Learned Embeddings from Deep Learning to Visualize and Predict Protein Sets
Christian Dallago, Christian Dallago, Burkhard Rost, Burkhard Rost, Konstantin Schütze, Konstantin Schütze, Michael Heinzinger, Michael Heinzinger, Tobias Olenyi, Tobias Olenyi, Maria Littmann, Maria Littmann, Amy X. Lu, Amy X. Lu, Kevin K. Yang, Kevin K. Yang, Seonwoo Min, Seonwoo Min, Sungroh Yoon, Sungroh Yoon, James T. Morton, James T. Morton
deep learning embeddings
machine learning
protein annotation pipeline
protein representations
protein visualization
Abstract
Models from machine learning (ML) or artificial intelligence (AI) increasingly assist in guiding experimental design and decision making in molecular biology and medicine. Recently, Language Models (LMs) have been adapted from Natural Language Processing (NLP) to encode the implicit language written in protein sequences. Protein LMs show enormous potential in generating descriptive representations (embeddings) for proteins from just their sequences, in a fraction of the time with respect to previous approaches, yet with comparable or improved predictive ability. Researchers have trained a variety of protein LMs that are likely to illuminate different angles of the protein language. By leveraging the bio_embeddings pipeline and modules, simple and reproducible workflows can be laid out to generate protein embeddings and rich visualizations. Embeddings can then be leveraged as input features through machine learning libraries to develop methods predicting particular aspects of protein function and structure. Beyond the workflows included here, embeddings have been leveraged as proxies to traditional homology-based inference and even to align similar protein sequences. A wealth of possibilities remain for researchers to harness through the tools provided in the following protocols. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
The following protocols are included in this manuscript:
Basic Protocol 1 : Generic use of the bio_embeddings pipeline to plot protein sequences and annotations
Basic Protocol 2 : Generate embeddings from protein sequences using the bio_embeddings pipeline
Basic Protocol 3 : Overlay sequence annotations onto a protein space visualization
Basic Protocol 4 : Train a machine learning classifier on protein embeddings
Alternate Protocol 1 : Generate 3D instead of 2D visualizations
Alternate Protocol 2 : Visualize protein solubility instead of protein subcellular localization
Support Protocol : Join embedding generation and sequence space visualization in a pipeline
INTRODUCTION
Protein sequences correspond to strings of characters, each representing an amino acid (referred to as residues when joined in a protein). While protein savants extrapolate a wealth of information from this representation, for machines this is as meaningless as any other text document. Finding meaningful, computable representations from protein sequences by converting text into vectors of numbers representing relevant features or descriptors of proteins is an important first step to find out properties of the protein with that sequence, e.g., what other proteins it resembles (sequence comparisons through alignments), what it looks like (membrane or water-soluble, regular globular or disordered), or what it does (enzyme or not, process involved in, molecular function, interaction partners).
Many approaches to generate knowledge and meaning from protein sequences have been proposed. Intuitive representations relied on what experts considered informative, e.g., converting sequences into numerical vectors representing polarity or hydrophobicity. More advanced ideas included substitution matrices (Henikoff & Henikoff, 1992), profiles of protein families (Stormo, Schneider, Gold, & Ehrenfeucht, 1982), and “evolutionary couplings” from events correlating the mutability at two or more residues (Morcos et al., 2011). Combining “evolutionary information” (Rost & Sander, 1993), along with global (entire protein) and local (only sequence fragment) features through machine learning (ML; Rost & Sander, 1993, 1994), led to the first breakthrough in protein structure prediction over two decades ago (Moult, Pedersen, Judson, & Fidelis, 1995; Rost & Sander, 1995). Combining more sophisticated tools from Artificial Intelligence (AI) to include even more protein evolutionary information have led to the most recent breakthrough by AlphaFold2 from DeepMind (Callaway, 2020).
Representations based on evolutionary information have improved remote homology detection (Steinegger et al., 2019) as well as the prediction of aspects of protein structure (Hopf et al., 2012; Rost, 1996) and protein function (Goldberg et al., 2014; Hopf et al., 2017). The amount of evolutionary information contained in these representations is proportional to the size and diversity of a protein family (Ovchinnikov et al., 2017; Rost, 2001); the generation of families relies on parameter-sensitive multiple sequence alignments (MSAs) that, due to growing databases, become increasingly computationally expensive, despite immense advances in method development (Steinegger & Söding, 2018; Steinegger, Mirdita, & Söding, 2019).
Deep learning−based Language Models (LMs) are a new class of machine learning devices learning the rules for semantics and syntax directly and autonomously from the statistics of text corpora. Modern LMs learn to represent language by being conditioned on either predicting the next word in a sentence given previous context, or by reconstructing corrupted text. In protein bioinformatics, these devices are trained on large sequence datasets, such as UniProt (The UniProt Consortium, 2019), through a process called “self-supervision”. LM representations (embeddings) have been used as input to other methods (a process referred to as transfer learning) to predict aspects of protein structure and function. Although embedding-based predictions tend to be less accurate than those using evolutionary information, they require less time (Heinzinger et al., 2019; Rao et al., 2019; Rives et al., 2019). By learning to represent sequence and background information, embeddings open the door to a completely new way of using protein sequences, successful enough to even compete with traditional remote homology detection and structural alignments (Biasini et al., 2014; Littmann, Heinzinger, Dallago, Olenyi, & Rost, 2021; Morton et al., 2020; Villegas-Morcillo et al., 2020).
Although embeddings derived from sequences contain substantially more information than raw sequences, one challenge for this new representation is to simplify its availability. This is one crucial objective of the bio_embeddings software resource, which collects tools to create and use protein embeddings. Basic Protocol 1 serves as a high-level overview of functionalities of the bio_embeddings pipeline. Basic Protocol 2 adds in-depth context for embeddings and details steps on how to extract embeddings from sequences. Through Basic Protocol 3 (and variations thereof in Alternate Protocols 1 and 2), embeddings are leveraged to plot sequence sets in combination with aspects of protein function, namely subcellular location and membrane-boundness. Finally, in Basic Protocol 4, the rich protein representations from a protein LM are used as input features to train a machine learning device to predict protein subcellular localization.
Basic Protocol 1: GENERIC USE OF THE bio_embeddings PIPELINE TO PLOT PROTEIN SEQUENCES AND ANNOTATIONS
This protocol serves as non-technical overview of what is available out-of-the-box through the bio_embeddings pipeline. The premise is simple: you will use software to plot protein sequences and color them by a property. For this purpose, we prepared three files for download: one containing about 100 protein sequences in FASTA format, a CSV file containing DisProt (Hatos et al., 2020) classifications for these sequences (whether their 3D structure presents mostly disorder or little disorder), and a configuration file that specifies parameters for the computation. Apart from downloading these files and the steps necessary to install the bio_embeddings software, executing the computation is a single step. The following basic protocols present greater detail about the technical aspects surrounding inputs, outputs and parameters of the pipeline.
The output obtained by us when executing this protocol is available for comparison at http://data.bioembeddings.com/disprot/disprot_sampled; the plot file resulting from executing the steps is available at http://data.bioembeddings.com/disprot/disprot_sampled/plotly_visualization/plot_file.html.
NOTE : This visualization is produced for a small sample of DisProt sequences; as such it is by no means representative of the power of the embeddings in distinguishing DisProt classes.
Materials
Hardware
- A modern computer (newer than 2012), with about 8 GB of available RAM, 2 GB of available disk space, and an Internet connection.
Software
- Windows users may need to install Windows Subsystem for Linux (https://docs.microsoft.com/en-us/windows/wsl). All users should have Python 3.7 or 3.8 installed (https://www.python.org/downloads).
Data
- You will need a FASTA sequence for some proteins in DisProt, which can be downloaded from http://data.bioembeddings.com/disprot/sequences.fasta; you will need annotations of disorder content, which can be downloaded from http://data.bioembeddings.com/disprot/disprot_annotations.csv; finally, you need a configuration file for the bio_embeddings pipeline, which can be downloaded from http://data.bioembeddings.com/disprot/config.yml.
1.Ensure that all software and hardware requirements are met (see Materials, above).
Install Python 3.7 or 3.8 on your system using https://www.python.org/downloads.
2.Download required files.
Through your browser, navigate to http://data.bioembeddings.com/disprot and download the files: sequences.fasta, config.yml, and disprot_annotations.csv.
If you prefer to use the terminal, run the following three commands:
wget http://data.bioembeddings.com/disprot/sequences.fasta
wget http://data.bioembeddings.com/disprot/config.yml
wget http://data.bioembeddings.com/disprot/disprot_annotations.csv
3.Create a project directory and move files into it.
Create a new directory called disprot on your computer and move the files downloaded in step 2 into this directory.
4.Open a new terminal window.
To open a terminal on MaxOS or Linux, search for the application “Terminal” and open it. On Windows, after having installed the Windows Subsystem for Linux (https://docs.microsoft.com/en-us/windows/wsl), search for and open the application called “bash” through the start menu.
5.Install bio_embeddings.
To install the pipeline and all of its dependencies, open a terminal window and type in the command:
pip install ---user "bio-embeddings[all]"
6.Navigate to the project directory from the terminal window.
If you called your project directory disprot inside the Downloads folder, the command to navigate to the directory through the MacOS and Linux Terminal is:
cd ∼/Downloads/disprot
7.Run the bio_embeddings pipeline.
To start running the bio_embeddings pipeline, type the following in your terminal window:
bio_embeddings config.yml
Then, press Enter.
This will start a job using parameters defined in the text configuration file (config.yml; detail about the parameters in the next protocols). Opening the file with a text editor will display the following content:
global:
sequences_file: sequences.fasta
prefix: disprot_sampled
protbert_embeddings:
type: embed
protocol: prottrans_bert_bfd
reduce: true
discard_per_amino_acid_embeddings: true
umap_projections:
type: project
protocol: umap
depends_on: protbert_embeddings
n_components: 2
plotly_visualization:
type: visualize
protocol: plotly
annotation_file: disprot_annotations.csv
display_unknown: false
depends_on: umap_projections
You should see output that resembles:
2020-11-09 20:37:13,753 INFO Created the prefix directory disprot_sampled
2020-11-09 20:37:13,756 INFO Created the file
disprot_sampled/input_parameters_file.yml
2020-11-09 20:37:13,970 INFO Created the file disprot_sampled/sequences_file.fasta
2020-11-09 20:37:14,118 INFO Created the file disprot_sampled/mapping_file.csv
…
8.Open the plot file.
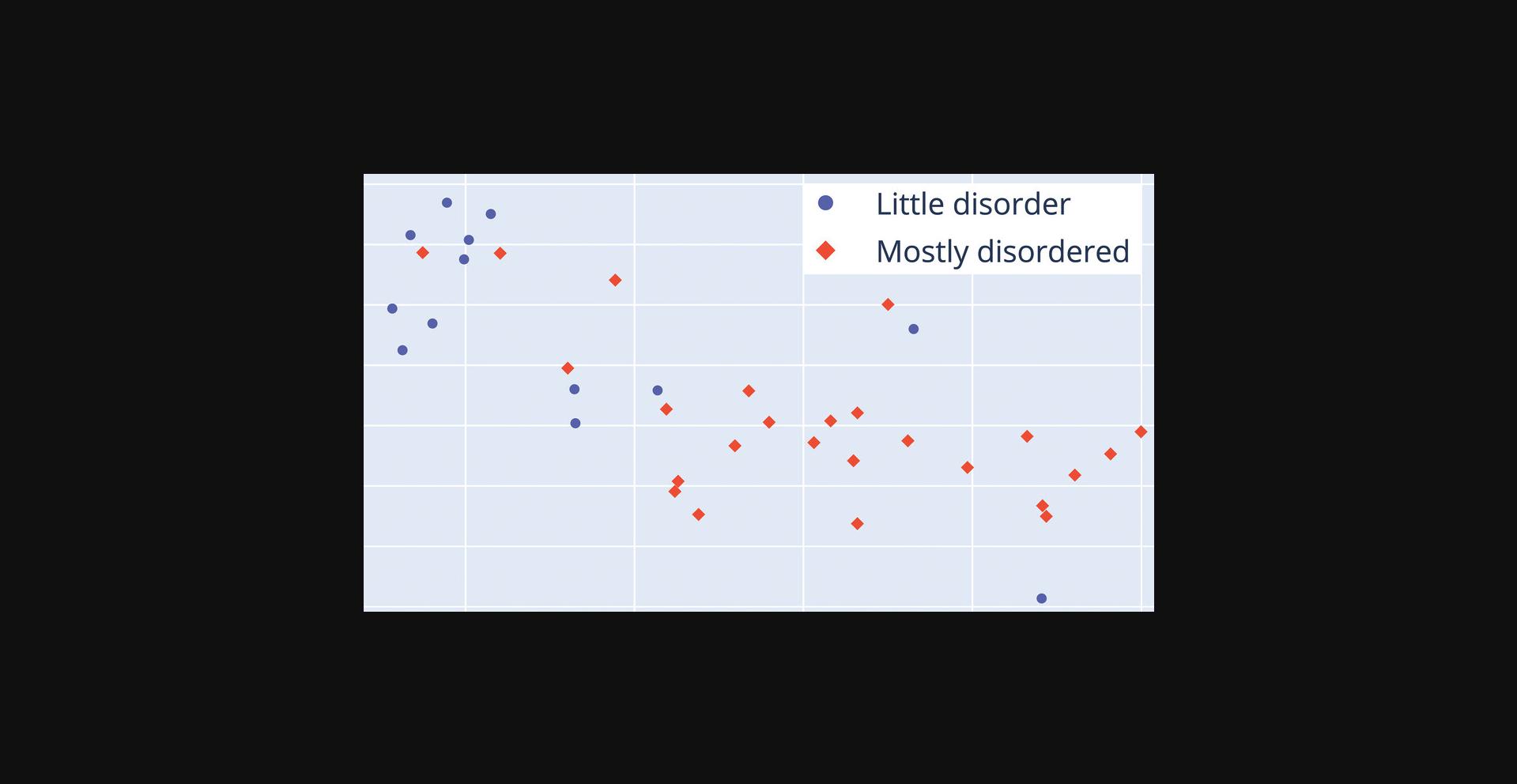
Basic Protocol 2: GENERATE EMBEDDINGS FROM PROTEIN SEQUENCES USING THE bio_embeddings PIPELINE
Through this protocol, you may generate machine-readable representations (embeddings) from a set of protein sequences using the “embed” stage of the bio_embeddings pipeline. The sequence file utilized for the example was written by the prediction program DeepLoc (Almagro Armenteros, Sønderby, Sønderby, Nielsen, & Winther, 2017), but you can also provide your own FASTA file. Embeddings constitute an abstract encoding of the information contained in protein sequences, and are the building block of the pipeline and its analytical tools. In this protocol, we use BERT (Devlin, Chang, Lee, & Toutanova, 2019) trained on BFD (Steinegger & Söding, 2018; Steinegger et al., 2019) to extract embeddings from protein sequences. This model is part of the ProtTrans protein LMs (Elnaggar et al., 2020), referred to as ProtBERT in text or prottrans_bert_bfd in the following code. You can find out how to choose a protein LM based on your requirements on our website (http://bioembeddings.com). The salient output of the embed stage are the embedding files. These come in two flavors: per-residue (embeddings_file.h5) and per-protein (reduced_embeddings.h5). While the per-residue embeddings are taken directly out of the LMs, per-protein embeddings are generated post-processing the information extracted by the LM through global average pooling (Shen et al., 2018) on all combined per-residue embeddings of a sequence. Per-residue embeddings are useful to analyze properties of residues in a protein (e.g., which residues bind ligands), while per-protein representations capture annotations describing entire proteins (e.g., native localization).
Materials
Hardware
-
Computer (newer than 2012), >8 GB of available RAM, ∼2 GB of available disk space
-
Optional : Graphical Processing Unit (GPU) with >4 GB of vRAM and supporting CUDA® 11.0
-
This will speed up the embedding process manyfold
-
Internet connection
Software (MacOS and Linux)
-
Python 3.7 or 3.8 (https://www.python.org/downloads)
-
Windows users : Windows Subsystem for Linux (https://docs.microsoft.com/en-us/windows/wsl)
-
Optional : CUDA® (https://developer.nvidia.com/cuda-downloads; at time of writing: version 11.1)
Data
-
DeepLoc (Almagro Armenteros et al., 2017): http://data.bioembeddings.com/deeploc/deeploc_data.fasta
-
DeepLoc (reduced sample) FASTA-formatted sequences: http://data.bioembeddings.com/deeploc/sampled_deeploc_data.fasta
-
NOTE : as input, we begin with two files containing protein sequences in a simplified FASTA format (first line begins with “>” followed by protein name, all subsequent lines contain the sequence in single-letter amino acid code).
1.Install bio_embeddings from pip.
To install the pipeline and all of its dependencies, open a terminal window and type in the command:
pip install --user "bio-embeddings[all]"
2.Create a project directory.
We suggest you create a new project directory on your disk. You can generate it through the terminal:
mkdir deeploc
Then, open the directory through the terminal:
cd deeploc
3.Download the DeepLoc FASTA file inside the project directory.
From the terminal (within the project directory):
wget http://data.bioembeddings.com/deeploc/deeploc_data.fasta
Alternatively, download the file using your browser, and move it to the project directory.
4.Create a configuration file.
A configuration file defines what the pipeline should do (files and parameters it should use and stages it should run). Many examples of configuration files are provided at http://examples.bioembeddings.com, including the one you will create here (called deeploc). To create the configuration file from the terminal:
nano config.yml
Then, type in the following and save the file (to save: press Ctrl+x, then “y”, then the Return key):
global:
sequences_file: deeploc_data.fasta
prefix: deeploc_embeddings
simple_remapping: True
prottrans_bert_embeddings:
type: embed
protocol: prottrans_bert_bfd
reduce: True
5.Run the bio_embeddings pipeline.
All that is left to do is to supply the configuration file to bio_embeddings and let the pipeline execute the job. To do so, type on the terminal:
bio_embeddings config.yml
You should see output that resembles:
2020-11-09 20:37:13,753 INFO Created the prefix directory deeploc_embeddings
2020-11-09 20:37:13,756 INFO Created the file deeploc_embeddings/input_parameters_file.yml
2020-11-09 20:37:13,970 INFO Created the file deeploc_embeddings/sequences_file.fasta
2020-11-09 20:37:14,118 INFO Created the file deeploc_embeddings/mapping_file.csv
…
6.Locate the embedding files.
Basic Protocol 3: OVERLAY SEQUENCE ANNOTATIONS ON A PROTEIN SPACE VISUALIZATION
The previous protocol generated embeddings from protein sequences in your dataset (here DeepLoc dataset). In Basic Protocol 3 you use functions from the bio_embeddings package to visualize “protein spaces” spanned by the embeddings extracted. These visualizations reveal whether or not the LM chosen for the “embed” stage (Basic Protocol 2) can roughly separate your data based on a desired property/phenotype. The property/phenotype in our example is subcellular location in 10 states. Alternate Protocol 2 uses the same data and similar steps to visualize protein solubility. While visualizations are useful, the discriminative power of embeddings can be boosted many times by training machine learning models on the embeddings to predict the desired property (Basic Protocol 4).
Between embedding generation and protein space visualization , another step has to be inserted. In the pipeline, we refer to this step as a “project” stage. Its purpose is to reduce the dimensionality of the embeddings (e.g., 1024 for ProtBERT) such that it can be visualized in either 2D or 3D. Here, we project embeddings onto 2D; Alternate Protocol 1 uses the same data and slight variations in parameters to 3D plots instead.
The final notebook constructed here is available at http://notebooks.bioembeddings.com as deeploc_visualizations.ipynb to be downloaded and executed locally, or executed directly online. The file also includes steps presented in Alternate Protocols 1 and 2.
The Support Protocol 1 explains how to integrate the final visualization options in a configuration file as instruction for the pipeline to manage the entire process—from sequences to visualizations. This is useful to enable colleagues to reproduce all your results from a few files.
Materials
Software
-
Jupyter Notebook (Kluyver et al., 2016)
-
Notebooks can be run locally, provided that the necessary dependencies are installed (python 3.7 and the Jupyter suite). Installation steps are described here : https://jupyter.org/install.
-
_Notebooks can be run on Google Colaboratory (_Bisong , 2019 ), without having to install software locally, given an internet connection and a Google account.
Data
-
DeepLoc embeddings input files, which you either calculated through Basic Protocol 2 or you can be download from http://data.bioembeddings.com/deeploc/reduced_embeddings_file.h5
-
Annotations of properties/phenotypes of the proteins; for DeepLoc, subcellular location annotations can be downloaded from http://data.bioembeddings.com/deeploc/annotations.csv
1.Create new Jupyter Notebook on Google Colaboratory (a) or locally (b).
-
We suggest running the following through Google Colaboratory. To open a new Google Colaboratory, navigate to:https://colab.research.google.com/\#create=true.
-
If you prefer to execute the steps on your local computer, through the terminal, navigate to thedeeplocfolder created previously, or to a new folder. Then, start a Jupyter notebook through the terminal:
jupyter-notebook
This should open a browser window. From the top-right drop-down menu called “new”, select “Python 3”.
2.Install bio_embeddings
- On Google Colaboratory paste in the following code in the first code block:
!pip3 install -U pip
!pip3 install -U "bio-embeddings[all]"
Then, press the play button on the left of the code cell. Given some version differences in Google Colaboratory, warnings may arise. These, however, can be ignored.
- If you already executed Protocol 1, you are set. Otherwise, open a new terminal window and type:
pip install --user "bio-embeddings[all]"
3.Download files.
- On Google Colaboratory, create a new code block (by pressing the “+ code” button). Then, paste in the following code:
!wget http://data.bioembeddings.com/deeploc/reduced_embeddings_file.h5
!wget http://data.bioembeddings.com/deeploc/annotations.csv
-
On your local computer, simply download the files listed in the Materials list for this protocol and move them into the folder in which the notebook was started (see step 1).
4.Import dependencies.
From here on, the execution steps are identical on Google Colaboratory and your local Jupyter notebook. You will now import the functions that allow you to open embedding files, reduce the dimensionality, and visualize scatter plots. To do so, in a new code block, type and execute the following:
import h5py
import numpy as np
from pandas import read_csv, DataFrame
from bio_embeddings.utilities import QueryEmbeddingsFile
from bio_embeddings.project import umap_reduce
from bio_embeddings.visualize import render_scatter_plotly
5.Read annotations file.
Assume that the original FASTA file, for which you generated embeddings, was the following:
>Q9H400-2
SEQVENCE
>P12962
SEQVVNCE
>P12686
MNQVENCE
You can define a set of annotations for the sequences in this set as a CSV file, containing minimally two columns called “identifier” and “label” such as:
identifier,label
Q9H400-2,Cell membrane
P12962,Cytoplasm
P12686,Mitochondrion
The identifiers have to match to the identifiers in the FASTA header of the protein sequences for which embeddings have been computed. They can, however, only contain a subset of identifiers with respect to the embeddings.
You can now load the annotations.csv file which we have created based on the DeepLoc data. These annotations contain experimentally validated subcellular location in 10 classes. To load them into the notebook, execute the following in a new code block:
annotations = read_csv(`annotations.csv')
6.Read the embeddings file.
In a new code block, type and execute the following:
identifiers = annotations.identifier.values
embeddings = list()
with h5py.File(`reduced_embeddings_file.h5', `r') as embeddings_file:
embedding_querier = QueryEmbeddingsFile(embeddings_file)
for identifier in identifiers:
embeddings.append(embedding_querier.query_original_id(identifier))
7.Project embeddings to 2D using UMAP (McInnes, Healy, & Melville, 2018).
In a new code block, type and execute the following:
options = {
`min_dist': .1,
`spread': 8,
`n_neighbors': 160,
`metric': `euclidean',
`n_components': 2,
`random_state': 10
}
projected_embeddings = umap_reduce(embeddings, **options)
8.Merge projected embeddings and annotations.
In a new code block, type and execute the following:
projected_embeddings_dataframe = DataFrame(
projected_embeddings,
columns=["component_0", "component_1"],
index=identifiers
)
merged_annotations_and_projected_embeddings = annotations.join(
projected_embeddings_dataframe, on="identifier", how="left"
)
Here, you create a DataFrame (similar to a table) from the projected embeddings. Rows are indexed by the “identifiers”, while the two columns contain the two components of te projected embeddings. In other words: you are constructing a table of coordinates for your protein sequences. Lastly, you merge these coordinates with the annotations. You can inspect the first five rows of the dataframe by typing the following into a new code block and executing it:
merged_annotations_and_projected_embeddings[:5]
| Identifier | Label | Component_0 | Component_1 |
|---|---|---|---|
| Q9H400 | Cell.membrane | 2.474637 | –8.919042 |
| Q5I0E9 | Cell.membrane | 32.507015 | 10.355012 |
| P63033 | Cell.membrane | 18.500378 | –0.299981 |
| Q9NR71 | Cell.membrane | 2.420154 | 18.161064 |
| Q86XT9 | Cell.membrane | –4.937888 | –1.767011 |
9.Plot the protein space spanned by the projected embeddings
In a new code block, type and execute the following:
figure = render_scatter_plotly(merged_annotations_and_projected_embeddings)
figure.show()
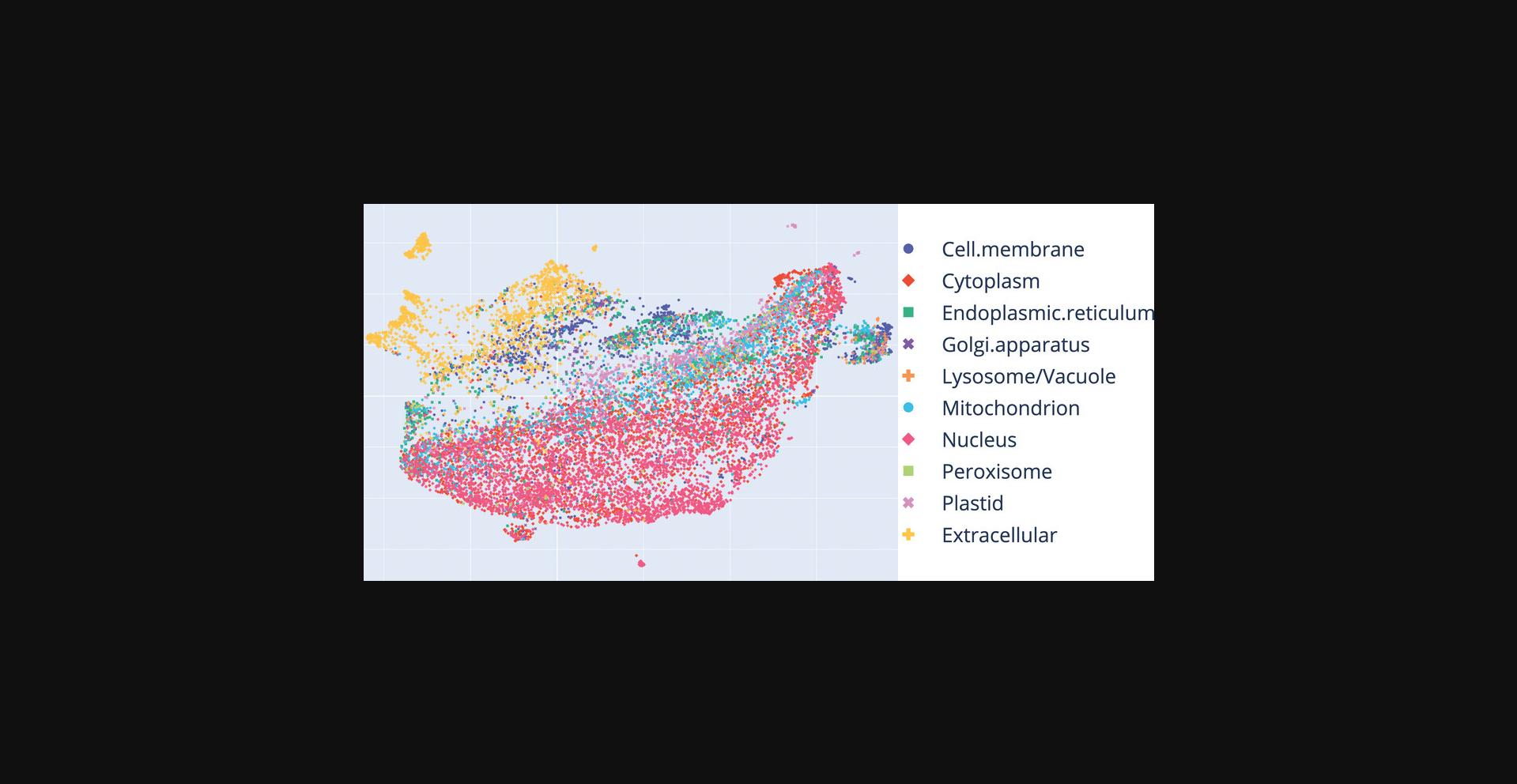
Basic Protocol 4: TRAIN A MACHINE LEARNING CLASSIFIER ON PROTEIN EMBEDDINGS
Basic Protocol 2 generated embeddings for proteins in DeepLoc (Almagro Armenteros et al., 2017). Basic Protocol 3 visualized the projected embeddings in a 2D plot and annotated the proteins in this 2D plot by colors signifying subcellular location. In the following steps, you will use the embeddings generated through the pipeline and the location annotations from DeepLoc to machine-learn the prediction of location from protein sequence embeddings. Once trained, you can apply this prediction method to annotate/predict location for any protein sequence. The simplest recipe to build a generic machine learning model is as follows:
- 1.Divide data into train and test sets (these should be sequence-non-redundant with respect to each other, i.e., no protein sequence in one should be more sequence-similar than some threshold to any protein in the other; what this threshold is depends on your task)
- 2.Split a subset from the train set to construct a validation set (non-redundant to split-off)
- 3.Evaluate some machine learning hyper-parameters using the validation set (e.g., which type of machine learning model—such as ANN, CNN, or SVM, what particular choice of parameters—such as number of hidden units/layers for ANN/CNN). Construct a leaderboard (i.e., a table keeping track of the relative performance of all the models/hyper-parameters).
- 4.Select the best model from the leaderboard, and evaluate on the test set (by NO MEANS apply all models to the test set and pick the best; instead, it is essential to choose the best using the validation set and to stick to that choice to avoid over-fitting).
- 5.Report performance for a diversity of relevant evaluation metrics for the final model using the test set (include estimates for standard errors)
The following steps explore this recipe using sci-kit learn (Pedregosa et al., 2011). You will produce a classifier which roughly separates the ten location classes from DeepLoc (Almagro Armenteros et al., 2017). The objective of this protocol is not to produce the best prediction method for subcellular location classification, which would require more parameter testing and tuning! Instead, the objective is to showcase the ease of going from data to prediction method when using embeddings. The final notebook constructed here is available at http://notebooks.bioembeddings.com as downloadable file called deeploc_machine_learning.ipynb.
Materials
- See Basic Protocol 3
1.Complete steps 1-5 of Basic Protocol 3.
2.Import additional dependencies.
Via a new code block, you will import a set of dependencies from the popular machine learning library scikit-learn (Pedregosa et al., 2011) in order to train and evaluate the machine learning model:
from sklearn.neural_network import MLPClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import accuracy_score
3.Split annotations into train and test sets
train_set = annotations[annotations.set == "train"]
test_set = annotations[annotations.set == "test"]
4.Load embeddings into train and test sets.
Once you have split the annotations into train and test sets, you need to create input and output for the machine learning model. The input will be the sequence embeddings (in the following, “training_embeddings”), while the output will be the subcellular location associated to those proteins (in the following, “training_labels”). In a new code block, type the following:
training_embeddings = list()
training_identifiers = train_set.identifier.values
training_labels = train_set.label.values
testing_embeddings = list()
testing_identifiers = test_set.identifier.values
testing_labels = test_set.label.values
with h5py.File(`reduced_embeddings_file.h5', `r') as embeddings_file:
embedding_querier = QueryEmbeddingsFile(embeddings_file)
for identifier in training_identifiers:
training_embeddings.append(embedding_querier.query_original_id(identifier))
for identifier in testing_identifiers:
testing_embeddings.append(embedding_querier.query_original_id(identifier))
5.Define basic machine learning architecture and parameters to optimize
In a new code block, type and execute the following:
multilayerperceptron = MLPClassifier(
solver=`lbfgs',
random_state=10,
max_iter=1000
)
parameters = {
`hidden_layer_sizes': [(30,), (20,15)]
}
6.Train classifiers and pick the best performing model.
In a new block of code, write and execute the following:
classifiers = GridSearchCV(
multilayerperceptron,
parameters, cv=3,
scoring="accuracy"
)
classifiers.fit(training_embeddings, training_labels)
classifier = classifiers.best_estimator_
7.Predict subcellular location for test set and calculate performance,
Lastly, to evaluate the performance of you final model, you predict the location for all proteins in the test set and calculate accuracy as follows:
predicted_testing_labels = classifier.predict(testing_embeddings)
accuracy = accuracy_score(
testing_labels,
predicted_testing_labels
)
print(f"Our model has an accuracy of {accuracy:.2}")
8.Optional : Embed a novel sequence and predict its subcellular location.
In this optional step, you generate the sequence embedding for an arbitrary sequence and use the classifier developed in the previous steps to predict its subcellular location. To do so, type and execute the following:
from bio_embeddings.embed import ProtTransBertBFDEmbedder
embedder = ProtTransBertBFDEmbedder()
sequence = "DDCGKLFSGCDTNADCCEGYVCRLWCKLDW"
per_residue_embedding = embedder.embed(sequence)
per_protein_embedding = embedder.reduce_per_protein(per_residue_embedding)
sequence_subcellular_prediction = classifier.predict([per_protein_embedding])[0]
print("The arbitrary sequence is predicted to be located in: "
f"{sequence_subcellular_prediction}")
Alternate Protocol 1: GENERATE 3D INSTEAD OF 2D VISUALIZATIONS
The following steps introduce minimal code changes with respect to the steps and code outlined in Basic Protocol 3 to visualize in 3D instead of 2D. We assume that the code from Basic Protocol 3 has been written in a Jupyter/Colab Notebook and highlight code changes in orange. Visit the docs at https://docs.bioembeddings.com to find out more about the functions of the bio_embeddings package.
The code from Basic Protocol 3 is available at http://notebooks.bioembeddings.com as downloadable file called deeploc_visualizations.ipynb. It includes the steps presented here in an alternate form.
Materials
- See Basic Protocol 3
1.Project embeddings onto 3D instead of onto 2D.
Take the code block written in Basic Protocol 3, step 7, and locate and change the line:
`n_components': 2
to:
`n_components':3
Then, re-run the code cell.
2.Import 3D scatter plot renderer instead of 2D.
Change the import of the visualization function from Basic Protocol 3, step 4, from:
from bio_embeddings.visualize import render_scatter_plotly
to:
from bio_embeddings.visualize importrender_3D_scatter_plotly
and execute the code block.
3.Add a third component to the projected embeddings DataFrame.
Change the number of components in the projected DataFrame defined in B.asic Protocol 3, step 8 from:
columns=["component_0", "component_1"],
to:
columns=["component_0", "component_1","component_2"],
and execute the code block.
4.Swap the plotting function with the 3D variant:
Lastly, swap out the plotting function name in the code block created in Basic Protocol 3, step 9, from:
figure = render_scatter_plotly(
merged_annotations_and_projected_embeddings
)
to:
figure =render_3D_scatter_plotly(
merged_annotations_and_projected_embeddings
)
and execute the code block.
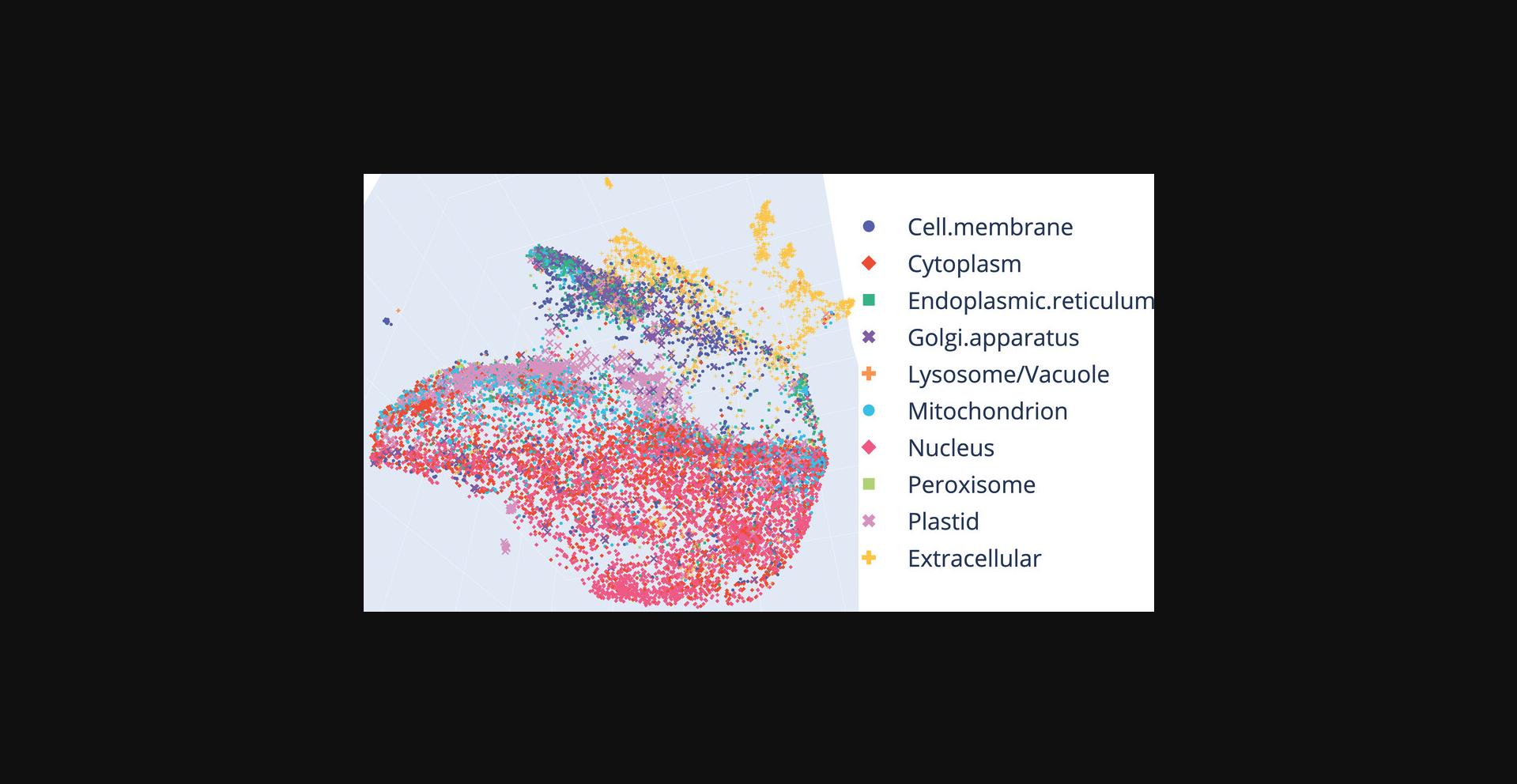
Alternate Protocol 2: VISUALIZE CLASSIFICATION INTO MEMBRANE/SOLUBLE INSTEAD OF PROTEIN SUBCELLULAR LOCATION
The following steps introduce minimal code changes with respect to the steps and code outlined in Basic Protocol 3 in order to visualize the classification into membrane/soluble proteins as annotated in DeepLoc (Almagro Armenteros et al., 2017) instead of location. We assume that the code from Basic Protocol 3 has been written up and highlights code changes in orange.
The code from Basic Protocol 3 is available at http://notebooks.bioembeddings.com as downloadable file called deeploc_visualizations.ipynb. It includes the steps presented here in an alternate form.
Materials
Software and Hardware
See Basic Protocol 3
Data
- DeepLoc solubility annotations: http://data.bioembeddings.com/deeploc/solubility_annotations.csv
1.Download additional file solubility_annotations.csv.
- On Google Colaboratory create a new code block (by pressing the “+ code” button). Then, paste in the following code:
!wget http://data.bioembeddings.com/deeploc/solubility_annotations.csv
-
On your local computer, simply download the file listed in the Materials list for this protocol and move into the folder in which the notebook was started (see Basic Protocol3, step 1).
2.Change the annotations file.
In the code block created in Basic Protocol 3, step 5, change the input file from:
annotations = read_csv(`annotations.csv')
to:
annotations = read_csv(`solubility_annotations.csv')
3.Re-run the subsequent code blocks.
Support Protocol: PUT EMBEDDING GENERATION AND SEQUENCE SPACE VISUALIZATIONS TOGETHER IN ONE PIPELINE
Basic Protocol 3 presents an explorative approach towards producing protein-space visualizations. In this Support Protocol, you will use the parameters chosen in Basic Protocol 3 to define a pipeline configuration file. These files allow reproducible workflows. You will do so by extending the bio_embeddings configuration presented in Basic Protocol 2, step 4, to also generate protein space visualizations. Noteworthy differences with previous files will be highlighted in orange.
Materials
Software and Hardware
See Basic Protocol 2
Data
-
DeepLoc FASTA file: http://data.bioembeddings.com/deeploc/deeploc_data.fasta
-
DeepLoc subcellular location annotations: http://data.bioembeddings.com/deeploc/annotations.csv
1.Execute steps 1 through 3 of Basic Protocol 2.
2.Download the annotations file into the project directory.
From the terminal (within the project folder):
wget http://data.bioembeddings.com/deeploc/annotations.csv
3.Define a configuration file to embed, project and visualize protein sequences.
Similarly to Basic Protocol 2, step 4, we define a text file (config.yml) that contains the following text:
global:
sequences_file: deeploc_data.fasta
prefix: deeploc_embeddings
simple_remapping: True
prottrans_bert_embeddings:
type: embed
protocol: prottrans_bert_bfd
reduce: True
discard_per_amino_acid_embeddings: True
umap_projections:
type: project
protocol: umap
depends_on: prottrans_bert_embeddings
min_dist: 0.1
spread: 8
n_neighbors: 160
metric: euclidean
n_components: 2
random_state: 10
plotly_visualization:
type: visualize
protocol: plotly
depends_on: umap_projections
annotation_file: annotations.csv
display_unknown: False
4.Run the bio_embeddings pipeline.
What remains is to supply the configuration file to bio_embeddings and let the pipeline execute the job. For that type into the terminal:
bio_embeddings-oconfig.yml
5.Locate the interactive figure file.
COMMENTARY
Background Information
Language Models (LMs) such as ELMo (Peters et al., 2018), BERT (Devlin et al., 2019), GPT-3 (Brown et al., 2020), and T5 (Raffel et al., 2020) improve over previous methods for learning to embed text (Bojanowski, Grave, Joulin, & Mikolov, 2017; Mikolov, Chen, Corrado, & Dean, 2013; Pennington, Socher, & Manning, 2014) by cleverly modeling context (“apple” company vs. fruit) and training on increasingly larger natural language corpora. They begin to suggest large models from artificial intelligence (AI) or machine learning (ML) to compete with human experts, at least for some tasks (Manning, 2011). They also help rising questions about current benchmarks (Heinzerling, 2020; McCoy, Pavlick, & Linzen, 2019) and the extent to which LMs truly understand language (Bender & Koller, 2020). Despite potential performance overestimates, LMs succeed to effectively translate natural language besting expert-based models, i.e., they captured the meaning in text automatically (Pires, Schlinger, & Garrette, 2019; Zhu et al., 2020).
Training LMs requires very large amounts of intrinsically structured, sequential data, making these approaches especially promising for ambitious attempts that try to automatically understand the language of life proxied by protein sequences (Heinzinger et al., 2019). In fact, the amount of data available for protein sequences is 500 times larger than the largest NLP data sets such as Google's Billion Word data (Chelba et al., 2014; Steinegger & Söding, 2018; Steinegger et al., 2019). With the increasing degree to which the speed of adding new protein sequences outpaces the improvement in computer hardware, experimental annotations—although also increasing exponentially—cannot keep track with this explosion. Therefore, the sequence-annotation gap, i.e., the gap between the number of proteins with known sequence and those with known annotation, continues to rise.
In analogy to natural languages, protein sequences are formed by tokens (proteins: amino acids, text: words) that have individual and context-dependent meaning through long- and short-range dependencies (proteins: inter-residue bonds, text: sentences). Thus, similarly to natural language, LMs trained on protein sequences (Alley, Khimulya, Biswas, AlQuraishi, & Church, 2019; AlQuraishi, 2019; Armenteros, Johansen, Winther, & Nielsen, 2020; Elnaggar et al., 2020; Heinzinger et al., 2019; Lu, Zhang, Ghassemi, & Moses, 2020; Madani et al., 2020; Min, Park, Kim, Choi, & Yoon, 2020; Rao et al., 2019; Rives et al., 2019) capture important meaning of the protein sequence language, as demonstrated by their ability to predict aspects of protein structure and function. For instance, SeqVec (Heinzinger et al., 2019) trained ELMo (Peters et al., 2018) on UniRef50 (The UniProt Consortium, 2019) and showed that the LM's representations clustered protein sequences by function (Heinzinger et al., 2019). In another analogy to NLP, protein LMs may be fine-tuned on specialized sequence sets (analogy to natural language: legal text vs. wikipedia articles) to encode for different protein properties (Armenteros et al., 2020).
Previously, machine-learning methods in computational biology leveraged data-driven protein representations such as substitution matrices, capturing biophysical features (Henikoff & Henikoff, 1992), family-specific profiles (Stormo et al., 1982), or evolutionary couplings (Morcos et al., 2011) that capture evolutionary features. Now, embeddings provide competitive results for many prediction tasks (Littmann et al., 2021; Rao et al., 2019, 2020). Protein LMs may even be combined with other representations to gain even better performance (Rives et al., 2019; Villegas-Morcillo et al., 2020). Protein sequence embeddings are generated in a fraction of the time it takes to generate MSAs (Heinzinger et al., 2019), and can thus be used on entire proteomes, where MSA-based approaches might be computationally prohibitive or even unavailable (e.g., small protein families).
The bio_embeddings pipeline, which is used throughout the manuscript to generate and leverage protein embeddings, is targeted to computational biologists and aims to abstract, via a uniform and standardized interface, the use of protein LMs. Embeddings can be used to train machine learning algorithms using “transfer learning” (Basic Protocol 4; Raina, Battle, Lee, Packer, & Ng, 2007), or for analytical purposes. The pipeline enables visual analysis of sequence sets by drawing protein spaces spawned by their embeddings (Basic Protocol 3). Users can create representations from a growing diversity of protein LMs, which at the time of writing include: SeqVec (Heinzinger et al., 2019), UniRep (Alley et al., 2019), ESM (Rives et al., 2019), ProtBERT, ProtALBERT, ProtXLNet, ProtT5 (Elnaggar et al., 2020), CPCProt (Lu et al., 2020), PLUS-RNN (Min et al., 2020). Via the “ extract ” stage, the pipeline incorporates supervised and unsupervised approaches for protein embeddings to further enhance analytical potential out-of-the-box. For instance, users can extract secondary structure in 3- and 8-states for embeddings from SeqVec (Heinzinger et al., 2019) and ProtBert (Elnaggar et al., 2020), or transfer GO annotations using embeddings of any available LM (Littmann et al., 2021). Pipeline runs are reproducible, as configurations are defined through files, and the output is stored in easily exchangeable formats, e.g., CSVs, FASTA, and HDF5 (The HDF Group, 2000).
For researchers contributing new protein LMs, bio_embeddings can provide a unified interface to distribute their work to the community, requiring minimal changes for pipeline consumers to make use of new protein LMs. For researchers contributing downstream uses of protein LMs [e.g., for the visualization of attention maps (Vig et al., 2020), which are most closely related to protein contact maps, or for the alignment of protein sequences (Morton et al., 2020)], the bio_embeddings pipeline provides a flexible approach to incorporate their work and directly extends it to all the LMs supported by bio_embeddings. In the future, as we expect more protein LMs to be developed, the bio_embeddings pipeline could be combined with the TAPE (Rao et al., 2019) evaluation system to provide an intuition for protein LM researchers about the best use of their new representations.
Critical Parameters
We strongly encourage users interested in generating their own sequence embeddings to do so on GPU-equipped machines, where the GPUs have at least 4 GB of vRAM and support CUDA® 11.0. While it is possible to generate embeddings via CPU computing, the slowdown with respect to GPU computing is significant and prohibitive for large sequence sets.
Differences in LM choice, sequence sets or parameters (e.g., UMAP) may lead to significantly different results than discussed in the protocols. While trying out the above steps on your own datasets is the ultimate goal, we encourage users to first try to execute the steps as laid out above to get a sense of the baseline behavior.
Troubleshooting
If you experience issues when installing the bio_embedding package, or when executing the steps laid out above, you may want to try to restart the Google Colab, or, if you are running the code locally, create a new python environment [e.g., by using Anaconda (“Anaconda Software Distribution,” 2020)]. In our experience, the most common issues are caused by installation problems, or limited computational resources. To address the former, you might want to consider using docker instead of python (this is available at the source code, see “Internet Resources”). To address the latter, you might want to discuss solutions with your local research computing facilities or try an online service (see “Internet Resources”).
Understanding Results
Basic Protocol 1
Through the steps outlined in this protocol, you generated an interactive plot of about 100 protein sequences with annotations of disorder content (either presenting high or low disorder content).
Basic Protocol 2
Through the steps outlined in this protocol, you generated embeddings for amino acids in sequences (embeddings_file.h5) and for sequences (reduced_embeddings_file.h5) from the DeepLoc sequence set. These files can be used on per-residue tasks (e.g., predict secondary structure) or per-protein tasks (e.g., predict subcellular location).
Basic Protocol 3
Through the steps outlined in this protocol, you generated interactive plots of sequence embeddings. You used color in plots to highlight annotated subcellular localization (from DeepLoc), and could test out different parameter choices (via Alternate Protocol 1) and annotations (via Alternate Protocol 2). You learned how to incorporate these steps in a bio_embeddings pipeline file to enable other researchers to reproduce your results (via the Support Protocol).
Basic Protocol 4
Through the steps outlined in this protocol, you trained a neural network on embeddings to predict subcellular localization of sequence embeddings.
Time Considerations
Basic Protocol 1
On a 2016 MacBook Pro with 16 GB of RAM, executing the pipeline took approximately 3 min. Considering installation of required software and download of necessary files, the overall execution time of the protocol should not exceed 20 min.
Basic Protocol 2
On an Nvidia 1080 GPU equipped with 8 GB of vRAM, embedding the whole DeepLoc dataset took ∼30 min. On a CPU (Intel i7-6700, 64 GB system RAM), embedding the sampled DeepLoc set took ∼2 min, while embedding the whole set took approximately 8 hr and 40 min. Executing the steps, not considering computation time, may take up to 30 min.
Basic Protocol 3
On Google Colab, the UMAP projection step (the most computationally expensive step) takes about 10 min. Writing the code and executing the steps, considering computation time, may take up to 1 hr.
Basic Protocol 4
On Google Colab, training various classifiers via grid search (the most computationally expensive step) takes about 15 min. Writing the code and executing the steps, considering computation time, may take up to 1 hr.
Acknowledgments
The authors thank Tim Karl (TUM) for help with hardware and software and Inga Weise (TUM) for support with many other aspects of this work. The authors thank Tom Sercu, Ali Madani, Daniel Berenberg, Alex Rives, Vladimir Gligorijevic, and Josh Meier for constructive discussions around protein language models and their use. The authors thank Roshan Rao, Neil Thomas, and Nicholas Bhattacharya for creating and maintaining TAPE. The authors also thank all those who deposited their experimental data in public databases, and those who maintain these databases. In particular, the authors thank Ioanis Xenarios (SIB, Univ. Lausanne), Matthias Uhlen (Univ. Upssala), and their teams at Swiss-Prot and HPA. This work was supported by the Deutsche Forschungsgemeinschaft (DFG), project number RO1320/4-1, by the Bundesministerium für Bildung und Forschung (BMBF), project number 031L0168, and by the BMBF through the program “Software Campus 2.0 (TU München)”, project number 01IS17049.
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Christian Dallago: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Visualization, Writing-original draft, Writing-review & editing, Konstantin Schütze: Methodology, Software, Writing-review & editing, Michael Heinzinger: Conceptualization, Investigation, Software, Writing-review & editing, Tobias Olenyi: Software, Writing-review & editing, Maria Littmann: Writing-original draft, Writing-review & editing, Amy X. Lu: Writing-original draft, Writing-review & editing, Kevin K. Yang, Seonwoo Min: Writing-original draft, Writing-review & editing, Sungroh Yoon: Writing-original draft, James T. Morton: Writing-original draft, Writing-review & editing, Burkhard Rost: Conceptualization, Funding acquisition, Supervision, Writing-original draft, Writing-review & editing
Conflicts of Interest
A.L. is employed at Insitro, South San Francisco, CA, 94080. Insitro had no involvement in the design or implementation of the work presented here.
Open Research
Data Availability Statement
The data that support the presented protocols are available at: https://github.com/sacdallago/bio_embeddings. These data were derived from the following resources available in the public domain: DisProt (https://www.disprot.org), DeepLoc (http://www.cbs.dtu.dk/services/DeepLoc).
Literature Cited
- Alley, E. C., Khimulya, G., Biswas, S., AlQuraishi, M., & Church, G. M. (2019). Unified rational protein engineering with sequence-based deep representation learning. Nature Methods , 16(12), 1315–1322. doi: 10.1038/s41592-019-0598-1.
- Almagro Armenteros, J. J., Sønderby, C. K., Sønderby, S. K., Nielsen, H., & Winther, O. (2017). DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics , 33(21), 3387–3395. doi: 10.1093/bioinformatics/btx431.
- AlQuraishi, M. (2019). End-to-end differentiable learning of protein structure. Cell Systems , 8(4), 292–301.e3. doi: 10.1016/j.cels.2019.03.006.
- Anaconda Software Distribution. (2020). In Anaconda Documentation (Vers. 2-2.4.0) [Computer software]. Anaconda Inc. Available at https://docs.anaconda.com/.
- Armenteros, J. J. A., Johansen, A. R., Winther, O., & Nielsen, H. (2020). Language modelling for biological sequences—curated datasets and baselines. BioRxiv , 2020.03.09.983585. doi: 10.1101/2020.03.09.983585.
- Bender, E. M., & Koller, A. (2020). Climbing towards NLU: On meaning, form, and understanding in the age of data. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics , 5185–5198. doi: 10.18653/v1/2020.acl-main.463.
- Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., … Schwede, T. (2014). SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research , 42, W252–W288. doi: 10.1093/nar/gku340.
- Bisong, E. (2019). Google colaboratory. In Building machine learning and deep learning models on google cloud platform (pp. 59–64). New York: Springer.
- Bojanowski, P., Grave, E., Joulin, A., & Mikolov, T. (2017). Enriching word vectors with subword information. ArXiv , 1607.04606 [Cs]. Available at http://arxiv.org/abs/1607.04606.
- Brown, T. B., Mann, B., Ryder, N., Subbiah, M., Kaplan, J., Dhariwal, P., … Amodei, D. (2020). Language models are few-shot learners. ArXiv , 2005.14165 [Cs]. Available at http://arxiv.org/abs/2005.14165.
- Callaway, E. (2020). ‘It will change everything’: DeepMind's AI makes gigantic leap in solving protein structures. Nature , 588(7837), 203–204. doi: 10.1038/d41586-020-03348-4.
- Chelba, C., Mikolov, T., Schuster, M., Ge, Q., Brants, T., Koehn, P., & Robinson, T. (2014). One billion word benchmark for measuring progress in statistical language modeling. ArXiv , 1312.3005 [Cs]. Available at http://arxiv.org/abs/1312.3005.
- Devlin, J., Chang, M.-W., Lee, K., & Toutanova, K. (2019). BERT: Pre-training of deep bidirectional transformers for language understanding. ArXiv , 1810.04805 [Cs]. Available at http://arxiv.org/abs/1810.04805.
- Elnaggar, A., Heinzinger, M., Dallago, C., Rehawi, G., Wang, Y., Jones, L., … Rost, B. (2020). ProtTrans: Towards cracking the language of life's code through self-supervised deep learning and high performance computing. In BioRxiv , 2020.07.12.199554. doi: 10.1101/2020.07.12.199554.
- Goldberg, T., Hecht, M., Hamp, T., Karl, T., Yachdav, G., Ahmed, N., … others (2014). LocTree3 prediction of localization. Nucleic Acids Research , 42(W1), W350–W355.
- Harris, C. R., Millman, K. J., van der Walt, S. J., Gommers, R., Virtanen, P., Cournapeau, D., … Oliphant, T. E. (2020). Array programming with NumPy. Nature , 585(7825), 357–362. doi: 10.1038/s41586-020-2649-2.
- Hatos, A., Hajdu-Soltész, B., Monzon, A. M., Palopoli, N., Álvarez, L., Aykac-Fas, B., … Piovesan, D. (2020). DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Research , 48(D1), D269–D276. doi: 10.1093/nar/gkz975.
- The HDF Group. (2000, 2010). Hierarchical data format version 5. Available at http://www.hdfgroup.org/HDF5.
- Heinzerling, B. (2020). NLP's clever Hans moment has arrived. Journal of Cognitive Science , 21(1), 159–167.
- Heinzinger, M., Elnaggar, A., Wang, Y., Dallago, C., Nechaev, D., Matthes, F., & Rost, B. (2019). Modeling aspects of the language of life through transfer-learning protein sequences. In BMC Bioinformatics , 20, 723. doi: 10.1186/s12859-019-3220-8.
- Henikoff, S., & Henikoff, J. G. (1992). Amino acid substitution matrices from protein blocks. Proceedings of the National Academy of Sciences , 89(22), 10915–10919. doi: 10.1073/pnas.89.22.10915.
- Hopf, T. A., Colwell, L. J., Sheridan, R., Rost, B., Sander, C., & Marks, D. S. (2012). Three-dimensional structures of membrane proteins from genomic sequencing. Cell , 149(7), 1607–1621. doi: 10.1016/j.cell.2012.04.012.
- Hopf, T. A., Ingraham, J. B., Poelwijk, F. J., Schärfe, C. P., Springer, M., Sander, C., & Marks, D. S. (2017). Mutation effects predicted from sequence co-variation. Nature Biotechnology , 35(2), 128–135.
- Kluyver, T., Ragan-Kelley, B., Pérez, F., Granger, B. E., Bussonnier, M., Frederic, J., … others (2016). Jupyter Notebooks-a publishing format for reproducible computational workflows. ELPUB , 87–90.
- Littmann, M., Heinzinger, M., Dallago, C., Olenyi, T., & Rost, B. (2021). Embeddings from deep learning transfer GO annotations beyond homology. Scientific Reports , 11(1), 1160. doi: 10.1038/s41598-020-80786-0.
- Lu, A. X., Zhang, H., Ghassemi, M., & Moses, A. (2020). Self-supervised contrastive learning of protein representations by mutual information maximization. BioRxiv , 2020.09.04.283929. doi: 10.1101/2020.09.04.283929.
- Madani, A., McCann, B., Naik, N., Keskar, N. S., Anand, N., Eguchi, R. R., … Socher, R. (2020). ProGen: Language modeling for protein generation. BioRxiv , 2020.03.07.982272. doi: 10.1101/2020.03.07.982272.
- Manning, C. D. (2011). Part-of-speech tagging from 97% to 100%: Is it time for some linguistics? In A. F. Gelbukh (Ed.), Computational Linguistics and Intelligent Text Processing (pp. 171–189). New York: Springer. doi: 10.1007/978-3-642-19400-9_14.
- McCoy, T., Pavlick, E., & Linzen, T. (2019). Right for the wrong reasons: Diagnosing syntactic heuristics in natural language inference. Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics , 3428–3448. doi: 10.18653/v1/P19-1334.
- McInnes, L., Healy, J., & Melville, J. (2018). UMAP: Uniform manifold approximation and projection for dimension reduction. ArXiv , 1802.03426 [Cs, Stat]. Available at http://arxiv.org/abs/1802.03426.
- Mikolov, T., Chen, K., Corrado, G., & Dean, J. (2013). Efficient estimation of word representations in vector space. ArXiv , 1301.3781 [Cs]. Available at http://arxiv.org/abs/1301.3781.
- Min, S., Park, S., Kim, S., Choi, H.-S., & Yoon, S. (2020). Pre-training of deep bidirectional protein sequence representations with structural information. ArXiv , 1912.05625 [Cs, q-Bio, Stat]. Available at http://arxiv.org/abs/1912.05625.
- Morcos, F., Pagnani, A., Lunt, B., Bertolino, A., Marks, D. S., Sander, C., … Weigt, M. (2011). Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proceedings of the National Academy of Sciences , 108(49), E1293–E1301. doi: 10.1073/pnas.1111471108.
- Morton, J. T., Strauss, C. E. M., Blackwell, R., Berenberg, D., Gligorijevic, V., & Bonneau, R. (2020). Protein structural alignments from sequence. BioRxiv , 2020.11.03.365932. doi: 10.1101/2020.11.03.365932.
- Moult, J., Pedersen, J. T., Judson, R., & Fidelis, K. (1995). A large-scale experiment to assess protein structure prediction methods. Proteins , 23, ii–iv.
- Ovchinnikov, S., Park, H., Varghese, N., Huang, P.-S., Pavlopoulos, G. A., Kim, D. E., … Baker, D. (2017). Protein structure determination using metagenome sequence data. Science , 355(6322), 294–298. doi: 10.1126/science.aah4043.
- Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., … Duchesnay, E. (2011). Scikit-learn: Machine learning in Python. Journal of Machine Learning Research , 12, 2825–2830.
- Pennington, J., Socher, R., & Manning, C. D. (2014). Glove: Global vectors for word representation. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) , 1532–1543. October 25-29, 2014, Doha, Qatar.
- Peters, M., Neumann, M., Iyyer, M., Gardner, M., Clark, C., Lee, K., & Zettlemoyer, L. (2018). Deep contextualized word representations. Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers) , 2227–2237. June 1-June 6, 2018, New Orleans, Louisiana. doi: 10.18653/v1/N18-1202.
- Pires, T., Schlinger, E., & Garrette, D. (2019). How multilingual is multilingual BERT? ArXiv , 1906.01502 [Cs]. Available at http://arxiv.org/abs/1906.01502.
- Raffel, C., Shazeer, N., Roberts, A., Lee, K., Narang, S., Matena, M., … Liu, P. J. (2020). Exploring the limits of transfer learning with a unified text-to-text transformer. ArXiv , 1910.10683 [Cs, Stat]. Available at http://arxiv.org/abs/1910.10683.
- Raina, R., Battle, A., Lee, H., Packer, B., & Ng, A. Y. (2007). Self-taught learning: Transfer learning from unlabeled data. Proceedings of the 24th International Conference on Machine Learning , 759–766. Bellevue, Washington. doi: 10.1145/1273496.1273592.
- Rao, R., Bhattacharya, N., Thomas, N., Duan, Y., Chen, P., Canny, J., … Song, Y. (2019). Evaluating Protein Transfer Learning with TAPE. In H. Wallach, H. Larochelle, A. Beygelzimer, F. Alché-Buc, E. Fox, & R. Garnett (Eds.), Advances in neural information processing systems 32 (pp. 9689–9701). Curran Associates, Inc. Available at https://static.yanyin.tech/literature/current_protocol/10.1002/cpz1.113/original_pdf/cpz1.113.pdf.
- Rao, R., Ovchinnikov, S., Meier, J., Rives, A., & Sercu, T. (2020). Transformer protein language models are unsupervised structure learners. BioRxiv , 2020.12.15.422761. doi: 10.1101/2020.12.15.422761.
- Reeb, J., Goldberg, T., Ofran, Y., & Rost, B. (2020). Predictive methods using protein sequences. In A. D. Baxevanis, G. D. Bader, & D. S. Wishart (Eds.) Bioinformatics ( 4th ed., p. 185).
- Rives, A., Goyal, S., Meier, J., Guo, D., Ott, M., Zitnick, C. L., … Fergus, R. (2019). Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. BioRxiv , 622803. doi: 10.1101/622803.
- Rost, B. (1996). PHD: Predicting one-dimensional protein structure by profile based neural networks. Methods in Enzymology , 266, 525–539.
- Rost, B. (2001). Protein secondary structure prediction continues to rise. Journal of Structural Biology , 134, 204–218.
- Rost, B., & Sander, C. (1993). Prediction of protein secondary structure at better than 70% accuracy. Journal of Molecular Biology , 232, 584–599.
- Rost, B., & Sander, C. (1994). Combining evolutionary information and neural networks to predict protein secondary structure. Proteins , 19, 55–72.
- Rost, B., & Sander, C. (1995). Progress of 1D protein structure prediction at last. Proteins , 23, 295–300.
- Saputro, D. R. S., & Widyaningsih, P. (2017). Limited memory Broyden-Fletcher-Goldfarb-Shanno (L-BFGS) method for the parameter estimation on geographically weighted ordinal logistic regression model (GWOLR). AIP Conference Proceedings , 1868(1), 040009. doi: 10.1063/1.4995124.
- Shen, D., Wang, G., Wang, W., Min, M. R., Su, Q., Zhang, Y., … Carin, L. (2018). Baseline needs more love: On simple word-embedding-based models and associated pooling mechanisms. Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) , 440–450. July 15 - 20, 2018, Melbourne, Australia. doi: 10.18653/v1/P18-1041.
- Steinegger, M., Meier, M., Mirdita, M., Vöhringer, H., Haunsberger, S. J., & Söding, J. (2019). HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics , 20(1), 473. doi: 10.1186/s12859-019-3019-7.
- Steinegger, M., Mirdita, M., & Söding, J. (2019). Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nature Methods , 16(7), 603–606. doi: 10.1038/s41592-019-0437-4.
- Steinegger, M., & Söding, J. (2018). Clustering huge protein sequence sets in linear time. Nature Communications , 9(1), 2542. doi: 10.1038/s41467-018-04964-5.
- Stormo, G. D., Schneider, T. D., Gold, L., & Ehrenfeucht, A. (1982). Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Research , 10(9), 2997–3011. doi: 10.1093/nar/10.9.2997.
- The UniProt Consortium. (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research , 47(D1), D506–D515. doi: 10.1093/nar/gky1049.
- Vig, J., Madani, A., Varshney, L. R., Xiong, C., Socher, R., & Rajani, N. F. (2020). BERTology meets biology: Interpreting attention in protein language models. ArXiv , 2006.15222 [Cs, q-Bio]. Available at http://arxiv.org/abs/2006.15222.
- Villegas-Morcillo, A., Makrodimitris, S., van Ham, R. C. H. J., Gomez, A. M., Sanchez, V., & Reinders, M. J. T. (2020). Unsupervised protein embeddings outperform hand-crafted sequence and structure features at predicting molecular function. Bioinformatics , 2020, btaa701. doi: 10.1093/bioinformatics/btaa701.
- Zhu, J., Xia, Y., Wu, L., He, D., Qin, T., Zhou, W., … Liu, T.-Y. (2020). Incorporating BERT into neural machine translation. ArXiv , 2002.06823 [Cs]. Available at http://arxiv.org/abs/2002.06823.
Internet Resources
bio_embeddings source code.
bio_embeddings Python documentation
Example bio_embeddings pipeline runs.
Notebooks for interactive bio-embeddings workflows.
For small FASTA files (<20000 residues in total) it is also possible to use the bio_embeddings web pipeline: https://api.bioembeddings.com. The web pipeline also allows execution of single sequences (<2000 residues) instantaneously, as utilized by PredictProtein (https://predictprotein.org) and https://embed.protein.properties.
Citing Literature
Number of times cited according to CrossRef: 24
- Chau Tran, Siddharth Khadkikar, Aleksey Porollo, Survey of Protein Sequence Embedding Models, International Journal of Molecular Sciences, 10.3390/ijms24043775, 24 , 4, (3775), (2023).
- Maha A. Thafar, Somayah Albaradei, Mahmut Uludag, Mona Alshahrani, Takashi Gojobori, Magbubah Essack, Xin Gao, OncoRTT: Predicting novel oncology-related therapeutic targets using BERT embeddings and omics features, Frontiers in Genetics, 10.3389/fgene.2023.1139626, 14 , (2023).
- Justin R. Randall, Cory D. DuPai, T. Jeffrey Cole, Gillian Davidson, Kyra E. Groover, Sabrina L. Slater, Despoina A. I. Mavridou, Claus O. Wilke, Bryan W. Davies, Designing and identifying β-hairpin peptide macrocycles with antibiotic potential, Science Advances, 10.1126/sciadv.ade0008, 9 , 2, (2023).
- Zhenjiao Du, Xingjian Ding, Yixiang Xu, Yonghui Li, UniDL4BioPep: a universal deep learning architecture for binary classification in peptide bioactivity, Briefings in Bioinformatics, 10.1093/bib/bbad135, 24 , 3, (2023).
- Maria Ninova, Hannah Holmes, Brett Lomenick, Katalin Fejes Tóth, Alexei A. Aravin, Pervasive SUMOylation of heterochromatin and piRNA pathway proteins, Cell Genomics, 10.1016/j.xgen.2023.100329, (100329), (2023).
- Shikha Mallick, Sahely Bhadra, CDGCN: Conditional de novo Drug Generative Model Using Graph Convolution Networks, Research in Computational Molecular Biology, 10.1007/978-3-031-29119-7_7, (104-119), (2023).
- Dimitri Boeckaerts, Michiel Stock, Bernard De Baets, Yves Briers, Identification of Phage Receptor-Binding Protein Sequences with Hidden Markov Models and an Extreme Gradient Boosting Classifier, Viruses, 10.3390/v14061329, 14 , 6, (1329), (2022).
- Konstantin Schütze, Michael Heinzinger, Martin Steinegger, Burkhard Rost, Nearest neighbor search on embeddings rapidly identifies distant protein relations, Frontiers in Bioinformatics, 10.3389/fbinf.2022.1033775, 2 , (2022).
- Dagmar Ilzhöfer, Michael Heinzinger, Burkhard Rost, SETH predicts nuances of residue disorder from protein embeddings, Frontiers in Bioinformatics, 10.3389/fbinf.2022.1019597, 2 , (2022).
- Samuel Goldman, Ria Das, Kevin K. Yang, Connor W. Coley, Machine learning modeling of family wide enzyme-substrate specificity screens, PLOS Computational Biology, 10.1371/journal.pcbi.1009853, 18 , 2, (e1009853), (2022).
- Katarzyna Stapor, Krzysztof Kotowski, Tomasz Smolarczyk, Irena Roterman, Lightweight ProteinUnet2 network for protein secondary structure prediction: a step towards proper evaluation, BMC Bioinformatics, 10.1186/s12859-022-04623-z, 23 , 1, (2022).
- David Hoksza, Hamza Gamouh, Exploration of protein sequence embeddings for protein-ligand binding site detection, 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 10.1109/BIBM55620.2022.9995025, (3356-3361), (2022).
- Michael Heinzinger, Maria Littmann, Ian Sillitoe, Nicola Bordin, Christine Orengo, Burkhard Rost, Contrastive learning on protein embeddings enlightens midnight zone, NAR Genomics and Bioinformatics, 10.1093/nargab/lqac043, 4 , 2, (2022).
- Valérie de Crécy-lagard, Rocio Amorin de Hegedus, Cecilia Arighi, Jill Babor, Alex Bateman, Ian Blaby, Crysten Blaby-Haas, Alan J Bridge, Stephen K Burley, Stacey Cleveland, Lucy J Colwell, Ana Conesa, Christian Dallago, Antoine Danchin, Anita de Waard, Adam Deutschbauer, Raquel Dias, Yousong Ding, Gang Fang, Iddo Friedberg, John Gerlt, Joshua Goldford, Mark Gorelik, Benjamin M Gyori, Christopher Henry, Geoffrey Hutinet, Marshall Jaroch, Peter D Karp, Liudmyla Kondratova, Zhiyong Lu, Aron Marchler-Bauer, Maria-Jesus Martin, Claire McWhite, Gaurav D Moghe, Paul Monaghan, Anne Morgat, Christopher J Mungall, Darren A Natale, William C Nelson, Seán O’Donoghue, Christine Orengo, Katherine H O’Toole, Predrag Radivojac, Colbie Reed, Richard J Roberts, Dmitri Rodionov, Irina A Rodionova, Jeffrey D Rudolf, Lana Saleh, Gloria Sheynkman, Francoise Thibaud-Nissen, Paul D Thomas, Peter Uetz, David Vallenet, Erica Watson Carter, Peter R Weigele, Valerie Wood, Elisha M Wood-Charlson, Jin Xu, A roadmap for the functional annotation of protein families: a community perspective, Database, 10.1093/database/baac062, 2022 , (2022).
- Matteo Manfredi, Castrense Savojardo, Pier Luigi Martelli, Rita Casadio, E-SNPs&GO: embedding of protein sequence and function improves the annotation of human pathogenic variants, Bioinformatics, 10.1093/bioinformatics/btac678, 38 , 23, (5168-5174), (2022).
- Emilio Fenoy, Alejando A Edera, Georgina Stegmayer, Transfer learning in proteins: evaluating novel protein learned representations for bioinformatics tasks, Briefings in Bioinformatics, 10.1093/bib/bbac232, 23 , 4, (2022).
- Felix Teufel, José Juan Almagro Armenteros, Alexander Rosenberg Johansen, Magnús Halldór Gíslason, Silas Irby Pihl, Konstantinos D. Tsirigos, Ole Winther, Søren Brunak, Gunnar von Heijne, Henrik Nielsen, SignalP 6.0 predicts all five types of signal peptides using protein language models, Nature Biotechnology, 10.1038/s41587-021-01156-3, 40 , 7, (1023-1025), (2022).
- Konstantin Weissenow, Michael Heinzinger, Burkhard Rost, Protein language-model embeddings for fast, accurate, and alignment-free protein structure prediction, Structure, 10.1016/j.str.2022.05.001, 30 , 8, (1169-1177.e4), (2022).
- Tobias Olenyi, Céline Marquet, Michael Heinzinger, Benjamin Kröger, Tiha Nikolova, Michael Bernhofer, Philip Sändig, Konstantin Schütze, Maria Littmann, Milot Mirdita, Martin Steinegger, Christian Dallago, Burkhard Rost, LambdaPP: Fast and accessible protein‐specific phenotype predictions, Protein Science, 10.1002/pro.4524, 32 , 1, (2022).
- Michal Gala, Gabriel Žoldák, Classifying Residues in Mechanically Stable and Unstable Substructures Based on a Protein Sequence: The Case Study of the DnaK Hsp70 Chaperone, Nanomaterials, 10.3390/nano11092198, 11 , 9, (2198), (2021).
- Seonwoo Min, HyunGi Kim, Byunghan Lee, Sungroh Yoon, Protein transfer learning improves identification of heat shock protein families, PLOS ONE, 10.1371/journal.pone.0251865, 16 , 5, (e0251865), (2021).
- Hannes Stärk, Christian Dallago, Michael Heinzinger, Burkhard Rost, Light attention predicts protein location from the language of life, Bioinformatics Advances, 10.1093/bioadv/vbab035, 1 , 1, (2021).
- Maria Littmann, Michael Heinzinger, Christian Dallago, Konstantin Weissenow, Burkhard Rost, Protein embeddings and deep learning predict binding residues for various ligand classes, Scientific Reports, 10.1038/s41598-021-03431-4, 11 , 1, (2021).
- Céline Marquet, Michael Heinzinger, Tobias Olenyi, Christian Dallago, Kyra Erckert, Michael Bernhofer, Dmitrii Nechaev, Burkhard Rost, Embeddings from protein language models predict conservation and variant effects, Human Genetics, 10.1007/s00439-021-02411-y, 141 , 10, (1629-1647), (2021).

