High-throughput Method for Novel Medium Development for Culture of Anaerobic Gut Bacteria
Kevin Lee, Kevin Lee, Kristina Wornell, Kristina Wornell, Bikiran Pardesi, Bikiran Pardesi, Svetlana Boycheva, Svetlana Boycheva, Anthony M. Roberton, Anthony M. Roberton, W. Lindsey White, W. Lindsey White
Abstract
Gut microbiota play important roles in the health of their host and detailed investigation of these organisms requires in vitro culture. Culturing strictly anaerobic bacteria can be a challenge as the gut environment they inhabit is nutritionally complex. Use of complex media containing nutritionally rich but undefined gut fluid reduces the accuracy of physiological and metabolomic studies. Here we present a high-throughput protocol for comparing growth rates of fastidiously anaerobic bacteria on different media. These protocols can be used to develop a solid medium made up of commercially sourced ingredients, providing replicable growth conditions for previously uncultured anaerobic bacteria. As many fastidious bacteria grow poorly in a liquid broth, these protocols measure bacterial growth rate on solid media. These protocols speed up and simplify the growth rate measurement process by using a multiwell format and equations in place of physical McFarland standards to calculate approximate cell density. Bacterial strains belonging to the families Erysipelotrichaceae and Lachnospiraceae (phylum Firmicutes) isolated from the hindgut of Kyphosus sydneyanus were used to demonstrate the efficacy of these protocols. Bacterial growth rates were compared between a nutritionally rich medium with gut fluid versus a novel replicable medium with mannitol. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
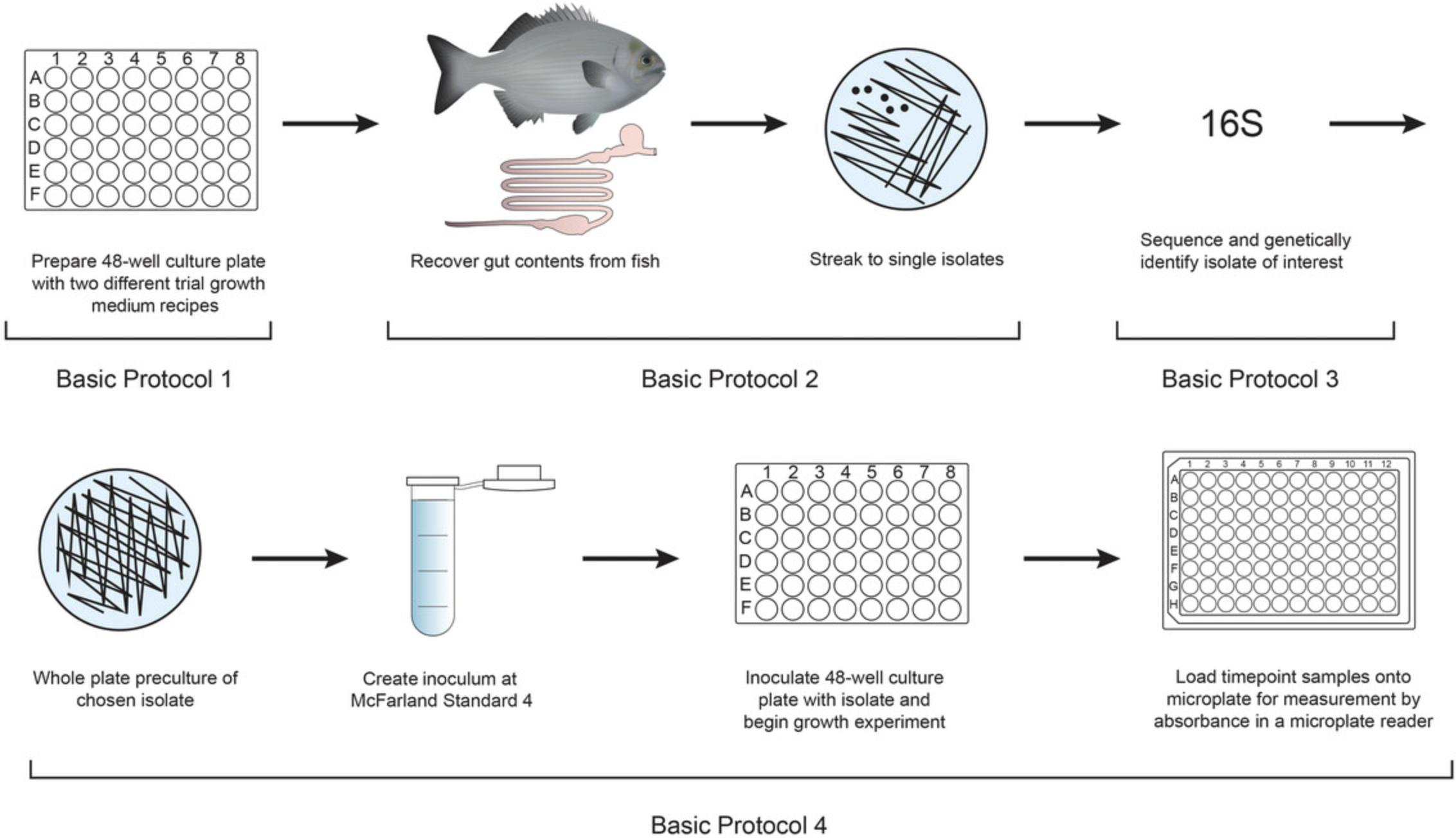
Basic Protocol 1 : Preparation of solid YCFA growth medium
Basic Protocol 2 : Collection of fish gut samples and plating to single isolates
Basic Protocol 3 : Genetic identification of single isolates with colony PCR and 16S rRNA gene sequencing
Basic Protocol 4 : Measurement of bacterial growth rates on solid media
INTRODUCTION
Culturing gut bacteria in vitro is a great challenge as the gut environment is anaerobic and nutritionally complex. Bacteria isolated from this environment are obligately anaerobic (Hungate, 1975), requiring the use of specialized techniques and equipment to ensure that all oxygen is removed from the system during in vitro culture. Gut bacterial isolates sourced from the same environment have varying and species-specific nutritional requirements (Tramontano et al., 2018). Predicting the likely essential growth factors of a gut bacterial isolate is difficult, and optimizing the growth rate of a chosen isolate on a replicable medium is a challenge with many possible limiting factors.
The difficulties in isolation and maintenance of viable gut bacteria cultures makes genomic sequencing the preferred alternative to in vitro analysis of these organisms. Genomic studies of environmentally isolated bacteria can suggest the presence of metabolic pathways (Béjà et al., 2000, 2002). However, to understand the physiology of novel microbes and the functional processes they engage in, cultivation is necessary (Stewart, 2012; Zengler et al., 2002). The protocols outlined in this study address the current challenge for microbiologists in achieving successful culture of environmentally isolated microorganisms in order to improve understanding of their metabolic properties and potential (Zengler et al., 2002). These protocols, outlined in Figure 1, are robust, reproducible, and easily adaptable.
Despite innovations in culture methods, many bacterial species are still classified as ‘viable non-culturable’ (Barer & Harwood, 1999), with sequencing studies revealing that less than 1% of extant bacterial species have been grown in the laboratory (Barer & Harwood, 1999; Bomar, Maltz, Colston, & Graf, 2011). This ‘great plate count anomaly’ is a decades-old problem in microbiological research, originally described by Staley and Konopka (1985). These as-yet uncultivated microorganisms represent a large unexplored reservoir of biological compounds, with great potential for structural diversity and use in novel drug development (Demain & Sanchez, 2009; Stott et al., 2008). However, the considerable time and energy required to produce primary bacterial cultures in vitro has led to a lack of interest by investigators and is one of the reasons for the large number of uncultivated bacteria (Lagier et al., 2015).
There is no standard process for isolating anaerobic microbes in vitro for the first time. Many gut bacteria of interest are still cultured in rich media with additions such as host gut fluid or blood (Lagier et al., 2015; Tramontano et al., 2018). These rich media recipes attempt to replicate the nutrient composition of the gut environment and are highly nutritionally complex, but problematically are chemically undefined. The use of media containing gut fluid reduces experimental reproducibility, as the nutrient profile varies among host organisms such as fish (Egerton, Culloty, Whooley, Stanton, & Ross, 2018), and thus is subject to variation from batch to batch. The use of rich media with gut fluid or blood limits the accuracy of metabolic and physiological investigations of microbes in culture (Tramontano et al., 2018).
Development of an alternative chemically defined culture medium requires trial-and-error experiments measuring the effect of nutrient additions on bacterial growth rates. For fastidious anaerobic microbes, this process is laborious and time consuming due to long incubation times and strict culture conditions. Obligate anaerobes isolated from the gut environment require specific bacteriological culture techniques (Bryant, 1972; Hungate, 1969) and media containing a variety of specific growth factors (Hensgens, Nienhuis-Kuiper, & Hansen, 1994; Jarrell & Kalmokoff, 1988; Scherer & Sahm 1981; Schönheit, Moll, & Thauer, 1979; Tanner & Wolf, 1988). Many gut bacteria species that have already been successfully cultured in vitro still take weeks to grow to detectable levels (Leadbetter, 2003). Unlike most bacterial growth, we have found that our fish gut bacterial isolates grow linearly rather than exponentially (Hall, Acar, Nandipati, & Barlow, 2013). This linear growth demonstrates the greater effect of limiting factors on the growth of these anaerobic bacteria, which cause reduced survival of the daughter cells of bacterial fission. The two most common ways to represent bacterial growth rate are doubling time (in hr) and first-order growth rate constant (α in min–1). However, these cannot be used in this case, as they are calculated based on an assumption of exponential bacterial growth. Therefore, in these protocols, we use the increase in colony forming units over time (CFU/hr) to represent bacterial growth.
Here we present a laboratory protocol for evaluating high-throughput microbial growth on a solid medium in strictly anaerobic conditions. This protocol begins with the isolation of bacteria from gut samples and preparation of an inoculum for multiwell cultures, and provides instructions for anaerobic culture maintenance and culture transfer to a microplate for growth rate measurement by optical density on a standard microplate reader. This approach is appropriate for efficient investigation of microbial growth rates on media with varying nutrient additions, with the outcome of developing a novel medium with defined ingredients, replicable in future work.
Importantly, these protocols have been designed for maximum recovery of viable anaerobic bacteria in in vitro culture. The results of Basic Protocols 1-3 are single bacterial isolates of known genetic identity, cultured on a rich medium containing host gut fluid. Solid medium is used in all protocols, as many anaerobic bacteria are unable to grow in liquid culture (see Current Protocols article Speers, Cologgi, & Reguera, 2009).
Basic Protocol 4 presents a method for high-throughput growth rate analysis on solid media. The usual method of measuring growth on solid media (colony counting) cannot be used here, as bacteria isolated from the gut may grow in mats or produce colonies that are invisible to the naked eye (Tramontano et al., 2018). This protocol is used to compare bacterial growth rates on novel replicable media to those on rich media and results in development of an optimal replicable medium for an isolate of interest. The protocol provides several calculations for converting absorbance readings to growth rates. If bacterial growth is linear, growth rates are calculated as an increase in colony-forming units over time (CFU/hr). If bacterial growth is exponential, growth rates are calculated as doubling time (in hr) or first-order growth rate constant (α in min–1).
These protocols describe how to isolate and identify bacteria from the fish gut while maintaining anaerobic culture conditions and use a high-throughput and replicable method for measuring the effect of various medium additions on growth rates. Results of these protocols may be used to develop an optimal replicable medium for gut bacteria isolates from fish or other organisms, including humans. This replicable medium may then be used during metabolomic analysis and to improve in vitro culture conditions of previously isolated and novel gut bacteria.
NOTE : All protocols involving live animals must be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must conform to government regulations for the care and use of laboratory animals.
STRATEGIC PLANNING
Equipment
Many bacteria isolated from the gut are strictly anaerobic and cannot tolerate exposure to oxygen (Hungate, 1975). Excluding all oxygen from an in vitro culture system requires specialized equipment and techniques. Detailed information on setting up a Hungate gassing apparatus (used in Basic Protocols 1, 2, and 4) is outlined in Hungate (1975) and Bryant (1972). This method requires the use of a Hungate gassing apparatus with the ability to deliver nitrogen, carbon dioxide, and hydrogen gas to the system. The technique, first introduced by Hungate (1969), enables aseptic and anaerobic transfer of ingredients between containers for generation of sterile anaerobic bacterial culture media. Gas is passed via a pressure-reducing valve through a hot reduced copper wire column, removing all trace oxygen present in the source gas (Moore, 1966). The gas passes out of this column through a glass Y tube, then through rubber tubing and out through 16- to 18-G bent gassing needles attached to 2-ml sterile cotton-plugged Luer-Lok syringe barrels (Bryant, 1972). A reducing agent (e.g., cysteine-HCl) is also added to the medium to ensure that all oxygen is removed (Bryant, 1972).
Other specialized equipment required to maintain in vitro culture of strictly anaerobic bacteria includes anaerobic jars, an anaerobic hood, and an incubator. The anaerobic jar, first introduced by Mcintosh, Fildes, and Bulloch (1916), has long been used for cultivating anaerobic microorganisms. The anaerobic jar used in this protocol (Fig. 2) was custom built for us by Mason Tool and Engineering Ltd. (Auckland, New Zealand). This anaerobic jar is filled with N2, CO2, and H2 (47.5%, 47.5%, 5% v/v/v) to 5 psi using the Hungate gassing apparatus (Fig. 3). Anaerobic hoods and incubators can be purchased from many suppliers and will interchangeably perform the same function. The anaerobic hood must be large enough to fit the anaerobic jar through the air lock. To ensure maintenance of anaerobic conditions in the hood, a catalyst (palladium) and a desiccant (calcium chloride) are used. To ensure maintenance of anaerobic conditions in the jars, a catalyst (palladium) and activated charcoal are present. Activated charcoal absorbs hydrogen sulfide produced by the bacteria, preventing it from affecting catalyst function. The catalyst, desiccant, and charcoal are activated by heating at 150°C for 1 hr. An anaerobic monitor allows quick detection of any oxygen entering the system. The incubator must also be large enough to fit the anaerobic jar and must have an adjustable temperature control that can be set to the body temperature of the host organism from which the gut bacteria were isolated. In our protocol, the gut bacteria are isolated from silver drummer (Kyphosus sydneyanus) and incubated at 20°C, which is within the 17°-23°C range of ambient seawater temperature used as K. sydneyanus body temperature in Mountfort, Campbell, and Clements (2002).


Bacteria of Interest
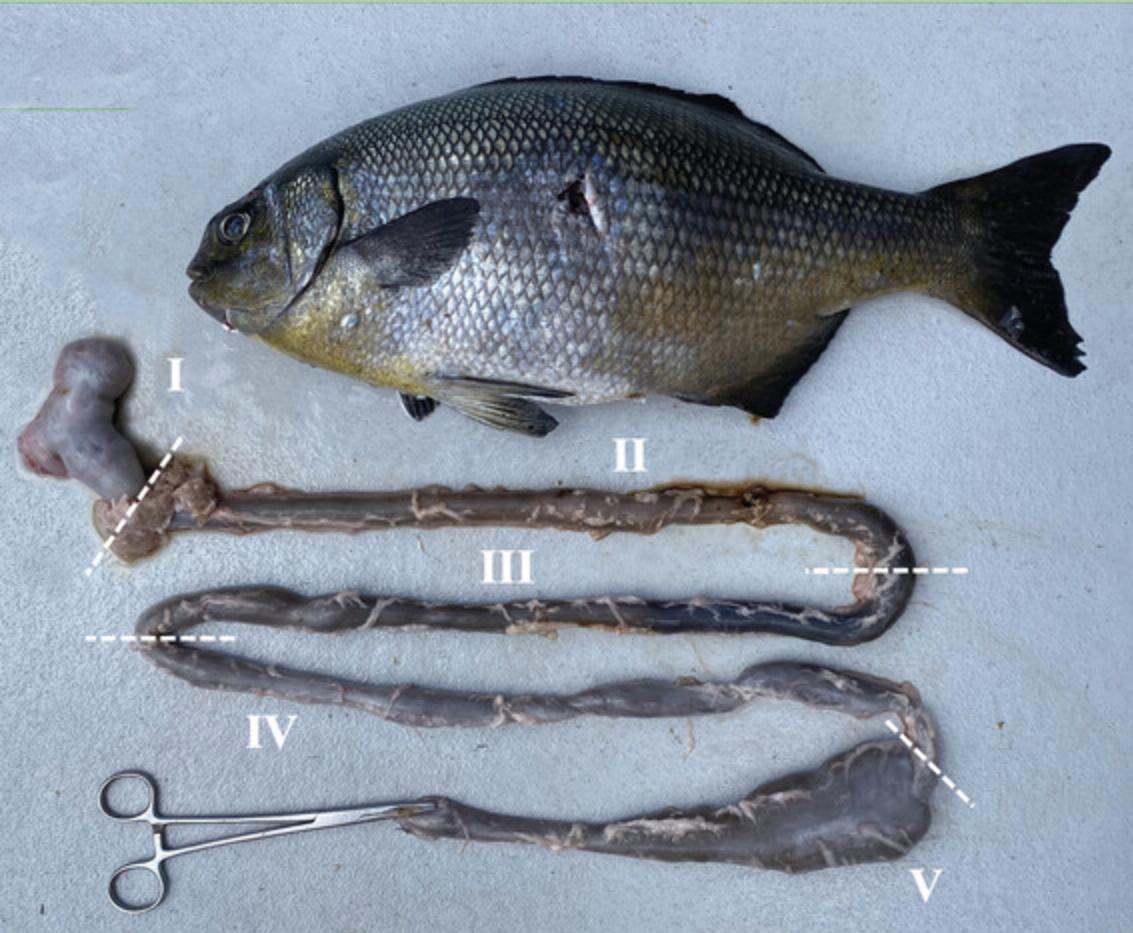
Proteobacteria, Firmicutes, and Bacteroidetes comprise 90% of the fish intestinal microbiota across species (Egerton et al., 2018; Ghanbari, Kneifel, & Domig, 2015). These protocols were verified using two bacterial isolates derived from section IV of the K. sydneyanus gut (Fig. 4). (1) Isolate BP52G in the Erysipelotrichaceae family, from the Firmicutes phylum. 16S rRNA gene sequence accessed from NCBI prokaryotic genome annotation pipeline (Tatusova et al., 2016). Accession number: MG827409. (2) Isolate BP5G in the Lachnospiraceae family, from the Firmicutes phylum. 16S rRNA gene sequence accessed from NCBI prokaryotic genome annotation pipeline (Tatusova et al., 2016). Accession number: CP092441.
Media
Yeast extract-casein hydrolysate-volatile fatty acids (YCFA) medium was designed by Dr. Flint's lab and the specific recipe is outlined in Duncan, Hold, Harmsen, Stewart, and Flint (2002). This work proved the effectiveness of YCFA as a base medium for the culture of anaerobic bacteria. Here we use an adapted version of YCFA medium with reduced yeast extract and tryptone (see Reagents and Solutions). Reducing nutrient richness reduces confounding variables during bacterial growth rate experiments, allowing the effect of nutrient additions to be seen clearly for all isolates. It also ensures that any novel medium developed is as minimal as possible.
The addition of gut fluid to this medium provides an unknown quantity and unknown variety of nutrients. Duncan et al. (2002) found that, when the older anaerobic M2GSC medium (Barcenilla et al., 2000; Miyazaki, Martin, Marinsek-Logar, & Flint, 1997) was used, gut fluid was necessary to cultivate the obligately anaerobic bacterium Fusobacterium prausnitzii from human feces. In contrast, they found that YCFA medium with added glucose contained all the nutrients required to culture F. prausnitzii in vitro. YCFA medium with K. sydneyanus gut fluid is used in these protocols as a standard against which the growth effects of alternative nutrient additions can be compared. Our research shows that YCFA with glucose alone does not support growth of strains BP5G and BP52G isolated from the hindgut of K. sydneyanus. This was not unexpected given the environment from which it was isolated. Members of the K. sydneyanus gut microbiota are likely under selection to use the abundant carbon substrates available in the host's gut. Mannitol is a common sugar alcohol present in brown algae, which is the main food source of this herbivorous (seaweed eating) fish (Clements & Choat, 1997). Also, mannitol concentration decreases along the gut of K. sydneyanus , correlating with an increase in gut microbiota density (White, Coveny, Robertson, & Clements, 2010), suggesting that mannitol is fermented by the gut microbiota. For these reasons, mannitol was chosen as the trial growth factor in these protocols. It was used at an overall 0.2% (w/v) or ∼10 mM concentration in YCFA, based on recommendations by Holdeman, Moore, and Cato (1977) and Browne et al. (2016), who used a 0.2% (w/v) concentration of sugars in their anaerobic media.
The growth rate of the chosen isolates (Erysipelotrichaceae and Lachnospiraceae) were measured on YCFA medium with mannitol (YCFA-M) and with gut fluid (YCFA-GF) in controlled conditions to allow comparison. Apart from gut fluid, the YCFA ingredients were commercially sourced. These protocols provide a pathway to develop a commercially sourced alternative to YCFA-GF for any gut bacteria isolate of interest.
Reproducibility
All qualitative and quantitative parameters in this experiment should be reported to ensure reproducibility. Each timepoint for one isolate in two conditions is replicated in triplicate on a single 48-well plate. Triplicate repeat timepoints are separately converted from absorbance readings to growth rates. These growth rates are averaged to generate a reliable average growth rate for each isolate on each medium.
Data Analysis
Reproducible and efficient data analysis methods are required for analysis of growth measurements from high-throughput methods. This protocol details the equations required to convert microplate reader absorbance measurements over time to approximate growth rates in colony forming units over time (CFU/hr) using McFarland standards (Casciato, Stewart, & Rosenblatt, 1975; McFarland, 1907). Analysis was conducted in R v. 4.1.2 (R Core Team, 2014), producing figures using the package ggplot2 v. 3.3.2 (Wickham, 2016).
Basic Protocol 1: PREPARATION OF SOLID YCFA GROWTH MEDIUM
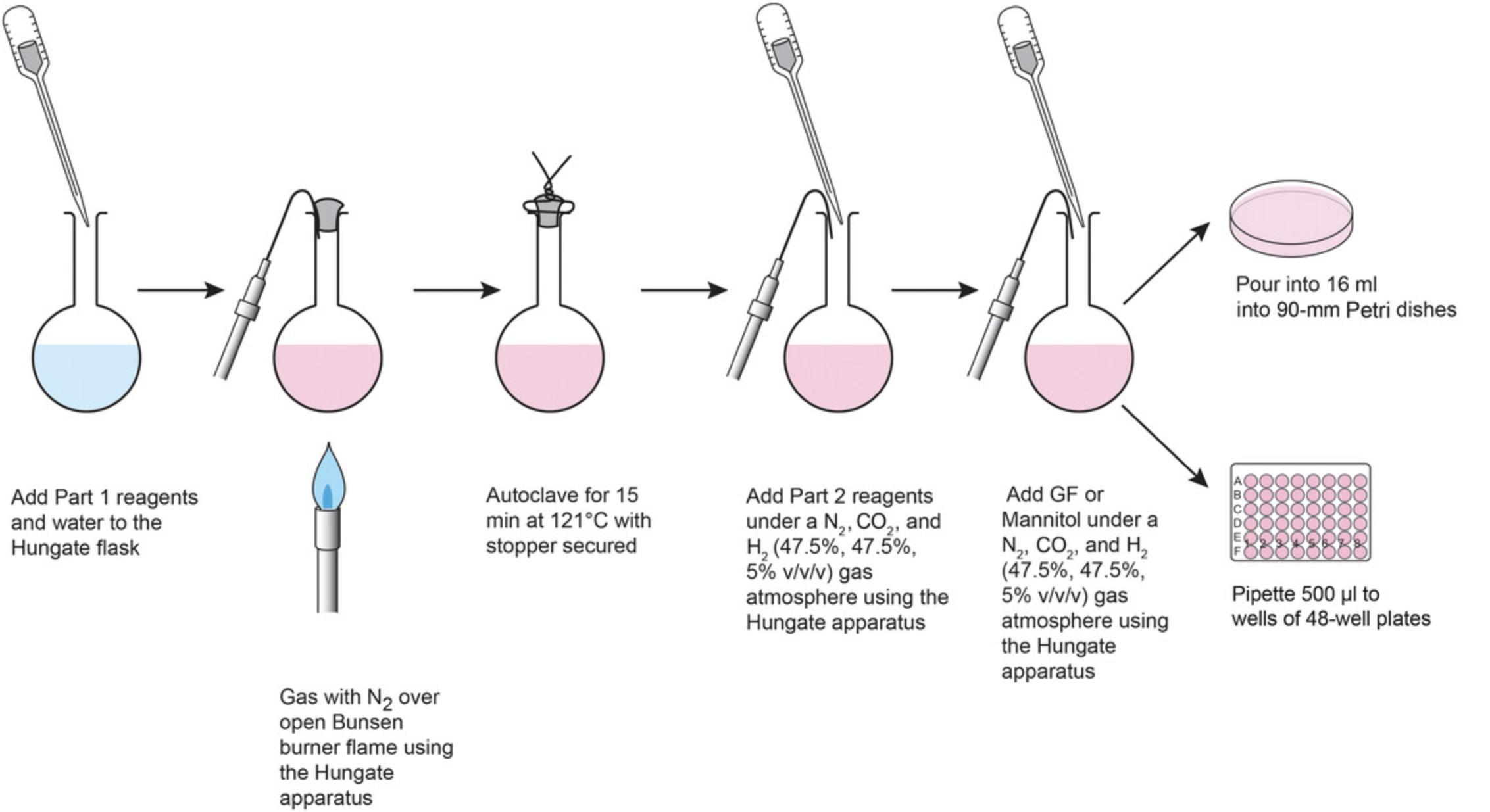
The protocol presented here is illustrated in Figure 5.The first section details the preparation of YCFA media adapted from Duncan et al. (2002), including the base medium and media supplemented with the desired growth factors to be compared. The second and third sections describe preparation of 90-mm Petri dishes (which will be used for plating to single isolates and for whole-plate streaking for growth rate experiments) and 48-well plates containing with two different media (for comparison in growth rate experiments). An example plate layout is provided for trial of a possible bacterial growth factor (in our case, mannitol in YCFA-M) against a standard of growth rate on YCFA medium with gut fluid (YCFA-GF). Both the 90-mm and 48-well plates are prepared in aerobic conditions, then depleted of any dissolved oxygen by storage in sealed anaerobic jars gassed with 47.5%, 47.5%, 5% (v/v/v) N2/CO2/H2 at 5 psi pressure and stored at 3-5°C. To ensure that new plates are completely depleted of oxygen, they should be stored in the anaerobic jar for at least 2 days.
Throughout these protocols, strict adherence to Hungate technique is required to maintain anaerobic conditions. The Hungate gassing apparatus, developed by Hungate (1969) and described by Bryant (1972), enables aseptic and anaerobic transfer of ingredients between containers outside of an anaerobic hood. This allows generation and manipulation of sterile anaerobic bacterial culture media without the restrictions associated with the use of an anaerobic hood. Details for setting up and using the Hungate apparatus are outlined in Bryant (1972). Three pure gas cylinders, separately containing N2, CO2, and H2, are attached to the Hungate apparatus prior to a copper column as outlined in Bryant (1972). The ratio of gasses provided through the apparatus to the gassing needles can be adjusted using the dials on the cylinders, which adjust the pounds per square inch (psi) released from each cylinder. The volume delivery of each gas to the gassing apparatus can further be checked using flow meters, which are used to measure the standard cubic centimeter per meter (sccm) for each gas. This allows a precise gas atmosphere of 47.5%, 47.5%, 5% (v/v/v) N2/CO2/H2 to be maintained.
Materials
-
Sterile water (deionized or autoclaved)
-
Bacto tryptone (BD, cat. no. 211705)
-
Yeast extract (BBL, BD, cat. no. 211929)
-
Agar (Difco, BD, cat. no. 21430)
-
0.6% (w/v) K2HPO4 in distilled water (store 100 ml stock in a Schott bottle for up to 30 days at 3-5°C)
-
Mineral mix (see recipe)
-
Trace elements (see recipe)
-
0.1% (w/v) resazurin in distilled water (store 100 ml stock in a foil-covered Schott bottle for up to 30 days at room temperature)
-
4% (w/v) NaHCO3 (see recipe)
-
2.5% (w/v) cysteine-HCl (see recipe)
-
Vitamin K3 + hemin mix (see recipe)
-
Volatile fatty acids mix (see recipe)
-
Gut fluid supernatant (see recipe)
-
5% (w/v) mannitol (Sigma, cat no. 63H01621) (store stock for up to 30 days at 3-5°C)
-
Stak-Pak Catalyst (palladium; Coy Laboratory Products, SKU: 6501000)
-
Stak-Pak Desiccant (Coy Laboratory Products, SKU: 6502000)
-
Charcoal, technical, activated for gas adsorption (Fisher Chemical, cat no. 7440-44-0)
-
Hungate gassing apparatus with (Fig. 3):
- 100-ml Hungate flasks (custom made by Mr. Alistair Mead, Senior Technician, Faculty of Science, University of Auckland)
- Butyl rubber stoppers (Thomas Scientific, cat no. 1208Z83)
- Anaerobic jar with pressure gauge (custom made by Mason Tool and Engineering Ltd, Auckland, New Zealand)
- N2, CO2, and H2 cylinders
- Materate flowmeters for air, bench standing (Glass Precision Engineering, cat no. MFA1001-BS)
-
Bunsen burner
-
Aluminum wire
-
60°C water bath
-
Sterile 90-mm Petri dishes
-
Sterile 48-well culture plates with lids
-
Laminar flow hood
-
10-ml serological pipettes
Prepare YCFA medium
These steps describe preparation of 100 ml medium. When comparing media (e.g., YCFA-M and YCFA-GF), 100 ml of each medium should be prepared.
1.Add the following to a 100-ml Hungate flask with 74 ml sterile water (Fig. 5, Part 1):
- 0.2 g Bacto tryptone
- 0.05 g yeast extract
- 1.7 g agar
- 3800 μl 0.6% (w/v) K2HPO4
- 3800 μl mineral mix
- 10 μl trace elements
- 100 μl 0.1% (w/v) resazurin
Deionized or autoclaved water can be used.
2.Gas the flask with N2 over an open flame until the medium turns pink and the agar has melted.
3.Remove the gassing needle and stopper the flask using the Hungate technique.
4.Secure the stopper in place with using two pieces of aluminum wire twisted around the stopper and the neck of the Hungate flask (Fig. 5).
5.Autoclave for 15 min at 121°C.
6.Immediately secure the 100 ml Hungate flask in a 60°C water bath and remove the aluminum wire.
7.Adjust the gas canisters of the Hungate apparatus to deliver a 47.5%, 47.5%, 5% (v/v/v) N2/CO2/H2 gas atmosphere.
8.Insert the needle of the Hungate gassing apparatus, hold needle in place with a butyl rubber stopper, perform this procedure using the Hungate technique.
9.Using the Hungate apparatus, add the following ingredients anaerobically under the N2/CO2/H2 atmosphere (Fig. 5, Part 2):
- 10,000 μl 4% (w/v) NaHCO3
- 2000 μl 2.5% (w/v) cysteine-HCl
- 1000 μl vitamin K3 + hemin mix
- 1000 μl volatile fatty acids mix
After each new ingredient addition, return the bung into place alongside the needle, maintaining anaerobic conditions.
10.Add the appropriate growth factor to make each desired medium for comparison:
- For YCFA-GF medium, add 10,000 μl gut fluid supernatant per 100 ml
- For YCFA-M medium, add 4000 μl of 5% (w/v) mannitol per 100 ml
- Swirl to ensure even distribution and keep in a 60°C water bath until ready topour plates.
YCFA media can be stored up to 90 days at 3-5°C.
Pour 90-mm Petri dishes
11.Pour 16 ml YCFA-GF into each 90-mm dish inside a sterile hood and leave in the hood for 30 min to cool and solidify.
12.Store anaerobically for at least 2 days (up to 90 days) at 3-5°C in sealed anaerobic jars gassed with 47.5%, 47.5%, 5% (v/v/v) N2/CO2/H2 gas at 5 psi.
Pour 48-well plates
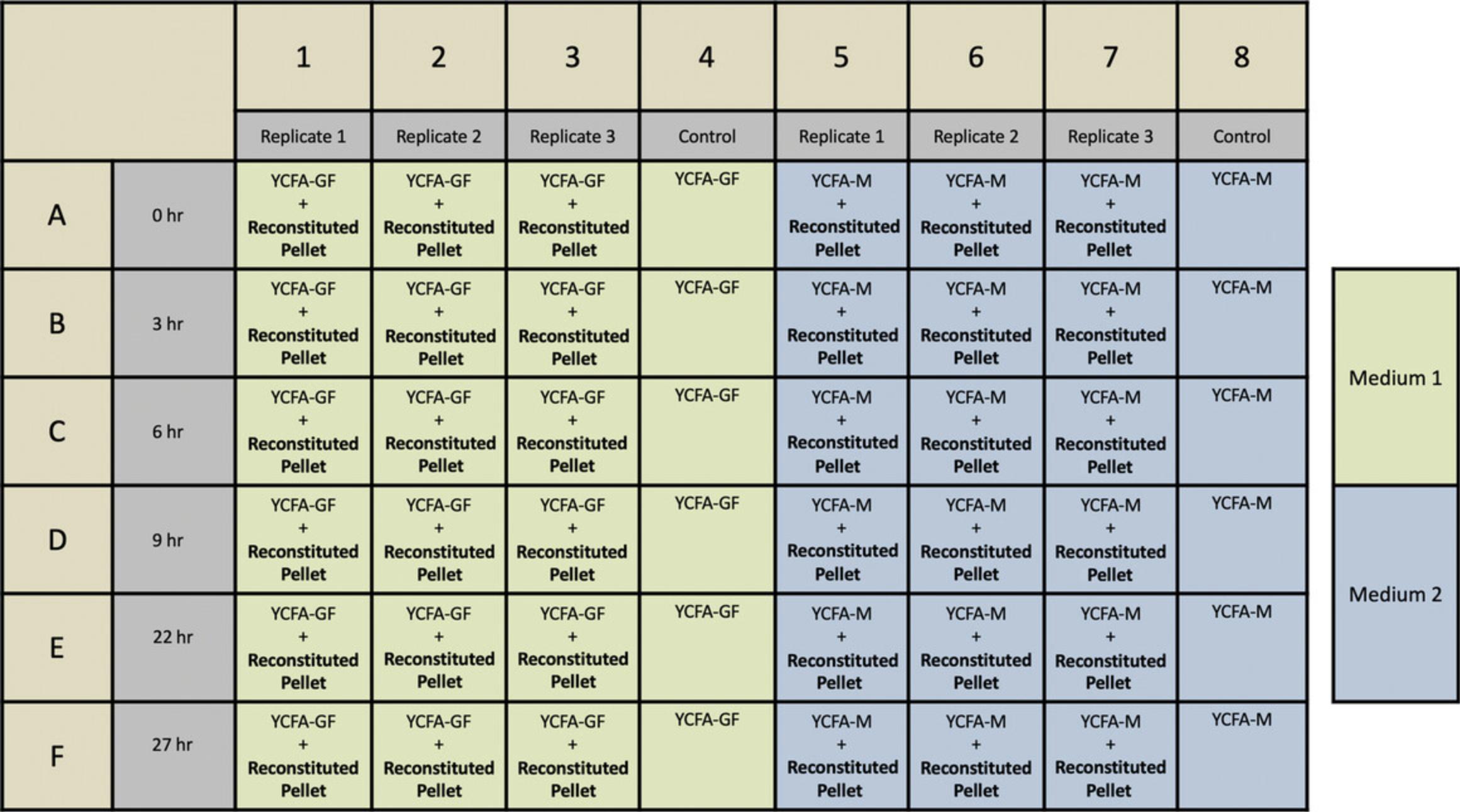
13.Design the layout of media to be poured in 48-well plates (for an example, see Fig. 6).
14.Prepare the required number of 48-well plates in a sterile hood.
15.Transfer the prepared YCFA medium variants (contained in Hungate flasks) from the 60°C water bath to the hood.
16.Use sterile 10-ml serological pipettes to transfer 500 μl prepared medium to each well of the plates inside the hood.
17.Leave poured plates in the hood for 30 min to cool and solidify.
18.Store anaerobically for at least 2 days as described in step 12.
Basic Protocol 2: COLLECTION OF FISH GUT SAMPLES AND PLATING TO SINGLE ISOLATES
For our experiments, bacteria were isolated from the hindgut of the silver drummer K. sydneyanus. Specimens were collected off the northeastern coast of New Zealand, where K. sydneyanus is a common herbivorous fish (Mountfort et al., 2002). The gut was removed by dissection and allocated into five sections (Fig. 4) following the guidelines of Johnson and Clements (2022). Microbial collection can take place from segments III, IV, and V, where microbial activity is highest. This was shown by Clements, Gleeson, & Slaytor (1994), where three herbivorous fish were found to have significantly higher concentrations of short-chain fatty acids in segments III-V compared to the earlier segments I and II. Increased short-chain fatty acid concentration is a sign of bacterial fermentation (Clements et al., 1994). Using this protocol, we sampled from gut sections IV and V and used novel bacterial species isolated from section IV for identification (Basic Protocol 3) and growth rate experiments (Basic Protocol 4). YCFA-GF was used to culture the bacteria to single isolates.
Materials
-
Freshly collected K. sydneyanus
-
Stak-Pak Catalyst (palladium, Coy Laboratory Products, SKU: 6501000)
-
Stak-Pak Desiccant (Coy Laboratory Products, SKU: 6502000)
-
Charcoal, technical, activated for gas adsorption (Fisher Chemical, cat. no. 7440-44-0)
-
90-mm YCFA-GF plates (see Basic Protocol 1)
-
Dissection equipment
-
Hungate gassing apparatus with:
- 10-ml Hungate tubes (glass test tubes)
- Butyl rubber stoppers for Hungate tubes (Thomas Scientific, cat no. 1208Z83)
- N2, CO2, and H2 cylinders
- Anaerobic jar with pressure gauge (custom made by Mason Tool and Engineering, Auckland, New Zealand)
- CAM-12 anaerobic monitor (Coy Laboratory Products, cat. no. 6250000)
-
Vinyl anaerobic chamber, type A glove box (Coy Laboratory Products, cat. no. 7000000)
-
Disposable inoculating loops (Thomas Scientific, cat no. 1230Z30)
-
20°C incubator
Collect fish hindgut contents
1.At the fish collection site, dissect gut from fish and divide into five sections (I-V) as in Figure 4.
2.Remove gut contents from sections IV and V by squeezing into separate Hungate tubes. Stopper tubes under anaerobic conditions and transport to the laboratory.
3.Once in the laboratory, place tubes inside an anaerobic chamber and allow solids to settle (5 min standing time) before moving to the next steps.
Culture to single isolates
4.Use a sterile disposable inoculating loop to streak 100 μl crude gut content onto a 90-mm YCFA-GF plate inside the anaerobic chamber.
5.Bring an anaerobic jar into the anaerobic chamber, place streaked plates inside the jar, and seal.
6.Remove anaerobic jar from the chamber and gas with mix of 47.5%, 47.5%, 5% (v/v/v) N2/CO2/H2 at 5 psi pressure using the Hungate gassing apparatus.
7.Place sealed anaerobic jar inside a 20°C incubator for 3 days or until single colonies are visible.
8.Pick well-isolated colonies and re-streak on YCFA-GF once they grow to a maximum cell number (∼3 days).
9.Re-streak two more subcultures of each colony of interest to produce pure single isolate cultures. Label cultures after each streak to ensure consistency in colony picking.
Basic Protocol 3: GENETIC IDENTIFICATION OF SINGLE ISOLATES USING COLONY PCR AND 16S SEQUENCING
This protocol describes how to genetically identify the single isolates cultured in Basic Protocol 2.It consists of two parts: (1) colony PCR amplification with purification and (2) 16S sequencing with analysis. Genetic identification of the isolates enables the results of Basic Protocol 4 to be analyzed in relation to previous studies on the same species or close genetic relatives.
Materials
-
16S primers for V3-V4 region (Klindworth et al., 2013):
- Forward primer (341F) 5′-CCTACGGGNGGCWGCAG-3′
- Reverse primer (785R) 5′-GACTACHVGGGTATCTAATCC-3′
-
REDTaq ReadyMix (Sigma, cat no. R2523-100RXN)
-
PCR-grade water (Roche, cat. no. 03315843001)
-
Single colony isolates to be tested (see Basic Protocol 2)
-
QIAquick PCR Purification Kit (Qiagen, cat no. 28104)
-
Vinyl anaerobic chamber, type A glove box (Coy Laboratory Products, cat. no. 7000000)
-
PCR tubes and lids
-
Sterile toothpicks
-
Permanent marker
-
PCR hood (Grant Instruments, cat. no. 7.970 866)
-
Thermal cycler (Thermo Fisher Scientific, cat no. 4375305)
-
IMPLEN NanoPhotometer N60 Microvolume UV-VIS Spectrophotometer (Fisher Scientific, cat. no. 15442203)
-
BLAST computer algorithm (Basic Local Alignment Search Tool, NCBI, nih.gov; Altschul, Gish, Miller, Myers, & Lipman, 1990)
-
Geneious Prime 2019.2.1 bioinformatics software (Kearse et al., 2012; www.geneious.com)
Perform colony PCR
1.Prepare the following PCR master mix (total 25 μl for each sample):
- 1 μl 25 μM primer F341 (final 1 μM)
- 1 μl 25 μM primer R806 (final 1 μM)
- 12.5 μl 2× REDTaq ReadyMix
- 10.5 μl H2O
2.Transfer the anaerobic jar containing the third subculture of single isolates from the incubator to the anaerobic chamber.
3.Place PCR tubes, lids, and toothpicks into the anaerobic chamber.
4.Touch a single colony with a toothpick and rub onto the wall of a PCR tube. Use a permanent marker to circle the colony on the bottom of the plate and label the circle to match the PCR tube.
5.Attach PCR lids and then transfer tubes from the anaerobic chamber and to a PCR hood.
6.Add 25 μl PCR mix to each tube and pipette up and down to mix.
7.Perform PCR using the following thermal cycler settings:
- 1 cycle: 95°C for 5 min
- 25 cycles:
- 95°C for 30 s
- 55°C for 30 s
- 72°C for 30 s
- 1 cycle: 72°C for 30 min
- Final step: 12°C (hold)
8.Use a QIAquick PCR Purification Kit to purify DNA and elute with 30 μl PCR-grade water.
9.Quantify the DNA on a nanospectrophotometer and adjust to an optimum concentration of 5-20 ng/μl using PCR-grade water.
Perform 16S sequence analysis
10.Send purified samples (along with F341 and R806 V3-V4 primers) for Sanger sequencing of the 16S rDNA gene at a sequencing facility.
11.Perform species identification by using BLAST to compare and align the 16S sequences to the NCBI public database.
12.Perform computational analysis of 16S sequencing data by using Geneious bioinformatics software to generate phylogenetic trees.
Basic Protocol 4: MEASUREMENT OF BACTERIAL GROWTH RATES ON SOLID MEDIA
This is a high-throughput protocol measuring the growth rates of fastidious anaerobic gut bacteria on different solid media. Growth effects of various nutrient additions can be compared to develop an optimal medium. In this protocol, YCFA (Duncan et al., 2002) with gut fluid (YCFA-GF) is used as a standard against which novel media are compared. YCFA-GF often produces optimal growth rates of bacteria in vitro , as gut fluid replicates the natural nutrient environment of gut bacteria in vivo. However, as the nutrient content of fish gut fluid is variable (Egerton et al., 2018), it is an unreplicable medium. This protocol allows a replicable medium with commercially sourced ingredients to be developed for any gut bacteria of interest. Importantly, strict anaerobic conditions are maintained at all times. Fish hindgut bacteria, cultured to single isolates and genetically identified using Basic Protocols 2 and 3, are tested for growth rate on two solid media: YCFA-GF and YCFA containing mannitol (YCFA-M). Mannitol was chosen as an ingredient of interest for reasons explained in Basic Protocol 1.
In this protocol, determination of bacterial growth rate on solid media is based on methods developed for anaerobic bacterial culture by Casciato et al. (1975). This group used water containing 0.1% gelatin (water-gel) to wash bacteria from the surface of solid medium in a Petri dish. Approximate cell density in the water-gel suspension was measured by converting the optical density at 600 nm (OD600) to approximate colony forming units per ml (CFU/ml) with the widely used McFarland standards (Casciato et al., 1975; McFarland, 1907). Using a single McFarland standard for all inocula within an experiment is important to maintain consistency and produce directly comparable results between isolates and conditions. Here, we have adjusted the inoculum to a cell density of McFarland standard 4 (∼1.24 × 109 CFU/ml) based on the growth success of our isolate in the preculture stage. However, any of the four McFarland standards can be chosen, and a lower McFarland standard can be used if the isolate of interest grows poorly.
This protocol applies principles from Casciato et al. (1975) to a high-throughput method with 48-well plates, allowing triplicate replicates and testing of multiple factors under the same conditions. Another novel development in this protocol is that there is no need for physical McFarland standards. These have been replaced with Equations 1 and 2, which we generated by graphing McFarland standards on an exponential curve against their approximate CFU/ml concentrations. These equations speed up and simplify the calculation of bacterial growth rates.
For isolates with exponential growth, equations are provided to calculate growth rates as either a doubling time (in hr) or a growth rate constant (α in min-1), two long-established ways to express bacterial growth rates (Hall et al., 2013). However, because previously uncultured anaerobic bacteria are often slow growing, the usual exponential growth profile may not apply. For isolates with linear growth, such as isolates BP5G and BP52G, bacterial growth rate can be calculated as an increase in colony forming units over time (CFU/hr). All of these growth rates will allow direct comparison of the growth effects of medium ingredients in order to develop an optimal medium.
Growth rates are estimates and interpreting them requires some judgement, as many confounding factors can affect absorbance readings and therefore growth rate calculations (Hall et al., 2013). Hall et al. (2013) recommend that results can be considered unreliable if the standard error is >3% of growth or if the correlation coefficient is <0.995.
Materials
-
Single colony isolates to be tested (see Basic Protocol 2)
-
90-mm YCFA-GF plates (see Basic Protocol 1)
-
Water-gel (see recipe)
-
48-well plate(s) with media to be tested (see Basic Protocol 1)
-
Anaerobic hood
-
Disposable sterile inoculating loops (Thomas Scientific, cat. no. 1230Z30)
-
Disposable sterile delta cell spreader (Fisher Scientific, cat. no. 50-751-5036, optional)
-
20°C incubator
-
2-ml microcentrifuge tubes (Eppendorf Safe-Lock tubes, Sigma-Aldrich, cat. no. EP022363344)
-
96-well flat-bottom plate (Sigma-Aldrich, cat. no. CLS3340)
-
Absorbance microplate reader (Parkin Elmer, EnVision 2104 Multilabel Plate Reader)
-
Additional equipment for maintaining anaerobic conditions using a Hungate gassing apparatus and anaerobic chamber (see Basic Protocol 2)
Prepare primary growth culture
1.Working in an anaerobic hood, use disposable inoculating loops to streak single colony isolates onto separate 90-mm Petri dishes containing solid YCFA-GF medium (one isolate per plate). Streak isolates over the entire plate.
2.Place inoculated dishes in an anaerobic jar inside an anaerobic chamber.
3.Remove sealed jar from the anaerobic chamber and gas with 47.5%, 47.5%, 5% (v/v/v) N2/CO2/H2 at 5 psi using the Hungate apparatus with copper tube inserted into the jar (Fig. 3).
4.Place sealed jar in a 20°C incubator for 3 days or to peak growth phase of the chosen isolates under these conditions.
5.Inside the anaerobic chamber, pipette 2000 μl water-gel onto each dish with an isolate culture. Resuspend bacteria in the water-gel by gently rubbing the surface with a disposable cell spreader or inoculating loop.
6.Tip the dish to gather all of the suspension at one side, then pipette the suspension from just above the agar layer and transfer to a 2-ml microcentrifuge tube.
7.Transfer 100 μl to a 96-well plate, then remove from the anaerobic chamber and measure the absorbance at 600 nm (OD600) in an absorbance microplate reader. Use 100 μl sterile water-gel in a second well as a blank reference.
8.Calculate the preculture cell concentration using Equation 1:
9.Calculate the amount of water-gel needed to reconstitute a cell pellet from the water-gel sample to McFarland standard 4 (∼12.4 × 108 CFU/ml) using Equation 2:
10.Inside the anaerobic chamber, centrifuge the 2 ml of water-gel suspension for 90 s at 1500 × g to pellet the bacteria in this sample.
11.Inside the anaerobic chamber, remove the supernatant, add the calculated volume of water-gel, and pipette gently up and down to resuspend the pellet.
Perform growth rate experiment
12.Inoculate 10 μl reconstituted pellet onto the solid medium in the appropriate wells of a prepared 48-well plate. Using the example layout in Figure 6, plate three replicates in columns 1-3 and 5-7.
13.Take the t = 0 hr timepoint by pipetting 100 μl water-gel onto the appropriate wells (wells A1-8 in Fig. 6), resuspending the bacteria by lightly scraping with a disposable inoculating loop, and transferring to a 96-well plate.
14.Measure the OD600. Use 100 μl sterile water-gel in a second well as a blank reference.
15.Repeat for at least four additional timepoints during the growth phase.
Calculate bacterial growth rates
16.For linear growth , calculate CFU/hr:
-
Use Equation1to convert absorbance readings to approximate cell density for each well (× 108CFU/ml).
-
Multiply these values by 0.1 ml (the sample volume for absorbance measurements) to convert cell density to the approximate cell number in each well (× 108CFU/well).
-
Plot CFU/well against time and apply a linear trend line. Determine the growth rate (in CFU/hr) as the gradient of this line.
17.For exponential growth , calculate the first-order growth rate constant (α):
-
Plot the natural log of the mean OD values (lnN) versus time (in hr) using a graphing program.
-
Use the software to generate a linear trendline between two points (at timestand0).
-
Use Equation3to calculate α:$$\begin{equation} {\rm{ln}}\frac{{{N_t}}}{{{N_0}}} = {\rm{;\alpha }}\left( {t - {\rm{;}}{t_0}} \right) \end{equation}$$
18.For exponential growth , also calculate the doubling time:
-
Plot mean OD values versus time (in hr) using a graphing program.
-
Apply an exponential trendline to the graph.
-
Use the equation associated with this exponential trendline to determine the κ coefficient as the exponential index.
-
Use the κ coefficient in Equation4to calculate the doubling time:$$\begin{equation} {\rm{Doubling;time;(hr)}} = ;\frac{{\rm l{n2}}}{{\rm \kappa ;coefficient}} \end{equation}$$
REAGENTS AND SOLUTIONS
Cysteine-HCl, 2.5% (w/v)
Dissolve 1 g cystine-HCl in 40 ml distilled water. Bring to a boil under a N2 atmosphere by gassing in a Hungate vial using the Hungate apparatus. Stopper the vial, secure using aluminum wire, and autoclave. Make 100 ml stock at a time and store in 10-ml aliquots in Hungate tubes under 100% N2 by gassing tubes using the Hungate technique. Store up to 30 days at 3-5°C.
Gut fluid supernatant
Filter K. sydneyanus hindgut lumen contents from sections IV and V (Fig. 4) through cheese cloth. Centrifuge filtrate 20 min at 9000 × g , 4°C. Collect supernatant and autoclave. Centrifuge supernatant 20 min at 13,500 × g , 4°C. Make 100 ml stock at a time. Divide between two 50-ml Falcon tubes and store frozen at −20°C for up to 9 months. Alternatively, prepare 10-ml aliquots in Hungate tubes under 100% N2 using the Hungate technique and store at 3-5°C for up to 30 days.
Mineral mix
- 0.6% (w/v) KH2PO4
- 0.6% (w/v) (NH4)2SO4
- 1.2% (w/v) NaCl
- 0.245% (w/v) MgSO4·7H2O
- 0.15% (w/v) CaCl2·2H2O
- Prepare 100 ml at a time
- Autoclave 15 min at 15 psi on liquid cycle
- Store in a Schott bottle up to 30 days at 3-5°C
NaHCO3, 4% (w/v)
Prepare a 4% (w/v) sodium carbonate solution and sterilize by autoclaving in an open flask. This turns to sodium hydroxide during heating. While hot, bubble with CO2 gas until cool, using a serological pipette attached to the Hungate apparatus to deliver the gas. The solution will have turned into sodium bicarbonate and carbonic acid when white flecks are visible on the end of the serological pipette. Make 100 ml stock at a time and stopper in 10-ml aliquots under CO2 gas. Store up to 30 days at 3-5°C.
Trace elements
- 4 mg CuSO4·4H2O
- 50 mg MnSO4·4H2O
- 0.2 mg Ni(II)Cl2
- 10 mg FeSO4
- 0.2 mg Co(II)(NO3)2·6H2O
- 0.2 mg H2MoO4
- Bring to 100 ml with distilled water
- Autoclave 15 min at 15 psi on liquid cycle
- Store in a Schott bottle up to 30 days at 3-5°C
Vitamin K + hemin mix
Dissolve 10 mg hemin in 0.2 ml 1 M NaOH. Add 20 ml distilled water and autoclave. Separately, add 100 mg sterile vitamin K1 or K3 to 20 ml of 95% ethyl alcohol. For the mixed stock, add 0.2 ml sterile vitamin K in ethyl alcohol to 20 ml hemin (final 0.005% [w/v] vitamin K, 0.05% [w/v] hemin). Store in 10-ml aliquots in Hungate tubes under 100% N2 by gassing tubes using the Hungate technique. Stored up to 30 days at 3-5°C.
Volatile fatty acids mix
- 17 ml acetic acid
- 6 ml propionic acid
- 1 ml n -valeric acid
- 1 ml isovaleric acid
- 1 ml isobutyric acid
- Bring to ∼50 ml with distilled water
- Neutralize with 5 M NaOH to pH 6.5
- Bring to 100 ml with water
Autoclave under a N2 atmosphere by gassing in a Hungate vial using the Hungate apparatus, then stoppering the vial, securing with an aluminum wire, and autoclaving. Make 100 ml at a time and prepare 10-ml aliquots in Hungate tubes under 100% N2 by gassing using the Hungate technique. Store up to 30 days at 3-5°C.
Water-gel, 0.1% (w/v) gelatin
Add 0.05 g gelatin from porcine skin (Sigma-Aldrich, cat. no. G2625) to 50 ml sterile water (deionized or autoclaved) in a 50-ml Falcon tube and vortex for 30 s. Ensure that the lid is only half way on (to allow pressure equalization) and then autoclave 15 min at 15 psi on liquid cycle. Transfer 10-ml aliquots to Hungate tubes using the Hungate technique and then gas tubes with 47.5%, 47.5%, 5% (v/v/v) N2/CO2/H2 using the Hungate apparatus. Leave to become anaerobic overnight inside the anaerobic chamber with bungs loose to allow gas exchange. Store up to 90 days at 3-5°C with bungs secure.
COMMENTARY
Background Information
The structure and function of gut microbiome communities and their relation to the host is an area of great research interest. However, few microorganisms isolated from environmental samples survive in in vitro culture (Barer & Harwood, 1999; Barns, Fundyga, Jeffries, & Pace, 1994; Giovannoni, Britschgi, Moyer, & Field, 1990; Kaeberlein, Lewis, & Epstein, 2002; Torsvik, Goksøyr, & Daae, 1990; Ward, Weller, & Bateson, 1990; Wayne et al., 1987). As culture methods were found to be inadequate for studying microbial community composition, there was an increased interest in sequence-based genetic techniques that bypassed the need for in vitro culture (Barns et al., 1994; Ward et al., 1990). These genetic techniques greatly expanded our knowledge of microbial biodiversity and related evolutionary processes (Barns et al., 1994). Although much progress has been made using a metagenomic cultivation-independent approach, the only secure and operational proof of organism viability is culturability (Barer & Harwood, 1999). Genomic 16S and rRNA studies of environmentally isolated bacteria can suggest the presence of metabolic pathways (Béjà et al., 2000, 2002), but cultivation is required in order to understand the physiology of novel microbes and the functional processes they engage in (Stewart, 2012; Zengler et al., 2002). It is a current challenge, therefore, for microbiologists to improve in vitro culture methods to allow a greater understanding of the metabolic properties and potentials of these organisms (Zengler et al., 2002).
Investigation of anaerobic bacteria in vitro requires specialized media containing all essential nutrients for growth. To improve the accuracy of bacterial function analysis, the medium should have reduced complexity with minimal undefined ingredients (Tramontano et al., 2018). The starting point of developing a replicable medium for novel bacteria is determining the essential elements for growth and their required concentrations. Some environmentally isolated bacteria grow in simple media with just a single carbon source, while others require addition of macroelements (C, H, O, N, P, and S) and metals (Ca, Cl, Fe, K, Mg, and Na), which together make up 95% of average microorganism dry weight (Plugge, 2005). Many bacteria remain unculturable even when all common essential elements have been added in sufficient amounts (Plugge, 2005). Such species require further nutrients, which are classified as growth factors and typically include amino acids, vitamins, or pyrimidines and purines (Plugge, 2005). For example, many Bacteroides strains require the presence of vitamin K1 and hemin for growth (Gibbons & Mc Donald, 1960).
Anaerobic bacteria require long incubation times; for routine clinical microbiology, anaerobic cultures should be incubated for a minimum of 5 days (Chapin & Murray, 2007; Lagier et al., 2015). Also, the gut environment is highly nutritionally complex, and bacterial species from the same host often have highly variable nutrient preferences (Egerton et al., 2018). These complications make developing an optimal medium for previously uncultured gut bacteria a long process that is nevertheless essential for the successful culture of many species.
Here, we present a high-throughput method for developing an optimal medium for novel anaerobic gut bacteria. This protocol uses solid media because fastidious anaerobic bacteria often do not grow in a liquid media (Speers et al., 2009). The use of solid media thus allows the protocol to be used on a broader range of gut bacteria. The key advantage of this technique its high-throughput setup, which allows three replicates of a potential growth factor to be tested over up to six timepoints on a single 48-well plate. If the anaerobic jar is large enough, two plates can be tested at one time. In this case, one experimental run can produce average growth rates of two isolates on two media (i.e., one with a test growth factor and one with gut fluid). To our knowledge, this protocol is the quickest way to determine precise nutrient requirements for a species of interest.
These protocols allow the trial of novel nutrient additions to media, and the trialing of various nutrient additions allows for discovery of novel growth factors. Furthermore, the growth effects of removing medium ingredients can be tested. Short-chain fatty acids and amino acids that promote bacterial growth of some species may inhibit the growth of other species (Tramontano et al., 2018). Removal of these inhibitors allows previously unculturable bacteria to be cultured in vitro for the first time.
A novel medium recipe developed using this technique can be exactly replicated in future work, reducing the effect of confounding variables on results. Most human gut bacteria have no growth characterization data available in replicable media (Tramontano et al., 2018), as they have only been cultured in media with gut fluid or other complex and undefined nutrient sources. A replicable medium recipe has the advantage of allowing metabolomic and proteomic studies to be carried out on bacteria in vitro. Moreover, gut bacteria must be cultured in a nutritionally defined medium to study the link between host diet, microbiota composition and dynamics, and interspecies interactions (Tramontano et al., 2018).
Other protocols for measuring growth rate in a high-throughput fashion are not suitable for strictly anaerobic bacteria. A high-throughput method developed by Hall, et al. (2013) uses microtiter plates to measure absorbance over the growth period aerobically in a plate reader. This method is not appropriate for strictly anaerobic species, as an anaerobic environment cannot be maintained in most plate readers. Another method by Tramontano et al. (2018) uses a microwell plate inside an anaerobic chamber to measure bacterial growth rates anaerobically, but it requires culture in liquid media (Tramontano et al., 2018), which is not suitable for many fastidious gut bacteria (Speers et al., 2009). In contrast, our protocol enables quick development of novel solid media for a broad range of fastidious anaerobic bacteria.
Critical Parameters
There are several critical parameters influencing bacterial growth that can be individually addressed to improve the performance of chosen isolates in these protocols. Maintenance of anaerobic conditions is crucial, hence oxygen must be completely excluded from the system. The Hungate technique must be strictly applied when making media to keep them anaerobic. The 48-well plates must be stored in an anaerobic jar gassed with standard gas mix of 47.5%, 47.5%, 5% (v/v/v) N2, CO2, and H2 at 5 psi at 3-5°C for at least 2 days before use to ensure that the solid medium is completely reduced. When gassing anaerobic jars, the pressure must not drop below 1 psi at any point. The gas must move in only one direction, away from the gas cylinders, through the jar, and out. The seals on the anaerobic jars must be verified for minimal loss of pressure over time. The water-gel must also be reduced in an anaerobic hood overnight before use. Oxygen levels in the anaerobic hood should be monitored with an anaerobic monitor to ensure that no oxygen has entered the system.
The amount of a trial growth factor added to YCFA medium must be decided based on previous work with similar compounds. Basic Protocol 1 explains that the concentration of mannitol in the YCFA-M medium used in our study was chosen based on recommendations by Holdeman et al. (1977) and Browne et al. (2016), who used a 0.2% (w/v) concentration of sugars in their anaerobic media.
Troubleshooting
See Table 1 for troubleshooting recommendations.
| Problem | Possible cause | Potential solutions |
|---|---|---|
| Too few bacterial CFU collected from Petri dish | Fastidious or slow growing isolate | Create two dishes for this isolate. To collect bacteria from both dishes, transfer bacteria suspended in water-gel from the first dish to the second |
| Bacteria adhere tightly to solid medium and cannot be removed by rubbing with loop | Isolate adheres strongly to nutrient source | Incubate at 20°C for a shorter time to harvest before strong adherence occurs |
| Bacteria do not grow on 48-well plate | Bacteria did not pellet | Increase centrifugation time (60-120 s at 1500 × g is recommended) |
| Bacteria were dead before inoculation | Reduce incubation time on Petri dishes | |
| Reduce centrifugation time to reduce sheer stress (60-120 s at 1500 × g is recommended) | ||
| Pipette gently to reduce sheer stress | ||
| Warm plates in hood for at least 20 min before streaking to reduce temperature shock | ||
| Bacterial growth was interrupted | Maintain strict anaerobic conditions at all points during growth | |
| Store anaerobic jars containing plates at 3-5°C to prevent drying of medium | ||
| Ensure palladium and charcoal are freshly activated before use (heat 1 hr at 150°C) | ||
| Increase gaps between timepoints for longer uninterrupted incubation | ||
| Increase length of growth period (some bacteria have long lag phases) |
Understanding Results
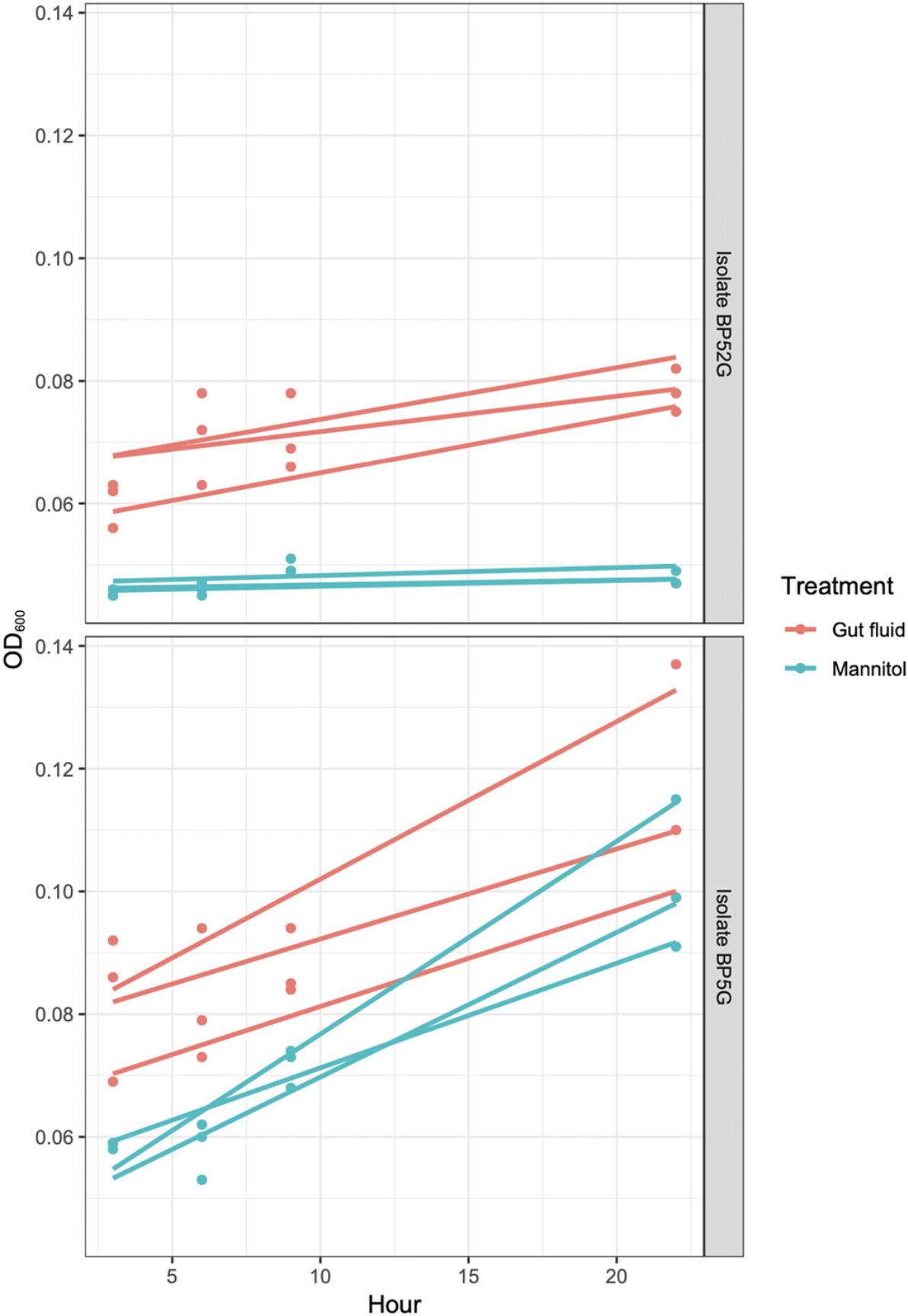
As an example of anticipated results, we conducted growth rate experiments on two isolates from K. sydneyanus gut section IV: BP5G and BP52G from the families Lachnospiraceae and Erysipelotrichaceae, respectively. Growth was measured by optical density at 600 nm in two different conditions: YCFA-GF and YCFA-M. Standard YCFA-GF (Duncan, et al., 2002) is used when culturing previously uncultured gut bacteria in vitro for the first time because it contains many of the nutrients present in the natural environment of these organisms. Mannitol is present at decreasing levels along the fish gut (White et al., 2010). As this decrease could be caused by fermentation by gut bacteria (White et al., 2010), this carbohydrate is a possible growth factor for these organisms. YCFA-M is a defined alternative to YCFA-GF, as all ingredients are industrially sourced, and thus is preferable to the complex and nutritionally undefined YCFA-GF for certain metabolic analyses (Tramontano et al., 2018). The OD600 was measured at four timepoints during the growth phase of the isolates, which was found in preliminary work to be between 3 and 27 hr. Three replicates were produced for each of the two trial media. Using the method described in Basic Protocol 4 for bacteria with linear growth, gradients of the linear regression lines applied to the data (Fig. 7) were used as approximate growth rates in CFU/hr. The individual growth rates of the three replicates were used to calculate average approximate growth rates (Table 2). Alternatively, growth rates of bacteria with exponential growth are calculated as doubling time (in hr) or first-order growth rate constant (α in min–1).
| Isolate | Medium | Replicate growth rate (× 105 CFU/hr) | Average growth rate (× 105 CFU/hr, ± SD) |
|---|---|---|---|
| BP5G (Lachnospiraceae) | YCFA-GF | 0.90 | |
| 0.80 | |||
| 1.60 | 1.10 (± 0.44) | ||
| YCFA-M | 1.30 | ||
| 1.80 | |||
| 0.90 | 1.30 (± 0.45) | ||
| BP52G (Erysipelotrichaceae) | YCFA-GF | 0.5 | |
| 0.3 | |||
| 0.4 | 0.40 (± 0.10) | ||
| YCFA-M | 0.04 | ||
| 0.05 | |||
| 0.06 | 0.05 (± 0.01) |
For the Lachnospiraceae isolate BP5G, YCFA-GF medium produced an average growth a rate of 1.1 × 105 (± 0.44) CFU/hr. This is used as the desired growth rate against which the alternative YCFA-M can be compared. The average growth rate on YCFA-M was 1.3 × 105 (± 0.45) CFU/hr. For a linear regression model of OD600 against time, growth on YCFA-GF (R 2 = 0.5574, p = 0.001386) and YCFA-M (R 2 = 0.9302, p = 6.82 × 10–9) were significant compared to no growth. No growth can be used as a comparison in this model, as no growth was seen in the no isolate (YCFA-GF or YCFA-M only) control wells. Also, the interaction effect (Hour:Media) was significant (p = 0.0302842), indicating a significant difference in growth rates of BP5G on YCFA-GF and YCFA-M. These results show that YCFA-M produced a significantly faster growth rate for this isolate than YCFA-GF. Based on these results, mannitol is a good replacement for fish gut fluid in YCFA medium for culture of BP5G, as it is a more effective growth factor and creates a defined and replicable medium.
In contrast, for the Erysipelotrichaceae isolate BP52G, YCFA-GF and YCFA-M media produced average growth rates of 0.4 × 105 (± 0.10) and 0.05 × 105 (± 0.01) CFU/ml, respectively. For a linear regression model of OD600 against time for this isolate, growth on YCFA-GF was significant compared to no growth (R 2 = 0.3678, p = 0.01653), whereas YCFA-M was not significant (p > 0.05). Although there was some growth of BP52G on YCFA-M (Supplementary material), it was poor and not statistically significant. Based on these results, mannitol cannot replace fish gut fluid in YCFA for this isolate as it leads to significantly inferior growth.
These results demonstrate the unique nutritional requirements of isolates from different families isolated from the same fish gut environment and even from the same gut section. This demonstrates that there is family-level variation in nutrient requirements within the fish gut microbiota. These same protocols can be used to search for other essential growth factors in nutritionally complex medium additions sourced from the natural environment. By evaluating the growth rate of anaerobic bacteria on different media, these protocols can be used to discover novel defined alternatives to YCFA-GF for in vitro gut bacteria culture.
Time Considerations
The preparation of multiwell anaerobic plates (Basic Protocol 1) takes 3 days. The initial isolation of fish gut bacteria from an environmental sample (Basic Protocol 2) takes ∼4 days, although this step (along with all other steps involving bacterial culture) may vary depending on the growth profile of the isolate. The time needed to reach saturation phase should be identified for an isolate in preliminary work before it is used in this protocol. The saturation phase should be reached by the end of incubation for all steps in Basic Protocol 2. Identification of isolates (Basic Protocol 3) takes 1 day of laboratory work; the total time depends on the sequencing facility used. Preculture preparation and growth rate experiments (Basic Protocol 4) take 4 days and at least 2 days, respectively. More time may be needed if isolates take more than 48 hr to reach saturation phase.
Acknowledgments
We thank Mason Tool and Engineering Ltd. (Auckland, New Zealand) for the custom-built anaerobic jar, and Mr. Alistair Mead (Senior Technician, Faculty of Science, University of Auckland) for the custom-built Hungate flasks. We thank Kristen Boxen (Auckland Genomics, DNA Sequencing Facility, School of Biological Sciences, University of Auckland) for Sanger sequencing services. We thank Vivian Ward for assistance with some illustrations. We also thank Kendall Clements for field assistance and helpful support and discussions. Funding for this project was provided by a grant to Kendall Clements and W. Lindsey White from the New Zealand Ministry of Business, Innovation and Employment (MBIE, grant No. UOAX1808).
Open access publishing facilitated by Auckland University of Technology, as part of the Wiley - Auckland University of Technology agreement via the Council of Australian University Librarians.
Author Contributions
Kristina Wornell : Conceptualization, data curation, formal analysis, investigation, methodology, validation, writing original draft; Bikiran Pardesi : Conceptualization, methodology, project administration, resources, supervision, writing review and editing; Kevin Lee : Formal analysis, writing review and editing; Svetlana Boycheva : Conceptualization, methodology; Anthony M. Roberton : Conceptualization, investigation, methodology, resources, writing review and editing; W. Lindsey White : Funding acquisition, project administration, resources, supervision, writing review and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supporting Information
| Filename | Description |
|---|---|
| cpz1463-sup-0001-SuppMat.docx14.2 KB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology , 215(3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
- Barcenilla, A., Pryde, S. E., Martin, J. C., Duncan, S. H., Stewart, C. S., Henderson, C., & Flint, H. J. (2000). Phylogenetic relationships of butyrate-producing bacteria from the human gut. Applied and Environmental Microbiology , 66(4), 1654–1661. doi: 10.1128/AEM.66.4.1654-1661.2000
- Barer, M. R., & Harwood, C. R. (1999). Bacterial viability and culturability. Advances in Microbial Physiology , 41, 93–137. doi: 10.1016/S0065-2911(08)60166-6
- Barns, S. M., Fundyga, R. E., Jeffries, M. W., & Pace, N. R. (1994). Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proceedings of the National Academy of Sciences , 91(5), 1609–1613. doi: 10.1073/pnas.91.5.1609
- Béjà, O., Aravind, L., Koonin, E. V., Suzuki, M. T., Hadd, A., Nguyen, L. P., … DeLong, E. F. (2000). Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science , 289(5486), 1902–1906. doi: 10.1126/science.289.5486.1902
- Béjà, O., Suzuki, M. T., Heidelberg, J. F., Nelson, W. C., Preston, C. M., Hamada, T., … DeLong, E. F. (2002). Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature , 415(6872), 630–633. doi: 10.1038/415630a
- Bomar, L., Maltz, M., Colston, S., & Graf, J. (2011). Directed culturing of microorganisms using metatranscriptomics. mBio , 2(2), e00012–11. doi: 10.1128/mBio.00012-11
- Browne, H. P., Forster, S. C., Anonye, B. O., Kumar, N., Neville, B. A., Stares, M. D., … Lawley, T. D. (2016). Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature , 533(7604), 543–546. doi: 10.1038/nature17645
- Bryant, M. P. (1972). Commentary on the Hungate technique for culture of anaerobic bacteria. American Journal of Clinical Nutrition , 25(12), 1324–1328. doi: 10.1093/ajcn/25.12.1324
- Casciato, D. A., Stewart, P. R., & Rosenblatt, J. E. (1975). Growth curves of anaerobic bacteria in solid media. Applied Microbiology , 29(5), 610–614. doi: 10.1128/am.29.5.610-614.1975
- Chapin, K. C., & Murray, P. R. (2007). Principles of stains and media. In P. R. Murray & E. J. Baron et al. (Eds.), Principles of clinical microbiology (pp, 182–191). Washington: American Society for Microbiology. doi: 10.1086/524076
- Clements, K. D., & Choat, J. H. (1997). Comparison of herbivory in the closely-related marine fish genera Girella and Kyphosus. Marine Biology , 127(4), 579–586. doi: 10.1007/s002270050048
- Clements, K. D., Gleeson, V. P., & Slaytor, M. (1994). Short-chain fatty acid metabolism in temperate marine herbivorous fish. Journal of Comparative Physiology B , 164(5), 372–377. doi: 10.1007/BF00302552
- Demain, A. L., & Sanchez, S. (2009). Microbial drug discovery: 80 years of progress. Journal of Antibiotics , 62(1), 5. doi: 10.1038/ja.2008.16
- Duncan, S. H., Hold, G. L., Harmsen, H. J., Stewart, C. S., & Flint, H. J. (2002). Growth requirements and fermentation products of Fusobacterium prausnitzii , and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. International Journal of Systematic and Evolutionary Microbiology , 52(6), 2141–2146. doi: 10.1099/00207713-52-6-2141
- Egerton, S., Culloty, S., Whooley, J., Stanton, C., & Ross, R. P. (2018). The gut microbiota of marine fish. Frontiers in Microbiology , 9, 873. doi: 10.3389/fmicb.2018.00873
- Ghanbari, M., Kneifel, W., & Domig, K. J. (2015). A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture , 448, 464–475. doi: 10.1016/j.aquaculture.2015.06.033
- Gibbons, R. J., & Macdonald, J. B. (1960). Hemin and vitamin K compounds as required factors for the cultivation of certain strains of Bacteroides melaninogenicus. Journal of Bacteriology , 80(2), 164–170. doi: 10.1128/jb.80.2.164-170.1960
- Giovannoni, S. J., Britschgi, T. B., Moyer, C. L., & Field, K. G. (1990). Genetic diversity in Sargasso Sea bacterioplankton. Nature , 345(6270), 60–63. doi: 10.1038/345060a0
- Hall, B. G., Acar, H., Nandipati, A., & Barlow, M. (2013). Growth rates made easy. Molecular Biology and Evolution , 31(1), 232–238. doi: 10.1093/molbev/mst187
- Hensgens, C. M., Nienhuis-Kuiper, M. E., & Hansen, T. A. (1994). Effects of tungstate on the growth of Desulfovibrio gigas NCIMB 9332 and other sulfate-reducing bacteria with ethanol as a substrate. Archives of Microbiology , 162(1), 143–147. doi: 10.1007/BF00264388
- Holdeman, L. V., Moore, W. E. C., & Cato, E. P. (1977). Anaerobe laboratory manual. Blacksburg, VA: Virginia Polytechnic Institute and State University.
- Hungate, R. E. (1969). A roll tube method for cultivation of strict anaerobes. In Methods in microbiology (Vol. 3, pp. 117–132). Academic Press. doi: 10.1016/S0580-9517(08)70503-8
- Hungate, R. E. (1975). The rumen microbial ecosystem. Annual Review of Ecology and Systematics , 6(1), 39–66. doi: 10.1146/annurev.es.06.110175.000351
- Jarrell, K. F., & Kalmokoff, M. L. (1988). Nutritional requirements of the methanogenic archaebacteria. Canadian Journal of Microbiology , 34(5), 557–576. doi: 10.1139/m88-095
- Johnson, K. S., & Clements, K. D. (2022). Histology and ultrastructure of the gastrointestinal tract in four temperate marine herbivorous fishes. Journal of Morphology , 283(1), 16–34. doi: 10.1002/jmor.21424
- Kaeberlein, T., Lewis, K., & Epstein, S. S. (2002). Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science , 296(5570), 1127–1129. doi: 10.1126/science.1070633
- Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., … Drummond, A. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics , 28(12), 1647–1649. doi: 10.1093/bioinformatics/bts199
- Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., & Glöckner, F. O. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research , 41(1), e1. doi: 10.1093/nar/gks808
- Lagier, J. C., Edouard, S., Pagnier, I., Mediannikov, O., Drancourt, M., & Raoult, D. (2015). Current and past strategies for bacterial culture in clinical microbiology. Clinical Microbiology Reviews , 28(1), 208–236. doi: 10.1128/CMR.00110-14
- Leadbetter, J. R. (2003). Cultivation of recalcitrant microbes: Cells are alive, well and revealing their secrets in the 21st century laboratory. Current Opinion in Microbiology , 6(3), 274–281. doi: 10.1016/S1369-5274(03)00041-9
- McFarland, J. (1907). The nephelometer: An instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. Journal of the American Medical Association , 49(14), 1176–1178. doi: 10.1001/jama.1907.25320140022001f
- Mcintosh, J., Fildes, P., & Bulloch, W. (1916). A new apparatus for the isolation and cultivation of anaerobic microorganisms. The Lancet , 187(4832), 768–770. doi: 10.1016/S0140-6736(01)11835-0
- Miyazaki, K., Martin, J. C., Marinsek-Logar, R., & Flint, H. J. (1997). Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe , 3(6), 373–381. doi: 10.1006/anae.1997.0125
- Moore, W. E. C. (1966). Techniques for routine culture of fastidious anaerobes. International Journal of Systematic and Evolutionary Microbiology , 16(2), 173–190. doi: 10.1099/00207713-16-2-173
- Mountfort, D. O., Campbell, J., & Clements, K. D. (2002). Hindgut fermentation in three species of marine herbivorous fish. Applied and Environmental Microbiology , 68(3), 1374–1380. doi: 10.1128/AEM.68.3.1374-1380.2002
- Plugge, C. M. (2005). Anoxic media design, preparation, and considerations. Methods in Enzymology , 397, 3–16. doi: 10.1016/S0076-6879(05)97001-8
- R Core Team. (2014). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Retrieved from http://www.R-project.org/
- Scherer, P., & Sahm, H. (1981). Effect of trace elements and vitamins on the growth of Methanosarcina barkeri. Acta Biotechnologica , 1(1), 57–65. doi: 10.1002/abio.370010108
- Schönheit, P., Moll, J., & Thauer, R. K. (1979). Nickel, cobalt, and molybdenum requirement for growth of Methanobacterium thermoautotrophicum. Archives of Microbiology , 123(1), 105–107. doi: 10.1007/BF00403508
- Speers, A. M., Cologgi, D. L., & Reguera, G. (2009). Anaerobic cell culture. Current Protocols in Microbiology , 12(1), A.4F.1–A.4F.16. doi: 10.1002/9780471729259.mca04fs12
- Staley, J. T., & Konopka, A. (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annual Review of Microbiology , 39(1), 321–346. doi: 10.1146/annurev.mi.39.100185.001541
- Stewart, E. J. (2012). Growing unculturable bacteria. Journal of Bacteriology , 194(16), 4151–4160. doi: 10.1128/JB.00345-12
- Stott, M. B., Crowe, M. A., Mountain, B. W., Smirnova, A. V., Hou, S., Alam, M., & Dunfield, P. F. (2008). Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environmental Microbiology , 10(8), 2030–2041. doi: 10.1111/j.1462-2920.2008.01621.x
- Tanner, R. S., & Wolfe, R. S. (1988). Nutritional requirements of Methanomicrobium mobile. Applied and Environmental Microbiology , 54(3), 625–628. doi: 10.1128/aem.54.3.625-628.1988
- Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., … Ostell, J. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Research , 44(14), 6614–6624. doi: 10.1093/nar/gkw569
- Torsvik, V., Goksøyr, J., & Daae, F. L. (1990). High diversity in DNA of soil bacteria. Applied and Environmental Microbiology , 56(3), 782–787. doi: 10.1128/aem.56.3.782-787.1990
- Tramontano, M., Andrejev, S., Pruteanu, M., Klünemann, M., Kuhn, M., Galardini, M., … Patil, K. R. (2018). Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nature Microbiology , 3(4), 514–522. doi: 10.1038/s41564-018-0123-9
- Ward, D. M., Weller, R., & Bateson, M. M. (1990). 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature , 345(6270), 63–65. doi: 10.1038/345063a0
- Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., … Truper, H. G. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. International Journal of Systematic and Evolutionary Microbiology , 37(4), 463–464. doi: 10.1099/00207713-37-4-463
- White, W. L., Coveny, A. H., Robertson, J., & Clements, K. D. (2010). Utilisation of mannitol by temperate marine herbivorous fishes. Journal of Experimental Marine Biology and Ecology , 391(1-2), 50–56. doi: 10.1016/j.jembe.2010.06.007
- Wickham, H. (2016). ggplot2: Elegant graphics for data analysis. New York: Springer-Verlag. Retrieved from https://ggplot2.tidyverse.org
- Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F. O., Ludwig, W., Schleifer, K. H., … Rosselló-Móra, R. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nature Reviews Microbiology , 12(9), 635–645. doi: 10.1038/nrmicro3330
- Zengler, K., Toledo, G., Rappé, M., Elkins, J., Mathur, E. J., Short, J. M., & Keller, M. (2002). Cultivating the uncultured. Proceedings of the National Academy of Sciences , 99(24), 15681–15686. doi: 10.1073/pnas.252630999
Internet Resources
Geneious Prime 2019.2.1 bioinformatics software (Kearse et al., 2012). Used to place a 16S sequence within a phylogenetic tree of sequences in the NBCI database. These phylogeny trees can be used to compare relatedness of isolates and understand the genetic relationships among all isolates under investigation. Geneious Prime 2019.2.1. was used in this study.
NCBI BLAST (Altschul et al., 1990). Algorithm and program for comparing 16S sequence data with sequences in the NBCI database.
Citing Literature
Number of times cited according to CrossRef: 4
- Hai Qu, Wenjing Zhang, Jianghao Li, Qingshan Fu, Xiaoxia Li, Miaomiao Wang, Guangyu Fu, Jing Cui, A rapid and sensitive CRISPR-Cas12a for the detection of Fusobacterium nucleatum , Microbiology Spectrum, 10.1128/spectrum.03629-23, 12 , 2, (2024).
- Xinru Huang, Shaoping Nie, Xiaodan Fu, Shihao Nan, Xinmiao Ren, Rong Li, Exploring the prebiotic potential of hydrolyzed fucoidan fermented in vitro with human fecal inocula: Impact on microbiota and metabolome, International Journal of Biological Macromolecules, 10.1016/j.ijbiomac.2024.131202, 267 , (131202), (2024).
- Honoka Aida, Bei-Wen Ying, Efforts to Minimise the Bacterial Genome as a Free-Living Growing System, Biology, 10.3390/biology12091170, 12 , 9, (1170), (2023).
- Stanislav Sukhikh, Svetlana Ivanova, Olga Babich, Anastasia Davydova, Ranjna Sirohi, Philippe Michaud, Vyacheslav Dolganyuk, Olga Kriger, Study of parameters for bioethanol production from Baltic Sea macroalgae, Bioresource Technology Reports, 10.1016/j.biteb.2023.101683, 24 , (101683), (2023).

