Mammalian Reoviruses: Propagation, Quantification, and Storage
Kevin M. Coombs, Kevin M. Coombs
cesium chloride ultracentrifugation
double-stranded RNA
mouse cell culture
non-enveloped virus
plaque assay
RNA virus
Abstract
Mammalian reoviruses are pathogens that cause gastrointestinal and respiratory infections. In humans, the mammalian reoviruses usually cause mild or subclinical disease, and they are ubiquitous, with most people mounting immunity at a young age. Reoviruses are prototypic representations of the Reoviridae family, which contains many highly pathogenic viruses. This article describes techniques for culturing mouse fibroblast L929 cell lines, the preferred cell line in which most mammalian reovirus studies take place. In addition, mammalian reovirus propagation, quantification, purification, and storage are described. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Propagation of mammalian reoviruses in cell culture from virus stocks
Alternate Protocol 1 : Large-scale propagation (and purification) of mammalian reoviruses in cell culture from virus stocks
Basic Protocol 2 : Quantification of mammalian reoviruses by plaque assay with neutral red staining
Alternate Protocol 2 : Quantification of mammalian reoviruses by plaque assay with crystal violet staining
Basic Protocol 3 : Storage of mammalian reoviruses
Support Protocol 1 : Growth and maintenance of mouse L929 cells
Support Protocol 2 : Plating L929 cells
INTRODUCTION
Mammalian reoviruses are pathogens that present as gastrointestinal and respiratory infections. In humans, the mammalian reoviruses usually cause mild or subclinical disease, and they are ubiquitous, with most people mounting immunity at a young age. Reoviruses are prototypic representations of the Reoviridae family, which contains many highly pathogenic viruses. Studies with reoviruses can provide insight into these more dangerous members.
This article describes techniques on culturing mouse fibroblast L929 cell lines, in which most of the mammalian reovirus studies take place (Support Protocol 1), as well as propagating, quantifying, and storing reovirus. Basic Protocol 1 describes the propagation of reovirus in mouse L929 cell lines. The titration, or quantification, of the virus is described in Basic Protocol 2. Methods for virus storage are explained in Basic Protocol 3.
CAUTION : Mammalian reoviruses are Biosafety Level 2 (BSL-2) pathogens. Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms.
Basic Protocol 1: PROPAGATION OF MAMMALIAN REOVIRUSES IN CELL CULTURE FROM VIRUS STOCKS
Although mammalian reoviruses will grow in many different cell types, mouse L929 cells are preferred to use for in vitro cell culture and manipulation because the virus grows to high titer, and these cells are easily manipulated in suspension and monolayer forms. The propagation of reovirus stocks is performed by a series of passages. Typical virus passaging is described below.
Materials
-
Mouse L929 cells (ATCC #CCL-1)
-
Neutral red–stained plate containing virus plaques (see Basic Protocol 2)
-
Gel saline (see recipe)
-
25-cm2 tissue culture flask with pre-attached L929 cell monolayer (see Support Protocol 1)
-
Complete Joklik's Minimum Essential Medium (J-MEM) (see recipe)
-
Penicillin-streptomycin solution, 100× (see recipe)
-
Amphotericin B working solution, 1000× (see recipe)
-
75-cm2 tissue culture flask(s) with pre-attached L929 cell monolayer (see Support Protocol 1)
-
Cotton-plugged Pasteur pipets, sterile
-
1- or 2-dram glass vials, sterile
-
Additional reagents and equipment for obtaining neutral red–stained plates containing virus plaques (Basic Protocol 2)
NOTE : All solutions and materials that come into contact with cells must be sterile, with proper aseptic techniques used.
NOTE : All cell culture incubations utilize a 37°C incubator with 5% CO2 unless otherwise stated.
P0 (passage zero)
1.To pick a virus plaque from a neutral red–stained plate (see Basic Protocol 2):
-
Squeeze the rubber bulb on a sterile cotton-plugged Pasteur pipet to expel the air.
-
Insert the tip of the pipet through the agar overlay into the plaque area, until the pipet touches the cell monolayer.
-
Rotate the pipet while slowly releasing the rubber bulb to suck up the agar and cells in the selected area.
To ensure a pure virus clone, pick only plaques that are separated from other plaques on the agar plate by ≥1 cm (Fig.1).
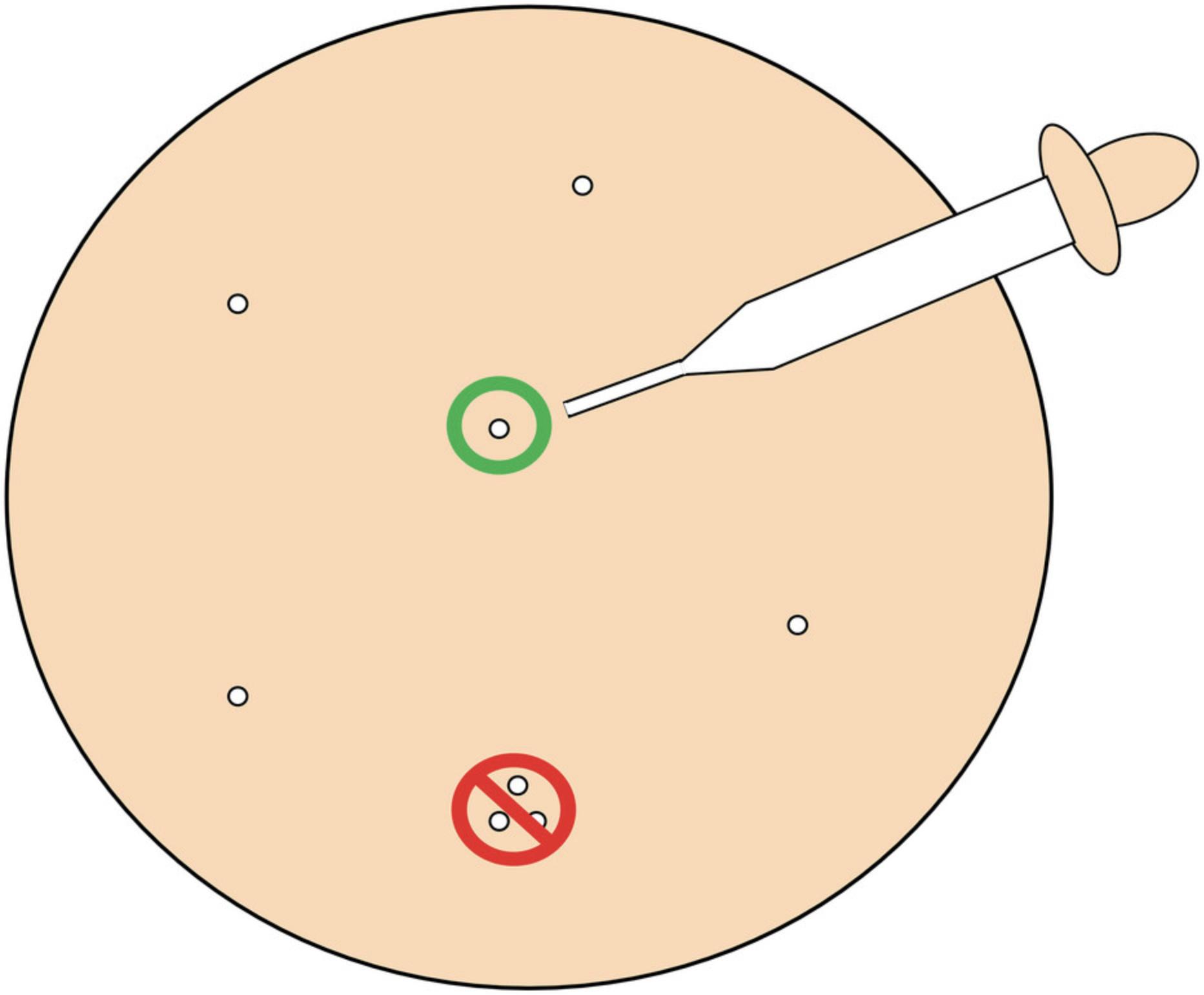
2.Expel the picked plaque into 1 ml of gel saline in a sterile 1- or 2-dram glass vial.
3.Store P0 virus stock at 4°C for ≥8 hr to allow virus to diffuse out of agar plug.
P1 (first passage)
4.Pour out medium from a 25-cm2 tissue culture flask that contains a pre-attached L929 cell monolayer.
5.Inoculate 0.5 ml of P0 virus stock into the flask.
6.Incubate the flask 1 hr at room temperature to allow virus to adsorb.
7.Add 4 ml of fresh complete J-MEM (supplemented with 40 µl 100× penicillin/streptomycin solution and 4 µl 1000× amphotericin B working solution) to the flask.
8.Incubate the flask at 37°C until ∼85% cytopathic effect (CPE).
9.Freeze-thaw three times by alternately placing the flask at less than –70°C and at room temperature for three complete cycles.
10.Transfer the sample into a labeled 2-dram vial and store at 4°C.
P2 (second passage)
11.Infect a pre-attached L929 cell monolayer in one or more 75-cm2 tissue culture flask(s) with 0.5 ml of P1 (from step 10). Use essentially the same procedure as for P1 passage, except add 11 ml of complete J-MEM per flask and store either in multiple 2-dram vials or in larger containers (i.e., small sterile media bottles).
Alternate Protocol 1: LARGE-SCALE PROPAGATION (AND PURIFICATION) OF MAMMALIAN REOVIRUSES IN CELL CULTURE FROM VIRUS STOCKS
To prepare large amounts of purified virus, a different method of amplification (compared to Basic Protocol 1), followed by purification of the virus can be used. This is demonstrated in the following protocol.
Materials
-
6.5 × 108 suspension-grown L929 cells (see Support Protocol 1)
-
Complete J-MEM (see recipe)
-
Mammalian reovirus P2 stock with titer > 108 PFU/ml (see Basic Protocol 1)
-
Penicillin-streptomycin solution, 100× (see recipe)
-
Amphotericin B working solution, 1000× (see recipe)
-
Bleach (Hypochlorite)
-
HO buffer (see recipe)
-
10% (w/v) sodium desoxycholate (DOC)
-
Vertrel-XF (Dupont Chemicals)
-
1.2 g/ml and 1.44 g/ml cesium chloride gradient(s) (see recipes)
-
Dialysis buffer (see recipe)
-
Glycerol (optional)
-
250-ml conical centrifuge bottles
-
Refrigerated low-speed (i.e., up to 2,000 rpm, or ∼850 × g) centrifuge with rotor for 250-ml conical centrifuge bottles
-
Glass roller bottle with sterile magnetic stir bar
-
Pipets and pipet tips
-
33°C spinning water bath (spinning water bath is set up by placing a large flat-bottom water-proof acrylic container on top of a multi-magnetic stirrer; the container should have sufficient room to hold a temperature-settable immersion heater and one or more glass roller bottles)
-
30-ml COREX centrifuge tubes
-
Ice bucket and ice
-
Sonicator with small probe (for ultrasonic disruption of cells)
-
Vortex
-
Parafilm
-
Refrigerated Super-speed (i.e., up to 15,000 rpm, or ∼21,000 × g) centrifuge with rotor and adaptors for 30-ml COREX tubes
-
SW-28 "ultra-clear"-type ultracentrifuge tubes
-
Refrigerated ultracentrifuge (i.e., up to 25,000 rpm, or ≥87,000 × g) with swinging-bucket rotor for SW-28 ultracentrifuge tubes
-
Dialysis tubing (e.g., Spectra/por 50,000 MWCO) and clips
-
UV spectrophotometer
-
Hemocytometer
NOTE : These protocols are for each 1-liter infection. They can be scaled up or down. For example, we have carried out 6-liter infections in 12-liter Florence flasks.
Perform virus propagation
1.Pour 6.5 × 108 suspension-grown L929 cells into 250-ml centrifuge bottles.
2.Spin cells in a refrigerated centrifuge 12 min at ∼350 × g , 4°C.
3.Pour 250 ml of the supernatant into the glass roller bottle (use supernatant as pre-adapted medium; pre-adapted medium = "conditioned" medium that contains growth factors and reduces costs), and discard the remaining supernatant.
4.Resuspend and pool the pelleted cells into fresh complete J-MEM so that after addition of virus P2 stock (next step) the concentration will be ≤2 × 107 cells/ml.
5.Add virus at an MOI of ∼5 PFU/cell.
6.Adsorb virus for 1 hr at room temperature with periodic swirling.
7.During adsorption, complete the glass roller bottle medium:
-
Add 700 ml fresh complete J-MEM, 11 ml 100× penicillin-streptomycin solution, and 1.5 ml 1000× amphotericin B working solution.
-
Place into a 33°C spinning water bath to equilibrate.
8.After the 1 hr adsorption, pour infected cell suspension into pre-equilibrated roller bottle.
9.Incubate, with spinning, ∼65 hr at 33°C.
10.After incubation, pellet cells using a refrigerated centrifuge in 250-ml centrifuge tubes 30 min at ∼500 × g.
11.Resuspend pelleted cells in 22 ml of HO buffer. Note the final volume of resuspended cells (should be 24 to 26 ml) and transfer into two 30-ml COREX tubes.
Perform virus purification
12.Thaw HO suspension if frozen, and place tubes on ice.
13.Sonicate suspension sample to disrupt cells.
14.Add 1/50th of the total volume recorded in step 11 of 10% DOC to each tube. Cover tubes with Parafilm and vortex the suspension 3 to 5 s.
15.Let sit for 5 min, then vortex 3 to 5 s. Let the tubes sit for an additional 25 min.
16.Add 2/5th of the total volume of Vertrel-XF to each tube and sonicate to mix the two layers and create an emulsion.
17.Add the same amount of Vertrel-XF as in step 16 to each tube and sonicate again at the same setting.
18.Centrifuge suspension 10 min at ∼9000 × g in a refrigerated Super-speed centrifuge.
19.Remove the top aqueous layers into fresh 30-ml COREX tubes with a plastic pipet, making sure to note the new volume.
20.Add 9/10th the total volume noted above of Vertrel-XF to each tube and re-emulsify.
21.Centrifuge again 10 min at ∼9000 × g to separate phases.
22.During final phase separation, prepare a 1.2 g/ml and 1.44 g/ml cesium chloride gradient(s) in an SW-28 ultracentrifuge tube.
23.Remove the aqueous phases from step 21 with a fresh pipet and layer contents of both COREX tubes onto a single 1.2-1.44 g/ml cesium chloride gradient.
24.Centrifuge gradient ≥5 hr at ≥87,000 × g in an ultracentrifuge at ∼5°C.
25.Collect the lower virus band by top, bottom, or side puncture into a sterile tube.
26.Pipet collected virus band into prepared dialysis tubing and dialyze against three sets of dialysis buffer (D-buffer or 2× SSC) for ≥1 hr, then ≥3 hr, then ≥6 hr at ∼5°C.
27.Collect the virus into an appropriate vessel and store at 4°C, or add glycerol to 15% and freeze at –80°C.
Basic Protocol 2: QUANTIFICATION OF MAMMALIAN REOVIRUSES BY PLAQUE ASSAY WITH NEUTRAL RED STAINING
Mammalian reoviruses cause cytopathic effect (CPE) and eventual death of host cells. Because of this, infected monolayers will show zones of lysed cells (called plaques), which are essentially areas of infected cells. A plaque assay is used to quantify the amount of infectious virus in a sample, indicated by the number of plaques formed on a monolayer of mouse L929 cells. If the dilution of the virus sample is high enough, the plaque represents a single infectious particle that has infected a cell and continued to spread to the adjacent cells. Using this method of quantification, a virus titer can be calculated to units per milliliter. This particular plaque assay uses a neutral red stain to enhance visibility for counting purposes. The neutral red assay technique is also used to replenish P0 virus stocks when these stocks run low (see Basic Protocol 1). Repeated passages should be performed at low multiplicities of infection to avoid generation of defective interfering particles.
Materials
-
Pre-attached L929 cell monolayers in 12- or 6-well plates or 60-mm dishes (see Support Protocol 2)
-
Gel saline (see recipe)
-
Mammalian reovirus stock/sample to be titrated (the virus may be initially obtained from the ATCC or clinical samples)
-
2% (w/v) agar (Difco Bacto)
-
Complete 2× Medium 199 (M-199) (see recipe)
-
Amphotericin B working solution, 1000× (see recipe)
-
2% (w/v) neutral red solution (in dH2O)
-
2× PBS (Current Protocols, 2006)
-
Penicillin-streptomycin solution, 100× (see recipe), optional
-
Dilution tubes
-
Micropipettors
-
100- to 1000-µl pipet tips
-
20- to 200-µl pipet tips
-
10-ml pipets
-
Vortex
-
Autoclavable waste container
-
Microwave
-
62°C water bath
Day 0
1.Set up L929 cells in appropriate well plates or dishes and incubate overnight at 37°C (see Support Protocol 2).
Day 1
2.Prepare a set of six dilution tubes for each sample by pipetting 900 µl of gel saline into each tube.
3.Make six serial 1:10 dilutions of the samples to be titered in gel saline as follows:
-
Vortex the sample, then pipet 100 µl of the sample into the first dilution tube, giving a 1 × 10–1dilution.
-
Discard the used pipet tip into an autoclavable waste container.
-
Vortex the 1 × 10–1dilution tube, then with a fresh pipet tip, pipet 100 µl of the 1 × 10–1dilution into the next dilution tube, giving a 1 × 10–2dilution.
-
Perform this for all dilution tubes for each sample (Fig.2), using a fresh pipet tip for each dilution.
For high-titered samples, start with a single 1:100 dilution, giving a 1 × 10–2dilution, followed by five 1:10 dilutions. In this case, the first dilution tube contains 10 µl of sample and 990 µl of gel saline.
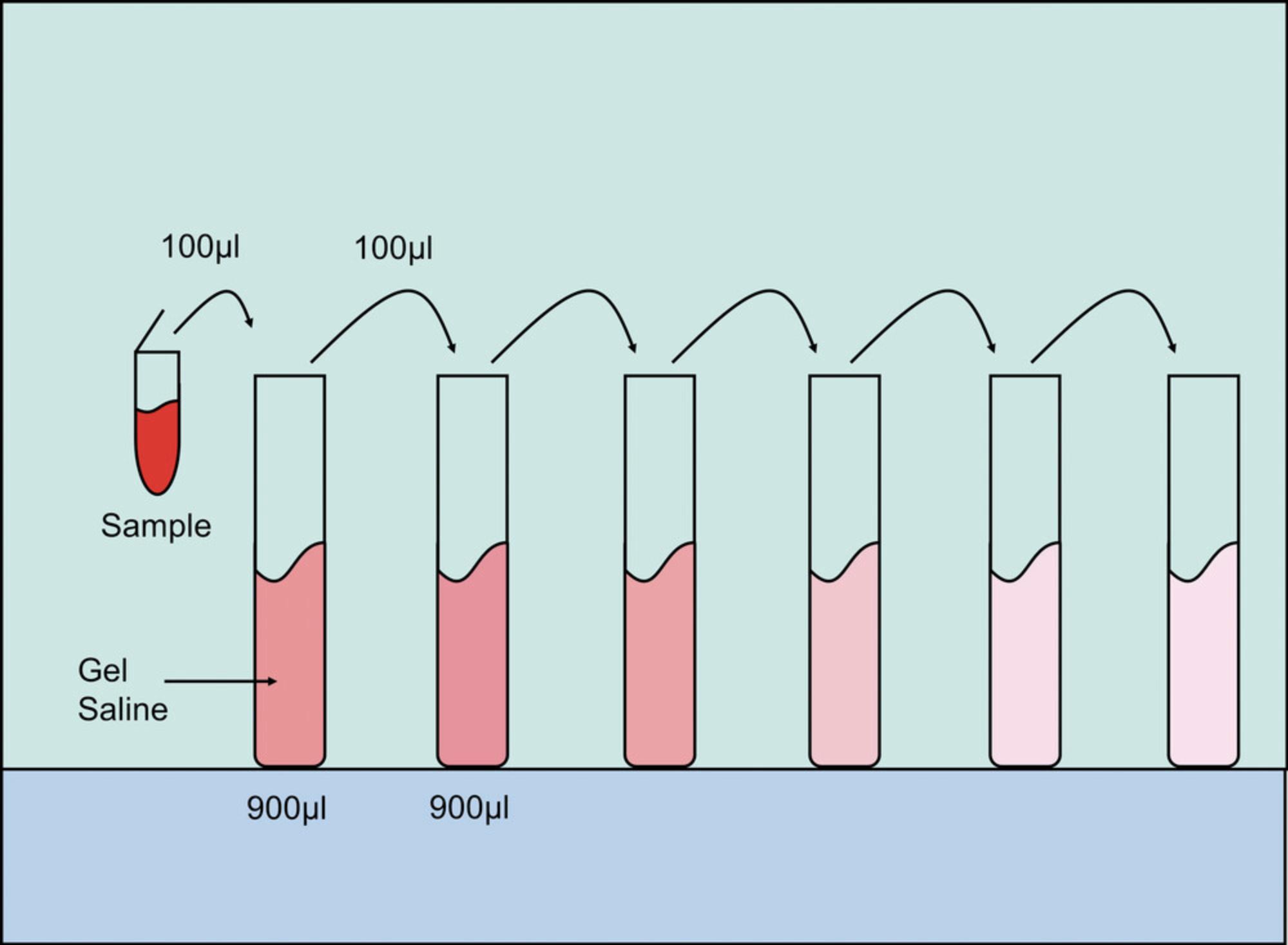
4.Dump out medium from the 6-well (and/or 12-well) plates that contain preformed L929 cell monolayers and inoculate 100 µl of the three highest dilutions onto cells, in duplicate, making sure to vortex each dilution tube first. Label wells with dilution factor used.
5.Allow virus to adsorb on cell monolayer by incubating 1 hr at room temperature.
6.While waiting for virus to adsorb, melt the 2% agar in a microwave and place in a 62°C water bath.
7.Allow the agar to cool to 62°C in the water bath.
8.Just before 1 hr adsorption time is up, prepare plaque assay overlay by adding an equal volume of room temperature 2× complete M-199 (with 6% FBS) into the bottle containing 62°C agar. Swirl to mix.
9.Add 1/1000th the total volume of 1000× amphotericin B working solution to the agar/199 mixture.
10.Overlay each well with 3 ml of complete agar/199 mixture for 6-well plates, or 1.25 ml for each well in a 12-well plate. Allow agar to solidify at room temperature for ∼10 min.
11.Put plates in a 37°C incubator. Incubate plates for 3 days.
Day 4
12.Prepare fresh agar/M-199 overlay mixture, as described above.
13.“Feed” assay by adding 2 ml of overlay into each well of a 6-well plate, or 0.75 ml of overlay into each well of a 12-well plate.
14.Return plates to a 37°C incubator. Incubate plates for 3 days.
Day 7
15.Prepare neutral red staining solution by melting the 2% agar in a microwave and placing in a 62°C water bath, as described above.
16.Allow agar to cool to 62°C.
17.Add 2 ml of 2% neutral red solution per 50 ml agar into warm agar.
18.Add an equal volume (to agar) of 2× PBS into agar/neutral red solution.
19.If planning to pick plaques for P0 virus stock (see Basic Protocol 1), add 1/100th the total volume of 100× penicillin/streptomycin solution and 1/1000th the total volume of 1000× amphotericin B working solution to the agar/PBS/neutral red solution.
20.Stain assay by adding 2 ml of staining solution into each well of a 6-well plate, or 0.75 ml into each well of a 12-well plate.
21.Return plates to a 37°C incubator.
Day 8
22.Count plaques within 18 to 24 hr of adding the staining solution.
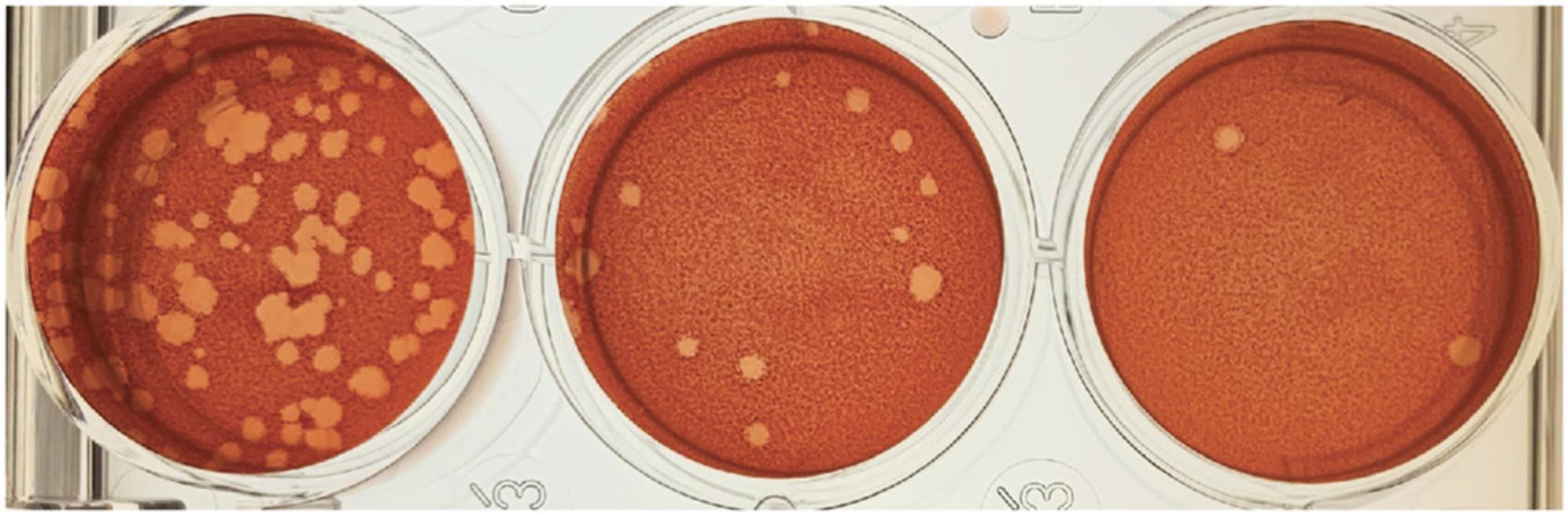
23.Count each plaque as one “plaque-forming unit” or PFU.
24.Calculate virus titer by taking into account the dilution factor.
Alternate Protocol 2: QUANTIFICATION OF MAMMALIAN REOVIRUSES BY PLAQUE ASSAY WITH CRYSTAL VIOLET STAINING
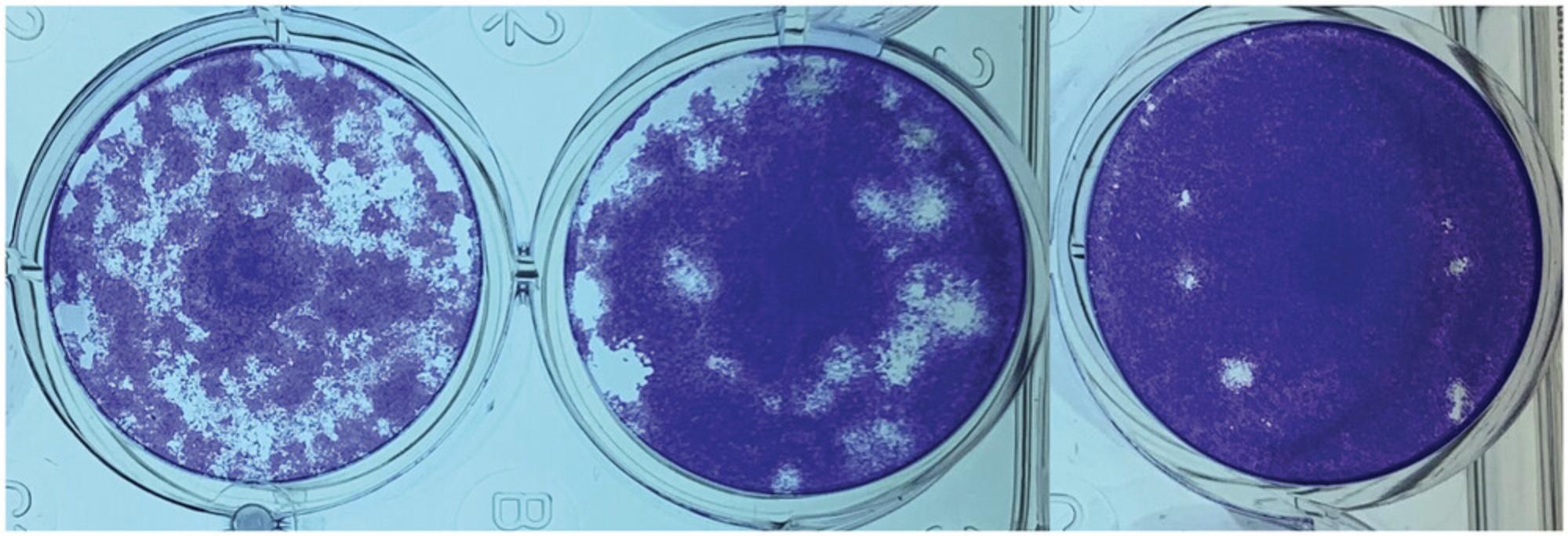
Plaques should be counted within 24 hr after staining with the neutral red staining technique (see Basic Protocol 2). However, a crystal violet staining method can be used instead for a stable assay that can be kept indefinitely (Fig. 4). This method kills both cells and virus, and it can therefore not be used when plaques need to be picked for replenishing P0 virus stocks.
Additional Materials (also see Basic Protocol 2)
-
2% (v/v) formaldehyde solution (37% formaldehyde in ddH2O)
-
Distilled water
-
2% (w/v) crystal violet stain (in methanol)
-
Small metal scoop
-
Paper towels
Days 0 to 4
1.Perform steps 1 to 14 of Basic Protocol 2.
Day 7
2.Fix cells in each well by pipeting 2% formaldehyde solution to cover agar. Leave on for > 1 hr (or overnight).
3.Gently scoop out agar plug from each well with a metal scoop.
4.Re-fix cells by pipeting 2% formaldehyde solution to cover cell monolayer. Leave on for ∼10 min.
5.Dump out formaldehyde solution, shaking once or twice to expel residual solution from the wells.
6.Gently wash wells once with distilled water.
7.Shake out remaining water from wells and add enough crystal violet solution to cover the bottom of each well.
8.Let crystal violet solution sit for at least 30 min.
9.Pipet out the crystal violet stain, putting it back into the stock solution for re-use.
10.Rinse off excess crystal violet stain from the wells under tap water, at an angle.
11.Place plates inverted onto paper towels, and leave to dry before counting plaques.
Basic Protocol 3: STORAGE OF MAMMALIAN REOVIRUSES
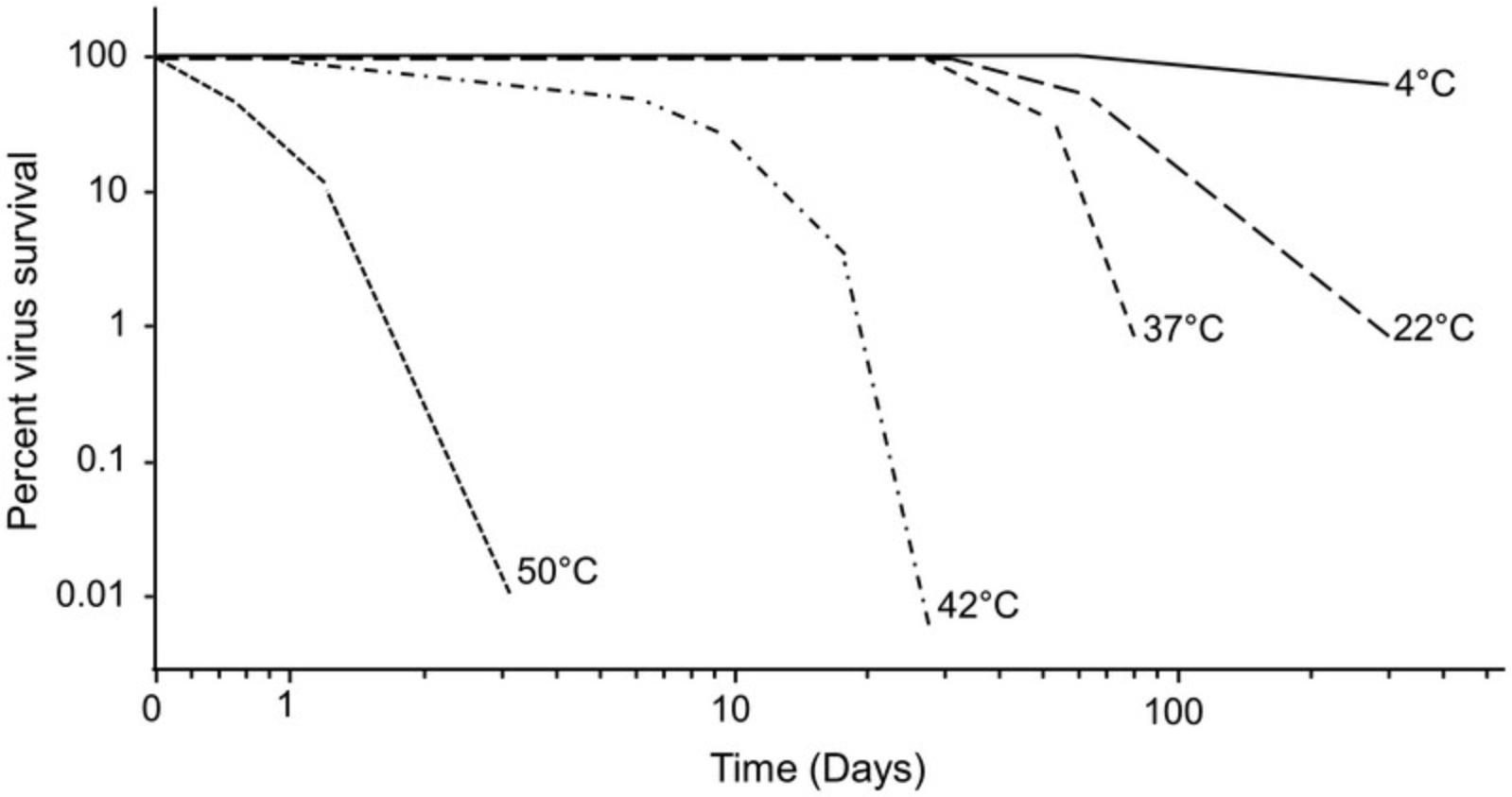
Mammalian reoviruses are fairly stable at 4°C, and they can be kept there for many months with little, if any, decline in infectivity (see Fig. 5). The virus also maintains high infectivity at room temperature (∼22°C) for about a month, but it loses infectivity more rapidly at higher temperatures (Fig. 5). Large stocks of virus may also be divided into aliquots and kept frozen (less than –70°C) for years.
Support Protocol 1: GROWTH AND MAINTENANCE OF MOUSE L929 CELLS
Although mammalian reoviruses may infect many different types of cell lines, mouse L929 cells are the preferred cell line used to efficiently amplify and quantify the virus. This protocol describes how to maintain the L929 cell line for use in Basic Protocols 1, 2, and 3 in either a monolayer or suspension form.
Materials
-
Mouse L929 cells grown in either 25-, 75-, or 150-cm2 tissue culture flask, or in a suspension culture flask (see Basic Protocol 1)
-
PBS/EDTA (see recipe)
-
Trypsin (see recipe)
-
Complete J-MEM (see recipe)
-
5-ml and 10-ml pipets
-
500- to 1000-ml glass flat-bottom “Florence” flask (for suspension culture)
-
Additional reagents and equipment for counting cells using a hemacytometer (Stevenson, 2006)
NOTE : All solutions and materials coming into contact with cells must be sterile, with proper aseptic techniques used.
NOTE : All cell culture incubations utilize a 37°C incubator at 5% CO2, except the suspension cultures, which are maintained in a 37°C non-CO2 incubator.
Prepare monolayer cultures
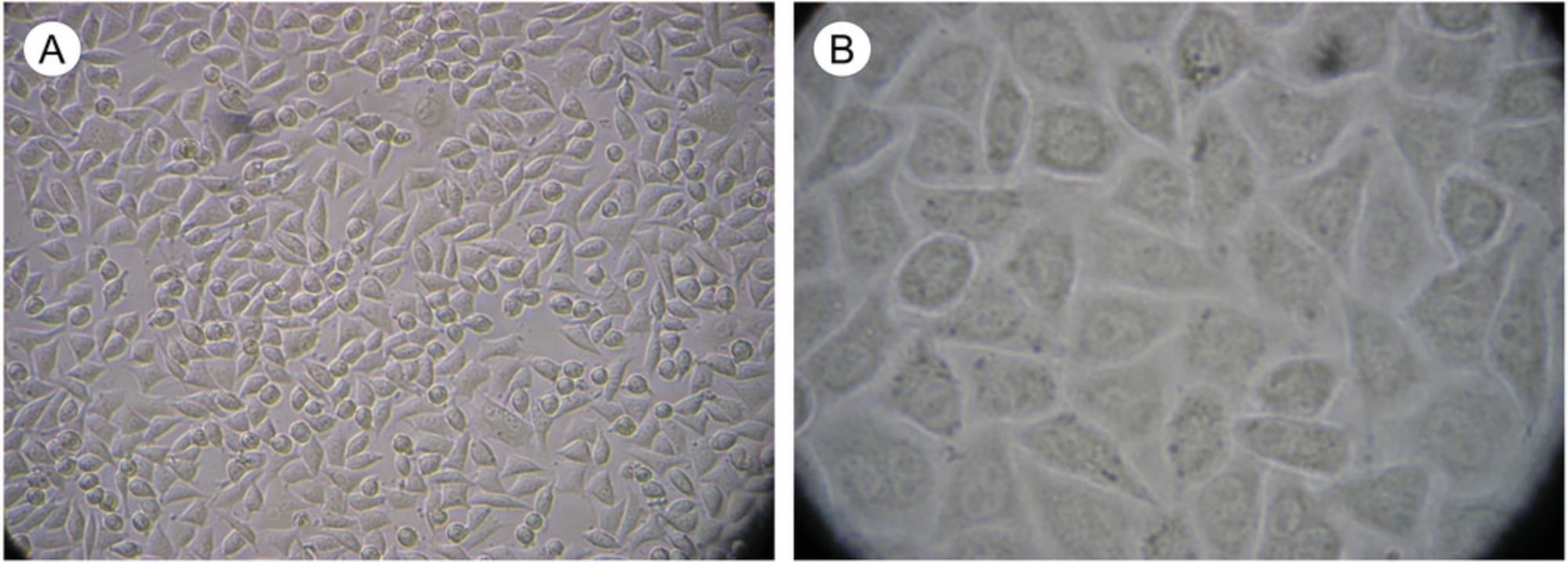
The steps that follow are based on treating a single 150-cm2 monolayer tissue culture flask so that it regains confluency in 2 days. We routinely split L929 cells 1:5 on Mondays and Wednesdays, and 1:10 on Fridays. The single 150-cm2 flask may be scaled up or down as anticipated needs dictate. In general, cells can be split ∼1:2.5 to be nearly confluent (∼90% to 95% confluent; optimal for virus growth; see Fig. 6) the next day; 1:5 to be confluent in ∼2 days; or 1:10 to be confluent in ∼3 days.
1a. Pour (or pipet) out old medium from confluent tissue culture flask, saving ∼10 ml in a sterile tube as “pre-adapted” medium.
2a. Rinse monolayer with ∼10 ml of PBS/EDTA. Pipet out PBS/EDTA.
3a. Add 2 ml of fresh PBS/EDTA/trypsin (at a final trypsin concentration of 2.5 mg/ml) into flask. Rock flask back and forth 2 to 3 times to completely cover bottom with trypsin mixture. Remove trypsin mixture.
4a. Put flask into a 37°C incubator for ∼10 min. After the time has elapsed, check for loosening of cells under a microscope.
5a. Pipet 5 ml of pre-adapted J-MEM (from step 1a) into trypsinized flask and titurate to wash cells off the bottom of the flask and break up clumps. Tituration is performed by placing the pipet against the flat bottom of the flask and expelling liquid to break up large cell clumps.
6a. Remove all but 1 ml of cell suspension.
7a. Add the remaining 5 ml of pre-adapted completed J-MEM (from step 1a) plus 24 ml of fresh medium.
8a. Place the flask back into the incubator.
Prepare suspension cultures
The steps that follow are based on 500 ml of cells kept in a 1000-ml flat-bottom glass “Florence” flask. These cells are checked each day and split back to a concentration of ∼5 × 105 cells/ml as follows:
1b. With a sterile pipet, remove ∼1 ml of cell suspension from the flask containing mouse L929 cells.
2b. Count the cell concentration using this 1 ml cell suspension in a hemacytometer (Stevenson, 2006).
3b. Calculate required amount of cell suspension needed to reduce stock solution to 5 × 105 cells/ml.
4b. Pour out extra cells into a sterile container to either discard or plate for use in further applications (see Support Protocol 2).
5b. Add fresh complete J-MEM into the flask to return the volume to 500 ml and the cells to a concentration of ∼5 × 105 cells/ml. Place the flask back into the incubator.
Support Protocol 2: PLATING L929 CELLS
This protocol describes plating L929 cells for quantification, purification, or amplification.
Materials
-
L929 cells in suspension (see Support Protocol 1)
-
Complete J-MEM (see recipe)
-
5- and 10-ml pipets
-
Appropriate-sized vessels
1.For use the next day, dilute cells to 4.2–4.5 × 105 cells/ml with complete J-MEM.
2.Pipet cells depending on their use:
| To titer virus stocks/samples: | 2.5 ml/well in 6-well cluster dishes |
| 1.25 ml/well in 12-well cluster dishes | |
| To pick plaques: | 5 ml/60-mm tissue culture dish |
| To set up virus growths: | 25-cm2 flask—5 ml |
| 75-cm2 flask—15 ml |
3.Incubate plates/flasks overnight for use the next day.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps.
Amphotericin B (1 mg/ml), 1000× (working solution)
- 1 ml of 20,000× amphotericin B solution (see recipe)
- Using aseptic techniques, bring to 20 ml with sterile ddH2O
- Divide into 10-ml aliquots into sterile tubes and store for several years at –20°C
Amphotericin B (20 mg/ml), 20,000× (stock solution)
- 100 mg amphotericin B solubilized powder, γ-irradiated (Sigma, cat. No. A9528)
- Using aseptic technique, reconstitute amphotericin B power in 5 ml sterile ddH2O
- Divide into 1-ml aliquots into sterile cryovials and store for several years at –20°C
Cesium chloride gradient solutions
1.2 g/ml :
- 33.3 g CsCl
- 1 ml 1 M Tris·Cl, pH 7.4 (Current Protocols, 2006), dissolved in 100 ml ddH2O
- Adjust refractive index to 1.3533 with 10 mM Tris·Cl, pH 7.4
1.44 g/ml :
- 67 g CsCl
- 1 ml 1M Tris·Cl, pH 7.4 (Current Protocols, 2006), dissolved in 100 ml ddH2O
- Adjust refractive index to 1.378 with 10mM Tris·Cl, pH 7.4
- Filter sterilize with a 0.2-µm membrane
- Store for several years at room temperature
Dialysis buffer
- 30 ml 5 M NaCl (150 mM)
- 15 ml 1 M MgCl2 (15 mM)
- 10 ml 1 M Tris·Cl, pH 7.4 (Current Protocols, 2006)
- Bring to 1 L with ddH2O
- Store up to 6 months at room temperature
Gel saline
- 8 g NaCl
- 0.03 g CaCl2
- 0.17 g MgCl2·6H2O
- 1.2 g H3BO3
- 0.05 g Na2B4O7·10H2O
- 3 g gelatin, type A
- Heat and stir to dissolve in 1 L ddH2O
- Autoclave
- Store for several years at 4°C
L-Glutamine (100×)
- 14.6 g L-glutamine
- Bring to 500 ml with ddH2O
- Filter sterilize with a 0.2-µm membrane
- Aliquot into sterile 100-ml bottles, and cool to 4°C overnight
- Place bottles at –20°C and swirl bottles every 30 min until frozen
- Store for several years at –20°C
HO buffer
- 1 ml 1 M Tris·Cl, pH 7.4 (10 mM final concentration) (Current Protocols, 2006)
- 5 ml 5 M NaCl (250 mM final concentration)
- 67 µl 2-mercaptoethanol (10 mM final concentration)
- Bring solution to 100 ml with ddH2O and filter sterilize with a 0.2-µm membrane
- Store for several years at 4°C
J-MEM (for culturing L929 cells), complete
- 940 ml 1 × S-MEM (Joklik's S-MEM; e.g., SAFC Biosciences, cat. No. 56449C)
- 50 ml tissue-culture grade fetal bovine serum (FBS)
- 10 ml L-glutamine stock solution (see recipe)
- Store up to 1 year at 4°C
Alternatively, if using a 2× S-MEM solution, use only 500 ml and add sterile ddH2O to final volume of 1 L.
Medium 199 (M-199) (6% FBS); 2×, complete
- 900 ml 2× 199 medium (made at double concentration; Medium 199 Modified; SAFC Biosciences, cat. No. 51312C or Invitrogen, cat no. 31100-019)
- 60 ml fetal bovine serum (FBS)
- 20 ml L-glutamine stock solution (see recipe)
- 20 ml penicillin-streptomycin stock solution (see recipe)
- Store up to 1 year at 4°C
PBS/EDTA
- 8 g NaCl
- 0.2 g KCl
- 1.15 g Na2HPO4
- 0.2 g KH2PO4
- 0.2 g EDTA
- Dissolve in 1 L of ddH2O
- Autoclave
- Store for several years at room temperature
Penicillin-streptomycin solution, 100×
- 1.2 g penicillin G
- 2.0 g streptomycin sulfate
- Bring solution to 200 ml with ddH2O and filter sterilize with a 0.2-µm membrane
- Aliquot in 100-ml bottles and store for several years at –20°C
Trypsin (25 mg/ml), 10×
- 1.25 g trypsin (1:250; porcine pancreas, Sigma)
- 0.425 g NaCl
- Bring to 50 ml with 1× PBS (Current Protocols, 2006) and filter sterilize with a 0.2-µm membrane
- Aliquot into desired amounts and store for several years at –20°C
COMMENTARY
Background Information
The mammalian reoviruses (MRV) are the prototypical members of the virus family Reoviridae. Members of this family have a genome of 9 to 12 segments of double-stranded (ds) RNA that are surrounded by 2 to 3 concentric, non-enveloped protein capsids (Dermody et al., 2013). The Reoviridae family contains the only group of dsRNA viruses (out of seven current dsRNA virus families) that infect mammals. The Reoviridae family currently consists of 15 recognized genera [NCBI Taxonomy Browser: Taxonomy browser (Reovirales), nih.gov], and a variety of other newly discovered viruses that share key features (dsRNA segmented genome, non-enveloped, and capacity to infect mammals) supports inclusion of additional currently unclassified agents (i.e., Cimodo virus). In addition to the genus Orthoreovirus, the Reoviridae includes Rotavirus , agents responsible for a significant amount of viral gastroenteritis and numerous deaths annually worldwide, the economically important insect-vectored Orbivirus , and a variety of other viruses that infect animals, fungi, and plants.
The Orthoreovirus genus is divided into three subgroups: non-syncytia-inducing mammalian reovirus (subgroup 1), avian reovirus and Nelson Bay virus (subgroup 2), and baboon reovirus (subgroup 3). The Orthoreoviruses have a genome of 10 segments of dsRNA surrounded by 2 concentric protein capsids. MRV are represented by three reovirus serotypes: type 1 Lang (T1L), type 2 Jones (T2J), and type 3 Dearing (T3D). A variety of other strains are placed into one of these three serotypes, based primarily upon hemagglutination characteristics. The complete genomic sequences of all 10 genes of all 3 prototype MRV have been determined. The orthoreovirus genome consists of 3 large segments (L1, L2, L3) ranging in size from ∼3.8 to 4.0 kilobase pairs (kbp), 3 medium segments (M1, M2, M3) ranging in size from ∼2.0 to 2.3 kbp, and 4 small segments (S1, S2, S3, S4) ranging in size from ∼1.0 to 1.6 kbp (total genome size ∼23.5 kbp) (Breun et al., 2001; Wiener et al., 1989; Yin et al., 2004).
The L genome segments encode large proteins [usually named lambda (λ)], which are found in the inner virus capsid (called core) and which have a variety of enzymatic roles associated with the generation of mRNA from the enclosed genome (Dryden et al., 2008; Sandekian & Lemay, 2015). Proteins λ1 and λ2 have major structural roles; λ1 constitutes the core shell and protein λ2 forms pentameric “turrets” at each of the core's icosahedral vertices. Protein λ3 serves as the viral RNA-dependent RNA polymerase (RdRp) and is located under each turret (Zhang et al., 2004). The core remains intact throughout the viral replicative cycle, which takes place in the infected cell's cytoplasm. Thus, the RdRp, which is located inside the virus capsid along with the genomic dsRNA, must have sufficient freedom of movement to read the template dsRNA to transcribe mRNA. This also implies that the core capsid must have holes in it to allow transport of nucleotides and other components into the core interior. In addition to the three λ proteins, orthoreovirus cores are composed of two additional proteins. These are a medium protein [named mu (µ); specifically µ2] and a small protein [named sigma (σ); specifically σ2]. The σ2 protein appears to serve as a “clamp” to help hold the core capsid together. The µ2 core protein appears to serve as an RdRp cofactor since expressed RdRp has little, if any, polymerase activity by itself (Starnes & Joklik, 1993). The core is surrounded by an outer capsid made up of three additional proteins. The major outer capsid protein is µ1 which, as a trimeric aggregate, forms the icosahedral T = 13 lattice. The µ1 outer capsid lattice is decorated with three copies of another major outer capsid protein (σ3) associated with each µ1 trimer. The outer capsid also contains a smaller number of another σ protein (σ1), which serves as the cell attachment protein. The related avian orthoreoviruses possess the same proteins, although they are given different names.
The development and refinement of a “Reverse genetics” system (Kobayashi et al., 2007; 2010) for reovirus has allowed further refinement of molecular signals for numerous functions performed by various reovirus proteins. These include tagging viral proteins for the visualization of “viral factories” (Bussiere et al., 2017), determining amino acid residues responsible for plaque size and oncolytic potential (Mohamed et al., 2020), and determining amino acid residues responsible for the temperature-sensitive phenotype of certain mutants (Glover et al., 2021).
Reoviruses infect a wide range of cells, both in vitro and in vivo. The virus usually infects specialized intestinal epithelial cells (M cells) that overlie Peyer's patches in vivo. The virus then migrates between and/or through the M cells into mucosal mononuclear cells in the Peyer's patch, and subsequently into a large number of extraintestinal sites, including heart, liver, and the central nervous system. The onset of cell infection is initiated by the interaction of the σ1 cell-attachment protein with an appropriate cell surface protein. Evidence points to junction adhesion molecules and/or sialyated proteins as possible receptors. Once bound, the virus is internalized. For reoviruses, this seems to involve receptor-mediated, low-pH-driven proteolysis, which serves to remove the σ3 outer capsid protein, and also results in clipping of the µ1 outer capsid protein into two peptides [called delta (δ) and phi (φ)]. The resulting particle [called Intermediate (or Infectious) Subviral Particle (ISVP)] can also be produced in the laboratory and is capable of directly penetrating membranes. Once exposed to the cellular milieu, the ISVP loses the rest of its outer capsid proteins and peptides to generate the core particle. Nascent mRNA produced from cores serves both for protein translation and also as a template to regenerate progeny dsRNA genomes. The progeny dsRNA genomes are assembled by an efficient process into sets of the 10 different genes, which then become progressively coated by newly translated viral proteins to produce progeny virus. Progeny virus is released when the infected cells lyse, a process that generally occurs 24 to 72 hr post-infection.
MRV infect a wide range of animals, including humans. Serologic evidence indicates >90% of tested human populations have antibodies to MRV by the time they reach adulthood. In humans, MRV infection is usually asymptomatic. There are sporadic reports of cold-like symptoms and more serious illnesses, including hepatic, and possibly neuronal, involvement (Hermann et al., 2004; Johansson et al., 1996; Tyler et al., 2004). The segmented nature of the viral genome allows genetic mixing when two different strains of the same virus infect the same cell. This genetic mixing (analogous to influenza virus genetic shift) leads, as in the case of influenza virus, to the generation of antigenically distinct virus clones, which may not initially be recognized by the human immune system.
Critical Parameters and Troubleshooting
Mammalian reoviruses generally are easily grown and achieve high titers. They also are among the most stable of known viruses, which, when combined with their general safety, makes their manipulation generally straightforward. As indicated earlier, they are stable when stored at refrigerator temperatures. The avian reoviruses are less stable than the mammalian viruses. Reoviruses also are stable under acidic conditions, and they will survive passage through the gastrointestinal system, their normal route of infection. However, the viruses are sensitive to alkaline conditions and will lose infectivity rapidly at pH values >9.Thus, solutions and media require buffering.
Virus growth is dependent upon healthy cells. Although the precise cell cycle stage of cells is not critical, the virus grows best in actively growing cells. Thus, optimum results are obtained with cells that are 90% to 98% confluent. Viruses will grow (and plaque) in cells that have just become confluent, but yields (and plaquing capacity) are reduced if cell monolayers have been confluent for >12 hr. Similarly, since optimal cell health is necessary, great care should be taken during plaque assays that the agar overlay is not too hot. We have found that mixing equivalent volumes of 62°C 2% agar with room-temperature (∼22°C) 2× medium, by adding the medium into the bottle that contains the agar, results in a solution that is 43° to 45°C. Be sure to “wrist test” the mixture's temperature, and, if necessary, allow it to cool sufficiently so as to not “cook” the cells. Plaque assays performed in 12-well plates are especially sensitive to the volume of agar/medium added. The amounts specified in the above protocols should not be exceeded. If they are, plaque development and staining suffer markedly.
Like other RNA viruses, reoviruses have the capacity to mutate more rapidly than many DNA viruses. In addition, defective interfering particles may arise after repeated high multiplicity infections. Thus, it generally is a good idea not to exceed four passages of virus amplification. Low passage stocks may be replenished by picking fresh plaques. Common problems, and their solutions, are indicated in Table 1.
| Problem | Possible cause | Solution |
|---|---|---|
| No visible plaques | Dilution series not appropriate | Use a wider range of dilutions, from 100 – 10-9 |
| No visible plaques | Agar overlay too hot | “Wrist test” to confirm solution not too hot |
| Plaques too large or too small | Incubation time too long or too short | Adjust incubation tme |
| Plaques appear as “comets” instead of being round | Plates moved before agar set | Overlay plates with agar and set down immediately without disturbing for at least 5 min |
| VertrelXF does not emulsify | Too much VertrelXF | Ensure the volume of VertrelXF is less than the volume of solution being extracted; keep sonicated sample chilled |
Understanding Results
Plaque assays
Plaques will present as clear or “milky” circular areas in the cell monolayers. They may be visible unstained, but enumeration is easier if the cell monolayer has been stained. Plaques normally contain ∼105 infectious viruses; thus, there is sufficient virus in a picked plaque (passage zero; P0) to amplify as described herein.
Virus passaging
The titer of virus usually increases 10× to 100× with each of the first two passages. Final titers obtained are dependent upon the virus strain. In our hands, MRV T1L regularly grows to titers of ∼109 PFU/ml, T2J to titers of ∼4 × 107 PFU/ml, and T3D to titers of 3-8 × 108 PFU/ml in L929 cells.
Virus purification
Mammalian reovirus grows well in L929 cells grown either as monolayers or as suspension cultures. For large-scale growth, it is easier to work with suspension cultures of 1 L or more. The amount of virus obtained is also strain-dependent; several milligrams of purified virus can usually be obtained from 1 L of infected cells. The centrifugation medium (cesium chloride) should be removed by dialysis before the virus is used.
Virus storage
Reoviruses are among the most stable of viruses. Nevertheless, their titer will decrease with time. The rate of infectivity decrease is temperature-dependent (see Fig. 5). Virus stocks will lose detectable infectivity if stored at 4°C for many years. Similarly, frozen virus stocks will also lose some infectivity after prolonged storage. Thus, periodic stock replenishment is needed.
Time Considerations
Plaque assays
The standard MRV plaque assay in 6-well plates takes ∼1 week when performed at 37°C. Temperature-sensitive clones (see Coombs, 1998b) take ∼50% longer if performed at a lower “permissive” temperature of 31°C to 33°C. When these assays are performed in 12-well plates, plaques develop slightly faster, and they may be visualized ∼1 day earlier.
Virus passaging
Completion of the P1 passage normally takes 5 to 10 days for sufficient CPE (>85%) to develop. Because the titer increases with each passage, P2 usually develop sufficient CPE within 3 to 5 days. The process of freeze-thawing flasks 3 times to aid release of virus from infected cells normally takes a minimum of an additional day.
Virus purification
Growth of virus takes ∼65 hr. Processing to obtain a cesium chloride–purified virus band will take most of an additional day, and dialyzing the virus to remove the cesium chloride will take 1 to 2 additional days.
Acknowledgments
The author's work has been supported by funding from the Canadian Institutes of Health Research and by the National Science and Research Engineering Council of Canada. The author also thanks the many individuals who trained in his laboratory and contributed to manuscripts and procedures cited herein.
Author Contributions
Kevin M. Coombs : Conceptualization, formal analysis, funding acquisition, investigation, methodology, project administration, supervision, visualization, writing – original draft, writing – review & editing.
Conflict of Interest
The author declares no conflict of interest.
Open Research
Data Availability Statement
All data are contained herein
Literature Cited
- Breun, L. A., Broering, T. J., McCutcheon, A. M., Harrison, S. J., Luongo, C. L., & Nibert, M. L. (2001). Mammalian reovirus L2 gene and Lambda2 core spike protein sequences and whole-genome comparisons of reoviruses type 1 Lang, type 2 Jones, and type 3 Dearing. Virology , 287, 333–348. https://doi.org/10.1006/viro.2001.1052
- Bussiere, L. D., Choudhury, P., Bellaire, B., & Miller, C. L. (2017). Characterization of a replicating mammalian orthoreovirus with tetracysteine-tagged µNS for live-cell visualization of viral factories. Journal of Virology , 91, e01371–17. https://doi.org/10.1128/JVI.01371-17
- Coombs, K. M. (1998a). Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology , 243, 218–228. https://doi.org/10.1006/viro.1998.9061
- Coombs, K. M. (1998b). Temperature-sensitive mutants of reovirus. Current Topics in Microbiology and Immunology , 233, 69–107. https://doi.org/10.1007/978-3-642-72092-5_4
- Current Protocols. (2006). Commonly Used Reagents. Current Protocols in Microbiology , 00, A.2A.1–A.2A.15. https://doi.org/10.1002/9780471729259.mca02as00
- Dermody, T. S., Parker, J. S. L., & Sherry, B. (2013). Orthoreoviruses. In D. M. Knipe & P. M. Howley (Eds.), Fields virology (pp. 1304–1346). Lippincott, Williams & Wilkins.
- Dryden, K. A., Coombs, K. M., & Yeager, M. (2008). The structure of orthoreoviruses. In J. T. Patton (Ed.), Segmented double-stranded RNA viruses: Structure and molecular biology (pp. 3–25), Horizon Press.
- Glover, K. K. M., Sutherland, D. M., Dermody, T. S., & Coombs, K. M. (2021). A single point mutation, Asn16 – Lys, dictates the temperature-sensitivity of the reovirus tsG453 mutant. Viruses , 13, 289. https://doi.org/10.3390/v13020289
- Hermann, L., Embree, J., Hazelton, P., Wells, B., & Coombs, K. (2004). Reovirus type 2 isolated from cerebrospinal fluid. The Pediatric Infectious Disease Journal , 23, 373–375. https://doi.org/10.1097/00006454-200404000-00026
- Johansson, P. J., Sveger, T., Ahlfors, K., Ekstrand, J., & Svensson, L. (1996). Reovirus type 1 associated with meningitis. Scandinavian Journal of Infectious Diseases , 28(2), 117–120. https://doi.org/10.3109/00365549609049060
- Kobayashi, T., Antar, A. A., Boehme, K. W., Danthi, P., Eby, E. A., Guglielmi, K. M., Holm, G. H., Johnson, E. M., Maginnis, M. S., Naik, S., Skelton, W. B., Wetzel, J. D., Wilson, G. J., Chappell, J. D., & Dermody, T. S. (2007). A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host & Microbe, 1, 147–157. https://doi.org/10.1016/j.chom.2007.03.003
- Kobayashi, T., Ooms, L. S., Ikizler, M., Chappell, J. D., & Dermody, T. S. (2010). An improved reverse genetics system for mammalian orthoreoviruses. Virology , 398, 194–200. https://doi.org/10.1016/j.virol.2009.11.037
- Mohamed, A., Clements, D. R., Gujar, S. A., Lee, P. W., Smiley, J. R., & Shmulevitz, M. (2020). Single amino acid differences between closely related reovirus T3D lab strains alter oncolytic potency in vitro and in vivo. Journal of Virology , 94, e01688–19. https://doi.org/10.1128/JVI.01688-19
- Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory.
- Sandekian, V., & Lemay, G. (2015). A single amino acid substitution in the mRNA capping enzyme λ2 of mammalian orthoreovirus increases interferon sensitivity. Virology , 483, 229–235. https://doi.org/10.1016/j.virol.2015.04.020
- Smith, R. E., Zweerink, H. J., & Joklik, W. K. (1969). Polypeptide components of virions, top component and cores of reovirus type 3. Virology , 39, 791–810. https://doi.org/10.1016/0042-6822(69)90017-8
- Starnes, M. C., & Joklik, W. K. (1993). Reovirus protein lambda 3 is a poly©-dependent poly(G) polymerase. Virology , 193, 356–366. https://doi.org/10.1006/viro.1993.1132
- Stevenson, B. (2006). Common Bacterial Culture Techniques and Media. Current Protocols in Microbiology , 00, A.4A.1–A.4A.8. https://doi.org/10.1002/9780471729259.mca04as00
- Tyler, K. L., Barton, E. S., Ibach, M. L., Robinson, C., Campbell, J. A., O'Donnell, S. M., Valyi-Nagy, T., Clarke, P., Wetzel, J. D., & Dermody, T. S. (2004). Isolation and molecular characterization of a novel type 3 reovirus from a child with meningitis. Journal of Infectious Diseases , 189, 1664–1675. https://doi.org/10.1086/383129
- Wiener, J. R., Bartlett, J. A., & Joklik, W. K. (1989). The sequences of reovirus serotype 3 genome segments M1 and M3 encoding the minor protein mu 2 and the major nonstructural protein mu NS, respectively. Virology , 169, 293–304. https://doi.org/10.1016/0042-6822(89)90154-2
- Yin, P., Keirstead, N. D., Broering, T. J., Arnold, M. M., Parker, J. S. L., Nibert, M. L., & Coombs, K. M. (2004). Comparisons of theM1 genome segments and encoded µ2 proteins of different reovirus isolates. Virology Journal , 1, 6. https://doi.org/10.1186/1743-422X-1-6
- Zhang, X., Walker, S. B., Chipman, P. R., Nibert, M. L., & Baker, T. S. (2004). Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 angstrom. Nature Structural & Molecular Biology, 10, 1011–1018. https://doi.org/10.1038/nsb1009

