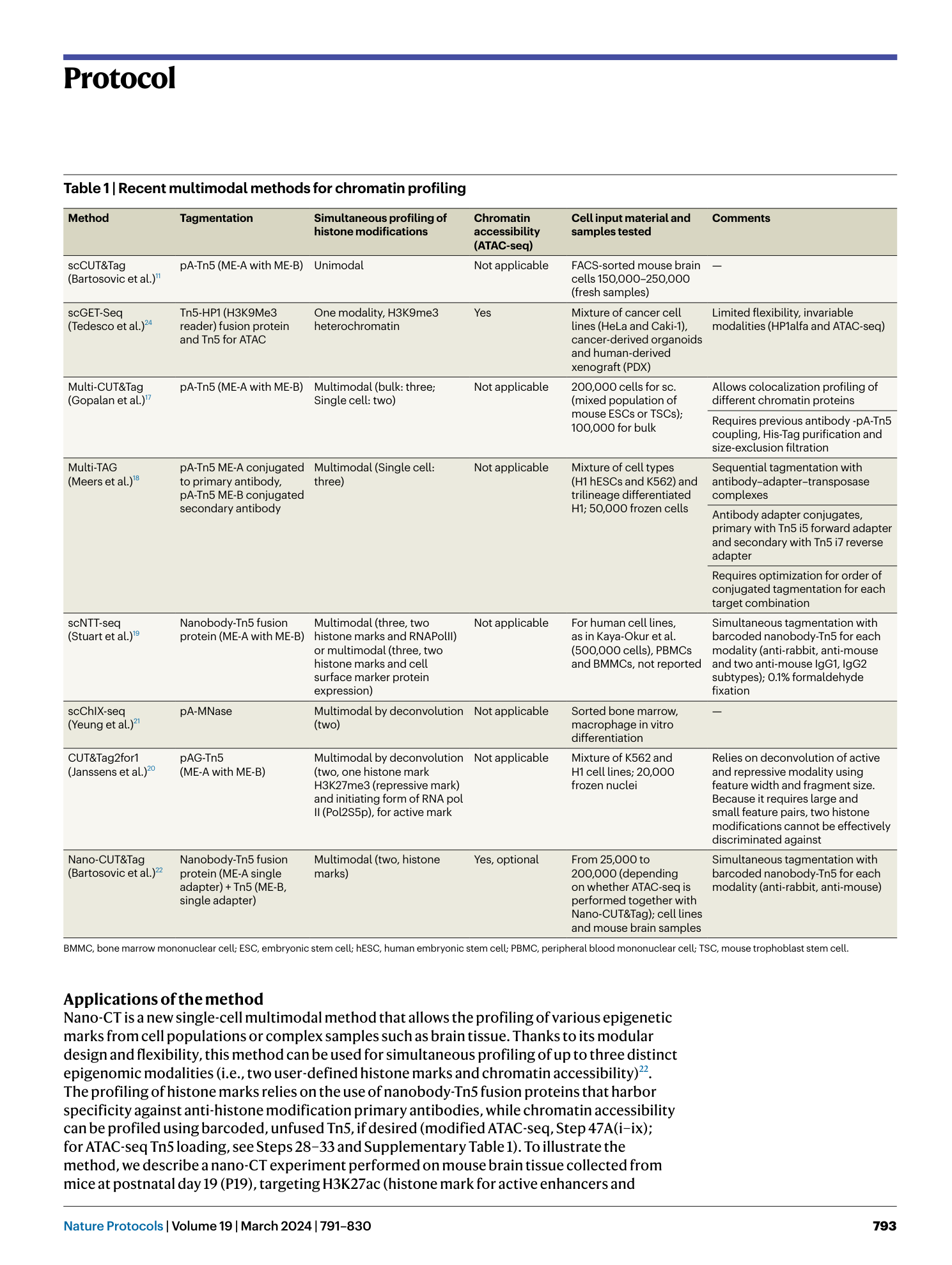
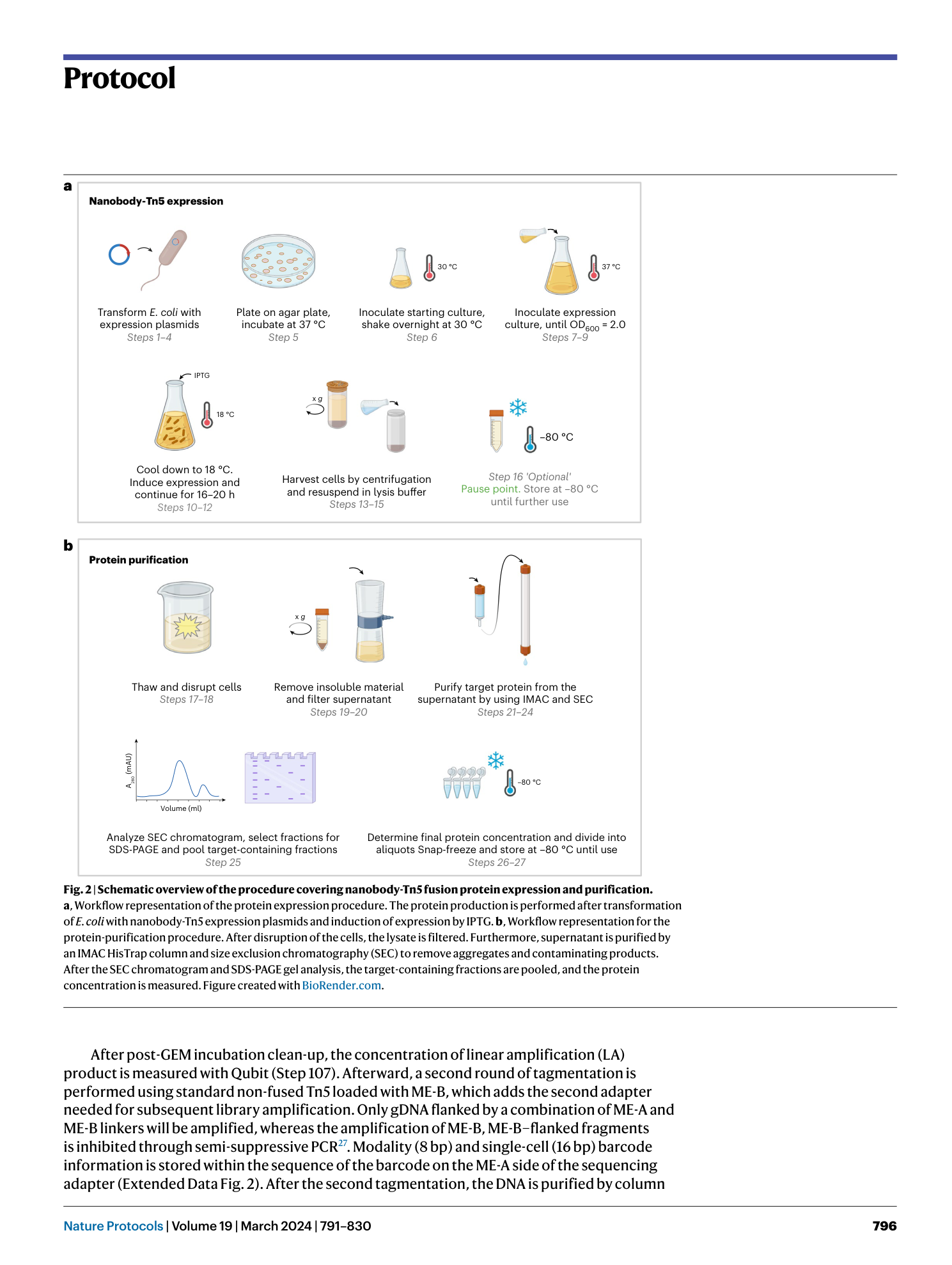
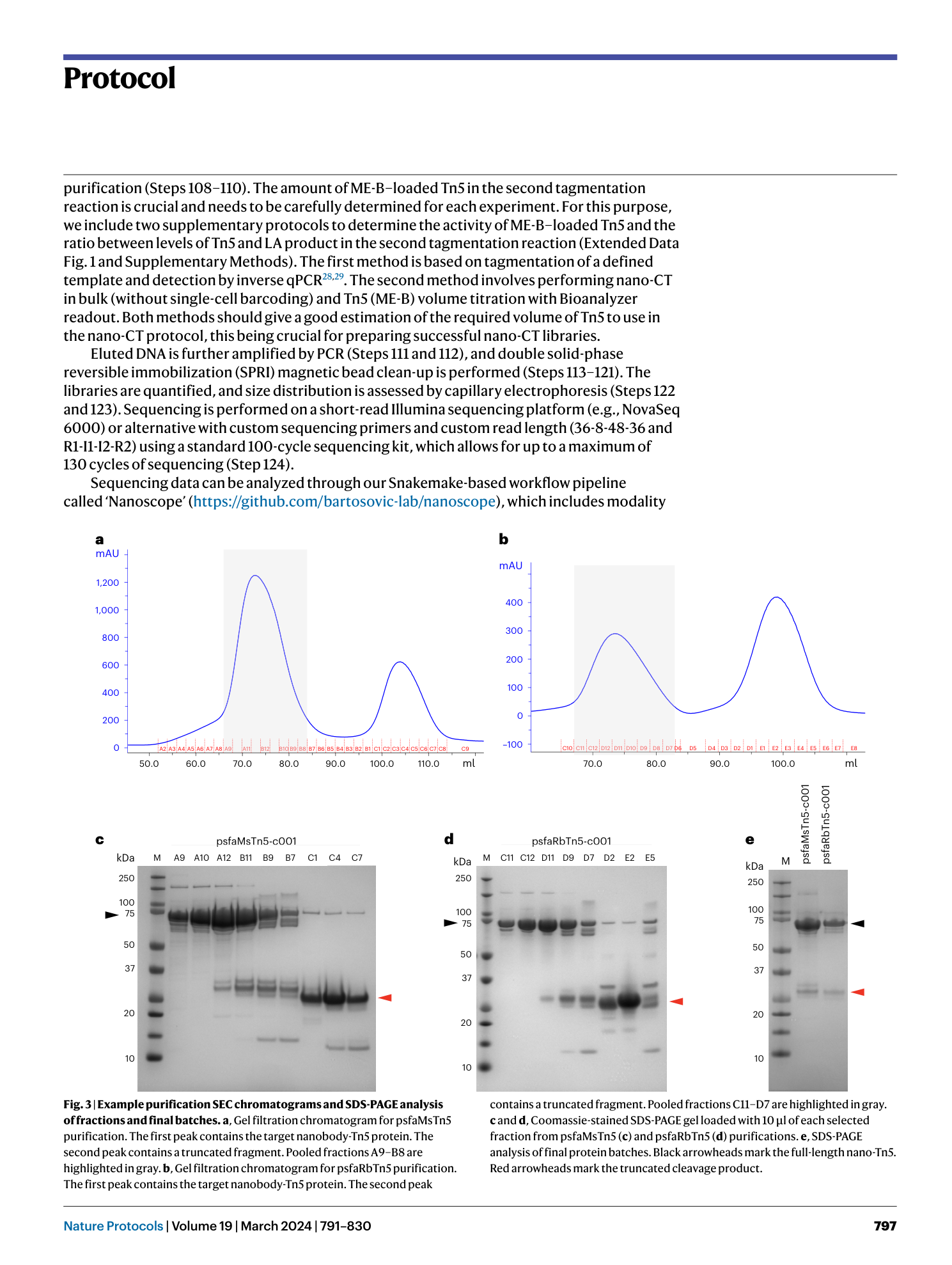
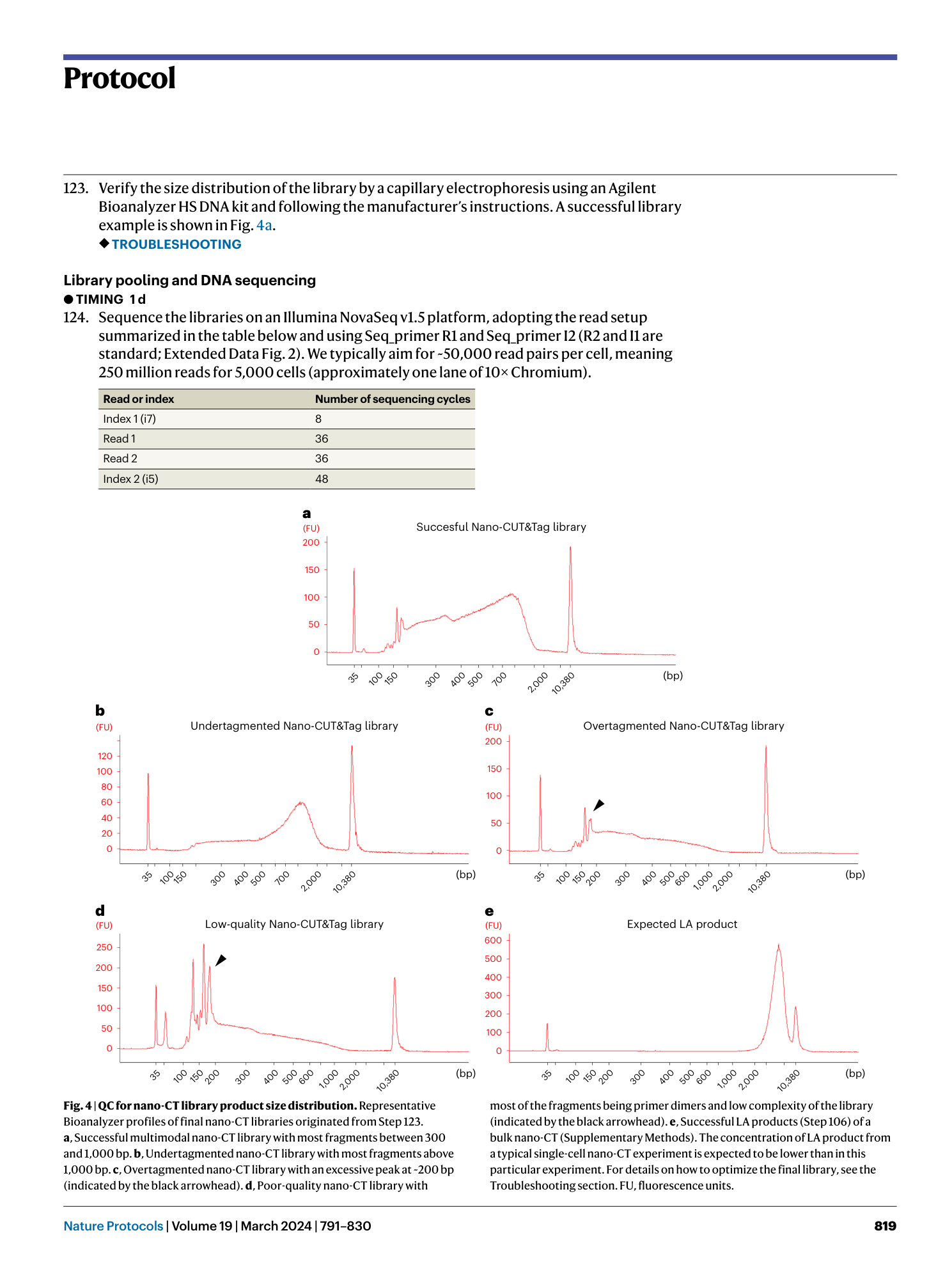
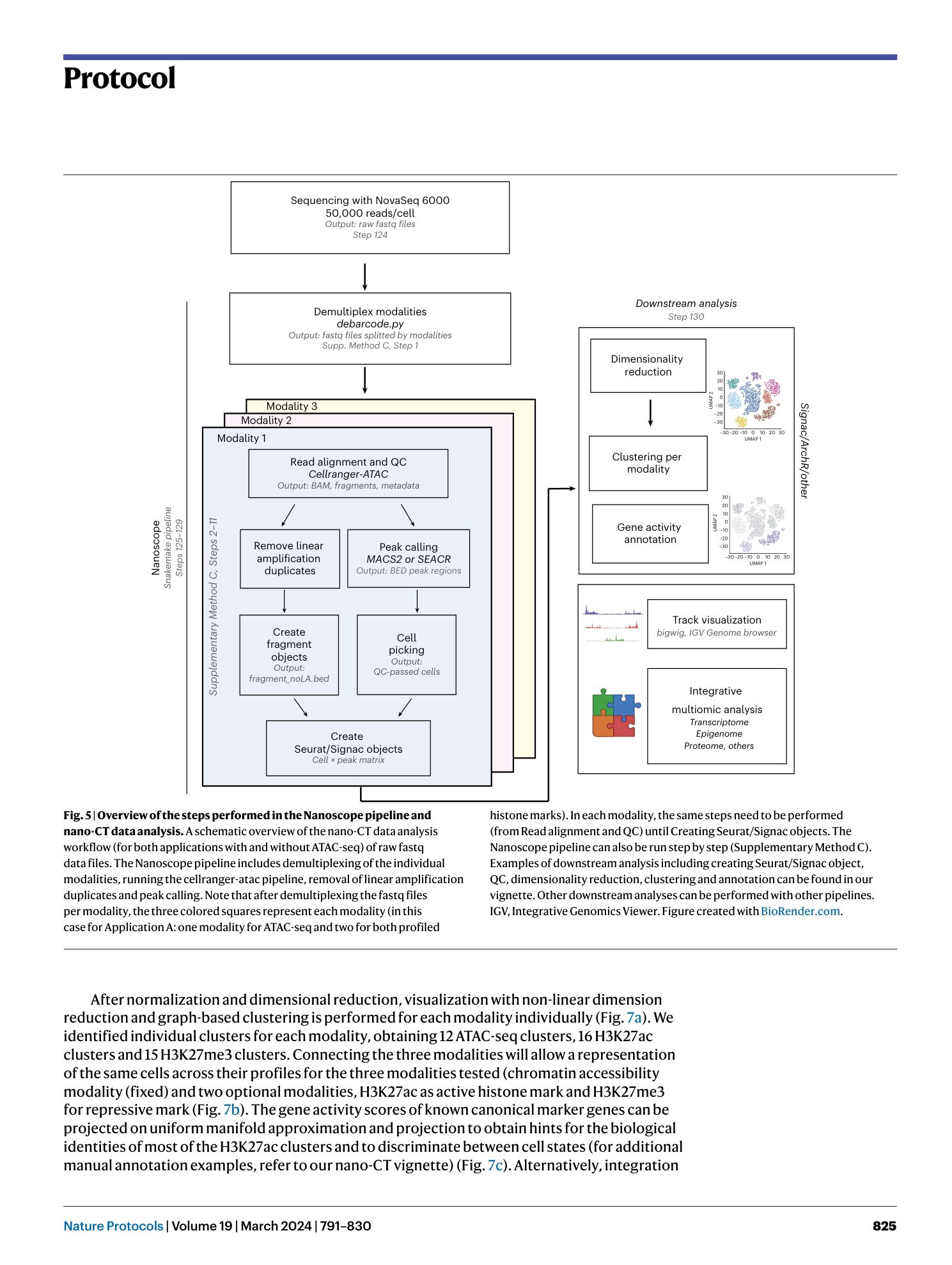
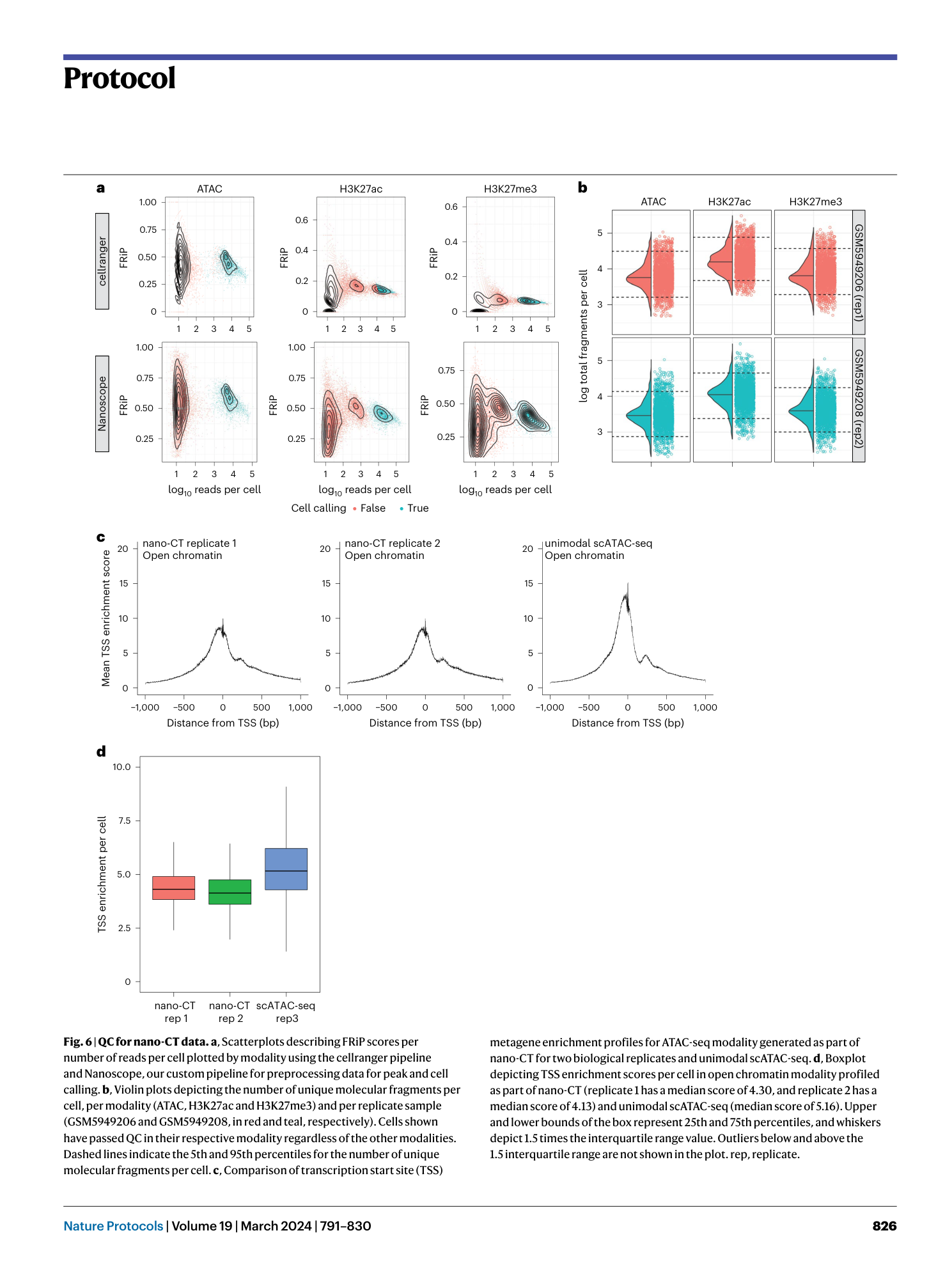
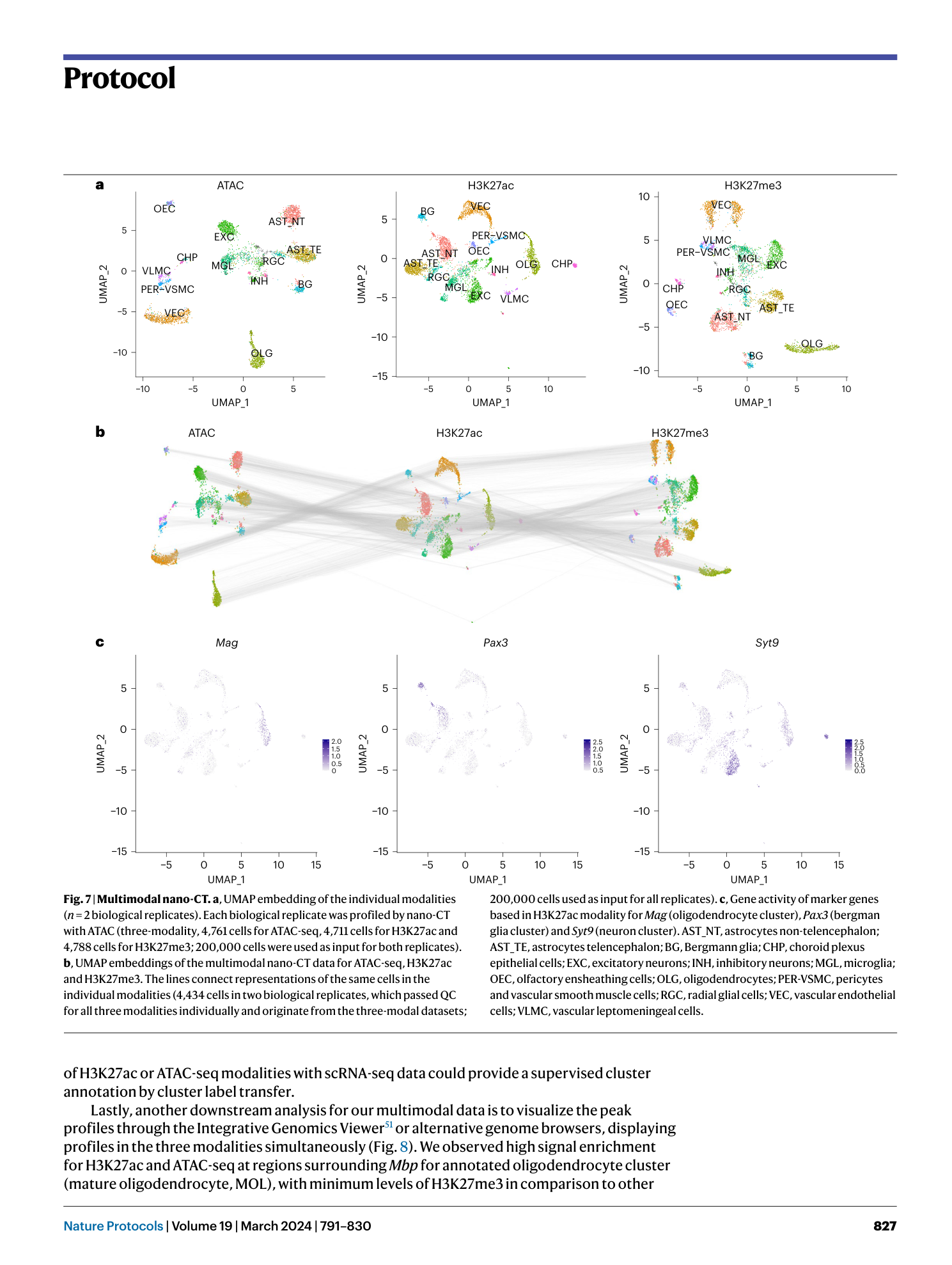
Nano-CUT&Tag for multimodal chromatin profiling at single-cell resolution
José Ramón Bárcenas-Walls, Federico Ansaloni, Bastien Hervé, Emilia Strandback, Tomas Nyman, Gonçalo Castelo-Branco, Marek Bartošovič
































Extended
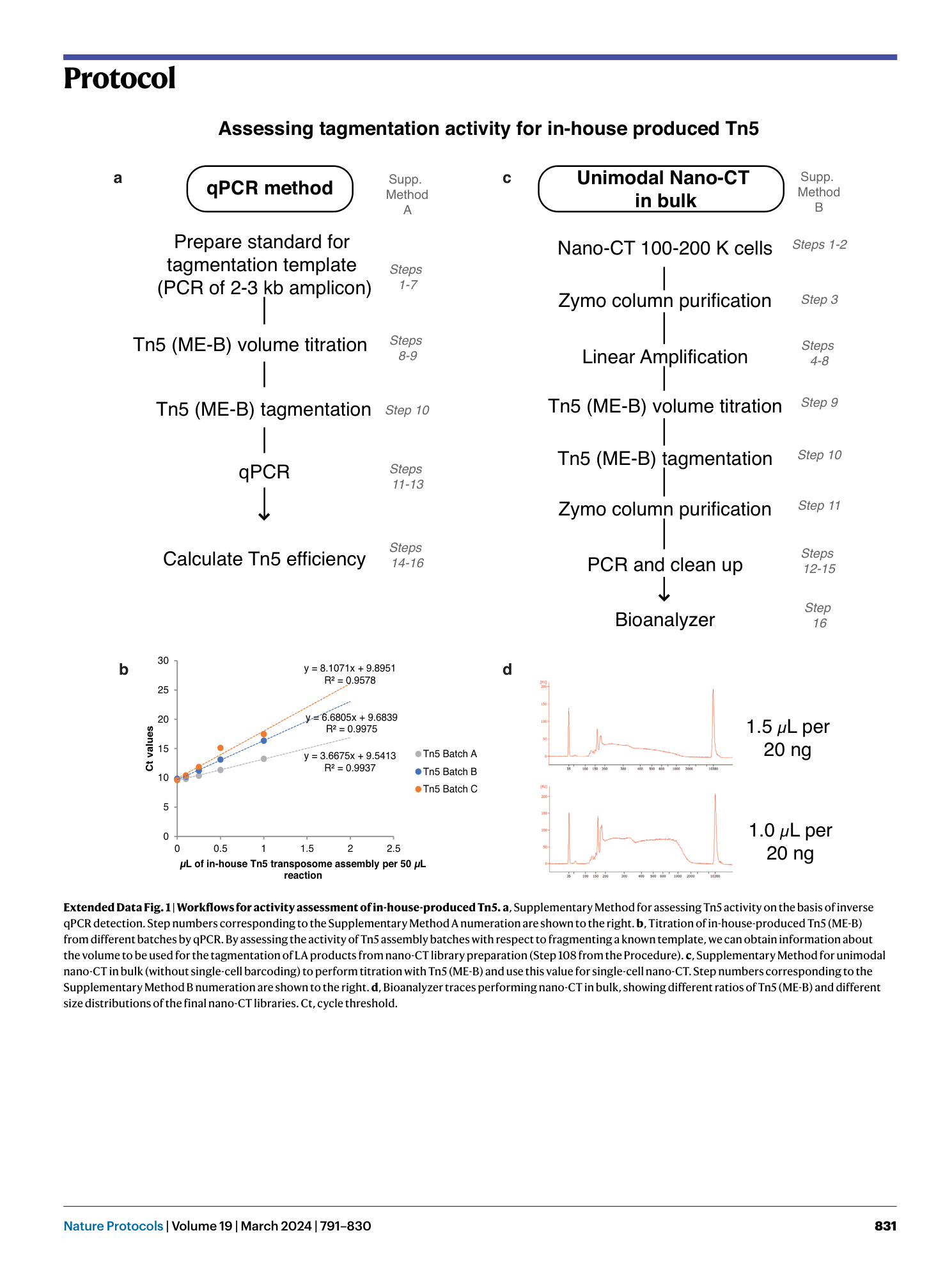
Extended Data Fig. 1 Workflows for activity assessment of in-house-produced Tn5.
a , Supplementary Method for assessing Tn5 activity on the basis of inverse qPCR detection. Step numbers corresponding to the Supplementary Method A numeration are shown to the right. b , Titration of in-house-produced Tn5 (ME-B) from different batches by qPCR. By assessing the activity of Tn5 assembly batches with respect to fragmenting a known template, we can obtain information about the volume to be used for the tagmentation of LA products from nano-CT library preparation (Step 108 from the Procedure). c , Supplementary Method for unimodal nano-CT in bulk (without single-cell barcoding) to perform titration with Tn5 (ME-B) and use this value for single-cell nano-CT. Step numbers corresponding to the Supplementary Method B numeration are shown to the right. d , Bioanalyzer traces performing nano-CT in bulk, showing different ratios of Tn5 (ME-B) and different size distributions of the final nano-CT libraries. Ct, cycle threshold.
Extended Data Fig. 2 Nano-CT library structure.
a and b , Full amplification sequence with Illumina P5 ( a ) and P7 ( b ) sequences for the nano-CT library. Note that both a and b correspond to the same nano-CT library molecule and are divided by genomic insert sequences. The custom primers required are for Read 1 and for Index 2 (shown in purple). Importantly, the ‘Modality barcode (8 nt)’ will contain the sequence from oligo adapters used during transposome complex loading (barcodes A, B and C in Steps 28–32). Library sequences will be demultiplexed by modalities by the Nanoscope pipeline. c , Schematic representation of the nano-CT library structure.
