Evolution of Viral Genomes: Interplay Between Selection, Recombination, and Other Forces
Stephanie J. Spielman, Steven Weaver, Stephen D. Shank, Brittany Rife Magalis, Michael Li, Sergei L. Kosakovsky Pond
Virus evolution
Molecular evolution
Recombination
Positive selection
Relaxed selection
Phylogenetics
Codon models
Abstract
Natural selection is a fundamental force shaping organismal evolution, as it both maintains function and enables adaptation and innovation. Viruses, with their typically short and largely coding genomes, experience strong and diverse selective forces, sometimes acting on timescales that can be directly measured. These selection pressures emerge from an antagonistic interplay between rapidly changing fitness requirements (immune and antiviral responses from hosts, transmission between hosts, or colonization of new host species) and functional imperatives (the ability to infect hosts or host cells and replicate within hosts). Indeed, computational methods to quantify these evolutionary forces using molecular sequence data were initially, dating back to the 1980s, applied to the study of viral pathogens. This preference largely emerged because the strong selective forces are easiest to detect in viruses, and, of course, viruses have clear biomedical relevance. Recent commoditization of affordable high-throughput sequencing has made it possible to generate truly massive genomic data sets, on which powerful and accurate methods can yield a very detailed depiction of when, where, and (sometimes) how viral pathogens respond to various selective forces.
Here, we present recent statistical developments and state-of-the-art methods to identify and characterize these selection pressures from protein-coding sequence alignments and phylogenies. Methods described here can reveal critical information about various evolutionary regimes, including whole-gene selection, lineage-specific selection, and site-specific selection acting upon viral genomes, while accounting for confounding biological processes, such as recombination and variation in mutation rates.
Steps
3.1 How to Run a Selection Analysis
To execute a selection analysis locally, the following steps will need to be taken:
Prepare your coding sequence alignment. In general, any duplicate sequences should be removed before analysis. Most importantly, it is imperative that the sequence alignment be in the correct reading frame, meaning that alignment must be performed with codon structure in mind. A common approach to ensure this criterion is met is to generate the alignment using translated amino-acid data and then back-translate to the original nucleotide sequences.
Prepare a phylogenetic tree from the multiple sequence alignment. Note that certain analyses may require a labeled phylogenetic tree, as indicated within each subsequent tutorial. Keep in mind that for most selection analyses, a tree topology is a nuisance parameter. Hence, while it is advisable to use good practices when inferring trees, minor errors in tree inference tend to have minor effects on gene- and site-level inference. A notable exception occurs when lineage-specific selection is investigated; in this case, ensuring high-quality tree topologies is important.
An essential and strongly recommended step before analyzing data for selection is to screen sequences for recombination. If recombinant sequences are naively analyzed without an appropriate phylogenetic correction, inference results are likely to be biased (Posada et al. [33]) (see the section on Screening sequences for recombination later in this chapter).
Prepare your data (alignment and phylogeny) for input to HyPhy. There are three ways to provide a dataset for HyPhy analysis, each of which will trigger a different analysis prompt at runtime:
- Two separate files containing the alignment and phylogeny, respectively. In this circumstance, HyPhy issues two successive prompts: the first for the file containing the alignment, and the second for the file containing the tree.
- A single file containing an alignment in one of the formats supported by HyPhy (FASTA, MEGA, and PHYLIP), with a Newick-formatted phylogeny included at the bottom of this file. In this circumstance, HyPhy issues two successive prompts: the first for the file containing the alignment, and the second asking whether to accept the tree found in the file (provide the affirmative response, e.g., “y,” to accept it).
- A NEXUS file containing both the alignment and phylogeny. In this circumstance, HyPhy automatically accepts the provided phylogeny and therefore only issues a single prompt for the file containing the alignment. This is also the format that can be used to specify partitioned data, which is necessary to account for recombination.
Execute the appropriate method in HyPhy, selecting options suitable for the specific analysis.
3.2 BUSTED
To run BUSTED, open a terminal session and enter HYPHYMP from the command line to launch the HyPhy analysis menu. Enter 1 (Selection Analyses) and then 5 to reach the BUSTED analysis menu, and supply values for the following prompts:
- Choose genetic code. This option tells HyPhy which translation table to use for codon-level analyses. Enter 1 to use the Universal genetic code.
- Select a coding sequence alignment file. Provide the full path to the dataset of interest: /path/to/data/ksr2.fna .
- A tree was found in the data file…Would you like to use it (y/n)? Enter “y” to use the tree.
- Choose the set of branches to test for selection. Enter 1 to test all branches for selection.
BUSTED will now run to completion, printing status indicators to screen while it runs. For an example of how this output will look when rendered into HTML (or similarly, PDF), see this link: http://bit.ly/2vsRZrh.
Listing 1 Partial BUSTED screen output
### Branches to test for selection in the BUSTED analysis
* Selected 15 branches to test in the BUSTED analysis: ‘HUM, PAN, Node6, GOR, Node5, PON, Node4, GIB, Node3, MAC, BAB, Node12, Node2, MAR, BUS‘
### Obtaining branch lengths and nucleotide substitution biases under the nucleotide GTR model
* Log ( L ) = -5768.01, AIC - c = 11582.06 (23 estimated parameters)
### Obtaining the global omega estimate based on relative GTR branch lengths and nucleotide substitution biases
* Log ( L ) = -5342.48, AIC - c = 10745.17 (30 estimated parameters)
* non - synonymous / synonymous rate ratio for *test* = 0.0342
### Improving branch lengths , nucleotide substitution biases, and global dN/dS ratios under a full codon model
* Log ( L ) = -5333.46, AIC - c = 10727.13 (30 estimated parameters)
* non - synonymous / synonymous rate ratio for *test* = 0.0307
### Performing the full ( dN / dS > 1 allowed) branch-site model fit
* Log ( L ) = -5319.67, AIC - c = 10707.62 (34 estimated parameters)
* For * test * branches , the following rate distribution for branch-site combinations was inferred
| Selection mode | dN/dS |Proportion, %| Notes |
|----------------------------------|--------------|-------------|------------------|
| Negative selection | 0.024 | 99.151 | |
| Negative selection | 0.085 | 0.812 | |
| Diversifying selection | 118.143 | 0.037 | |
### Performing the constrained (dN/dS > 1 not allowed) model fit
* Log ( L ) = -5326.18, AIC - c = 10718.63 (33 estimated parameters)
* For * test * branches under the null (no dN/dS > 1 model), the following rate distribution for branch-site combinations was inferred
| Selection mode | dN/dS |Proportion, %| Notes |
|-----------------------------|--------------|-------------|-------------------------|
| Negative selection | 0.000 | 10.598 | |
| Negative selection | 0.000 | 86.086 | Collapsed rate class |
| Neutral evolution | 1.000 | 3.316 | |
----
## Branch - site unrestricted statistical test of episodic diversification [BUSTED]
Likelihood ratio test for episodic diversifying positive selection, **p = 0.0015**.

3.3 RELAX
To run RELAX, open a terminal session and enter HYPHYMP from the command line to launch the HyPhy analysis menu. Enter 1 (Selection Analyses) and then 7 to reach the RELAX analysis menu, and supply values for the following prompts:
- Choose genetic code. Enter 1 to use the Universal genetic code.
- Select a coding sequence alignment file. Provide the full path to the dataset of interest: /path/to/data/pb2.fna .
- A tree was found in the data file…Would you like to use it (y/n)? Enter “y” to use the tree.
- Choose the set of branches to test for selection. This option asks you to specify the label inside your tree used to specify the test lineages. You can either select all unlabeled branches, or HyPhy will show all labels it found in the tree you provided. Enter1to select the branches labeled as “test” as the test set in RELAX analysis. Note that when multiple labels are present in your tree, HyPhy will issue an additional prompt to choose the set of Reference branches, in the event that some branches should remain Unclassified .
- Analysis type. This option asks you to specify the scope of RELAX analysis. Selecting “Minimal” will run the RELAX hypothesis test, and selecting “All” will run hypothesis testing and fit two additional descriptive models, described earlier. Here, we will perform only hypothesis testing to determine whether the data shows evidence for a relaxation or intensification of selection intensity between the test and reference lineages. Enter the option 2 to run the “Minimal” analysis.
RELAX will now run to completion, printing status indicators to screen while it runs.
Listing 2 Partial RELAX screen output
### Obtaining branch lengths and nucleotide substitution biases under the nucleotide GTR model
* Log ( L ) = -16755.26, AIC - c = 33660.66 (75 estimated parameters)
### Obtaining the global omega estimate based on relative GTR branch lengths and nucleotide substitution biases
* Log ( L ) = -14410.97, AIC - c = 28988.46 (83 estimated parameters)
* non - synonymous / synonymous rate ratio for *Reference* = 0.0401
* non - synonymous / synonymous rate ratio for *Test* = 0.0604
### Improving branch lengths , nucleotide substitution biases, and global dN/dS ratios under a full codon model
* Log ( L ) = -14354.67, AIC - c = 28875.86 (83 estimated parameters)
* non - synonymous / synonymous rate ratio for *Reference* = 0.0358
* non - synonymous / synonymous rate ratio for *Test* = 0.0609
### Fitting the alternative model to test K != 1
* Log ( L ) = -14337.22, AIC - c = 28849.02 (87 estimated parameters)
* Relaxation / intensification parameter (K) = 0.73
* The following rate distribution was inferred for **test** branches
| Selection mode | dN/dS |Proportion, %|Notes |
|---------------------------------|--------------|-------------|--------------------|
| Negative selection | 0.031 | 94.752 | |
| Negative selection | 0.086 | 2.951 | |
| Diversifying selection | 1.406 | 2.297 | |
* The following rate distribution was inferred for **reference** branches
| Selection mode | dN/dS |Proportion, %| Notes |
|---------------------------------|--------------|-------------|--------------------|
| Negative selection | 0.009 | 94.752 | |
| Negative selection | 0.035 | 2.951 | |
| Diversifying selection | 1.591 | 2.297 | |
### Fitting the null ( K := 1) model
* Log ( L ) = -14342.33, AIC - c = 28857.22 (86 estimated parameters)
* The following rate distribution for test/reference branches was inferred
| Selection mode | dN/dS |Proportion, %|Notes |
|---------------------------------|--------------|-------------|--------------------|
| Negative selection | 0.010 | 94.149 | |
| Negative selection | 0.021 | 3.391 | |
| Diversifying selection | 1.735 | 2.460 | |
----
## Test for relaxation ( or intensification) of selection [RELAX]
Likelihood ratio test ** p = 0.0014**.
> Evidence for * relaxation of selection* among **test** branches _relative_ to the **reference** branches at P<=0.05
----
3.4 aBSREL
To run aBSREL, open a terminal session and enter HYPHYMP from the command line to launch the HyPhy analysis menu. Enter 1 (Selection Analyses) and then 6 to reach the aBSREL analysis menu, and supply values for the following prompts:
- Choose genetic code. This option tells HyPhy which translation table to use for codon-level analyses. Enter 1 to use the Universal genetic code.
- Select a coding sequence alignment file. Provide the full path to the dataset of interest: /path/to/hiv1_transmission.fna .
- A tree was found in the data file…Would you like to use it (y/n)? Enter “y” to use the included tree.
- Choose the set of branches to test for selection. You can now select on which branches aBSREL should conduct a formal hypothesis test for positive selection. Enter 1 to test all branches for selection.
aBSREL will now run to completion, printing status indicators to screen while it runs (some output abbreviated).
Listing 3 Partial aBSREL screen output
### Obtaining branch lengths and nucleotide substitution biases under the nucleotide GTR model
* Log ( L ) = -5524.50, AIC - c = 11153.08 (52 estimated parameters)
### Fitting the baseline model with a single dN/dS class per branch, and no site-to-site variation.
* Log ( L ) = -5402.40, AIC - c = 11009.72 (102 estimated parameters)
* Branch - level non - synonymous / synonymous rate ratio distribution has median 0.66, and 95% of the weight in 0.00--5.41
### Determining the optimal number of rate classes per branch using a step up procedure
| Branch | Length | Rates | Max. dN/dS | Log(L) | AIC-c |Best AIC-c so far|
|-----------|-------|-----|------------------|------------|---------|-----------------|
| 0564 _22| 0.01 | 2 | 1.96 (52.27%) | -5402.41 | 11013.78 | 11009.72 |
| 0564 _7 | 0.01 | 2 | 0.74 ( 5.19%) | -5402.40 | 11013.76 | 11009.72 |
| Separator | 0.01 | 2 | 197.32 ( 3.95%) | -5397.53 | 11004.02 | 11004.02 |
| Separator | 0.01 | 3 | 180.22 ( 4.08%) | -5397.53 | 11008.06 | 11004.02 |
| 0564 _4 | 0.01 | 2 | 29.79 ( 2.15%) | -5394.37 | 11001.74 | 11001.74 |
| 0564 _4 | 0.01 | 3 | 29.78 ( 2.15%) | -5394.37 | 11005.78 | 11001.74 |
| 0564 _3 | 0.01 | 2 | 126.86 ( 3.14%) | -5388.59 | 10994.22 | 10994.22 |
| 0564 _3 | 0.01 | 3 | 135.96 ( 3.05%) | -5388.59 | 10998.25 | 10994.22 |
| 0564 _9 | 0.01 | 2 | 10.01 ( 8.61%) | -5388.37 | 10997.82 | 10994.22 |
...
| Node53 | 0.00 | 2 | 1.00 (100.00%) | -5371.63 | 10976.46 | 10971.76 |
| 0557 _6 | 0.00 | 2 | 27.66 (100.00%) | -5371.32 | 10975.83 | 10971.76 |
| 0557 _21 | 0.00 | 2 | 0.25 ( 1.96%) | -5371.30 | 10975.80 | 10971.76 |
| 0557 _7 | 0.00 | 2 | 0.25 ( 1.96%) | -5371.30 | 10975.80 | 10971.76 |
### Rate class analyses summary
* 38 branches with **1** rate classes
* 6 branches with **2** rate classes
### Improving parameter estimates of the adaptive rate class model
* Log ( L ) = -5370.66, AIC - c = 10970.49 (114 estimated parameters)
### Testing selected branches for selection
| Branch | Rates | Max. dN/dS | Test LRT |Uncorrected p-value|
|-------------------|---------|----------------------|----------|--------------------|
| 0564 _22 | 1 | 1.22 (100.00%) | 0.11 | 0.43015 |
| 0564 _7 | 1 | 0.61 (100.00%) | 0.00 | 1.00000 |
| Separator | 2 | 197.72 ( 3.95%) | 14.13 | 0.00029 |
| 0564 _4 | 2 | 28.89 ( 2.15%) | 4.81 | 0.03281 |
| 0564 _3 | 2 | 127.66 ( 3.14%) | 14.06 | 0.00030 |
| 0564 _9 | 1 | 0.72 (100.00%) | 0.00 | 1.00000 |
| 0564 _1 | 1 | 1.07 (100.00%) | 0.01 | 0.48208 |
...
| 0557 _21 | 1 | 1.00 (100.00%) | 0.00 | 1.00000 |
| 0557 _7 | 1 | 1.00 (100.00%) | 0.00 | 1.00000 |
----
### Adaptive branch site random effects likelihood test
Likelihood ratio test for episodic diversifying positive selection at Holm-Bonferroni corrected _p = 0.0500_ found **3** branches under selection among **44** tested.
* Node35 , p - value = 0.00018
* Separator , p - value = 0.01251
* 0564 _3 , p - value = 0.01266

3.5 Site-Level Selection: MEME, FEL, SLAC, and FUBAR
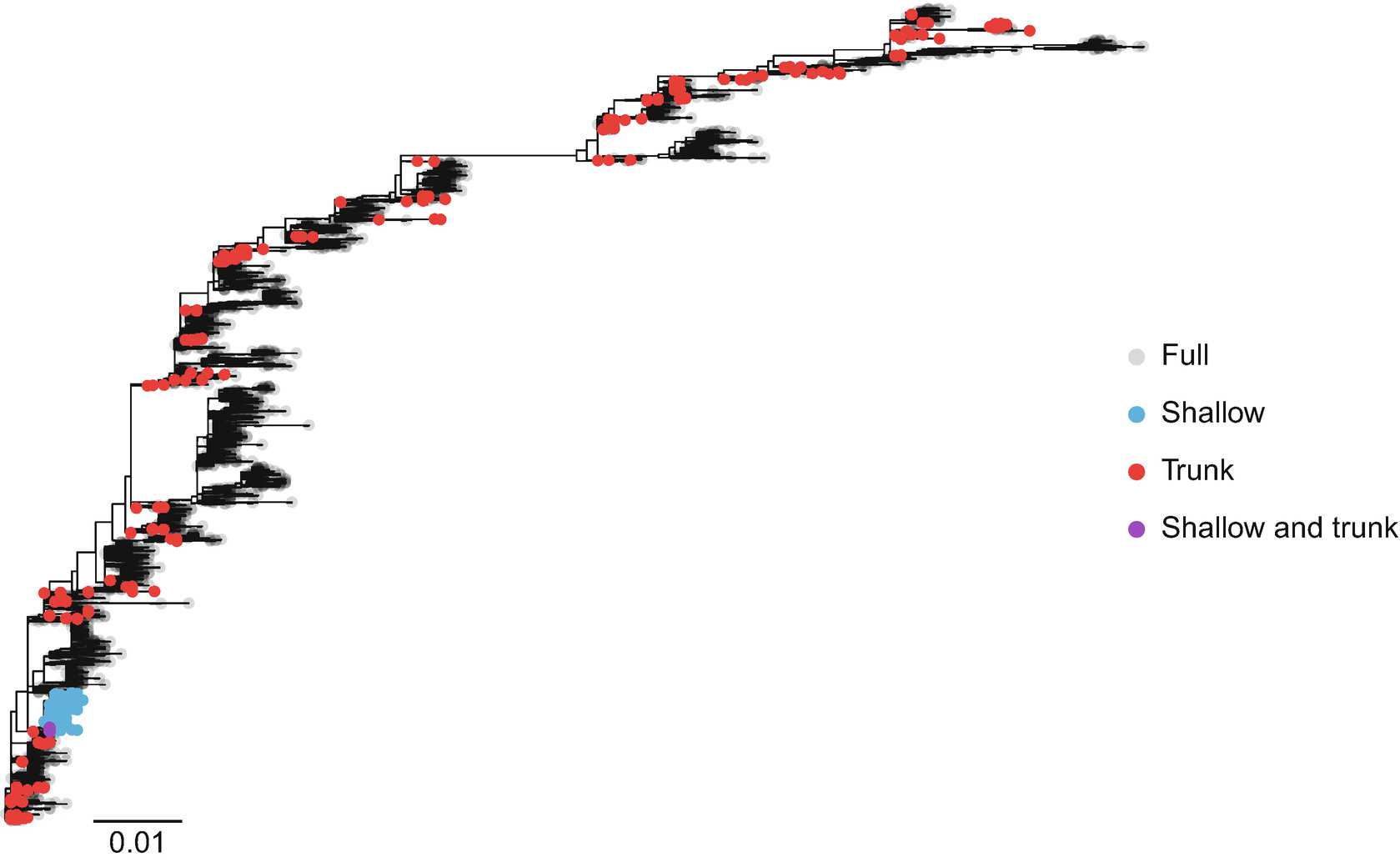

FEL: Launch HyPhy from the command line, and enter options 1 (Selection Analyses) and then 2 to reach the FEL analysis menu, and supply values for the following prompts:
- Choose genetic code . Enter 1 to use the Universal genetic code.
- Select a coding sequence alignment file . Provide the full path to the dataset of interest: /path/to/data/h3_trunk.fna .
- A tree was found in the data file…Would you like to use it (y/n)? . Enter “y” to use the tree.
- Choose the set of branches to test for selection . This option allows you to specify which branches along which site-level inference should be performed. Enter 1 to test all branches for selection.
- Use synonymous rate variation? . This option asks you to specify whether the dS parameter in the codon model should be allowed to vary across sites (“Yes”) or be fixed to 1 at all sites (“No”). Enter1to use a model with synonymous rate variation.
- Select the P -value used to perform the test at (permissible range = [0,1], default value = 0.1) . Provide the default threshold of 0.1 .
FEL will now run to completion and print status indicators to the screen, including results for any site found to be under selection (either positive or negative). Abbreviated results are shown below.
Listing 4 Partial FEL screen output
### Obtaining branch lengths and nucleotide rates under the GTR model
* Log ( L ) = -7506.06
### Obtaining the global omega estimate based on relative GTR branch lengths and nucleotide substitution biases
* Log ( L ) = -7302.10
* non - synonymous / synonymous rate ratio for *test* = 0.2923
### Improving branch lengths , nucleotide substitution biases, and global dN/dS ratios under a full codon model
* Log ( L ) = -7289.65
* non - synonymous / synonymous rate ratio = 0.2598
### For partition 1 these sites are significant at p <=0.1
| Codon | Partition | alpha | beta | LRT |Selection detected?|
|:-----------:|:------------:|:----------:|:--------:|:-----------:|:-------------------:|
...
| 146 | 1 | 3.818 | 0.000 | 7.336 | Neg. p = 0.0068 |
| 152 | 1 | 1.968 | 0.000 | 3.634 | Neg. p = 0.0566 |
| 154 | 1 | 0.000 | 3.912 | 4.652 | Pos. p = 0.0310 |
| 159 | 1 | 4.413 | 0.716 | 2.972 | Neg. p = 0.0847 |
| 164 | 1 | 2.082 | 0.000 | 2.713 | Neg. p = 0.0995 |
| 176 | 1 | 1.659 | 0.000 | 2.986 | Neg. p = 0.0840 |
| 177 | 1 | 6.393 | 0.000 | 8.421 | Neg. p = 0.0037 |
| 181 | 1 | 1.928 | 0.000 | 3.286 | Neg. p = 0.0699 |
| 190 | 1 | 2.085 | 0.000 | 2.715 | Neg. p = 0.0994 |
| 201 | 1 | 1.645 | 0.000 | 3.370 | Neg. p = 0.0664 |
| 208 | 1 | 0.000 | 3.625 | 4.668 | Pos. p = 0.0307 |
...
### ** Found _3_ sites under pervasive positive diversifying and _115_ sites under negative selection at p <= 0.1**
Listing 5 Partial SLAC screen output
...
### For partition 1 these sites are significant at p <=0.1
| Codon | Partition | S | N | dS | dN |Selection detected?|
|:---------:|:--------:|:---------:|:-------:|:--------:|:--------:|:-----------------:|
...
| 146 | 1 | 3.000 | 0.000 | 3.000 | 0.000 | Neg. p = 0.037 |
| 154 | 1 | 0.000 | 8.000 | 0.000 | 4.000 | Pos. p = 0.039 |
| 177 | 1 | 3.000 | 0.000 | 4.038 | 0.000 | Neg. p = 0.020 |
| 208 | 1 | 0.000 | 6.000 | 0.000 | 2.994 | Pos. p = 0.089 |
...
### Ancestor sampling analysis
> Generating 100 ancestral sequence samples to obtain confidence intervals
Resampling results for partition 1
|Codon|Part.|S [median,IQR]|N [median, IQR]|dS [median, IQR]|dN [median, IQR]|p-value [median, IQR]|
|:----:|:----:|:---------:|-----------:|----------:|-----------:|---------------------:|
...
| 146 | 1 | 3.00 [3.00-3.00]| 0.00 [0.00-0.00]| 3.00 [3.00-3.00] | 0.00 [0.00-0.00] | 0.04 [0.04-0.04].|
|154|1|0.00 [0.00-0.00]| 8.00 [8.00-8.00]| 0.00 [0.00-0.00]| 4.00 [4.00-4.00]| 0.04 [0.04-0.04] |
| 177 | 1 | 3.00 [3.00-4.00]| 0.00 [0.00-0.00]| 4.04 [4.04-5.38]| 0.00 [0.00-0.00]| 0.02 [0.01-0.02] |
| 208 | 1| 0.00 [0.00-0.00]| 6.00 [6.00-6.00]| 0.00 [0.00-0.00]| 2.99 [2.99-2.99]| 0.09 [0.09-0.09]|
...
Listing 6 Partial MEME screen output
...
| Codon | Partition | alpha | beta+ | p+ | LRT |Episodic selection detected?|# branches|
|:------:|:-------:|:----:|:----:|:---:|:---:|:-----------------------:|:--------:|
| 64 | 1 | 0.000 | 14.717 | 0.204 | 3.512 | Yes, p = 0.0816 | 5 |
| 154 | 1 | 0.000 | 35.302 | 0.145 | 5.334 | Yes, p = 0.0317 | 8 |
| 171 | 1 | 0.000 | 45.005 | 0.017 | 5.753 | Yes, p = 0.0256 | 1 |
| 208 | 1 | 0.000 | 59.749 | 0.089 | 5.554 | Yes, p = 0.0283 | 6 |
| 242 | 1 | 1.839 | 34.114 | 0.216 | 4.273 | Yes, p = 0.0549 | 7 |
| 402 | 1 | 0.000 | 10.476 | 0.091 | 3.493 | Yes, p = 0.0824 | 2 |
### ** Found _6_ sites under episodic diversifying positive selection at p <= 0.1**
FUBAR: To run FUBAR, launch HyPhy from the command line, and enter options 1 (Selection Analyses) and then 4 to reach the FUBAR analysis menu, and supply values for the following prompts ( see footnote 5):
- Choose genetic code . Enter 1 to use the Universal genetic code.
- Select a coding sequence alignment file . Provide the full path to the dataset of interest: /path/to/data/h3_trunk.fna .
- A tree was found in the data file…Would you like to use it (y/n)? . Enter “y” to use the tree.
- Number of grid points per dimension. This option controls how fine the FUBAR analysis is by setting the range of possible dN and dS values that can be inferred, along an N × N grid. We will use the default value of 20 (leading to a 20 × 20 grid of dN ∕ dS ratios). FUBAR will now pre-compute likelihoods for each value in the grid.
- Number of MCMC chains to run . This option determines the number of Markov Chain Monte Carlo chains to run during Bayesian inference of evolutionary rates. Enter the default value of 5 to run 5 chains.
- The length of each chain . This option controls for how long each MCMC chain should be run. Enter the default value of 2000000 to run each chain for two million generations (thus obtaining two million samples).
- Use this many samples as burn-in . This option determines how many initial samples drawn from the MCMC chain should be discarded as burn-in, as is standard in Bayesian analyses. Enter the default value of 1000000 , leading to a final value of one-million draws per chain.
- How many samples should be drawn from each chain . This option determines the final number of samples to draw from the full set of one-million draws per chain. Enter the default value of 100 .
- The concentration parameter of the Dirichlet prior . This option controls the shape of the Dirichlet prior distribution. Enter the default value of 0.5 .
Listing 7 Partial FUBAR screen output
...
### Tabulating site - level results
| Codon | Partition | alpha | beta | N.eff |Posterior prob for positive selection|
|:-------:|:--------:|:----:|:-----:|:--------:|:------------------------------------:|
| 61 | 1 | 0.753 | 4.365 | 64.549 | Pos. posterior = 0.9262 |
| 64 | 1 | 0.753 | 3.920 | 77.106 | Pos. posterior = 0.9095 |
| 69 | 1 | 0.730 | 4.447 | 64.182 | Pos. posterior = 0.9325 |
| 154 | 1 | 0.637 | 6.595 | 53.312 | Pos. posterior = 0.9826 |
| 208 | 1 | 0.622 | 5.908 | 55.794 | Pos. posterior = 0.9731 |
| 242 | 1 | 2.215 | 12.055 | 1489.879 | Pos. posterior = 0.9131 |
----
## FUBAR inferred 6 sites subject to diversifying positive selection at posterior probability >= 0.9
Of these , 0.36 are expected to be false positives (95% confidence interval of 0-2 )
3.6 Screening Sequences for Recombination
3.7 GARD
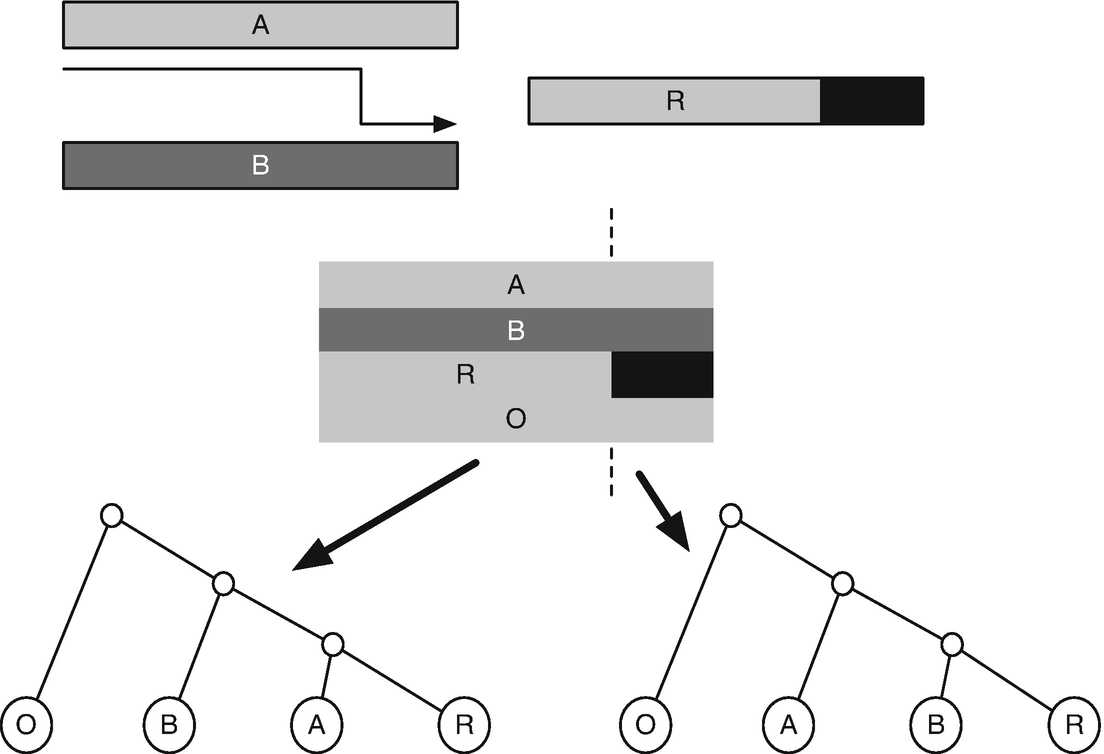
To run GARD, open a terminal session and start HYPHYMPI in the appropriate MPI environment (e.g., MPIRUN in OpenMPI) from the command line to launch the HyPhy analysis menu. Enter 12 (Recombination) and then 1 to reach the GARD analysis menu, and supply values for the following prompts:
- Nucleotide file to screen : Provide the full path to the dataset of interest: /path/to/data/cvf.fna .
- Please enter a 6-character model designation (e.g., 010010 defines HKY85) . This option controls which nucleotide substitution model is to be used for analysis, using PAUP notational shorthand. The six-character shorthand allows the user to specify the entire spectrum from F81 (000000) to GTR (012345), which we recommend as default option. Provide the value 012345 for this prompt.
- Rate variation options . This option determines how site-to-site rate variation should be modeled. The option None will discount site-to-site rate variation, allowing the analysis to run several times faster than other options but also creating the risk of mistaking rate heterogeneity for recombination. As such, we can only recommend this option for extremely small alignments (i.e., 3–5 sequences). The option General Discrete (the default) models rate variation using an N bin general discrete distribution, and option Beta-Gamma models rate variation using an adaptively discretized distribution, a more flexible version of the standard Gamma+4 model. Enter option 2 to select the General Discrete model.
- How many distribution bins [2–32]? . If rate variation was selected in the previous step, this option allows the user to decide how many different rate classes should be included in the model. We recommend using 3 rate classes by default, as both General Discrete and Beta-Gamma distributions are flexible enough to reliably capture rate variability in the majority of alignments with only a few rate classes. Therefore, enter the value 3 .
- Save results to . For this option, provide a full path to the output file to which you would like GARD to write results. The supplied file name will ultimately contain an HTML-formatted summary of the analysis. HyPhy will generate several other files with names obtained by appending suffixes (as in
_suffix) to the main result file. In particular, the _finalout file stores the original alignment in NEXUS format with inferred non-recombinant sections of the alignment saved in the ASSUMPTIONS block and trees inferred for each partition in the TREES block. This NEXUS file can be input into many recombination-aware analyses in HyPhy and other programs that can read NEXUS. The _ga_details file contains two lines of information about each model examined by the genetic algorithm: its AICc score and the location of breakpoints in the model. Finally, the _ga_splits file stores information about the location of breakpoints and trees inferred for each alignment region under the best model found by the GA.
GARD will now run to completion, printing status indicators to screen while it runs.
Listing 8 Partial GARD output
Fitting a baseline nucleotide model...
Done with single partition analysis. Log(L) = -5921.9511901113, c-AIC = 11914.85153276497
Starting the GA ...
GENERATION 2 with 1 breakpoints (~0% converged)
Breakpoints c - AIC Delta c - AIC [BP 1]
0 11914.85
1 11804.56 110.291 1393
GA has considered 92/ 328 (92 over all runs) unique models
Total run time 0 hrs 0 mins 2 seconds
Throughput 46.00 models/second
Allocated time remaining 999 hrs 59 mins 58 seconds (approx. 165599908 more models.)
...
GENERATION 52 with 4 breakpoints (~100% converged)
Breakpoints c - AIC Delta c - AIC [BP 1] [BP 2] [BP 3] [BP 4]
0 11914.85
1 11804.56 110.291 1445
2 11783.92 20.638 617 1490
3 11778.94 4.978 587 962 1475
4 11778.94 0.000 587 962 1475
GA has considered 268/ 473490550 (1356 over all runs) unique models
Total run time 0 hrs 4 mins 2 seconds
Throughput 5.60 models/second
Allocated time remaining 999 hrs 55 mins 58 seconds (approx. 20170544.82644628 more models.)
Performing the final optimization...
3.8 Accounting for Synonymous Rate Variation
4 Tips
5 Exercises
Earlier, we performed a BUSTED analysis without designating a specific subset of test lineages. For this exercise, we will analyze the HIV-1 transmission dataset with BUSTED in two different ways: testing all branches, and testing only recipient-derived HIV-1 sequences. The input data for this exercise, with an appropriately labeled phylogeny, is available in exercises/hiv1_transmission_exercise1.fna . For select branches labeled All or test as the test lineages.
- Is there evidence (compare model fits using the small sample AIC) that test branches have a different selective regime than the rest of the tree?
- The entire dataset should provide evidence for episodic diversification, but the recipient only analysis should return a negative result. What does this mean biologically, i.e., where does the selection signal come from?
Investigate the effect of recombination of site-specific inference of episodic selection using MEME. Run MEME on exercises/cvf.fna (single partition data, i.e., assuming no recombination), and then on the same dataset screened for recombination using GARD exercises/cvf_gard.nexus , testing for selection on all branches, with P=0.1 . Compare the list of sites detected to be under selection by the two analyses.
- Which analysis generated more positive results?
- Do you think these results are true or false positives? How does this compare to the FEL analysis we described in the text?
- Compare site-wise estimates of substitution rates (e.g., α ) between the two analyses. Is there a discernible bias introduced by not accounting for recombination?
When analyzing intraspecies or intrahost data, dN ∕ dS estimates may be inflated due to the fact that not all observed sequence variation are due to substitutions, but some are simply mutations that have not yet been filtered by selection [17, 23, 31, 35]. In other words, dN ∕ dS may be elevated by intraspecies/intrahost polymorphism that should not necessarily be attributed to positive selection. One simple approach to mitigating this undesirable effect is to restrict site-specific analyses to Internal branches only. Internal branches are less likely to contain spurious polymorphic variants because they encompass at least one process on which selection can act (i.e., transmission and/or multiple rounds of replication). Apply MEME and FEL to an intrahost sample of HIV-1 sequences, found in exercises/JS1774.nex , from an infected individual analyzed in Lorenzo-Redondo et al. [19] first choosing to test All branches, and next choosing Internal branches.
Compare the lists of selected sites between All/Internal analyses. How different are they?
Use RELAX to formally test whether or not selective regimes ( dN ∕ dS distributions) are different between terminal and internal branches in exercises/JS1774.nex .

