Human-Induced Pluripotent Stem Cell Culture Methods Under cGMP Conditions
Teresa Rivera, Teresa Rivera, Yuanyuan Zhao, Yuanyuan Zhao, Yuhui Ni, Yuhui Ni, Jiwu Wang, Jiwu Wang
Abstract
The discovery of induced pluripotent stem cells (iPSCs) revolutionized the approach to cell therapy in regenerative medicine. Reprogramming of somatic cells into an embryonic-like pluripotent state provides an invaluable resource of patient-specific cells of any lineage. Implementation of procedures and protocols adapted to current good manufacturing practice (cGMP) requirements is critical to ensure robust and consistent high-quality iPSC manufacturing. The technology developed at Allele Biotechnology for iPSC generation under cGMP conditions is a powerful platform for derivation of pluripotent stem cells through a footprint-free, feeder-free, and xeno-free reprogramming method. The cGMP process established by Allele Biotechnology entails fully cGMP compliant iPSC lines where the entire manufacturing process, from tissue collection, cell reprogramming, cell expansion, cell banking and quality control testing are adopted. Previously, we described in this series of publications how to create iPSCs using mRNA only, and how to do so under cGMP conditions. In this article, we describe in detail how to culture, examine and storage cGMP-iPSCs using reagents, materials and equipment compliant with cGMP standards. © 2020 The Authors.
Basic Protocol 1 : iPSC Dissociation
Support Protocol 1 : Stem cell media
Support Protocol 2 : ROCK inhibitor preparation
Support Protocol 3 : Vitronectin coating
Basic Protocol 2 : iPSC Cryopreservation
Basic Protocol 3 : iPSC Thawing
INTRODUCTION
The arrival and development of induced pluripotent stem cells (iPSCs), thanks to Dr. Yamanaka and other stem cell researchers (Takahashi & Yamanaka, 2006), provided new possibilities to cell therapy and regenerative medicine. iPSCs are derived from somatic cells that are reprogrammed back into an embryonic-like pluripotent state by forced expression of key transcription factors (Takahashi et al., 2007; Yu et al., 2007). iPSCs have the capacity to self-renew and the potential to differentiate into all cell lineages of the three germ layers. Streamlined cGMP cell reprogramming allows generation of patient-specific differentiated cells to replace diseased or damaged tissues. In addition to regenerative medicine, iPSCs have numerous biomedical applications in drug screening, toxicological studies, and disease modeling (Shi, Inoue, Wu, & Yamanaka, 2017).
Prior to 2007, derivation of pluripotent stem cells from pre-implantation embryos, known as embryonic stem cells (ESC), opened new avenues for the development of treatments of many degenerative diseases. However, generation of human ESC lines raises ethical and political concerns as it requires the destruction of human embryos (Aach, Lunshof, Iyer, & Church, 2017). In addition, the use of differentiated cells from ESC for clinical applications suffers from the concerns of immune rejection after allogenic cell transplantation (de Almeida, Ransohoff, Nahid, & Wu, 2013). Therefore, the use of iPSC in regenerative medicine holds great promise as a therapeutic product circumventing the ethical problems associated with the use of human embryos and immune rejection after transplantation.
The development of directed differentiation protocols using human iPSC into various cell types has allowed the initiation of several cell-therapy human trials based on iPSCs. The first reported clinical trial involving iPSC-derived retinal pigment epithelial cells was designed to treat the Age-Related Macular Degeneration (AMD) (Mandai, Kurimoto, & Takahashi, 2017). In 2018, the Kyoto University announced the first clinical trial to treat Parkinson's disease using iPSC-derived dopaminergic neurons to be transplanted into human patients (Takahashi, 2019). iPSC-derived cells are also used in clinical trials for cancer immunotherapy to treat advanced solid tumors (ClinicalTrials.gov Identifier: NCT02923375). Recently, a clinical trial using iPSC-derived cardiomyocytes has been approved for heart diseases (Cyranoski, 2018).
As any therapeutic material to be applied to human, iPSC-derived clinical products should be well defined for the safety and efficacy of the therapeutic applications, and development of clinical-grade iPSC lines suitable for various applications as a starting material for cell therapy is a priority to the whole field.
This article describes the procedures for human iPS cell culture and handling techniques compliant with cGMP regulations in order to generate high quality iPSC for use in clinical applications. This protocol has been tested and validated using multiple cGMP-grade iPSC lines generated with Allele reprogramming core technology at Allele's GMP facility in San Diego, California. To adhere to cGMP regulations for cell manufacturing, reagents, materials, and equipment compliant with cGMP standards were used and the best practices for cell culture and quality control were adopted. In addition to the implementation of this protocol for iPSC culture, a cGMP-compliant facility that meets the FDA regulations and requirements imposed by institutional review boards (IRBs) is required to successfully obtain cGMP compliant iPSC.
Basic Protocol 1 covers iPSC dissociation procedures during iPSC expansion and selection. cGMP stem cell media is described in Support Protocol 1. ROCK inhibitor reconstitution is detailed in Support Protocol 2 and coating of culture vessels in Support Protocol 3. How the cGMP iPSC lines are cryopreserved is described in Basic Protocol 2 and finally, how the iPSC frozen stocks are thawed is outlined in Basic Protocol 3.
STRATEGIC PLANNING
Manufacturing of high-quality iPSCs using a robust cGMP-compliant process is essential for iPS cell-based therapies, suitable methods for every step throughout iPS cell manufacturing should be established to assure the desired high quality. Using cGMP-grade and xeno-free reagents is required to ensure defined and consistent culture conditions for iPSC manufacturing. Well-characterized and quality-controlled solutions and reagents must be obtained from reliable sources and the corresponding Certificate of Analysis should be acquired. Routine iPSC cell culture manipulations should be performed inside a Class II biological safety cabinet (BSC). All reagents and materials described in this protocol are sterile and should be disinfected with proper aseptic techniques before introducing into BSC. During iPSC culture under cGMP conditions there are critical factors and components that require detailed evaluation as discussed below.
Critical Factors of iPSC Culture
Cell morphology
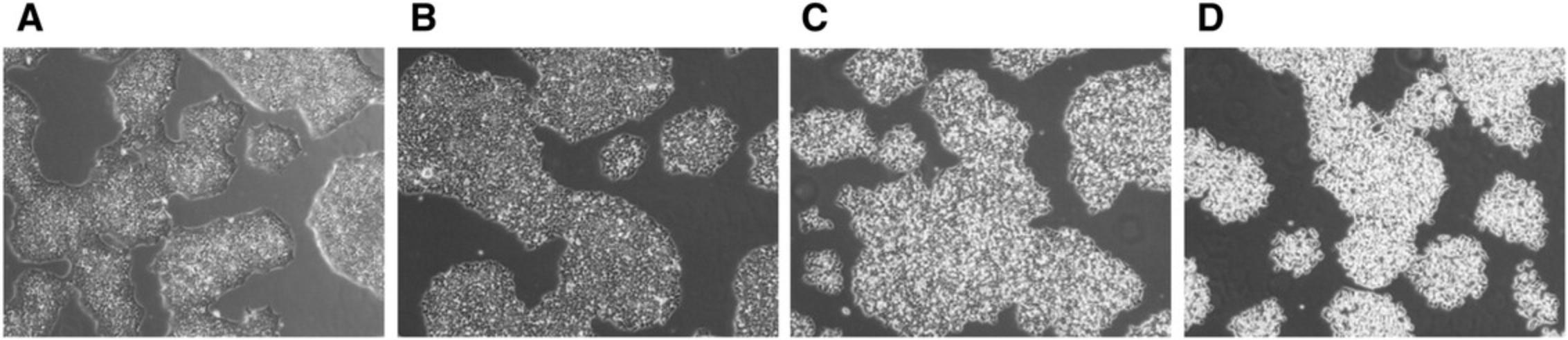
Evaluation of the culture process is critical to ensure manufacturing of high-quality iPSCs. It is, therefore, essential to examine iPS cell culture every day under a microscope to check for iPSC-like morphology, presence of any differentiated cells and cell culture confluency. Cell morphology of human iPSC has been reported to resemble that of human ESC (Takahashi et al., 2007), these characteristics include a high nucleus:cytoplasm ratio, prominent nucleoli, scant cytoplasm, and formation of compact colonies with clear and smooth colony edges (Yu et al., 2007) (Fig. 1).
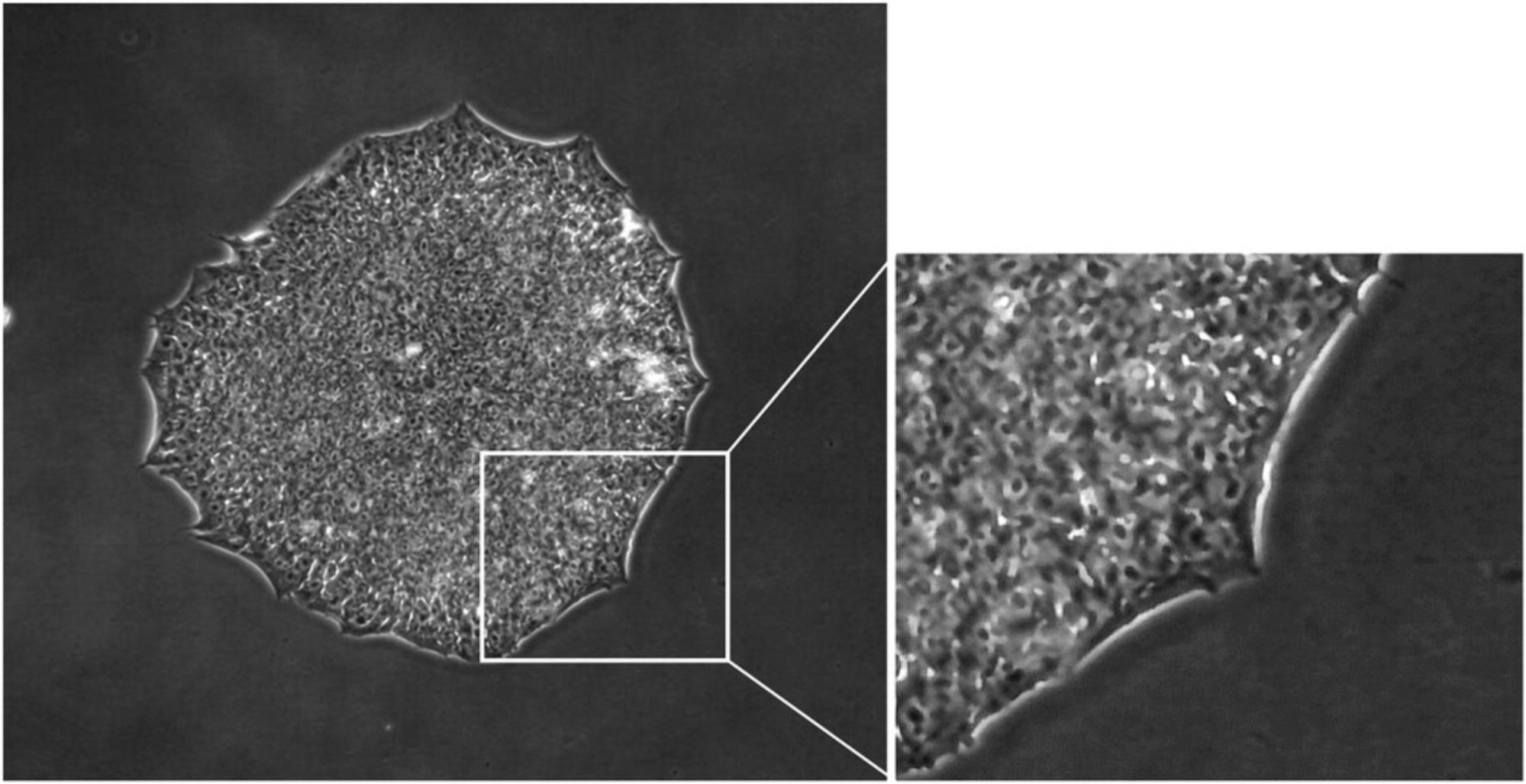
High-quality iPS cells are attainable only when optimal cell culture conditions are achieved. In order to maintain cell growth in exponential phase it is important that iPSCs be sub-cultured every 4-5 days according to cell density described below; if the density of the culture is too high, iPSCs tend to grow as 3D-like aggregates and cell viability decreases. Representative images of iPSC cultures grown to different density are shown in Figure 2.
Cell proliferation
Maintenance of indefinite self-renewal and unlimited expansion potential are fundamental hallmarks of successful stem cell culture. ESCs and iPSCs present a short cell cycle and their doubling time is approximately 16-20 hr (Hanna et al., 2010), allowing for very fast cell division, which supports pluripotency. When pluripotent stem cells differentiate, the cell cycle duration lengthens to >24 hr, which is the typical cell cycle of a differentiated somatic cell (Becker, Stein, Lian, van Wijnen, & Stein, 2010). Several reports showed that elongated or arrested cell cycle results in spontaneous differentiation (Neganova, Zhang, Atkinson, & Lako, 2009; Ruiz et al., 2011). Accordingly, analysis of cell proliferation and stem cell renewal was used as a test method to evaluate robustness of iPS cell culture.
Cell proliferation is determined by analyzing the doubling time of the cell culture. The information required for calculation of the doubling time includes: date, starting and ending time of the culture, as well as initial and final cell number.
Doubling time is calculated as follows: 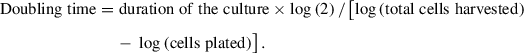
We have established and validated a quality control test to perform the “Cell Counting and Viability” assay of iPSC cultures. This assay is used to determine iPS cell number, viability, and proliferation. We have incorporated a cGMP-compliant automated cell counter into our iPSC culture platform that allows determination of the viability and cell concentration of aggregating cells such as iPSCs. The cell counter includes fluorescent dyes to determine the total cell number and dead cell population of a given sample using image analysis. Since it allows a cell count of aggregated cells, no single-cell dissociation of the iPSC culture is required to perform the analysis.
Critical Components of iPSC Culture
Stem cell medium
The quality and consistency of the stem cell medium is crucial to the maintenance and expansion of iPSCs. Essential 8TM (E8) Medium is a fully defined, xeno-free, and feeder-free medium formulated for the growth and expansion of human pluripotent stem cells. It was originally developed by Chen et al. (2011) and showed to support iPSC growth for >50 passages.
cGMP-compliant E8 medium (GibcoTM, ThermoFisher, Waltham, MA) was evaluated to support iPSC growth during our Process Development phase. We have observed high proliferation rates (doubling time around 20 hr) and typical iPSC morphology in cultures grown in cGMP-E8 medium. Consequently, we established cGMP-E8 as a suitable medium for iPSC manufacture.
Essential 8™ Medium has been extensively tested and is proven to be able to maintain pluripotency in multiple pluripotent stem cells lines. Unlike most feeder-free media, Essential 8™ medium does not require the presence of BSA (bovine serum albumin) or HSA (human serum albumin) that contributes to lot-to-lot variability.
Cell culture matrix
We aimed for and have established a defined iPSC culture system that is completely xeno-free, including both the medium and vessel coating material. We previously showed that the synthetic peptide-based surface-coating Synthemax II-SC (Corning), certifiable under cGMP, could be used to support iPSC growth (Ni, Zhao, Warren, Higginbotham, & Wang, 2016). We have further explored the use of other extracellular matrices (ECM) manufactured under cGMP guidelines.
We evaluated Vitronectin (VTN) as a xeno-free ECM to culture iPSC under cGMP conditions. VTN is a recombinant human protein used as ECM substrate, which was shown to support human pluripotent stem cell sell-renewal (Braam et al., 2008). The vitronectin variant, VTN-N, lacks the N-terminal SMB functional domain of the native vitronectin and has been shown to support human pluripotent attachment and survival better than wild-type vitronectin (Chen et al., 2011).
A cell culture system based on the above-described cGMP components provides an optimal system for the in vitro maintenance of iPSC, supporting self-renewal and high proliferative rates in a way that is highly consistent and practical for cGMP operations. Pluripotent stem cells maintained on CTSTM-VTN-N matrix appear to be homogeneous of typical iPSC colonies with no spontaneous differentiation observed even after long-term culturing.
Cell culture conditions/oxygen
iPSC cultures should be maintained in a multi-gas incubator to provide a stable environment of 37°C, 5% CO2, 5% O2 and relative humidity of about 95%. CO2 and O2 sensors should be calibrated regularly according to cGMP Equipment and Operation Qualification schedules to ensure accurate measurements.
These parameters are defined based on the culture requirements of iPSC. E8 Base Medium is suitable for 5% CO2 conditions providing a pH of 7.4. Moreover, regulation of oxygen concentration towards physiological levels (2%-5%), which is natural to pluripotent stem cells, is critical for reducing spontaneous differentiation and chromosomal abnormalities while supporting stem cell renewal and increasing cell proliferation (Dakhore, Nayer, & Hasegawa, 2018). During our Process Development, low oxygen tension at 3%-5% was found to be optimal.
Cell density
Maintaining a proper colony density is another critical aspect of growing human iPS cells in vitro. Frequent dissociation of the iPSC colonies can cause cell stress and poor cell survival; on the other hand, chromosomal abnormalities and spontaneous differentiation may arise in overgrown iPSC cultures. Therefore, it is essential to find the proper cell density in order to maintain cultures of high quality iPSCs.
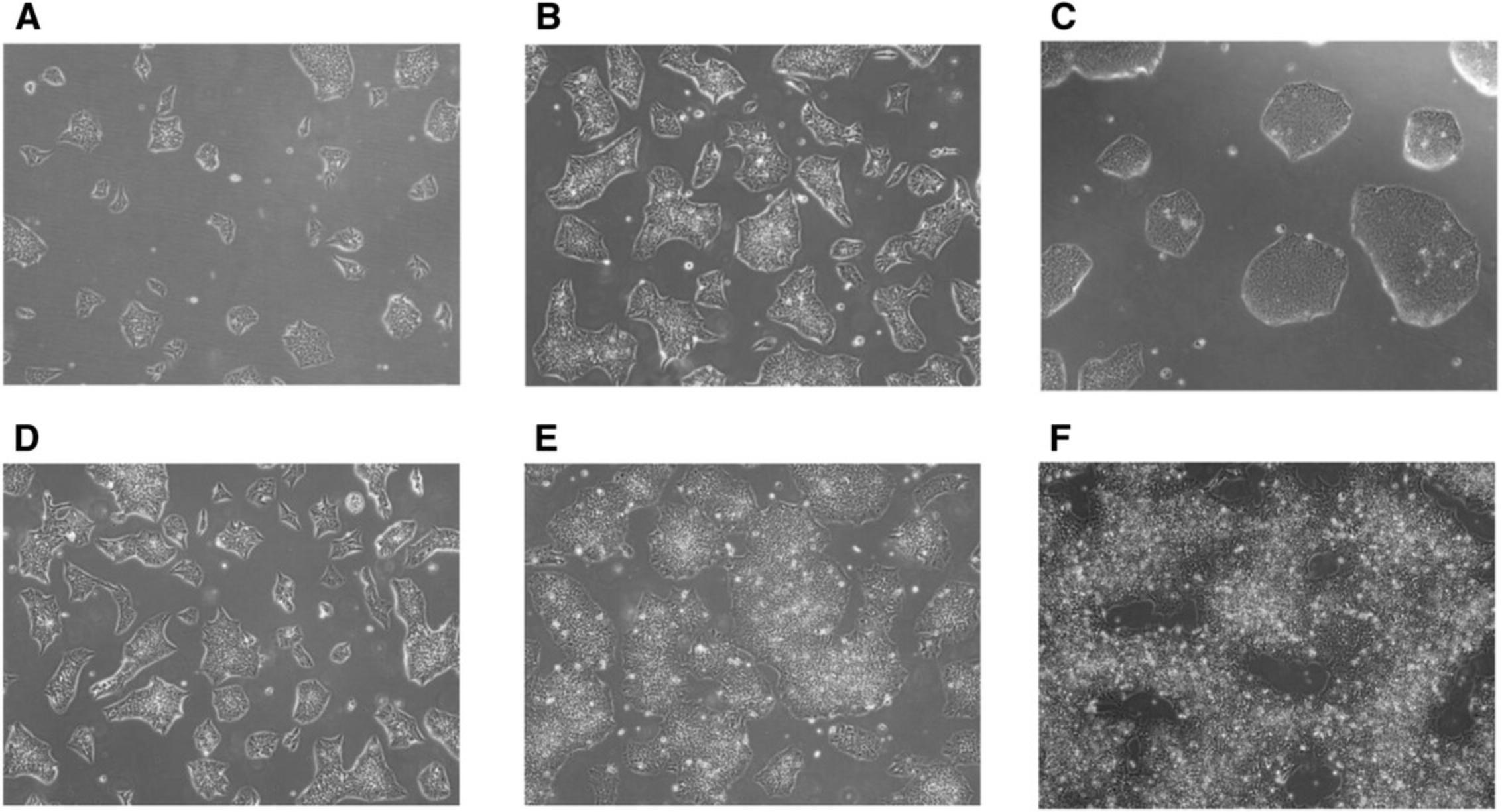
The criteria used in assessing the timing for iPSC dissociation was based on colony size or overall confluency of the culture (Fig. 2A-F). We have determined that iPSCs should be passaged when the colonies have reached medium-to-large colony size (Fig. 2B and C) or when culture reaches approximately 60% to 75% confluency (Fig. 2E) to ensure healthy high proliferative cultures. Microscopic observation is therefore essential to evaluate cell culture confluency. Overgrown cultures can be easily detected as increased number of non-viable cells remain floating in the media and the center of the colonies appear to be densely packed, compromising cell viability of the culture (Fig. 2F).
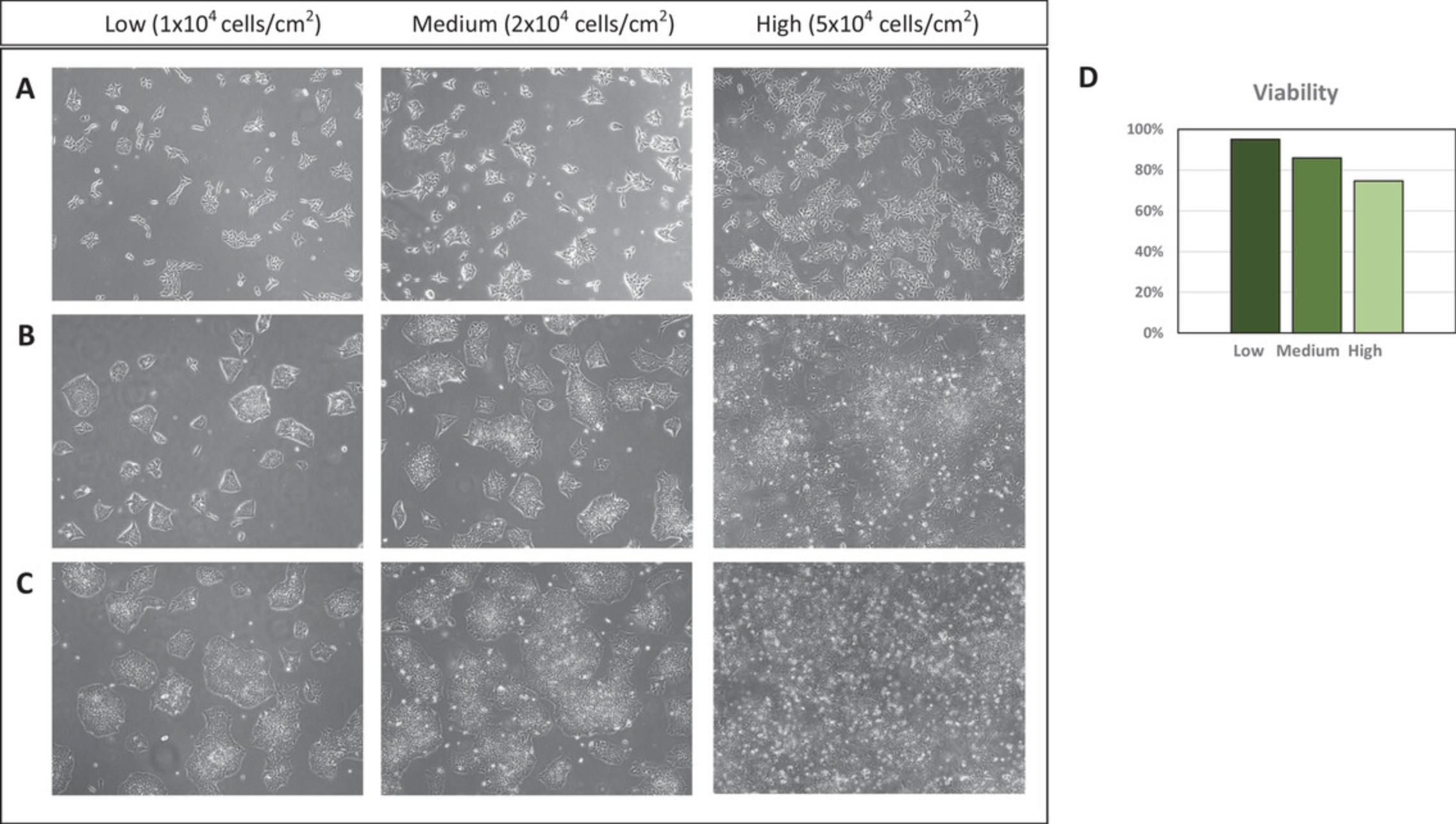
To reliably and consistently achieve the accuracy of cell density, we perform three cell counts every time and calculate their average. We have evaluated different cell seeding numbers and tested cell viability under these conditions. When cell seeding was high, reduced cell viability was found and frequent cell dissociation was required. Thus, in order to avoid frequent dissociations of the colonies that can lead to poor cell survival, we recommend cell seeding numbers ranging from 1 × 104 to 2 × 104 cells/cm2 (Fig. 3). When seeding cells near the lower limit of the optimal range, typically a 4-day dissociation/splitting interval can be established for fast dividing cell populations.
Basic Protocol 1: iPSC DISSOCIATION
Culture conditions, including cell passaging methods, affect the genetic and epigenetic stability of iPSCs; importantly, enzymatic passaging of iPSCs is associated with increased genomic instability of iPSCs (Garitaonandia et al., 2015). To circumvent this problem, enzyme-free dissociation methods, such as the EDTA-based passaging procedure, have been developed to successfully derive and expand iPSC (Beers et al., 2012).
CRITICAL STEP : During iPSC selection and expansion, cell survival is the priority. Procedures related to iPSC dissociation should be performed quickly to avoid keeping cell cultures outside of incubator for long periods of time. We recommend completing this procedure in approximately 30 min or less.
For iPSC expansion, we recommend treatment with a dissociation reagent (Allele Biotechnology) for 2 to 5 min at 37°C to preserve iPSC clusters. Microscope observation can be performed every 2 min to evaluate the dissociation of the colonies. iPSCs are ready for dissociation when cells become rounded and separated throughout the colony, usually after 3 min of treatment (Fig. 4C), and small clusters of iPSCs are dissociated from the original tissue culture vessel. Tapping of the tissue culture vessel after treatment was found to induce dissociation of the colonies into single cells, therefore gentle rocking, instead of tapping, is recommended to preserve iPSC clusters.
Materials
- E8: Essential 8™ Medium, cGMP grade (GibcoTM, ThermoFisher, cat. no. A1517001)
- ROCK inhibitor (ROCKi): 1 mM ROCK inhibitor solution in water (see Support Protocol 2)
- DPBS with no calcium and magnesium, cGMP grade (GibcoTM, ThermoFisher, cat. no. 14190144)
- iPSC culture
- 70% USP-grade isopropanol wipes, Contec™ PROSAT™ Presaturated Knitted Polynit Wipes (Fisher Scientific, cat. no. 19-120-817)
- Dissociation reagent, EDTA-containing solution, (Allele Biotechnology, cat. no. ABP-SC-DR100)
- Multi-gas Incubator (Panasonic MCO, CO2/O2 Incubator)
- Vitronectin- (VTN) coated flasks (see Support Protocol 1)
- Conical tubes (ThermoFisherTM, NuncTM, 15 ml and 50 ml, cat. no. 339653 and 339651)
- Microscope (Live Technologies EVOSTM XL Core Imaging System, cat no. AMEX1000)
- Automated Cell Counting instrument (ChemoMetec NucleoCounter® NC-200TM System)
Dissociation of iPSCs from VTN-coated flasks
1.Prior to cell dissociation, reagents including: DPBS (without calcium and magnesium), dissociation reagent, and E8 should be pre-warmed at 37°C from 30 min to 2 hr.
2.Prepare the amount of E8 + ROCKi required for each procedure according to Table 1.
| Flask size | DPBS without calcium and magnesium | Dissociation reagent | E8+ROCKi | iPSC seeding number |
|---|---|---|---|---|
| 12.5 cm2 | 3 ml | 1 ml | 3 ml | <5E5 |
| 25 cm2 | 5 ml | 3 ml | 5 ml | 5E5∼1E6 |
| 225 cm2 | 50 ml | 15 ml | 50 ml | 1E6∼5E6 |
3.Retrieve the iPSC culture from the incubator, disinfect the outside with 70% isopropanol wipes, and transfer into BSC.
4.Aspirate medium from iPSC culture that is about to be dissociated.
5.Add pre-warmed DPBS (with no calcium and magnesium) to rinse the cells. Incubate for 1 min at room temperature in the BSC.
6.Aspirate DPBS from the flask.
7.Add pre-warmed dissociation solution and spread gently by rocking the flask side to side. Incubate for 1 min at room temperature in BSC.
8.Carefully remove most of dissociation solution without drying out the cells. Incubate the flask in the incubator for 2-5 min.
9.Transfer the required volume of E8+ROCKi into the flask (3 ml for 12.5-cm2 and 25-cm2 flasks and 15 ml for 225-cm2 flasks), pipette cell suspension up and then gently allow it to flow back down over the cell culture surface to help release remaining iPSC colonies. Transfer the entire cell suspension to conical tube and gently pipette up and down to suspend iPSCs without breaking apart the colonies.
Plating iPSCs on VTN-coated flask
10.Disinfect with 70% isopropanol wipes and transfer a new VTN-coated flask into BSC. Aspirate the coating solution, transfer the required volume of cell suspension and E8+ROCKi into new flask according to Table 1.
11.Shake the flask back and forth and side to side to distribute the cells evenly. Place the flask in the incubator (settings: 37°C, 5% CO2, 5% O2).
12.On the next day, observe the cells under the microscope to confirm cell attachment and confluency.
13.Monitor the cell culture daily and evaluate readiness to cell dissociation according to cell density.
Support Protocol 1: STEM CELL MEDIA
Complete Essential 8™ Medium is prepared by adding Essential 8™ Supplement (50×, provided with the medium) to Essential 8™ Basal Medium. Once E8 complete medium is prepared, it should be stored up to 2 weeks at 2°C-8°C.
To ensure adequate nutrition levels in a highly proliferative iPSC culture and avoid spontaneous differentiation, media change every day is recommended. Cell proliferation is optimal at 37°C whereas low temperatures can slow down cell growth and negatively impact the quality of cells; therefore, media used for daily change should be pre-warmed at 37°C. On the other hand, previous studies have shown that growth factors in stem cell media are sensitive to proteolysis and thermal denaturation and degradation of some of these factors are observed after 24 hr at 37°C (Levenstein et al., 2006). Thus, long-term exposure to 37°C of E8 medium should be avoided prior to use. During our process development work, we have found optimal performance of cGMP-E8 medium can be achieved by warming the medium between 30 and 120 min.
Support Protocol 2: ROCK INHIBITOR PREPARATION
Human embryonic stem cells are prone to cell death after individualization during cell dissociation treatments. Rho-associated protein kinase (ROCK) inhibitor has been shown to increase cell survival of dissociated cells by preventing dissociation-induced apoptosis in human embryonic stem cells (Watanabe et al., 2007). Therefore, we recommend adding ROCK inhibitor (ROCKi) to E8 medium at concentrations of 5 or 10 μM after cell dissociation, which is not necessary during daily iPSC culture medium change without passaging.
ROCK inhibitor (Y-27632, Sigma Aldrich, cat. no. SCM075) is supplied as a solid powder in 5 mg vials. For reconstitution, add 14.8 ml sterile cell culture grade water to make 1 mM stock solution. Dividing into small volume aliquots is recommended (30- and 250-µl aliquots are used in this protocol). Aliquots are stored up to 1 year at −80°C per Allele's cGMP protocol. We have evaluated working ROCKi concentrations of 5 and 10 μM in E8 medium and demonstrated that both concentrations efficiently inhibit apoptosis of dissociated hiPSCs and promote cell survival without affecting pluripotency. We routinely add ROCKi to E8 medium at 5 μM. Whenever relevant in the following protocols, ROCKi from Sigma Aldrich will be included at such a concentration. Note that cGMP grade ROCKi is now also available from Tocris Bioscience and can be incorporated into our protocol described below once it passes the Quality Control Testing in our cGMP system.
Support Protocol 3: VITRONECTIN COATING
We have tested cell therapy-grade Vitronectin (CTSTM-VTN-N, GibcoTM, ThermoFisher Scientific) for iPSC culture and observed consistent support of pluripotent stem cell renewal. VTN-N (GibcoTM, ThermoFisher) is provided in 1-ml aliquots at 0.9 mg/ml by the manufacturer with a recommended coating concentration range of 0.1-1.0 μg/cm2 on the culture vessel surface. We recommend using a solution concentration at 9 μg/ml with a minimum amount of 0.9 μg/cm2.
Basic Protocol 2: iPSC CRYOPRESERVATION
The quality of iPSC can be severely influenced by culture conditions and culture manipulation techniques, with cryopreservation being a major potential obstacle to maintaining iPSC quality. Human embryonic stem cells typically suffer from very low post-thaw recoveries. Similarly, cryopreservation is one of the most critical steps after iPSC expansion. Suboptimal cryopreservation methods lead to poor recovery of iPSCs after thawing, resulting in low survival rate of their population. The low cryopreservation efficiency of human pluripotent stem cells often leads to the arising of abnormal cells as well. To counter this, we (as described below) and others (Liu & Chen, 2014) have extensively tested and achieved technical improvements in cryopreservation methods.
Cryopreservation parameters, including components of cryopreservation medium and methods for processing samples during cryopreservation, were thoroughly evaluated in our Process Development:
- -Methods: Optimization of cryopreservation procedure begins with good training of operators to ensure robustness and consistency to provide uniform cell population for downstream clinical applications. Critical steps during cryopreservation should be performed in a timely and efficient manner to preserve cell viability, especially when generating Master Cell Banks (MCB). The protocol shown here for cryopreservation of iPSC has been validated for large cGMP banking and, importantly, provides a robust method and easily performed by different operators.
- -Cell freezing containers: The CoolCell® freezing containers available from BioCision (San Rafael, California) have already been adopted for use in cGMP cleanrooms. These containers provide highly reproducible, alcohol-free −1°C/min controlled rate freezing when transferred to a −80°C freezer. The −80°C freezer must follow the cGMP requirements for temperature-controlled storage equipment.
- -Cryopreservation medium: A critical component of the cryopreservation medium is the cryopreservant dimethyl sulfoxide (DMSO), we established that DMSO concentration at 10% in E8 medium provides high cryopreservation efficiency.
Protein-rich medium has been shown to increase cryopreservation efficiency; thus, serum is commonly added to cryomedium in most research labs. However, medium containing serum is not recommended to manufacture cGMP-grade iPSC. Human serum albumin (HSA) had been widely used as replacement of serum for human pluripotent stem cell cultivation (Ludwig et al., 2006). Along this line, we investigated cryopreservation efficiencies using cryomedium containing different concentrations of HSA and found high cryopreservation efficiency in medium containing 1% or 2.5% HSA. Post-thaw cell viability of iPSC lines cryopreserved under these conditions was generally higher than 90% (Fig. 5).
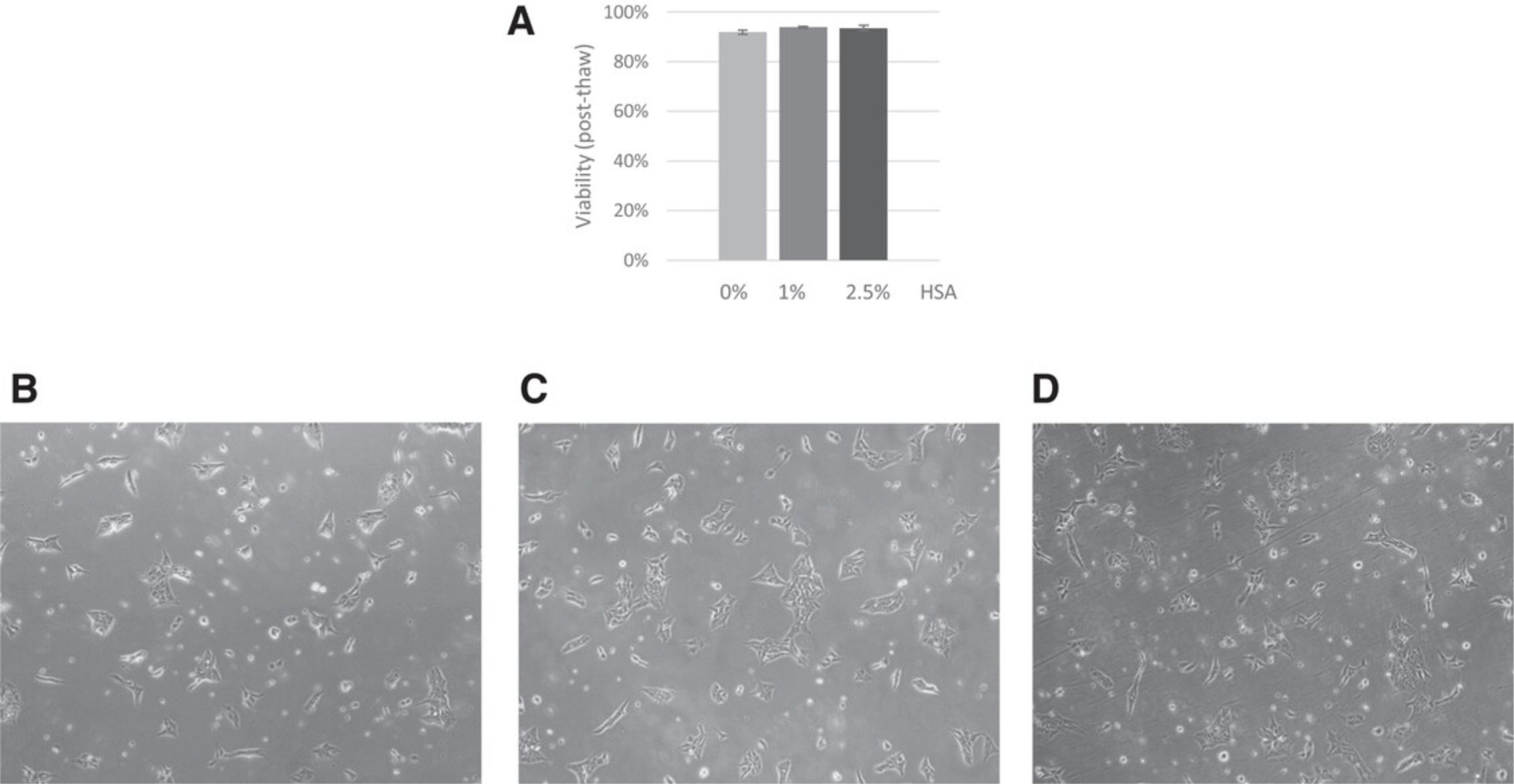
Therefore, to ensure high cryopreservation efficiencies and good cell survival (>90% viability) we recommend cryopreserving iPSCs in E8 medium containing 10% DMSO and HSA at concentrations ranging from 1% to 2.5%. We routinely use cryomedium containing 1% HSA but this concentration can be adapted according to each cell line's growth conditions.
The following procedure describes cryopreservation of iPSC at a concentration of 1 × 106 cells /ml in 1 ml of cryopreservation medium.
Volume of cryopreservation medium and number of cryogenic vials to prepare are determined by the results of live cell number of iPSCs obtained during cell count of the flask being cryopreserved.
If only a fraction of the iPSCs are to be cryopreserved, calculate the volume of cryopreservation medium accordingly, but maintain concentration of 1 × 106 cells/ ml to preserve high survival rate.
Materials
-
70% USP-grade isopropanol wipes, Contec™ PROSAT™ Presaturated Knitted Polynit Wipes (Fisher Scientific, cat. no. 19-120-817)
-
DMSO: Dimethyl Sulfoxide, USP grade (Sigma Aldrich, cat. no. D2438)
-
HSA: Human Serum Albumin (100 mg/ml), USP grade (Irvine Scientific, cat. no. 9988)
-
E8: Essential 8™ Medium, cGMP grade (GibcoTM, ThermoFisher, cat. no. A1517001)
-
ROCK inhibitor (ROCKi): 1 mM ROCK inhibitor solution in water (see Support Protocol 2)
-
1.2-ml Cryogenic vials (Corning® External Thread Cryogenic Vials, cat. no. 430658)
-
60-ml Reagent bottle (Thermo Scientific, cat. no. 3420200060)
-
Cell freezing container, CoolCell® BioCision
-
Automated cell counting instrument, ChemoMetec NucleoCounter® NC-200TM System
-
Ultra-low freezer, Panasonic MDF-U76VC-PA
Collecting iPSCs and preparing cryopreservation medium
1.Perform iPSC dissociation and cell count as described in the Basic Protocol 1, steps 1 to 9.
2.Calculate volume of cryopreservation medium according to Table 2.
| Required volume | Abbreviation | Formula |
|---|---|---|
| Cryopreservation medium (ml) | Vol. cryo | Vol. cryo = Total live cell numbera/1 × 106 |
| E8 (ml) | Vol. E8 | Vol. E8 = (Vol. cryo × 0.8) – Total volume of cell suspensionb (ml) |
| ROCK inhibitor | Vol. ROCKi | Vol. ROCKi = Vol. E8 (ml)/200 |
| DMSO | Vol. DMSO | Vol. DMSO = Vol. cryo (ml) × 0.1 |
| HSA | Vol. HSA | Vol. HSA = Vol. cryo (ml) × 0.1 |
- a
Total live cell number: obtained from cell count and viability test.
- b
Total volume of cell suspension: volume of cell suspension in E8+ROCKi in which cells were collected from flask after iPSC dissociation.
3.Prepare cryopreservation medium as follows:
-
Disinfect with 70% isopropanol wipes and transfer into BSC: cryogenic vials, one reagent bottle, consumables and reagents including, E8 medium aliquot, ROCKi aliquot(s), DMSO vial(s), HSA aliquot(s).
-
Add corresponding volume of E8 and ROCKi (according to Table2) to the reagent bottle.
-
Transfer the required volume of DMSO (according to Table2) to the reagent bottle. Swirl the bottle gently while adding DMSO to help mix evenly.
-
Transfer the required volume of HSA (according to Table2) to the reagent bottle.
-
Mix the reagents by gently pipetting up and down at least 5 times. Label the reagent bottle as Cryopreservation Medium (CM).
Cryopreserving iPSCs
4.Mix dissociated cells by gently pipetting and transfer the required cell suspension into the reagent bottle containing CM. Carefully mix the cell suspension in CM.
5.Transfer 1 ml of cells in CM into each cryotube.
6.Upon completion of vialing, immediately place cryogenic vials into cell freezing container and transfer into ultra-low temperature freezer.
7.Transfer cryogenic vials into liquid nitrogen for long-term storage after 24 hr of cryopreservation.
Basic Protocol 3: iPSC THAWING AND SEEDING
The following steps describe the thawing procedure of one cryogenic vial of iPSC and cell seeding into 75- cm2 VTN-coated flask.
In our protocols, thawing of cells in automated cell thawing instruments replaces common methods of cell thawing such as water baths to ensure consistency, as rapid and reproducible thawing reduces operation variability and the risk of contamination. Cryovials are retrieved from the liquid nitrogen tank or −80°C freezer and thawed in BioCision ThawSTAR® (BioCision LLC, Larkspur, CA) automated cell thawing instrument according to manufacturer's instructions.
Materials
-
Cryopreserved iPSC (see Basic Protocol 2)
-
70% USP-grade isopropanol wipes, Contec™ PROSAT™ Presaturated Knitted Polynit Wipes (Fisher Scientific, cat. no. 19-120-817)
-
E8: Essential 8™ Medium, cGMP grade (GibcoTM, ThermoFisher, cat. no. A1517001)
-
ROCK inhibitor (ROCKi): 1 mM ROCKi solution in water (see Support Protocol 2)
-
Automated Cell Thawing instrument, BioCisionTM ThawSTAR®
-
Automated Cell Counting instrument, ChemoMetec NucleoCounter® NC-200TM System
-
1.5-ml Microcentrifuge tubes (AxygenTM Scientific cat. no. SCT150SSCS)
-
15- and 50-ml Conical tubes (ThermoFisherTM, NuncTM, cat. no. 339653 and 339651)
-
Centrifuge, Eppendorf ®Centrifuge 5810/5810R
-
VTN-coated flask: one 75-cm2 VTN-flask per line (see Support Protocol 3)
-
Multi-gas incubator, Panasonic MCO, CO2/O2 Incubator
-
Microscope, Live Technologies EVOSTM XL Core Imaging System (cat. no. AMEX1000)
Thawing cryopreserved iPSCs
1.Retrieve cryovials with cryopreserved iPSC from liquid nitrogen.
2.Thaw the cells in an automated cell thawing instrument according to manufacturer's instructions.
3.Once the cell thawing program is completed, hold the vial and turn the vial upside down to check for the disappearance of ice.
4.Disinfect the vial with 70% isopropanol wipes and transfer into BSC.
5.Transfer a small volume of cell suspension (100 µl) into a microcentrifuge tube to perform cell count and viability test according to procedure determined by the automated cell counter.
6.Disinfect with 70% isopropanol wipes and transfer of E8 medium and ROCKi aliquot of appropriate quantity into BSC.
7.Combine pre-warmed E8 medium and ROCKi in a 50-ml conical tube (recommended volume is 30 ml of E8 medium and 150 µl ROCKi) and mix by pipetting.
8.Transfer approximately 9 ml of E8+ROCKi into a new 15-ml conical tube and add cell suspension from the cryogenic vial; gently mix by pipetting up and down 5 times.
9.Place the conical tube in the centrifuge and pellet the cells for 2 min at 300 × g , room temperature.
Seeding iPSCs in VTN-coated flask
10.Disinfect with 70% isopropanol wipes the conical tube and VTN-coated 75-cm2 flask with 70% isopropanol wipes and transfer into BSC.
11.Discard VTN coating solution from flask.
12.Carefully remove the supernatant from the conical tube in step 9 without disturbing the cell pellet.
13.Transfer approximately 5 ml of E8+ROCKi from step 7 into conical tube and gently pipette up and down to resuspend cell pellet.
14.Transfer the entire cell suspension from step 13 into VTN-coated flask.
15.Add approximately an additional 10 ml of E8+ROCKi from step 7 into VTN-coated flask. Close the flask and move the flask back and forth and side to side to distribute cells across the entire cell culture surface.
16.Perform microscope observation to ensure that cell suspension is evenly distributed.
17.Place the flask into the incubator (settings: 37°C, 5% CO2, 5% O2).
UNDERSTANDING RESULTS
When establishing an iPSC culture from a cryopreserved cell population, a 24-48 hr “recovery period” is normal since cell proliferation could be slower than expected, but exponential cell growth should resume after this period. This recovery stage should be taken into consideration when calculating the first post-thawing doubling time. However, when high levels of apoptosis and cellular debris are observed 24 hr post-thaw, it might be indicative of suboptimal cryopreservation storage or thawing procedures. An extended post-thaw recovery time can be considered in this situation and media change can be skipped the next day to allow more cells to attach. Alternatively, cells in suspension can be re-seeded 24 hr post-thaw into a new coated vessel to rescue some of the cell population that would otherwise be lost.
Lines with slow proliferative rates will require adaptation to the culture method described here, in which case cell seeding densities should be adjusted accordingly in order to gain healthy cell cultures. Daily evaluation of the culture under a microscope is essential to determine the readiness for cell dissociation and to detect appearance of spontaneously differentiated cells. For morphology evaluation, it is important to know that 24-48 hr after cell dissociation, iPSCs might initially exhibit elongated cell morphology and loose colony appearance. However, they should become more compact and uniform of typical pluripotent stem cell shapes once the colonies grow bigger, normally in 1 or 2 more days.
iPSC lines generated and expanded using Allele's system under cGMP standards have exhibited: cell doubling times similar to those of human ES cells (approximately 18-20 hr) (Hanna et al., 2010), homogeneous pluripotent gene expression profile, high and broad differentiation potential, and genomic stability over 100 passages in culture. Some iPSC lines are already available and have been adopted for future clinical applications. This set of protocols has been tested with multiple iPSC lines derived from a variety of cell types from donors across different ages, genders, and disease conditions. When implemented properly, our system's consistency will benefit any cGMP iPSC operation.
Time Considerations
The method for iPSC culture shown in this protocol has been optimized to minimize operation time when working under cGMP conditions. Improvements at each step of the process, such as iPSC expansion, cryopreservation, and thawing, allows processing of multiple iPSC lines in a more confined time frame while preserving cell viability and quality.
iPSCs cultures established after thawing one cryovial of cells containing 1 × 106 cells should give enough cells to seed a 75-cm2 flask, which will reach 60%-70% confluency within 3-4 days. At such cell confluence, iPSC yield will be approximately 5 × 106 cells. At this point, they can be further expanded into 225-cm2 flask and grown for another 3-4 days to obtain yields between 20 × 106 and 30 × 106 cells. At this stage, cells can be used for differentiation into a desired cell type or to generate an iPS cell bank. If large batches of iPSCs are required, cells collected form 225-cm2 flasks can be expanded for another 3-4 days. Following our method of dissociation regimen and a cell seeding density ranging from 1 × 104–2 × 104 cells/cm2, approximately 8 × 107–18 × 107 iPSCs can be obtained within 10 to 15 days of culture from 1 × 106 cells of a starting population.
This cell culture procedure enables the production of a working cell bank (WCB) of 100 vials of 1 × 106 iPSCs each within 10-12 days from a cryopreserved vial of iPSC of a master cell bank (MCB). It is optimized for manufacturing large numbers of cells at reduced cost by allowing industrial scale production of iPSCs in a time-efficient manner.
COMMENTARY
Background Information
Generation of iPSC for clinical applications
Genetic reprogramming of human somatic cells to a pluripotent state was first achieved in 2007 by overexpression of four pluripotent factors (Takahashi et al., 2007; Yu et al., 2007) using viral vectors that integrate into the genome. Since then, numerous methods have been developed to generate iPSCs and substantial efforts have been made to improve reprogramming efficiencies and to optimize iPSC culture conditions to obtain high-quality iPSCs that are comparable to human ES cells with respect to colony morphology, cell behavior, gene expression, epigenetic status, and differentiation potential both in vitro and in vivo (Malik & Rao, 2013).
Genomic alterations represent an obstacle for therapeutic applications, as random integration of exogenous DNA can lead to insertional mutagenesis with unpredictable effects on the quality of the cells and the potential safety after transplantation (Hacein-Bey-Abina et al., 2003). Therefore, integration-free methods for iPSC derivation are desired for manufacturing clinical-grade cells. Several non-integrating methods, including recombinant proteins, DNA plasmids, and Sendai virus, have been developed to transiently express transcription factors to induce reprogramming (Okita et al., 2011; Seki et al., 2010; Zhou et al., 2009). However, these methods show low efficiency, lack of consistency, and require the removal of vector-positive cells in DNA plasmid and Sendai virus-based cell reprogramming methods. Overcoming these obstacles, the mRNA-based reprogramming technology provides a powerful platform for generating “footprint-free” iPSC. This system allows the ectopic expression of transcription factors with no risk of genomic integration, no requirement of cleanup phase to purge residual traces of vectors, and allows dose-titratable expression of any desired factor (Warren, Ni, Wang, & Guo, 2012). mRNA-based cell reprogramming has also been shown to be the most efficient technology (Ni et al., 2016), and, more importantly, the most suitable method to ensure genome integrity of the iPSCs. A detailed comparison of the genetic integrity of iPSCs derived by the three different integration-free methods showed extremely low rates of aneuploidy and very low karyotype abnormalities detected in RNA-derived iPSCs compared to Sendai virus or episomal-derived iPSCs (Schlaeger et al., 2015). Therefore, mRNA technology represents a very powerful tool and is justifiably the method of choice to generate high-quality cGMP iPSCs for clinical applications.
Initial studies showed that the reprogramming process itself compromises genomic integrity of iPSCs (Laurent et al., 2011). However, increasing evidence lately suggested that most genetic variations observed in iPSC clones were observed in the parental cell population they were reprogrammed from (Letourneau et al., 2014). Interestingly, reprogramming has been shown to actually rectify chromosomal abnormalities present in somatic cells (Bershteyn et al., 2014). Optimization of iPSCs derivation and expansion to promote genetic stability is a priority during iPSC manufacturing to avoid spontaneous chromosomal abnormalities.
The mRNA reprogramming technology developed by Allele Biotechnology has demonstrated to be extremely efficient, consistent, and reproducible. It allows generation of iPSCs from multiple cell types with a faster kinetics than other methods based on our comparisons. Moreover, mRNA-derived iPSCs have been shown to better recapitulate the molecular signatures of human ESCs than virally derived iPSCs (Warren et al., 2010). Differentiation potential of iPSC lines generated by Allele Biotechnology has been tested on more than a dozen different cell types successfully by us and by several teams in the industry with minimal line-to-line variations.
Critical Parameters
cGMP-iPSC
Derivation of iPSC for clinical use has been proposed to be either manufactured under cGMP requirements or converted from research-grade conditions to cGMP quality standards. However, rigorous tests need to be performed on the converted lines to verify that the cells are free of contamination since they were not under full custody of a cGMP operation all the time. Allele's platform for iPSC derivation and expansion relies on the first method where every procedure from tissue collection from the donor to iPSC generation and banking is performed under cGMP requirements.
In order to ensure consistency and uniformity of the cGMP-iPSCs, strict standards are required to develop and validate manufacturing processes. Several principles of cGMP guidelines need to be established regarding quality control and assurance, design of the facility and the equipment, adequate and precise documentation, personnel training and certification, and environmental monitoring.
Allele Biotechnology has followed well-documented quality control principles and established a quality assurance system so that the entire manufacturing operation fulfills expected requirements from the initial informed consent to the final iPSC bank. All procedures are documented in the form of standard operating procedures (SOPs), procedure descriptions, registries, records, training documents, manuals, lists, and others to assure the good management, functionality, and traceability of the cell bank.
Allele cGMP-facility is built with ISO 7/Class cleanrooms, ISO 5/Class biosafety cabinets, and ISO 8/Class Facility Support Areas. Every procedure is performed in dedicated cleanrooms equipped with qualified biosafety cabinets, incubators, freezers, refrigerators, and centrifuges for iPSC generation. All the equipment is calibrated periodically by certified vendors to ensure accuracy. To acquire quality assurance to the maximum level, we have incorporated certified materials, reliable suppliers, qualified equipment, detailed SOPs, and rigid process validations at each step. Environmental monitoring program and procedures for the manufacturing suite are conducted to assure compliance of the manufacturing environments according to the 21 Code of Federal Regulation (CFR). The GMP Manufacturing suite is monitored for non-viable airborne particulate, viable airborne particulate, and viable surface according to ISO 14644-1:2015. Personnel dedicated to processing iPSCs have been thoroughly trained and manufacturing operations are monitored and recorded daily. In addition, the Quality Control-Designated Laboratory supports all product testing.
Quality control of iPSC
The critical quality attributes in respect to clinical-grade iPSCs are still under development by Allele Biotechnology and the entire field. The International Stem Cell Banking Initiative is currently providing general guidance regarding standards to qualify clinical grade iPSC lines. There is a general agreement in the field, as manifested through workshop discussions and publications, of the critical quality attributes that include cell identity, microbiological sterility, genetic fidelity and stability, viability, characterization, and differentiation potency. The combination of these data should provide enough quality control information at the clinical stage to determine the quality and safety of the iPSC line. However, the specific parameters, assays, and standards to validate iPSC lines still need to be defined to acquire such data. Allele Biotechnology is taking a lead in this development.
As will be described elsewhere, strict standards and methods for quality control have been adopted by Allele Biotechnology to qualify cGMP-iPSC lines. Sterility testing is performed routinely following FDA guidance for biological products, including mycoplasma and endotoxin tests specified under title 21 of the CFR. Purity of cell population is determined by expression of pluripotent markers using flow cytometry analysis and RNA sequencing. Genome integrity is evaluated by high-throughput sequencing of the donor cells and reprogrammed cells, genetic identity matching with starting material by short tandem repeat (STR) analysis, and karyotypic analysis is routinely performed to evaluate genome stability. Finally, careful analysis of differentiation potential is performed to confirm potency.
iPSC Culture under cGMP conditions
Material
Selecting GMP-grade or animal-origin-free reagents is extremely important to provide defined and consistent iPSC manufacturing. In addition, quality control methods need to be performed to demonstrate that the generated iPSCs meet quality standards. For Allele's cGMP-iPSC culture, cGMP-grade reagents and materials were used whenever possible; otherwise, strict quality control tests have been developed and implemented to obtain quality assurance.
In the protocol published in 2016 (Ni et al., 2016), we provided a detailed method to generate iPSC under cGMP conditions using the mRNA technology; here we present a protocol to culture and expand cGMP-iPSC. We provide information regarding where to acquire reagents, vessels, and other materials that are compliant with cGMP standards and regulations.
Extensive optimization of cell culture media has been performed to develop a completely defined medium that efficiently supports derivation and culturing of human iPSCs (Chen et al., 2011). Multiple matrix proteins and synthetic surfaces have been shown to support human ES cell growth, but extensive evaluation is required to find the matrix that ensures efficient cell attachment and survival of pluripotent stem cell in defined medium (Chen et al., 2011). To find optimal cell culture conditions for iPSC under cGMP standards, we have evaluated parameters related to stem cell media, cell culture matrix, and subculture reagents in the Process Development phase. Reagents, materials, and qualified vendors are detailed in each section of the protocol.
Cell culture methods
iPSC adherent cultures have been considered labor-intensive and time-consuming; however, the protocol presented here provides a set of methods to culture iPSC in a time-efficient manner, which allows the establishment of a reliable platform for cGMP manufacturing. Our optimized cell culture techniques enable more efficient iPSC culturing by reducing the time of manufacturing operations and increasing reproducibility and quality.
Several changes in iPSC culture conditions and adoption of optimal manipulation of iPS cells have been proposed to reduce genetic aberrations during reprogramming and subsequent iPSC expansion (Lee, Tang, Rao, Weissman, & Wu, 2013). Culture conditions at physiologic oxygen levels are among the most important factors that could prevent genomic instability of iPSC lines. Several lines of evidence support this notion. For instance, reprogrammed cells have been shown to present accelerated cell growth and division when changing from oxidative respiration to oxidative glycolysis, which increases the potential for leakage of reactive oxygen species (ROS) into the iPSC cytoplasm. ROS may oxidize nucleotides, leading to the mutagenic 7,8 dihydro-8-oxoguanine, capable of generating single- and double-strand DNA breaks (Turinetto, Orlando, & Giachino, 2017). Hence, finding optimal culture conditions for reprogrammed cells is critical to reduce genetic alterations associated to adaptation in culture and also to avoid lengthy culture time, particularly when large master or working cell banks are being generated. For this purpose, low oxygen environment has been incorporated into our cGMP platform for both generating and expanding iPSCs in order to ensure genomic stability throughout the whole manufacturing process.
During iPSC expansion, harvesting of iPSC clusters using non-enzymatic passaging method affords high controllability of cluster size while preserving cell viability. Moreover, EDTA-based cell dissociation does not require pretreatment of the culture with ROCK inhibitor or enzyme neutralization as posttreatment, which are recommended for enzymatic methods (Beers et al., 2012). In addition, EDTA treatment preferentially dissociates iPSCs over any differentiated cells, thereby at the same time providing a suitable method for iPSCs selection from heterogeneous populations when spontaneous differentiation occurs, avoiding the need of manual colony picking (Beers et al., 2012). The protocol we provide in this article for iPSC dissociation integrates a time-efficient procedure and cGMP grade reagents. Importantly, our optimized method eliminates the need of centrifugation of the cells prior to seeding to a new culture vessel; thus, reduces cellular stress.
This method for iPS cell culture provides high viability after dissociation as well as a consistent cell stability after cryopreservation, resulting in reduced cost and time to meet iPS cell banking requirements at various expansion stages. Cell stability of our cell bank has been validated using cell recovery and viability assays after thawing, reaching post-thaw recovery rates of over 90%.
Overall, this fully tested protocol for iPSC culture has been optimized for efficient iPSC proliferation and stemness. It allows scalable manufacturing operations that are fully compliant with cGMP standards; it can be adopted by any cGMP facility, providing a valuable platform to generate iPSCs for clinical applications. Moreover, this platform is designed to allow an easy transition to closed cell culture systems towards automation, which facilitates scalability and controllability and reduces operator-introduced variability, a direction of development Allele is actively engaged in.
Troubleshooting
There are a number of key parameters that could impact the quality of iPSC cultures. Establishment of vigorous cGMP level protocols provides the best route to quality control of iPSC through the entire manufacturing process. During our Process Development stage, we addressed the factors that could potentially affect the quality of the iPSC culture.
Cell culture handling can be a source of severe disruption of iPSC clusters or damages to cells can be caused by excessive pipetting of cell suspension, causing low cell viability after dissociation. In addition, overconfluence of iPSC cultures should be avoided to enhance cell viability. Cell dissociation schedule should be carefully determined and consistently executed. Through early experimentation, we determined that 4 days was the optimal dissociation regimen when following our criteria for cell seeding density.
iPSC could undergo spontaneous differentiation in vitro, which is commonly characterized by the presence of colonies without distinct borders, flattened cells, or colonies with less compacted areas. Major causes for differentiation are (1) overconfluent cultures : Therefore, it is essential that colonies are passaged before reaching full confluence; (2) poor quality of ECM coating : This can be prevented by QC testing of each lot of coated vessels prior to transferring into manufacturing; alternatively, increasing the volume of coating solution could reduce the risk of coating failure; (3) high oxygen levels We suggest adjustment to low oxygen tension of 3%-5% O2 for all pluripotent stem cell culturing; (4) cell culture exposed to high fluctuations of temperature : This can occur when cell culture is kept for extended periods of time outside of the incubator; thus, the execution of the protocol should be studied and evaluated by the managers to reduce operation time. Moreover, addition of pre-warmed reagents into cultures is recommended but prolonged exposure of stock media to 37°C should be limited to keep the growth factors from losing activities. Removal of differentiated cells can be achieved during the dissociation step by performing short incubation times with EDTA-based dissociation reagent since iPSCs will be preferentially harvested and differentiated cells will remain attached to the current culture surface.
If poor cell recovery rates or low cell attachment after cryopreservation is detected, it is advisable that the thawing procedure should be carried out more quickly and proper concentration of ROCKi added into the media at the time of thawing.
To avoid spontaneous chromosomal abnormalities in cultured iPSCs, several precautionary steps may be implemented: (1) make sure that oxygen tension is maintained at pluripotent stem cell-appropriate physiological levels, (2) careful selection of extracellular matrices that best maintain the normal karyotypes of pluripotent stem cells, such as human vitronectin or laminin-521 (Braam et al., 2008; Rodin et al., 2010), (3) use only enzyme-free methods for cell dissociation to prevent passage-induced mutations during prolonged culturing (Beers et al., 2012).
Author Contribution and Acknowledgments
YN, YZ, and TR lead the cGMP team in developing the protocols; TR and JW wrote the manuscript; YN reviewed the manuscript. We thank Lisa Stewart, Hemangiben Mehta, Chuanpit Boonchitsirikul, Francis Bauzon and the whole Allele cGMP operation team; quality and regulatory staff, Drs. Kathrin Copley, Joseph Chuang; and equipment and facility management, David Dale, Shirlee Smhart for their support and contribution to work reported in this article.
Literature Cited
- Aach, J., Lunshof, J., Iyer, E., & Church, G. M. (2017). Addressing the ethical issues raised by synthetic human entities with embryo-like features. Elife , 6, e20674. doi: 10.7554/eLife.20674
- Becker, K. A., Stein, J. L., Lian, J. B., van Wijnen, A. J., & Stein, G. S. (2010). Human embryonic stem cells are pre-mitotically committed to self-renewal and acquire a lengthened G1 phase upon lineage programming. Journal of Cellular Physiology , 222(1), 103–110. doi: 10.1002/jcp.21925
- Beers, J., Gulbranson, D. R., George, N., Siniscalchi, L. I., Jones, J., Thomson, J. A., & Chen, G. (2012). Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nature Protocols , 7(11), 2029–2040. doi: 10.1038/nprot.2012.130
- Bershteyn, M., Hayashi, Y., Desachy, G., Hsiao, E. C., Sami, S., Tsang, K. M., … Wynshaw-Boris, A. (2014). Cell-autonomous correction of ring chromosomes in human induced pluripotent stem cells. Nature , 507(7490), 99–103. doi: 10.1038/nature12923
- Braam, S. R., Zeinstra, L., Litjens, S., Ward-van Oostwaard, D., van den Brink, S., van Laake, L., … Mummery, C. L. (2008). Recombinant vitronectin is a functionally defined substrate that supports human embryonic stem cell self-renewal via alphavbeta5 integrin. Stem Cells , 26(9), 2257–2265. doi: 10.1634/stemcells.2008-0291
- Chen, G., Gulbranson, D. R., Hou, Z., Bolin, J. M., Ruotti, V., Probasco, M. D., … Thomson, J. A. (2011). Chemically defined conditions for human iPSC derivation and culture. Nature Methods , 8(5), 424–429. doi: 10.1038/nmeth.1593
- Cyranoski, D. (2018). ‘Reprogrammed’ stem cells approved to mend human hearts for the first time. Nature , 557(7707), 619–620. doi: 10.1038/d41586-018-05278-8
- Dakhore, S., Nayer, B., & Hasegawa, K. (2018). Human Pluripotent Stem Cell Culture: Current Status, Challenges, and Advancement. Stem Cells International , 2018, 7396905. doi: 10.1155/2018/7396905
- de Almeida, P. E., Ransohoff, J. D., Nahid, A., & Wu, J. C. (2013). Immunogenicity of pluripotent stem cells and their derivatives. Circulation Research , 112(3), 549–561. doi: 10.1161/CIRCRESAHA.111.249243
- Garitaonandia, I., Amir, H., Boscolo, F. S., Wambua, G. K., Schultheisz, H. L., Sabatini, K., … Laurent, L. C. (2015). Increased risk of genetic and epigenetic instability in human embryonic stem cells associated with specific culture conditions. Plos One , 10(2), e0118307. doi: 10.1371/journal.pone.0118307
- Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M. P., Wulffraat, N., Leboulch, P., … Cavazzana-Calvo, M. (2003). LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science , 302(5644), 415–419. doi: 10.1126/science.1088547
- Hanna, J., Cheng, A. W., Saha, K., Kim, J., Lengner, C. J., Soldner, F., … Jaenisch, R. (2010). Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proceedings of the National Academy of Sciences , 107(20), 9222–9227. doi: 10.1073/pnas.1004584107
- Laurent, L. C., Ulitsky, I., Slavin, I., Tran, H., Schork, A., Morey, R., … Loring, J. F. (2011). Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell , 8(1), 106–118. doi: 10.1016/j.stem.2010.12.003
- Lee, A. S., Tang, C., Rao, M. S., Weissman, I. L., & Wu, J. C. (2013). Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nature Medicine , 19(8), 998–1004. doi: 10.1038/nm.3267
- Letourneau, A., Santoni, F. A., Bonilla, X., Sailani, M. R., Gonzalez, D., Kind, J., … Antonarakis, S. E. (2014). Domains of genome-wide gene expression dysregulation in Down's syndrome. Nature , 508(7496), 345–350. doi: 10.1038/nature13200
- Levenstein, M. E., Ludwig, T. E., Xu, R. H., Llanas, R. A., VanDenHeuvel-Kramer, K., Manning, D., & Thomson, J. A. (2006). Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells , 24(3), 568–574. doi: 10.1634/stemcells.2005-0247
- Liu, W., & Chen, G. (2014). Cryopreservation of human pluripotent stem cells in defined medium. Current Protocols in Stem Cell Biology , 31, 1C 17 11–13. doi: 10.1002/9780470151808.sc01c17s31
- Ludwig, T. E., Levenstein, M. E., Jones, J. M., Berggren, W. T., Mitchen, E. R., Frane, J. L., … Thomson, J. A. (2006). Derivation of human embryonic stem cells in defined conditions. Nature Biotechnology , 24(2), 185–187. doi: 10.1038/nbt1177
- Malik, N., & Rao, M. S. (2013). A review of the methods for human iPSC derivation. Methods in Molecular Biology , 997, 23–33. doi: 10.1007/978-1-62703-348-0_3
- Mandai, M., Kurimoto, Y., & Takahashi, M. (2017). Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. New England Journal of Medicine , 377(8), 792–793. doi: 10.1056/NEJMc1706274
- Neganova, I., Zhang, X., Atkinson, S., & Lako, M. (2009). Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene , 28(1), 20–30. doi: 10.1038/onc.2008.358
- Ni, Y., Zhao, Y., Warren, L., Higginbotham, J., & Wang, J. (2016). cGMP Generation of Human Induced Pluripotent Stem Cells with Messenger RNA. Current Protocols in Stem Cell Biology , 39(1), 4A 6 1–4A 6 25. doi: 10.1002/cpsc.18
- Okita, K., Matsumura, Y., Sato, Y., Okada, A., Morizane, A., Okamoto, S., … Yamanaka, S. (2011). A more efficient method to generate integration-free human iPS cells. Nature Methods , 8(5), 409–412. doi: 10.1038/nmeth.1591
- Rodin, S., Domogatskaya, A., Strom, S., Hansson, E. M., Chien, K. R., Inzunza, J., … Tryggvason, K. (2010). Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nature Biotechnology , 28(6), 611–615. doi: 10.1038/nbt.1620
- Ruiz, S., Panopoulos, A. D., Herrerias, A., Bissig, K. D., Lutz, M., Berggren, W. T., … Izpisua Belmonte, J. C. (2011). A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Current Biology , 21(1), 45–52. doi: 10.1016/j.cub.2010.11.049
- Schlaeger, T. M., Daheron, L., Brickler, T. R., Entwisle, S., Chan, K., Cianci, A., … Daley, G. Q. (2015). A comparison of non-integrating reprogramming methods. Nature Biotechnology , 33(1), 58–63. doi: 10.1038/nbt.3070
- Seki, T., Yuasa, S., Oda, M., Egashira, T., Yae, K., Kusumoto, D., … Fukuda, K. (2010). Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell , 7(1), 11–14. doi: 10.1016/j.stem.2010.06.003
- Shi, Y., Inoue, H., Wu, J. C., & Yamanaka, S. (2017). Induced pluripotent stem cell technology: A decade of progress. Nature Reviews Drug Discovery , 16(2), 115–130. doi: 10.1038/nrd.2016.245
- Takahashi, J. (2019). Preparing for first human trial of induced pluripotent stem cell-derived cells for Parkinson's disease: An interview with Jun Takahashi. Regenerative Medicine , 14(2), 93–95. doi: 10.2217/rme-2018-0158
- Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., & Yamanaka, S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell , 131(5), 861–872. doi: 10.1016/j.cell.2007.11.019
- Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell , 126(4), 663–676. doi: 10.1016/j.cell.2006.07.024
- Turinetto, V., Orlando, L., & Giachino, C. (2017). Induced Pluripotent Stem Cells: Advances in the Quest for Genetic Stability during Reprogramming Process. International Journal of Molecular Sciences , 18(9), 1952. doi: 10.3390/ijms18091952
- Warren, L., Manos, P. D., Ahfeldt, T., Loh, Y. H., Li, H., Lau, F., … Rossi, D. J. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell , 7(5), 618–630. doi: 10.1016/j.stem.2010.08.012
- Warren, L., Ni, Y., Wang, J., & Guo, X. (2012). Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Scientific Reports , 2, 657. doi: 10.1038/srep00657
- Watanabe, K., Ueno, M., Kamiya, D., Nishiyama, A., Matsumura, M., Wataya, T., … Sasai, Y. (2007). A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology , 25(6), 681–686. doi: 10.1038/nbt1310
- Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., … Thomson, J. A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science , 318(5858), 1917–1920. doi: 10.1126/science.1151526
- Zhou, H., Wu, S., Joo, J. Y., Zhu, S., Han, D. W., Lin, T., … Ding, S. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell , 4(5), 381–384. doi: 10.1016/j.stem.2009.04.005
Internet Resources
Reference of clinical trial using iPSC-derived cells for cancer immunotherapy.
Key References
- Ni et al. (2016). See above.
Describes a protocol to generate iPSCs through an established cGMP process.
Citing Literature
Number of times cited according to CrossRef: 34
- Xi Lu, Eli Perr, Tahmina Naqvi, David Galitz, Marnelle Andersen, David Grabowski, Anthony Person, Alex Kalyuzhny, Kevin C. Flynn, A Novel Recombinant Vitronectin Variant Supports the Expansion and Differentiation of Pluripotent Stem Cells in Defined Animal-Free Workflows, Cells, 10.3390/cells13181566, 13 , 18, (1566), (2024).
- Ratchapong Netsrithong, Laura Garcia-Perez, Maria Themeli, Engineered T cells from induced pluripotent stem cells: from research towards clinical implementation, Frontiers in Immunology, 10.3389/fimmu.2023.1325209, 14 , (2024).
- Pengfei Yu, Carol Christine Bosholm, Hainan Zhu, Zhongping Duan, Anthony Atala, Yuanyuan Zhang, Beyond waste: understanding urine’s potential in precision medicine, Trends in Biotechnology, 10.1016/j.tibtech.2024.01.009, 42 , 8, (953-969), (2024).
- Hannah W. Song, Jennifer N. Solomon, Fernanda Masri, Amanda Mack, Nisha Durand, Emmanuelle Cameau, Noushin Dianat, Arwen Hunter, Steve Oh, Brianna Schoen, Matthew Marsh, Christopher Bravery, Cenk Sumen, Dominic Clarke, Kapil Bharti, Julie G. Allickson, Uma Lakshmipathy, Bioprocessing considerations for generation of iPSCs intended for clinical application: perspectives from the ISCT Emerging Regenerative Medicine Technology working group, Cytotherapy, 10.1016/j.jcyt.2024.05.024, 26 , 11, (1275-1284), (2024).
- Gamal A. Atia, Fatema Rashed, Ehab S. Taher, Ssang-Goo Cho, Ahmed Abdal Dayem, Magdalen M. Soliman, Hany K. Shalaby, Nourelhuda A. Mohammed, Noha Taymour, Mohamed El-Sherbiny, Elturabi Ebrahim, Mahmoud M. Ramadan, Afaf Abdelkader, Mohamed Abdo, Ahmed A. Aldarmahi, Ahmed M. Atwa, Duaa A. Bafail, Ahmed Abdeen, Challenges of therapeutic applications and regenerative capacities of urine based stem cells in oral, and maxillofacial reconstruction, Biomedicine & Pharmacotherapy, 10.1016/j.biopha.2024.117005, 177 , (117005), (2024).
- Saumey Jain, Dimitrios Voulgaris, Surangrat Thongkorn, Rick Hesen, Alice Hägg, Mohsen Moslem, Anna Falk, Anna Herland, On‐Chip Neural Induction Boosts Neural Stem Cell Commitment: Toward a Pipeline for iPSC‐Based Therapies, Advanced Science, 10.1002/advs.202401859, 11 , 25, (2024).
- Miguel Mendivil-Perez, Carlos Velez-Pardo, Francisco Lopera, Kenneth S. Kosik, Marlene Jimenez-Del-Rio, PSEN1 E280A Cholinergic-like Neurons and Cerebral Spheroids Derived from Mesenchymal Stromal Cells and from Induced Pluripotent Stem Cells Are Neuropathologically Equivalent, International Journal of Molecular Sciences, 10.3390/ijms24108957, 24 , 10, (8957), (2023).
- Yoshiki Nakashima, Hiroki Iguchi, Eiko Shimizu, Minh N.T. Le, Kenta Takakura, Yuta Nakamura, Teruhiko Yanagisawa, Rutvi Sanghavi, Satoshi Haneda, Masayoshi Tsukahara, Improved Production of Induced Pluripotent Stem Cells Using Dot Pattern Culture Plates, Tissue Engineering Part C: Methods, 10.1089/ten.tec.2023.0068, 29 , 9, (410-423), (2023).
- Elke Gabriel, Walid Albanna, Giovanni Pasquini, Anand Ramani, Natasa Josipovic, Aruljothi Mariappan, Maria Giovanna Riparbelli, Giuliano Callaini, Celeste M. Karch, Olivier Goureau, Argyris Papantonis, Volker Busskamp, Toni Schneider, Jay Gopalakrishnan, Generation of iPSC-derived human forebrain organoids assembling bilateral eye primordia, Nature Protocols, 10.1038/s41596-023-00814-x, 18 , 6, (1893-1929), (2023).
- Roger A. Barker, Melissa Carpenter, Catriona H.M. Jamieson, Charles E. Murry, Graziella Pellegrini, Rajesh C. Rao, Jihwan Song, Lessons learnt, and still to learn, in first in human stem cell trials, Stem Cell Reports, 10.1016/j.stemcr.2022.11.019, 18 , 8, (1599-1609), (2023).
- Fei-Chien Chang, Matthew Michael James, Abdullah Mohammed Qassab, Yang Zhou, Yoshiki Ando, Min Shi, Miqin Zhang, 3D chitosan scaffolds support expansion of human neural stem cells in chemically defined condition, Matter, 10.1016/j.matt.2023.08.014, 6 , 10, (3631-3660), (2023).
- Valeria Peli, Mario Barilani, Araceli Rivera-Ordaz, Paolo Manzini, Francesco Rusconi, Cristiana Lavazza, Silvia Cimoni, Lorenza Lazzari, Validation of an automated viable cell counting assay for GMP manufacturing of human induced pluripotent stem cells, Biochemical Engineering Journal, 10.1016/j.bej.2023.108953, 196 , (108953), (2023).
- Jinhu Li, Yurou Wu, Xiang Yao, Yao Tian, Xue Sun, Zibo Liu, Xun Ye, Chunjie Wu, Preclinical Research of Stem Cells: Challenges and Progress, Stem Cell Reviews and Reports, 10.1007/s12015-023-10528-y, 19 , 6, (1676-1690), (2023).
- Pelin Kilic, Begum Cosar, An Optimized Protocol for piggyBac-Induced iPSC Generation from hPBMCs by Automatic Electroporation, Stem Cells and Lineage Commitment, 10.1007/7651_2023_500, (193-205), (2023).
- Dosh Whye, Delaney Wood, Wardiya Afshar Saber, Erika M. Norabuena, Nina R. Makhortova, Mustafa Sahin, Elizabeth D. Buttermore, A Robust Pipeline for the Multi‐Stage Accelerated Differentiation of Functional 3D Cortical Organoids from Human Pluripotent Stem Cells, Current Protocols, 10.1002/cpz1.641, 3 , 1, (2023).
- Shashank Pandey, Michal Jirásko, Jan Lochman, Alexandr Chvátal, Magdalena Chottova Dvorakova, Radek Kučera, iPSCs in Neurodegenerative Disorders: A Unique Platform for Clinical Research and Personalized Medicine, Journal of Personalized Medicine, 10.3390/jpm12091485, 12 , 9, (1485), (2022).
- Markus Uhrig, Fernando Ezquer, Marcelo Ezquer, Improving Cell Recovery: Freezing and Thawing Optimization of Induced Pluripotent Stem Cells, Cells, 10.3390/cells11050799, 11 , 5, (799), (2022).
- Marta Włodarczyk, Beata Pyrzynska, CAR-NK as a Rapidly Developed and Efficient Immunotherapeutic Strategy against Cancer, Cancers, 10.3390/cancers15010117, 15 , 1, (117), (2022).
- Kalaiselvaan Thanaskody, Amirah Syamimi Jusop, Gee Jun Tye, Wan Safwani Wan Kamarul Zaman, Sylvia Annabel Dass, Fazlina Nordin, MSCs vs. iPSCs: Potential in therapeutic applications, Frontiers in Cell and Developmental Biology, 10.3389/fcell.2022.1005926, 10 , (2022).
- Zhiyao Ma, Marcelo Augusto Szymanskide Toledo, Paul Wanek, Mohamed H. Elsafi Mabrouk, Francis Smet, Rock Pulak, Simon Pieske, Tobias Piotrowski, Werner Herfs, Christian Brecher, Robert H. Schmitt, Wolfgang Wagner, Martin Zenke, Cell Cluster Sorting in Automated Differentiation of Patient-specific Induced Pluripotent Stem Cells Towards Blood Cells, Frontiers in Bioengineering and Biotechnology, 10.3389/fbioe.2022.755983, 10 , (2022).
- Lay Shuen Tan, Juin Ting Chen, Lillian Yuxian Lim, Adrian Kee Keong Teo, Manufacturing clinical‐grade human induced pluripotent stem cell‐derived beta cells for diabetes treatment, Cell Proliferation, 10.1111/cpr.13232, 55 , 8, (2022).
- Amanda Cooper, Adam Sidaway, Abishek Chandrashekar, Elizabeth Latta, Krishnendu Chakraborty, Jingyou Yu, Katherine McMahan, Victoria Giffin, Cordelia Manickam, Kyle Kroll, Matthew Mosher, R. Keith Reeves, Rihab Gam, Elisa Arthofer, Modassir Choudhry, Tom Henley, Dan H. Barouch, A genetically engineered, stem-cell-derived cellular vaccine, Cell Reports Medicine, 10.1016/j.xcrm.2022.100843, 3 , 12, (100843), (2022).
- Paolo Manzini, Valeria Peli, Araceli Rivera-Ordaz, Silvia Budelli, Mario Barilani, Lorenza Lazzari, Validation of an automated cell counting method for cGMP manufacturing of human induced pluripotent stem cells, Biotechnology Reports, 10.1016/j.btre.2022.e00708, 33 , (e00708), (2022).
- Jianjian Shi, Lei Wei, Rho Kinases in Embryonic Development and Stem Cell Research, Archivum Immunologiae et Therapiae Experimentalis, 10.1007/s00005-022-00642-z, 70 , 1, (2022).
- Pankhi Vatsa, Sadaf Jahan, Uzair Ahmad Ansari, Andleeb Khan, Shabir Ahmad Mir, Bader Alshehri, Ranjay Kumar Choudhary, Arif Jamal Siddiqui, Stem Cell Safety and Sterility Testing: A Promising Approach in Regenerative Medicine, Stem Cell Production, 10.1007/978-981-16-7589-8_9, (205-232), (2022).
- Daniela Lisini, Simona Frigerio, Sara Nava, Simona Pogliani, Stem Cell Production: Processes, Practices, and Regulation, Stem Cell Production, 10.1007/978-981-16-7589-8_6, (125-158), (2022).
- Jason P. Acker, Mykola Bondarovych, Ricarda Brunotte, Iryna A. Buriak, Barry J. Fuller, Birgit Glasmacher, Anatoliy M. Goltsev, Jiří Gregor, Oleksandr Gryshkov, Kieran Herrity, Barbora Honegrová, Charles J. Hunt, Miroslava Jandová, Brian H. Johnstone, Peter Kilbride, Miriam Lánská, Jennifer Mann, Pavel Měřička, Kelsey G. Musall, Vitalii Mutsenko, Olga Mykhailova, Yuriy Petrenko, Jakub Radocha, Aubrey M. Sherry, Glyn Nigel Stacey, Lubomír Štěrba, Doris Vokurková, Nishaka William, Erik J. Woods, Preservation and Storage of Cells for Therapy: Current Applications and Protocols, Cell Engineering and Regeneration, 10.1007/978-3-319-37076-7_68-2, (1-69), (2022).
- Jason P. Acker, Mykola Bondarovych, Ricarda Brunotte, Iryna A. Buriak, Barry J. Fuller, Birgit Glasmacher, Anatoliy M. Goltsev, Jiří Gregor, Oleksandr Gryshkov, Kieran Herrity, Barbora Honegrová, Charles J. Hunt, Miroslava Jandová, Brian H. Johnstone, Peter Kilbride, Miriam Lánská, Jennifer Mann, Pavel Měřička, Kelsey G. Musall, Vitalii Mutsenko, Olga Mykhailova, Yuriy Petrenko, Jakub Radocha, Aubrey M. Sherry, Glyn Nigel Stacey, Lubomír Štěrba, Doris Vokurková, Nishaka William, Erik J. Woods, Preservation and Storage of Cells for Therapy: Current Applications and Protocols, Cell Engineering and Regeneration, 10.1007/978-3-319-37076-7_68-1, (1-69), (2022).
- Fei‐Chien Chang, Yang Zhou, Matthew Michael James, Hadi M. Zareie, Yoshiki Ando, Jihui Yang, Miqin Zhang, Effect of Degree of Deacetylation of Chitosan/Chitin on Human Neural Stem Cell Culture, Macromolecular Bioscience, 10.1002/mabi.202200389, 23 , 1, (2022).
- Dosh Whye, Delaney Wood, Kristina H. Kim, Cidi Chen, Nina Makhortova, Mustafa Sahin, Elizabeth D. Buttermore, Dynamic 3D Combinatorial Generation of hPSC‐Derived Neuromesodermal Organoids With Diverse Regional and Cellular Identities, Current Protocols, 10.1002/cpz1.568, 2 , 10, (2022).
- Peter J. Koch, Saiphone Webb, Jessica A. Gugger, Maddison N. Salois, Maranke I. Koster, Differentiation of Human Induced Pluripotent Stem Cells into Keratinocytes, Current Protocols, 10.1002/cpz1.408, 2 , 4, (2022).
- Giuseppe Scesa, Raffaella Adami, Daniele Bottai, iPSC Preparation and Epigenetic Memory: Does the Tissue Origin Matter?, Cells, 10.3390/cells10061470, 10 , 6, (1470), (2021).
- Ishnoor Sidhu, Sonali P. Barwe, Kristi L. Kiick, E. Anders Kolb, Anilkumar Gopalakrishnapillai, A 3-D hydrogel based system for hematopoietic differentiation and its use in modeling down syndrome associated transient myeloproliferative disorder, Biomaterials Science, 10.1039/D1BM00442E, 9 , 18, (6266-6281), (2021).
- Dusko Ilic, Mirjana Liovic, Industry Updates from the Field of Stem Cell Research and Regenerative Medicine in July 2020, Regenerative Medicine, 10.2217/rme-2020-0122, 15 , 11, (2253-2260), (2020).

