Extraction and PCR for animal samples using the Thermofisher Scientific TaqPath COVID-19 Combo Kit
Brianna Stenger
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
To describe the steps and materials needed to perform a multiplex real-time reverse transcriptase PCR test intended for the qualitative detection of nucleic acid from SARS-CoV-2, the causative agent of COVID-19, in animal respiratory swab samples.
This document includes the information and procedures to comply with the ThermoFisher Scientific TaqPath™ COVID-19 Combo Kit under the Food and Drug Administration’s (FDA) Emergency Use Authorization (EUA). The TaqPath™ COVID-19 Combo Kit includes a Real Time PCR assay multiplex that contains three primer/probe sets specific to the SARS-CoV-2 and a primer probe set for bacteriophage MS2 as an internal extraction control.
Sections of this procedure are taken directly from the TaqPath™ COVID-19 Combo Kit Instructions for Use, Publication MAN0019181, Version G.0. For additional information, please refer to these instructions.
Steps
SARS-CoV-2 RNA Extraction
Prepare working areas by cleaning with ~10% bleach or ~70% alcohol, followed by a rinse and use of another solution to reduce the presence of RNases such as RNaseZAP or Eliminase.
Prepare Binding Beads/Lysis mix on each day of use.
Vortex the Total Nucleic Acid Magnetic Beads to ensure that the bead mixture is homogeneous.
For the number of required reactions, prepare the Binding Bead/Lysis Mix according to Table 1
Table 1: Binding Bead/Lysis Mix
| A | B | C |
|---|---|---|
| Component | Volume per RNA Sample or Control | Volume for 150 rxns |
| Binding Solution | 530 μL | 79.5 mL |
| Nucleic Acid Magnetic Beads | 20 μL | 3.0 mL |
| Proteinase K | 10 μL | 1.5 mL |
| MS2 Phage Control | 10 μL | 1.5 mL |
| Total Reaction Mix volume | 570 μL | 85.5 mL |
***Highly recommend more than a 10% overage.
Mix well by inversion, then store at room temperature.
Prepare Plates for extraction following table 2.
Table 2: Plate setup.
| A | B | C | D | E |
|---|---|---|---|---|
| Plate Position | Plate ID | Plate Type | Reagent | Volume per Well |
| 1 | Sample Plate | KingFisher™ Deepwell 96 Plate or Equivalent | 400 µLSample* + 570 µL Bead/lysis mix | |
| 2 | Wash 1 | Wash Buffer | 1000 µL | |
| 3 | Wash 2 | 80% Ethanol | 1000 µL | |
| 4 | Elution Plate | Elution Solution | 50 µL | |
| 5 | Tip Comb | Place a KingFisher™ 96 tip comb for DW magnets in a KingFisher™ 96 KF microplate |
*Add sample to plate in a biological safety cabinet.
Invert the Binding bead Mix (from 2.2) five times gently to mix, then add 570µL (binding solution + beads + PK + MS2 phage control) to each sample well and negative control well in the sample plate.
Add samples and controls to the appropriate wells:
Samples: 400µLof sample to a well.
Negative Control: L of Nuclease-free Water 400µLL of Nuclease-free Water (not DEPC-Treated) to the Negative Control
Seal sample plate with film. Wipe outside of plate with disinfectant and place in transport container. Wipe/disinfect transport container and carefully bring to BSC in Extraction Room.
Select the program MVP_2Wash_400_Flex on the KingFisher™ Flex Magnetic Particle Processor with 96 Deep-Well Head
Start the run and load prepared plates in positions when prompted by the instrument.
Immediately remove the elution plate from the instrument and cover with film/foil. Run lasts ~24 minutes.
Seal immediately to avoid evaporation and store up to 48hrs at 4°C, -20°C, or -70°C.
SARS-CoV-2 PCR
If frozen, thaw the reagents on ice. Gently vortex the reagents, then centrifuge briefly to collect liquid at the bottom of the tubes.
Prepare a Master Mix of the PCR reagents containing enough for all samples, controls, plus extra. Master Mix is prepared manually using the volumes in Table 3.
Table 3. PCR Reagent Volumes.
| A | B |
|---|---|
| Nuclease-free Water | 12.5 µL |
| 4X TaqPath 1-Step Multiplex MMX (No ROX) | 6.25 µL |
| COVID-19 Real Time PCR Assay Multiplex | 1.25 µL |
| 20 µL MMX per rxn |
Pipette 20µL of master mix into each well.
Add 5µL of extraction RNA or negative control to corresponding well. Total reaction volume will be 25µL
Add 2µL of diluted positive control +3µL of nuclease-free water for positive control well.
Diluted positive control is made by:
Pipet 98µL of TaqPath™ COVID‑19 Control Dilution Buffer into a microcentrifuge tube, then add 2µL of TaqPath™ COVID‑19 Control. Mix well, then centrifuge briefly
Pipet 87.5µL of TaqPath™ COVID‑19 Control Dilution Buffer into a second microcentrifuge tube, then add 12.5µL of the dilution created in substep 13.1. Mix well, then centrifuge briefly.
Once sample and controls have been added, seal plate thoroughly.
Vortex plate at high speed for 10-30 seconds. Recommend a plate holder attachment to avoid damaging wells.
Centrifuge the reaction plate for 2 minutes (~600 x g) to remove bubbles and collect liquid to the bottom of the reaction plate.
Setting up Thermal Cycler
This protocol used the ABI 7500 Fast or ABI 7500 DX thermal cycler.
Set up thermal cycler according to manufacture instruction manuals along with table 4 and 5.
Refer to https://www.fda.gov/media/136112/download in reference sections for more information
Table 4. Target, dyes, and quencher.
| A | B | C |
|---|---|---|
| Target | Reporter dye | Quencher |
| ORF1ab | FAM | None |
| N gene | VIC | None |
| S gene | ABY | None |
| MS2 | JUN | None |
Table 5. Thermal cycler conditions.
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Number of cycles |
| UNG incubation | 25 C | 2 min | 1 |
| Reverse transcription | 53 C | 10 min | 1 |
| Activation | 95 C | 2 min | 1 |
| Denaturation | 95 C | 3 sec | 40 |
| Anneal/extension | 60 C | 30 sec |
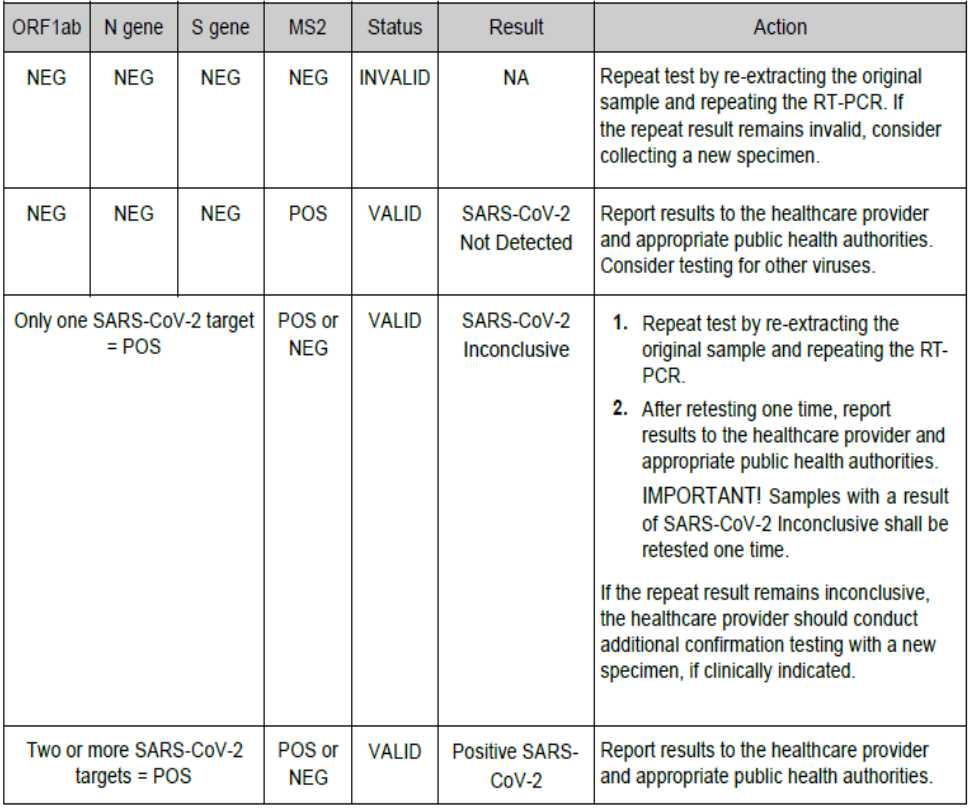
Interpretation of Results
A minimum of one Negative Control and one Positive Control must be present for each run. Additional Negative Control wells must be run for each extraction that is represented on a real-time RT-PCR plate. All control wells must pass for the real-time RT-PCR plate to be considered valid.