Establishing an In Vitro 3D Spheroid Model to Study Medulloblastoma Drug Response and Tumor Dissemination
Sophie J. Roper, Sophie J. Roper, Beth Coyle, Beth Coyle
Abstract
Medulloblastoma is the most common malignant pediatric brain tumor. Current treatment involves surgery, chemotherapy, and craniospinal radiotherapy, and these are associated with a significant reduction in quality of life. Metastatic dissemination at diagnosis is found in up to 30% of medulloblastoma cases and, alongside therapy resistance, is a significant feature in determining poor outcome. Development of new therapeutic approaches requires models where drug resistance and migration can be readily quantified and that are representative of patient disease. 3D medulloblastoma (3D-MB) spheroids are a simple yet effective means of bridging the gap between 2D culture and in vivo methods, providing users with highly reproducible in vitro models that more accurately recapitulate tumor morphology, drug response, and migration from a tumor mass. Unlike other cancer types, medulloblastoma spheroids fail to grow in their different standard cell culture media; instead, each cell line requires the same stem cell–enriching conditions. This requirement, however, has the advantage that it allows direct comparison of growth and response between cell lines in the absence of any potential media bias. In addition, spheroids can be used to model the initial stages of metastatic dissemination, something that cannot be achieved in 2D culture, providing insight into key changes occurring in migratory cells. Here, we provide protocols that detail the initial generation and maintenance of 3D-MB spheroids from sonic-hedgehog, Group 3, and Group 4 medulloblastoma subgroups, as well as describing functional assays to study drug response and cell migration across hyaluronan matrices, which represent the extracellular matrix backbone of the brain parenchyma. Through application of these simple yet highly representative models, it will be possible to test novel therapeutics targeting metastasis and drug resistance, as well as to develop insights into the mechanistic processes driving relapse in this malignant pediatric brain tumor. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Generation and maintenance of 3D medulloblastoma (3D-MB) spheroids
Support Protocol 1 : Measuring spheroid size for coefficient-of-variation analysis
Basic Protocol 2 : Assessing drug response in 3D-MB spheroids
Support Protocol 2 : 384-well 3D-MB spheroid generation
Basic Protocol 3 : Immunohistochemical staining of 3D-MB spheroids
Basic Protocol 4 : Modeling metastatic dissemination using 3D-MB migration models
Support Protocol 3 : RNA extraction from 3D-MB spheroids
INTRODUCTION
Medulloblastoma, a grade IV embryonal tumor of the cerebellum, is the most common malignant pediatric brain tumor, accounting for around a fifth of all childhood brain cancers (Pui, Gajjar, Kane, Qaddoumi, & Pappo, 2011). Almost 20% to 30% of medulloblastoma patients present with metastatic disease at diagnosis, a finding that is associated with poor outcome (Kool et al., 2012). Advances in molecular and genetic profiling over the past decade have enabled the classification of medulloblastomas into one of four subgroups [i.e., WNT, sonic hedgehog (SHH), Group 3, or Group 4] based on factors including tumor location, histology, genetic alterations, and survival (Cavalli et al., 2017). Despite these advances, methods of studying medulloblastoma at the in vitro level have not really progressed. Standard 2D cell culture fails to recapitulate the multidimensionality and cell-cell/cell-matrix interactions found in solid patient tumors (Edmondson, Broglie, Adcock, & Yang, 2014). Animal models replicate some of these features, although there may be species differences. In addition, such models are costly and only used at the final stage, after targets have been validated in cell culture models. Hence, more realistic in vitro models that consider physiological gradients and tumor microenvironmental factors are needed to bridge the gap between cellular and animal models.
Spheroid models, in which cell aggregates are grown in suspension in a 3D conformation, have been widely adopted by cancer researchers for drug evaluation studies and investigations of cell migration and invasion (Friedrich, Seidel, Ebner, & Kunz-Schughart, 2009; Vinci, Box, Zimmermann, & Eccles, 2013; Zanoni et al., 2016). Previous studies have shown that established cell lines of many tumor types, including glioblastoma multiforme, colorectal cancer, and breast cancer, produce 3D spheroids suitable for functional analyses when cultured under their standard conditions in ultra-low-attachment (ULA) wells (Vinci et al., 2012). We have recently demonstrated that, unlike other cell types, medulloblastoma cell lines were unable to grow successfully as spheroids in their diverse types of standard culture medium, and thorough optimization for growth in 3D spheroid culture was required (Roper, Linke, Scotting, & Coyle, 2021). The resulting 3D medulloblastoma (3D-MB) spheroids were both highly reproducible and extremely suitable for use in drug response experiments and migration assays.
Here, we describe the optimal growth conditions necessary to generate and maintain 3D-MB spheroids for a panel of widely available medulloblastoma cell lines, representing SHH, Group 3, and Group 4 medulloblastoma. We also detail, for the first time, robust and easy-to-follow protocols for downstream applications, including drug response studies, immunohistochemical (IHC) analysis, and hydrogel-based tumor dissemination cell migration assays. Support protocols for coefficient-of-variation (CV) analysis, higher-throughput (384-well) spheroid culture suitable for large-scale drug studies, and RNA extraction methods suitable for RNA sequencing (RNA-Seq) analyses are also included. A summary of the protocols included is shown in Figure 1.
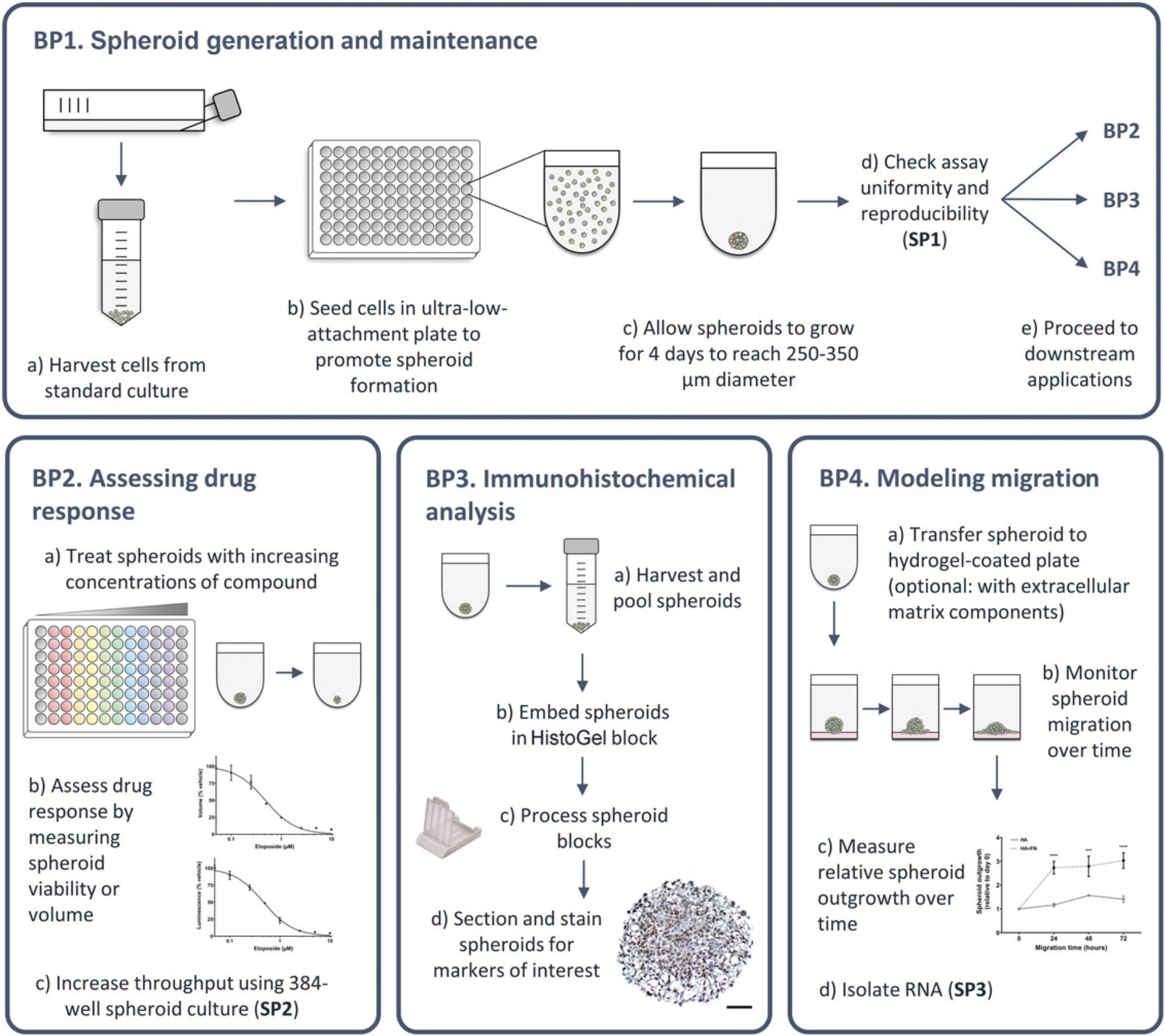
Given the potential of 3D-MB spheroids to bridge the gap between cell culture and animal studies, we have optimized culture conditions to produce actively growing 3D spheroids that are directly comparable between medulloblastoma cell lines, and we describe these in Basic Protocol 1. In Support Protocol 1, we detail CV analysis to validate spheroid uniformity. Next, in Basic Protocol 2, we describe simple drug compound screening and response analysis, with the potential to scale up to 384-well culture, if needed (Support Protocol 2). In Basic Protocol 3, we describe how to embed and section 3D-MB spheroids for IHC analyses, allowing target validation and visualization of protein expression gradients within the spheroid structure. Given that metastatic dissemination is a major factor in medulloblastoma progression, we also provide a protocol describing how this can be modeled using 3D-MB spheroids’ migration across a brain–extracellular matrix (ECM) hyaluronic hydrogel (Basic Protocol 4). Finally, we provide a support protocol describing RNA isolation methods suitable for RT-qPCR and high-throughput RNA-Seq (Support Protocol 3). In all, these protocols will allow users to implement our simple quantifiable approaches to measure and compare therapeutic response and dissemination between cell lines in closer-to-patient 3D cultures.
STRATEGIC PLANNING
The user will need to obtain medulloblastoma cell lines before commencing 3D-MB spheroid culture. Table 1 details clinical information (including patient demographics and metastatic status), source, and 2D versus 3D growth characteristics of some of these cell lines. Prior to 3D-MB spheroid generation, grow cells in their standard 2D culture conditions and medium, as detailed in Table 1. SHH cell lines (DAOY, ONS76, and UW228-3) grow adherently, Group 3 cell lines (HD-MB03, D283, D425, and D458) grow semi-adherently, and Group 4 cell lines (CHLA-01-MED and CHLA-01R-MED) grow in suspension. Manually inspect cells under a microscope before harvesting for 3D-MB spheroid generation. Ensure cells are free from infection and of around 70% to 80% confluence. One or two T-75 flasks at 70% to 80% confluence are usually sufficient to seed multiple 96- or 384-well spheroid plates.
- a 2D culture medium: 1: DMEM + 10% FBS; 2: RPMI 1640 + 10% FBS; 3: DMEM/F-12 + 15% FBS + 1% sodium pyruvate; 4: DMEM/F-12 + 2% B-27 supplement + 20 ng/mL EGF + 20 ng/mL FGF.
- b See recipe for 1.2% methyl cellulose stock.
- c,d Cell lines with matched primary and metastatic pairs.
NOTE : All solutions and equipment coming into contact with cells must be sterile, and proper sterile technique should be used accordingly.
NOTE : All culture incubations are performed in a 37°C, 5% CO2 incubator unless otherwise specified.
Basic Protocol 1: GENERATION AND MAINTENANCE OF 3D MEDULLOBLASTOMA (3D-MB) SPHEROIDS
Here, users will use ULA plates to generate 3D-MB spheroids from established medulloblastoma cell lines. ULA plates have a round bottom and a covalently bound hydrogel layer to inhibit cellular attachment. This structure promotes the formation of a single, highly reproducible spheroid in each well. The resulting spheroids can then be used in downstream applications, as described in Basic Protocols 2 to 4.
3D culture conditions, such as spheroid size, incubation periods, and culture medium, have been standardized across all of the medulloblastoma cell lines described in this protocol in order to allow direct comparisons between spheroids of different types. In addition, the cell seeding densities in this protocol have been optimized to produce 3D-MB spheroids of 250 to 350 µm in diameter following an initial 4-day formation period. This is the optimal size for establishment of pathophysiological oxygen gradients while still permitting drug perfusion throughout the spheroid (Vinci et al., 2013).
Use of a serum-free neurosphere medium, containing supplements and growth factors routinely used in the culture and maintenance of neural stem cells, is essential for sustained 3D-MB spheroid growth (Roper et al., 2021).
We also provide Support Protocol 1, which describes how to perform CV analysis, a method of calculating variation in 3D-MB spheroid size within and between plates, to validate the model's reproducibility. We advise that users perform CV analysis on day 4, following the initial spheroid formation period, before moving on to downstream applications (Basic Protocols 2 to 4).
Materials
-
Medulloblastoma cell lines (growing under conditions described in Strategic Planning and Table 1)
-
Hank's Balanced Salt Solution (HBSS; Gibco, 14170) or sterile phosphate-buffered saline (PBS)
-
Trypsin-EDTA (Sigma-Aldrich, T4174)
-
Culture medium, 37°C
-
Neurosphere medium (see recipe), 37°C
-
Trypan blue (Gibco, 15250061)
-
Brightfield microscope
-
1.5- and 15-ml tubes
-
Centrifuge (capable of holding 15-ml tubes and plates)
-
Hemocytometer (or automated cell-counting system)
-
P200 multichannel pipet (capable of pipetting volumes ranging from 50 to 200 µl)
-
Reagent reservoirs (Thermo Fisher Scientific, 8096-11, or equivalent)
-
ULA round-bottom 96-well plates (Corning, 7007)
-
Additional reagents and equipment to measure 3D-MB spheroid variability (see Support Protocol 1; optional)
Generation of 3D-MB spheroids
Timing: 2 hr of hands-on work followed by 4 days of incubation
1.Manually inspect medulloblastoma cell lines under a brightfield microscope to ensure that they are around 70% to 80% confluent before proceeding.
2.Remove culture medium and wash cells twice with 10 ml HBSS or sterile PBS.
3.Add 2 ml trypsin-EDTA and incubate at 37°C for 5 min to allow cell detachment.
4.When the cells have detached from the flask, wash with 8 ml culture medium and transfer cell suspension into a pre-labeled 15-ml tube.
5.Centrifuge cell suspension for 5 min at 180 × g at room temperature to form a cell pellet.
6.During centrifugation, prepare volume of neurosphere medium needed.
7.Following centrifugation, remove and discard supernatant and resuspend cell pellet in 2 to 3 ml neurosphere medium.
8.Perform a cell count to calculate number of cells/ml:
-
Prepare a 1:1 dilution of cells (10 µl) and trypan blue (10 µl) in a fresh 1.5-ml tube and then pipet up and down to mix.
-
Add 10 µl of the solution to a hemocytometer chamber and place under the microscope.
-
Count total number of cells in the four outer squares of the hemocytometer grid.
-
Once you have obtained the total count, calculate cell concentration using the following formula:FIGURE
9.Dilute cells in neurosphere medium to the appropriate seeding density for seeding in a volume of 200 µl/well.
10.Add 200 µl HBSS or sterile PBS to all outer wells of a ULA round-bottom 96-well plate.
11.Gently resuspend diluted cell suspension to ensure that the cells are well distributed.
12.Seed cells in the ULA plate in a volume of 200 µl/well using a P200 multichannel pipet and reagent reservoir.
13.For semi-adherent and suspension cell lines (HD-MB03, D283, D425, D458, CHLA-01-MED, and CHLA-01R-MED), centrifuge plate for 5 min at 100 × g after seeding to encourage spheroid compactness.
14.Visually inspect plate under the microscope to check that cells have been seeded in each well.
15.Transfer plate to the incubator and allow spheroid formation and growth for 4 days to achieve an optimal size of 250 to 350 µm in diameter.
16.On day 4, follow Support Protocol 1 to measure 3D-MB spheroid variability before proceeding with downstream applications (Basic Protocol 2, 3, or 4). Alternatively, continue growing 3D-MB spheroids following steps 17 to 21.
Maintenance of 3D-MB spheroids
Timing: Every 2 to 3 days following initial 4-day incubation period
17.Manually inspect ULA plate under the microscope to assess spheroid health.
18.Gently remove 100 µl neurosphere medium from each well using a multichannel pipet.
19.Pour fresh neurosphere medium into a reagent reservoir.
20.Add 100 µl fresh neurosphere medium to each well using a multichannel pipet.
21.If needed, place plate back in the incubator and continue to maintain spheroids by performing medium changes every 2 to 3 days following steps 17 to 20.
Support Protocol 1: MEASURING SPHEROID SIZE FOR COEFFICIENT-OF-VARIATION ANALYSIS
In this support protocol, we describe how to perform CV analysis by manually imaging each well (containing an individual spheroid) and measuring spheroid size using the macro developed by Ivanov et al. (2014) and the open-access software ImageJ. CV analysis is a useful method of determining assay uniformity by calculating variations in spheroid size within a plate. CV is commonly used to assess the reproducibility of in vitro models, and CV scores of <20% variation are considered acceptable (Sittampalam, Grossman, Brimacombe, Arkin, & Auld, 2004).
We advise that this protocol be performed on day 4 of spheroid formation (see Basic Protocol 1, step 16), when spheroids should have reached the optimal size of 250 to 350 µm in diameter, before moving on to downstream applications (Basic Protocols 2 to 4).
Materials
-
3D-MB spheroids (see Basic Protocol 1)
-
Imaging apparatus [e.g., brightfield microscope (Canon, DS126431, or equivalent) with attached camera (Olympus, CKX41) or automated imaging system]
-
Calibration slide (AmScope, A36CALM7-3PL)
-
Computer
-
ImageJ version 1.53n (https://imagej.nih.gov/ij/)
Timing: ≤1 hr
1.Image 3D-MB spheroids at 10× magnification with an imaging apparatus (there should be one spheroid per well). Use a calibration slide to determine scale of the images.
2.Transfer images to a computer.
3.Use ImageJ macro developed by Ivanov et al. (2014) and open-access software ImageJ to analyze spheroid size:
-
Download ImageJ and start program.
-
Using the image of the calibration slide from step 1, measure scale of images (number of pixels equal to 100 μm) at 10× magnification.
-
Download macro provided by Ivanov et al. (https://figshare.com/s/32f81784ee28e3fde015) and enter scale value after “distance=.”
-
Go to Plugins > Macros > Install and select amended macro text file.
-
Go to Plugins > Macros and select installed macro (typically listed at the bottom).
-
Select input folder containing the original spheroid images.
-
Select output folder (an empty folder that will include any processed images with the spheroid outlined).
-
Run macro.
This macro automatically calculates spheroid area and draws a blue outline of the detected spheroid.
-
Wait for macro to finish processing before analyzing the images (see steps 4 to 8).
4.Drag and drop output folder into ImageJ and assess blue outline of the detected spheroids. If the macro has failed to detect the correct spheroid circumference, manually outline spheroid and go to Analyze > Measure (or Ctrl + M) to calculate area.
5.Export spheroid measurements from ImageJ into a spreadsheet.
6.Using the area values, calculate spheroid diameter using the following equations:
7.Calculate mean and standard deviation of the diameter measurements for each plate (or by cell line if multiple lines have been seeded within the same plate).
8.Calculate CV for each plate/cell line using the following equation:
9.Proceed with downstream protocols (Basic Protocols 2 to 4) if CV is <20%.
Basic Protocol 2: ASSESSING DRUG RESPONSE IN 3D-MB SPHEROIDS
In this protocol, users will generate 3D-MB spheroids using an adaptation of Basic Protocol 1 that allows subjection of spheroids to a drug treatment of interest in a 96-well plate format. As opposed to in Basic Protocol 1, the 3D-MB spheroids here will be generated in opaque-walled ULA plates, thereby avoiding the need to transfer delicate drug-treated spheroids to an assay plate for viability assessment, which is the protocol's readout.
Vehicle-treated controls should always be used when assessing drug response, and treatment exposure and time points for measurements should be modified depending upon the compound's mechanism of action. The advantage of the approach described here is that all spheroids (regardless of the cell line from which they are derived) will have the same starting dimensions (250 to 350 µm) and are grown in the same medium, so any differences in response should be due to intrinsic properties of the cells themselves. Drug penetration and stability in media will also be the same, allowing direct comparisons between cell lines and compounds.
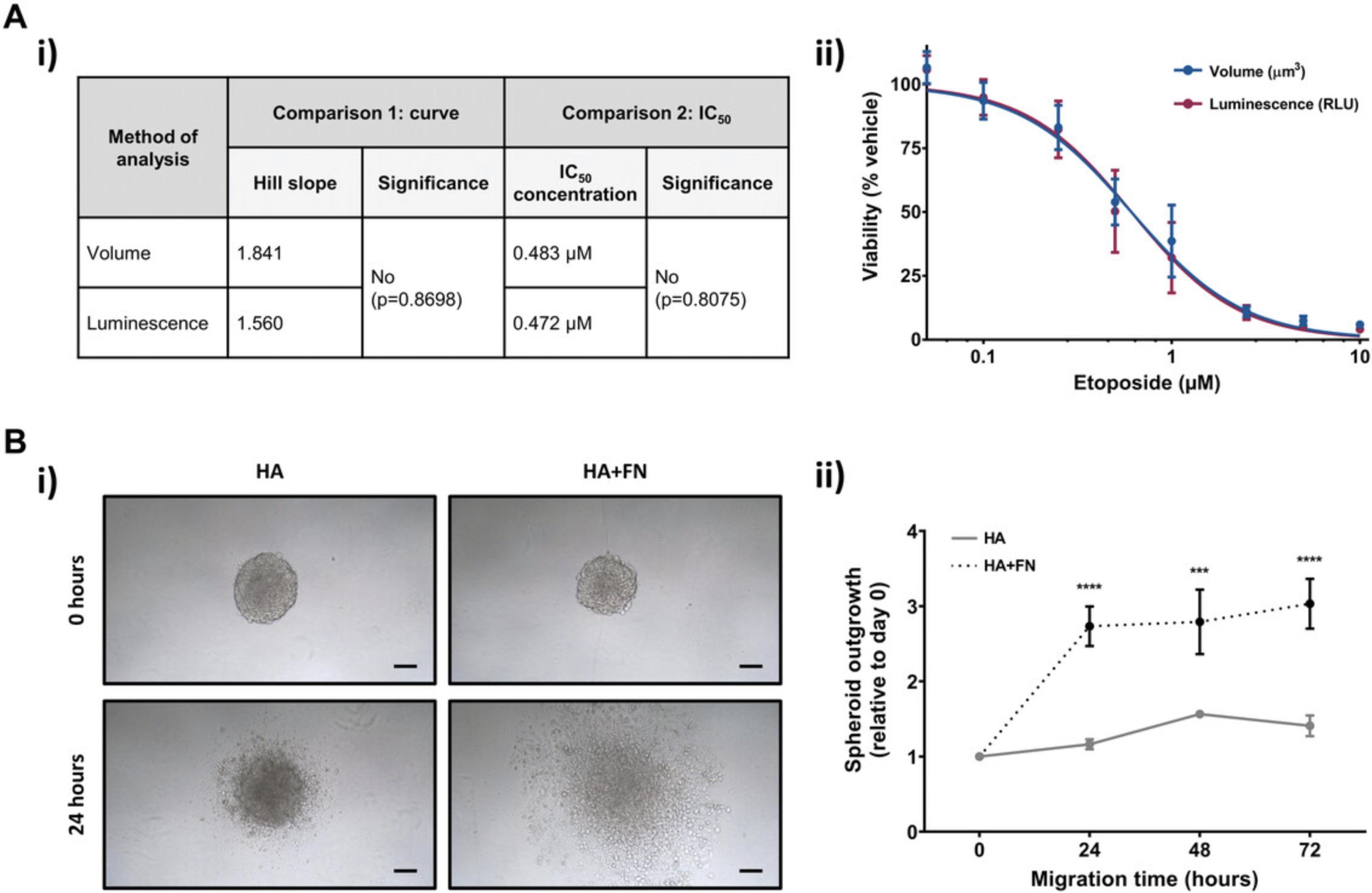
Drug response can be monitored using the CellTiter-Glo 3D cell viability assay to measure 3D-MB spheroid viability. This luciferase-based, commercially available assay has been optimized for use with 3D in vitro culture. The CellTiter-Glo reagent used penetrates the 3D spheroid structure and lyses cells to generate a luminescent signal proportional to the amount of adenosine triphosphate (ATP) present, indicative of the number of viable cells within the spheroid. Alternatively, drug response can be measured by assessing changes in spheroid size, and we describe the steps for both approaches. These two methods of analysis produce equivalent drug response curves and IC50 values (Fig. 3A).
This protocol is designed for 96-well plate cultures; however, this method can easily be scaled up to 384-well plate cultures with automated dispensing and imaging approaches for users requiring a higher throughput, for example, those wishing to assess large drug compound panels. Such adaptation is described in Support Protocol 2.
Materials
-
Drug of interest (e.g., etoposide, Selleckchem, S1225)
-
Vehicle control [e.g., dimethyl sulfoxide (DMSO), Sigma, D2650]
-
CellTiter-Glo 3D cell viability assay (Promega, G9683)
-
Opaque-walled ULA round-bottom 96-well plates (Corning, 4520)
-
Orbital incubator (Stuart, SI500, or equivalent)
-
Plate reader (FLUOstar Omega, BMG Labtech, or equivalent)
-
Statistical package of choice (GraphPad Prism version 9.2.0 or equivalent)
-
Additional reagents and equipment for preparing medulloblastoma cell lines to generate 3D-MB spheroids (see Basic Protocol 1) and for measuring 3D-MB spheroid uniformity (see Support Protocol 1)
Generation of 3D-MB spheroids
Timing: 2 hr of hands-on work followed by 4 days of incubation
1.Follow steps 1 to 9 of Basic Protocol 1.
2.Add 200 µl neurosphere medium to any three outer wells of an opaque-walled ULA round-bottom 96-well plate (to be used later as blank controls). Add 200 µl HBSS or sterile PBS to remaining outer wells.
3.Gently resuspend diluted cell suspension to ensure that the cells are well distributed.
4.Seed cells in neurosphere medium in a volume of 200 µl/well in the opaque-walled ULA plate using a P200 multichannel pipet and reagent reservoir.
5.For semi-adherent and suspension cell lines (HD-MB03, D283, D425, D458, CHLA-01-MED, and CHLA-01R-MED), centrifuge plates for 5 min at 100 × g after seeding to encourage spheroid compactness.
6.Visually inspect plate under a brightfield microscope to check that cells have been seeded in each well.
7.Transfer plate to the incubator and allow 3D-MB spheroid formation and growth for 4 days before proceeding to drug treatment.
Drug treatment
Timing: 1 to 2 hr
8.On day 4 of spheroid growth/formation:
- Visually inspect ULA plate under the microscope to assess spheroid health.
Examples of day 4 3D-MB spheroids are shown in Table1.
-
Follow Support Protocol1to measure CV to confirm 3D-MB spheroid uniformity on day 4 prior to addition of drug treatment.
9.Prepare 100 µl/well of solution of drug of interest at 2× concentration and vehicle control.
10.Gently remove 100 µl neurosphere medium from each well using a multichannel pipet.
11.Add 100 µl of 2× drug treatment or vehicle to each well.
12.Transfer plate to the incubator for the desired treatment length.
Measuring spheroid viability following drug treatment using the CellTiter-Glo 3D cell viability assay
Timing: 1 hr
13a. Following drug treatment, use a multichannel pipet to remove 100 µl medium from ULA plate.
14a. Dispense required volume of CellTiter-Glo 3D cell viability reagent (thawed at room temperature) into a reagent reservoir, with enough for all wells.
15a. Add 100 µl CellTiter-Glo 3D cell viability reagent per well using a multichannel pipet.
16a. Shake plate at 250 rpm for 5 min in an orbital incubator to promote cell lysis.
17a. Incubate plate for 25 min at room temperature in the dark.
18a. Insert plate into a plate reader and measure luminescence.
19a. Export luminescence measurements into a spreadsheet.
20a. Correct luminescence measurements against the blank (medium-only) controls included in step 2.
21a. Using the statistical package of choice, generate dose-response curves and calculate IC50 values (drug concentration required to inhibit growth by 50%) by nonlinear regression analysis.
Measuring changes in spheroid volume following drug treatment
Timing: 1 hr
13b. Following drug treatment, follow steps 1 to 5 of Support Protocol 1.
14b. Using the area values, calculate spheroid volume using the following equations:
15b. Calculate mean spheroid volume for each treatment condition.
16b. Using the statistical package of choice, generate dose-response curves and calculate IC50 values (drug concentration required to inhibit growth by 50%) by nonlinear regression analysis.
Support Protocol 2: 384-WELL 3D-MB SPHEROID GENERATION
In Basic Protocol 2, we describe how to generate 3D-MB spheroids that are suitable for drug response analysis in a 96-well plate format. However, 384-well culture is more amenable when higher throughput is required for drug screening studies, for example, when using compound libraries. Here, we describe 3D-MB spheroid culture in a 384-well format. Use of opaque-walled 384-well plates allows easy spheroid generation and analysis of luminescence-based viability assays.
Additional Materials (also see Basic Protocols 1 and 2)
- Opaque-walled ULA round-bottom 384-well plates (Corning, 3830)
Generation of 3D-MB spheroids
Timing: 2 hr of hands-on work followed by 4 days of incubation
1.Manually inspect cells under a brightfield microscope to ensure that they are around 70% to 80% confluent before proceeding.
2.Remove culture medium and wash cells twice with 10 ml HBSS or sterile PBS.
3.Add 2 ml trypsin-EDTA and incubate at 37°C for 5 min to allow cell detachment.
4.When the cells have detached from the flask, wash with 8 ml culture medium and transfer cell suspension into a pre-labeled 15-ml tube.
5.Centrifuge cell suspension for 5 min at 180 × g at room temperature to form a cell pellet.
6.During centrifugation, prepare volume of neurosphere medium needed.
7.Following centrifugation, remove and discard supernatant and resuspend cell pellet in 2 ml neurosphere medium.
8.Perform a cell count to calculate number of cells/ml according to Basic Protocol 1, step 8.
9.Dilute cells in neurosphere medium to the appropriate seeding density for seeding in a volume of 50 µl/well.
10.Gently resuspend diluted cell suspension to ensure that the cells are well distributed.
11.Seed cells in neurosphere medium in a volume of 50 µl/well in the opaque-walled ULA round-bottom 384-well plate using a P200 multichannel pipet and reagent reservoir. Include at least six wells as blank (medium-only) controls per plate to allow for luminescence correction later.
12.For semi-adherent and suspension cell lines (HD-MB03, D283, D425, D458, CHLA-01-MED, and CHLA-01R-MED), centrifuge ULA plates for 5 min at 100 × g at room temperature after seeding to encourage spheroid compactness.
13.Visually inspect plate under the microscope to check that cells have been seeded in each well.
14.Transfer plate to the incubator and allow spheroid formation and growth for 4 days before proceeding.
15.Continue to maintain the spheroids (see Basic Protocol 1, steps 17 to 21) or subject them to drug treatment (Basic Protocol 2, steps 8 to 12) before measuring viability. Reduce volumes to always achieve a final volume of 50 µl/well.
Measuring spheroid viability following drug treatment using the CellTiter-Glo 3D cell viability assay
Timing: 1 hr
16a. Dispense required volume of CellTiter-Glo 3D cell viability reagent (thawed at room temperature) into a reagent reservoir.
17a. Add 50 µl CellTiter-Glo 3D cell viability reagent per well using a multichannel pipet.
18a. Shake ULA plate at 250 rpm for 5 min in an orbital incubator to promote cell lysis.
19a. Incubate plate at for 25 min at room temperature in the dark.
20a. Insert plate into a plate reader and measure luminescence.
21a. Export luminescence measurements into a spreadsheet.
22a. Correct luminescence measurements against blank (medium-only) controls included in step 11.
23a. Using the statistical package of choice, generate dose-response curves and calculate IC50 values (drug concentration required to inhibit growth by 50%) by nonlinear regression analysis.
Measuring changes in spheroid volume following drug treatment
Timing: 1 hr
16b. Measure changes in spheroid size according to Basic Protocol 2, steps 13b to 16b.
Basic Protocol 3: IMMUNOHISTOCHEMICAL STAINING OF 3D-MB SPHEROIDS
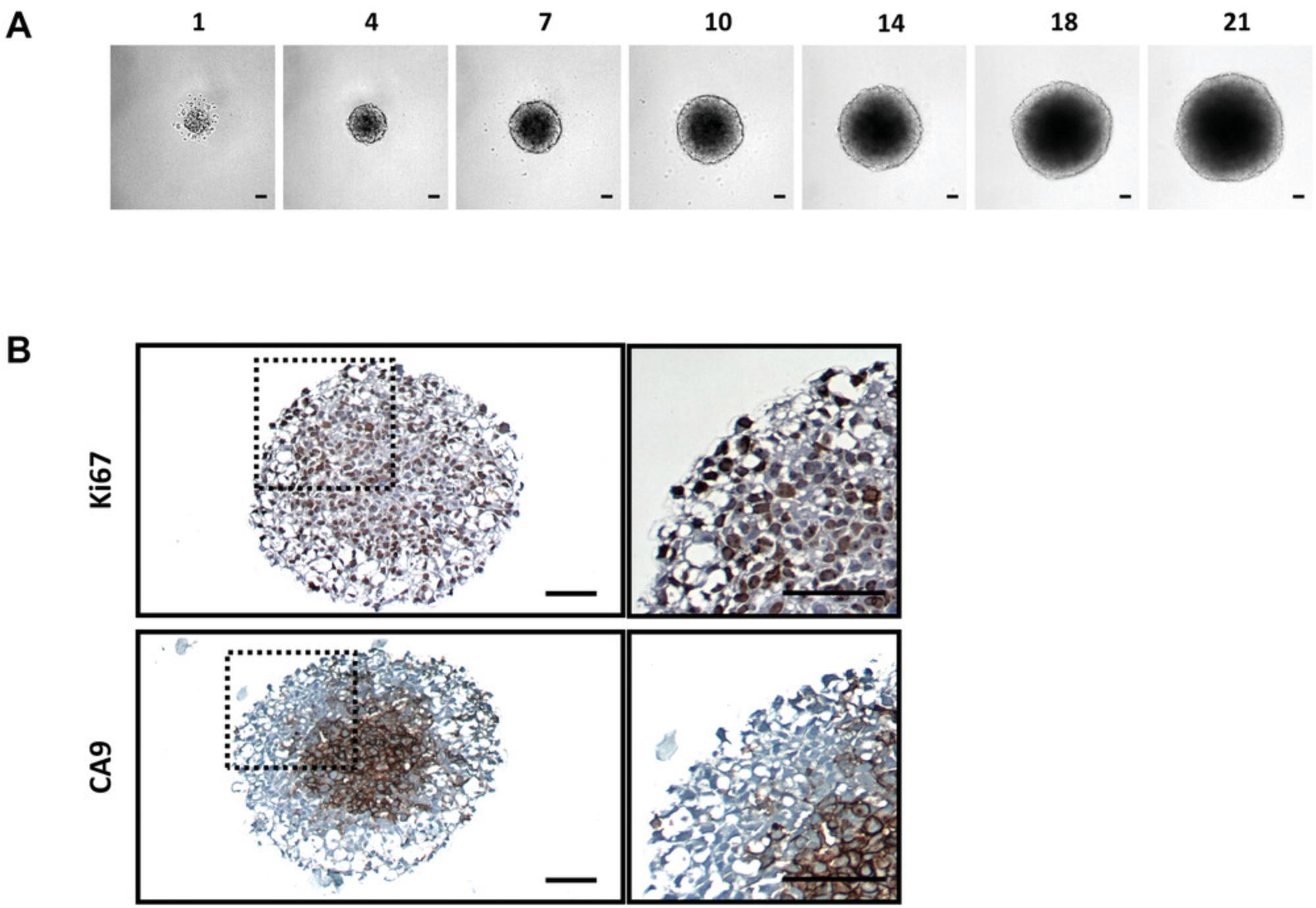
In this protocol, users will harvest 3D-MB spheroids generated from Basic Protocol 1 or drug-treated spheroids from Basic Protocol 2 for IHC staining. Multiple 3D-MB spheroids will be harvested and processed into histological blocks that, following standard fixation and embedding procedures, can be sectioned to produce slides for IHC staining. This protocol allows target validation and the visualization of protein expression patterns and gradients throughout spheroid sections, for example, for proliferation and hypoxia markers (Fig. 2B). It also suitable for the assessment of cytotoxic response following drug treatment. This method can be performed at multiple time points to allow the assessment of staining patterns as the 3D-MB spheroids mature or in response to drug exposure.
Materials
-
HistoGel (Thermo Fisher Scientific, HG-4000-012)
-
4% (v/v) paraformaldehyde (PFA) in PBS (Thermo Fisher Scientific, J19943.K2)
-
PBS (Thermo Fisher Scientific, J75889.AE)
-
Methanol
-
Xylene (Honeywell, 534056)
-
Paraffin
-
Sodium citrate buffer, pH 6 (see recipe)
-
70%, 80%, 95%, and 100% (v/v) ethanol
-
Peroxidase suppressor (Thermo Fisher Scientific, 35000)
-
Blocking buffer (Universal Blocker Blocking Buffer, Thermo Fisher Scientific, 1859332)
-
Primary antibody (e.g., Ki67, Cell Signaling Technology, 9449; CA9, Absolute Antibody, Ab00414-1.1)
-
HRP polymer–conjugated secondary antibody (e.g., goat anti-mouse, Abcam, ab214879)
-
SignalStain® DAB chromogen concentrate (Cell Signaling Technology, 11725)
-
SignalStain® DAB diluent (Cell Signaling Technology, 11724)
-
Distilled water
-
Hematoxylin (Harris Modified Hematoxylin, Thermo Fisher Scientific, 1859352)
-
Mounting medium (Thermo Fisher Scientific, 1859351)
-
60°C water bath (Leica, HI1210)
-
1.5-ml microcentrifuge tubes
-
Cryomolds (Tissue-Tek, Agar Scientific, AGG4581)
-
Tissue processing cassette (Simport, M490)
-
Beakers
-
Tissue processor (Leica, TP1020)
-
Wax embedding machine (Leica, EG1160)
-
Microtome (Leica, RM2235)
-
52°C water bath
-
Forceps
-
3-aminopropyltriethoxysilane (APES)-coated slides (BioWorld, 42763006-1)
-
60°C flattening table (Leica, HI1220)
-
60°C oven
-
Steamer (IHC World, IW-1102, or equivalent)
-
Humidified chamber (BioLegend, 926301)
-
Heat-proof containers (e.g., Tupperware)
-
Slide rack
-
Coplin jar (Fisher Scientific, 12858735)
-
Tissue paper
-
Hydrophobic pen (Vector Laboratories, H-4000)
-
Coverslips (AmScope, CS-24 × 50-100-3PL)
-
Brightfield microscope (Brunel Microscopes, N510)
-
Additional reagents and equipment for generating 3D-MB spheroids (see Basic Protocol 1)
Generation of 3D-MB spheroids
Timing: 2 hr of hands-on work followed by ≥4 days of incubation
1.Generate and maintain 3D-MB spheroids (10 spheroids per condition/time point) as described in Basic Protocol 1, steps 1 to 15.
Embedding spheroids in a HistoGel block
Timing: 3 hr
2.Pre-heat HistoGel to 60°C in a water bath.
3.At the desired time points, pool 10 spheroids using a pipet and gently transfer them to a 1.5-ml microcentrifuge tube.
4.Allow spheroids to settle at the bottom of the tube.
5.Carefully remove as much medium as possible and avoid disrupting spheroids.
6.Add 1 ml HBSS or sterile PBS to gently wash spheroids.
7.Carefully remove HBSS/PBS wash.
8.Resuspend spheroids in 500 µl HistoGel (pre-heated to 60°C in step 2).
9.Transfer spheroid/HistoGel suspension to a cryomold and place on ice.
10.When the spheroid block has set, transfer into a tissue processing cassette.
11.Place cassette in a beaker and cover with 4% PFA in PBS for 2 hr.
12.Transfer cassette to a clean beaker and cover with PBS for 10 min.
13.Transfer cassette to another clean beaker and cover with PBS for a further 10 min.
Processing of spheroid blocks
Timing: 18 hr
14.Process spheroid block overnight in a tissue processor with the following cycles: one bath each of 50%, 70%, and 90% methanol and four baths of 100% methanol (1 hr each), three baths of xylene (1 hr each), and two baths of molten paraffin (2 hr each, vacuum on).
15.The following day, embed spheroid block in paraffin using a wax embedding machine.
16.To section the spheroid block, place on ice before carefully aligning on a microtome.
17.Cut 4-µm sections and carefully place ribbons into a pre-heated 52°C water bath.
18.Separate each ribbon with forceps and place onto an APES-coated slide.
19.Leave slides to dry for ≥1 hr at 60°C on a flattening table.
20.Store slides at room temperature or proceed directly to the next stage.
IHC staining of spheroid sections
Timing: 1 day
21.Prepare for the IHC protocol:
- Place slides in an oven pre-heated to 60°C for ≥30 min.
Alternatively, slides can be incubated overnight.
-
Fill a heat-proof container with sodium citrate buffer (pH 6) and place in a steamer.
-
Pre-heat steamer for ≥20 min so that it reaches 90°C.
-
Prepare a humidified chamber by placing wet paper towels under the slide supports.
22.Place slides from step 20 in a slide rack and immerse in the following solutions for the indicated lengths of time:
-
Xylene (10 min).
-
Xylene (5 min).
-
100% ethanol (5 min).
-
100% ethanol (5 min).
-
95% ethanol (5 min).
-
95% ethanol (5 min).
-
80% ethanol (2 min).
-
70% ethanol (2 min).
23.Remove slides from the final 70% ethanol bath and wash slides briefly in running tap water.
24.To perform heat-induced antigen retrieval, place slide rack in the sodium citrate buffer bath in the pre-heated steamer (see step 21) for 40 min.
25.Turn off steamer and leave slides inside to cool for 10 min.
26.Place slides in a Coplin jar filled with PBS for 2 min.
27.With a tissue paper, gently wipe off excess PBS from slide and draw one large circle on slide around the spheroids with a hydrophobic pen.
28.Place slides in the humidified chamber from step 21d.
29.Add 150 µl peroxidase suppressor over spheroid sections on each slide and incubate for 30 min to quench endogenous peroxidase activity.
30.Wash slides by placing in a Coplin jar filled with PBS for 3 min.
31.Replace with fresh PBS and wash slides for an additional 3 min.
32.Add 150 µl blocking buffer over spheroid sections on each slide and incubate for 30 min.
33.Gently blot excess buffer from slide with a tissue paper.
34.Add 150 µl primary antibody diluted in blocking buffer (e.g., Ki67 1:400, CA9 1:800) over spheroid sections on each slide and incubate for 1 hr.
35.Wash slides by placing in a Coplin jar filled with PBS for 3 min.
36.Replace with fresh PBS and wash slides for an additional 3 min.
37.Add 150 µl HRP polymer–conjugated secondary antibody diluted in blocking buffer per slide and incubate for 1 hr.
38.Wash slides twice for 3 min each with PBS.
39.Prepare a 1× DAB solution by diluting 30 µl SignalStain® DAB chromogen concentrate in 1 ml SignalStain® DAB diluent.
40.Add 150 µl of 1× DAB solution and incubate until staining intensifies.
41.Wash slides by placing in a Coplin jar filled with PBS for 3 min.
42.Replace with fresh PBS and wash slides for an additional 3 min.
43.Rinse slides with distilled water and drain.
44.Add 150 µl hematoxylin over spheroid sections on each slide and incubate until desired staining is achieved.
45.Drain off excess hematoxylin and wash slides several times with distilled water.
46.Wash slides briefly in running tap water.
47.Place slides in a rack and wash in the following solution baths for 2 min each:
-
95% ethanol.
-
100% ethanol.
-
100% ethanol.
-
Xylene.
48.Place slides on a flat surface and apply 1 to 3 drops of mounting medium.
49.Apply a coverslip on top of the mounting medium and allow to set for 1 to 2 hr at room temperature.
50.Image stained spheroid sections under a brightfield microscope at 10× and 20× magnification to visualize protein expression patterns (Fig. 2B).
Basic Protocol 4: MODELING METASTATIC DISSEMINATION USING 3D-MB MIGRATION MODELS
Tumor spread involves the alteration of the cell's cytoskeleton, increased cell motility, and interaction with the surrounding ECM. Spheroid models are a useful tool to study tumor dissemination in vitro by modeling the migration of cells from a solid tumor (in this case, the 3D-MB spheroids) across the brain ECM (represented by a hyaluronan hydrogel). Here, we describe how users can transfer spheroids produced in Basic Protocol 1 onto a simple brain-specific matrix and measure migration of cells across the matrix over time (also called relative spheroid outgrowth). We refer to this as the hyaluronan hydrogel spheroid migration assay. The hyaluronan hydrogel that we use represents the backbone of the brain parenchyma, and we describe how this can be tailored by the addition of other ECM factors, such as fibronectin (Fig. 3B), to discern how these factors influence cell migration.
Materials
-
HyStem hydrogel kit (ESI-BIO, GS311), containing Glycosil, Extralink-Lite, and DG Water
-
Syringe (Scientific Laboratory Supplies, SYR6158)
-
Needle (Fisher Scientific, BD301750)
-
Rocker (Stuart, SRT6)
-
Tissue culture–treated 96-well plate (Corning, 10695951)
-
Statistical package of choice (GraphPad Prism version 9.2.0 or equivalent)
-
Additional reagents and equipment for generating 3D-MB spheroids (see Basic Protocol 1) and for imaging spheroids (see Support Protocol 1)
Generation of 3D-MB spheroids
Timing: 2 hr of hands-on work followed by 4 days of incubation
1.Prepare spheroids as described in Basic Protocol 1, steps 1 to 15, for use in the HA hydrogel spheroid migration assay.
2.Before proceeding with the setup of the migration model described below, visually inspect spheroids under a microscope to ensure that they are of optimal size and health (see Support Protocol 1).
Preparation of HA hydrogel layer
Timing: 2 hr
3.Thaw components of the HyStem hydrogel kit (Glycosil, Extralink-Lite, and DG Water) at room temperature.
4.Using a syringe and needle, add 1 ml DG Water to the thawed Glycosil.
5.Place vial horizontally on a rocker for 40 min to allow the solution to dissolve.
6.Using the syringe and needle, add 250 µl DG Water to the thawed Extralink-Lite. Invert several times to dissolve.
7.Prepare HA hydrogel at a 1:4 (Extralink-Lite to Glycosil) ratio by combining 250 µl Extralink-Lite and 1 ml Glycosil.
8.Add 50 µl of the HA hydrogel to each well of a tissue culture–treated 96-well plate.
9.Incubate hydrogel-coated plate at 37°C for 30 min to allow the gel layer to set.
Transfer of spheroids to HA hydrogel
Timing: 2 hr
10.Remove both hydrogel-coated plate (from step 9) and spheroid plate (from step 1) from the incubator.
11.Carefully position a P200 pipet tip at base of the spheroid plate and gently aspirate one spheroid in 100 µl neurosphere medium.
12.Transfer one spheroid into center of each well of the hydrogel-coated plate.
13.Repeat procedure for the remaining spheroids (a minimum of three repeats per experimental condition is recommended).
14.Add an additional 100 µl fresh neurosphere medium to each well.
15.Incubate spheroid migration plate at 37°C for 1 hr to allow the spheroids to settle on the matrix.
Assessment of spheroid migration
Timing: 1 hr per time point
16.Place spheroid migration plate under a brightfield microscope with an attached camera.
17.Image each well at both 4× and 10× magnification.
18.Transfer images to a computer.
19.Follow step 3 of Support Protocol 1 to calculate spheroid area.
20.Drag and drop output folder into ImageJ and assess blue outline of the migrated spheroid area. If the macro has failed to detect the correct spheroid circumference, manually outline migrated spheroid area and go to Analyze > Measure (or Ctrl + M) to calculate area.
21.Export spheroid measurements from ImageJ into a spreadsheet.
22.Using the area values, calculate relative spheroid outgrowth by normalizing the area covered by migrating cells over time to the initial spheroid size at 0 hr.
23.Using the statistical package of choice, generate spheroid outgrowth plots (see Fig. 3B-ii). Calculate statistical significance using two-way ANOVA analyses with Sidak's multiple-comparisons post hoc test.
Support Protocol 3: RNA EXTRACTION FROM 3D-MB SPHEROIDS
In addition to protein expression analysis by IHC (Basic Protocol 3), RNA analysis methods can also be used to investigate gene expression, either globally (RNA-Seq) or by candidate gene-based approaches (RT-qPCR). Here, we describe RNA extraction protocols for both (a) 3D-MB spheroids grown without a hydrogel matrix (as described in Basic Protocols 1 and 2) and (b) 3D-MB spheroids used in hydrogel-containing migration models (Basic Protocol 4). These allow the user to determine gene expression changes occurring in response to cytotoxic drugs (Basic Protocol 2) or to the influence of ECM components (Basic Protocol 4) in comparison to control samples (Basic Protocol 1). In each case, the purified RNA is suitable and sufficient for use in downstream applications, including RNA-Seq and RT-qPCR.
Materials
-
Liquid nitrogen (if snap-freezing; optional)
-
NucleoSpin RNA Plus Kit (Macherey-Nagel, 740984) or equivalent RNA extraction kit, including lysis buffer, gDNA removal column, binding solution, NucleoSpin RNA Plus column, Wash Buffers 1 and 2, 1.5-ml collection tube, and RNA-free water
-
Matrix D beads (2-ml tubes, MP Biomedicals, 11412420)
-
1.5-ml microcentrifuge tubes
-
Vortex
-
Microcentrifuge
-
Spectrophotometer (NanoDrop, Thermo Scientific, ND2000C)
-
Bioanalyzer (2100 Bioanalyzer, Agilent Technologies, G2939BA)
-
Tissue homogenizer (FastPrep-24 tissue homogenizer, MP Biomedicals, 12079310)
-
Additional reagents and equipment for generating 3D-MB spheroids (see Basic Protocol 1 or 2) and for imaging spheroids (see Support Protocol 1)
NOTE : Experiments involving RNA require careful technique to prevent contamination.
RNA extraction from 3D-MB spheroids (without hydrogel matrix)
Timing: 1 hr
1a. Prepare 3D-MB spheroids as described in Basic Protocol 1 or 2, with 10 spheroids per condition/time point of interest.
2a. Before proceeding, visually inspect spheroids under a microscope to ensure that they are of optimal size and health (see Support Protocol 1).
3a. At the desired time points, pool 10 spheroids into a 1.5-ml microcentrifuge tube.
4a. Allow spheroids to settle at bottom of the tube.
5a. Carefully remove as much medium as possible and avoid disrupting spheroids.
6a. Wash spheroids with 1 ml HBSS or sterile PBS.
7a. Carefully remove HBSS/PBS wash.
8a. At this stage, either snap-freeze spheroids in liquid nitrogen and store at –80°C (for RNA extraction later) or proceed directly to RNA extraction (step 9a).
9a. Extract RNA using a NucleoSpin RNA Plus Kit or equivalent RNA extraction kit:
- Add 350 µl lysis buffer to spheroids. Mix by pipetting and vortexing sample.
- ii. Transfer lysate to gDNA removal column. Centrifuge 30 s at 11,000 × g. Following centrifugation, discard column.
- Add 100 µl binding solution to flow-through and transfer to a NucleoSpin RNA Plus column. Centrifuge 15 s at 11,000 × g and discard flow-through.
- Add 200 µl Wash Buffer 1. Centrifuge 15 s at 11,000 × g and discard flow-through.
- Add 600 µl Wash Buffer 2. Centrifuge 15 s at 11,000 × g and discard flow-through.
- Add 250 µl Wash Buffer 2. Centrifuge 2 min at 11,000 × g to dry membrane.
- Transfer column to a 1.5-ml collection tube.
- Add 30 µl RNase-free water. Centrifuge 1 min at 11,000 × g .
10a. Determine RNA concentration using a spectrophotometer.
11a. Determine RNA integrity using a bioanalyzer.
12a. Store eluted RNA at –80°C until use in RT-qPCR or RNA-Seq.
RNA extraction from 3D-MB spheroids (with hydrogel matrix)
Timing: 1 hr
1b. Prepare 3D-MB spheroid migration models as described in Basic Protocol 4, with 10 spheroids per condition/time point.
2b. At the desired time points, gently remove medium from each well and use a pipet tip to move hydrogel layer containing migrated spheroids into a 1.5-ml microcentrifuge tube (pooling 10 wells).
3b. At this stage, either snap-freeze samples in liquid nitrogen and store at –80°C (for RNA extraction later) or proceed directly to RNA extraction (step 4b).
4b. Extract RNA using a NucleoSpin RNA Plus Kit or equivalent RNA extraction kit:
-
Transfer samples into tubes containing lysing Matrix D beads.
-
Add 350 µl lysis buffer and homogenize samples for 30 s using a FastPrep tissue homogenizer.
-
Transfer lysate to a new 1.5-ml microcentrifuge tube and centrifuge 5 min at 11,000 ×gto pellet hydrogel.
-
Transfer centrifuged lysate to gDNA removal column, avoiding the hydrogel pellet. Centrifuge 30 s at 11,000 ×g. Following centrifugation, discard column.
-
Add 100 µl binding solution to flow-through and transfer to a NucleoSpin RNA Plus column. Centrifuge 15 s at 11,000 ×gand discard flow-through.
-
Add 200 µl Wash Buffer 1. Centrifuge 15 s at 11,000 ×gand discard flow-through.
-
Add 600 µl Wash Buffer 2. Centrifuge 15 s at 11,000 ×gand discard flow-through.
-
Add 250 µl Wash Buffer 2. Centrifuge 2 min at 11,000 ×gto dry membrane.
-
Transfer column to a 1.5-ml collection tube.
-
Add 30 µl RNase-free water. Centrifuge 1 min at 11,000 ×g.
5b. Determine RNA concentration using a spectrophotometer.
6b. Determine RNA integrity using a bioanalyzer.
7b. Store eluted RNA at –80°C until use in RT-qPCR or RNA-Seq.
REAGENTS AND SOLUTIONS
Methyl cellulose stock, 1.2%
- Add 50 ml pre-heated DMEM/F-12 to 600 mg methyl cellulose (Sigma, M7027; pre-weighed and autoclaved)
- Stir solution for 30 min at room temperature
- Stir solution for further 2 hr at 4°C
- Centrifuge for 2 hr at 5,000 × g
- Remove clear, viscous supernatant
- Store ≤3 months at 4°C
Dilute 1.2% (w/v) stock to 0.24% (w/v) in neurosphere medium immediately before use.
Neurosphere medium
- 48.36 ml DMEM/F-12 (Gibco, 11320)
- 1 ml B-27 supplement (Gibco, 17504044)
- 500 µl N-2 supplement (Gibco, 17502048)
- 125 µl heparin (Stem Cell Technologies, 07980) (2 µg/ml final)
- 10 µl recombinant human epidermal growth factor (EGF; Gibco, PHG0315) (20 ng/ml final)
- 5 µl recombinant human basic fibroblast growth factor (bFGF; Gibco, PHG0266) (10 ng/ml final)
- Store ≤1 week at 4°C
Sodium citrate buffer, pH 6
- 2.1 g citric acid monohydrate
- ∼1 L distilled water
- ∼13 ml sodium hydroxide
- 0.5 ml Tween 20
- Store ≤3 months at room temperature
COMMENTARY
Background Information
To date, the majority of medulloblastoma in vitro experiments have been performed in 2D monolayer culture. Although reproducible and amenable to high-throughput drug screening studies (Zhang et al., 2019), 2D cultures fail to recapitulate the multidimensional growth, physiological gradients, and cell-cell interactions that exist in animal models and, more importantly, in patient tumors. Hence, this approach can result in an over/under-estimation of the therapeutic efficacy of test compounds. An improved understanding of medulloblastoma tumor biology and behavior requires adoption of a suitable culture system. In recent years, there have been significant improvements in in vitro technologies, allowing researchers to routinely grow and analyze cancer cells in 3D culture. 3D culture approaches have also been demonstrated to mimic many of the characteristics observed in patient tumors (Edmondson et al., 2014), providing a more predictive model of tumor biology and hence therapeutic efficacy.
3D culture models can be broadly categorized into four types: gel/matrix based, spheroids, transwell based, and tumor microvessels (reviewed in Katt, Placone, Wong, Xu, & Searson, 2016). Each model has its own advantages and disadvantages, and the choice depends upon the needs of the researcher. For example, gel/matrix-based models, in which cells are embedded within a gel or ECM, are ideal for studying matrix remodeling as well as cell migration and invasion. Transwell-based and tumor microvessel models can also be used to assess cell movement; however, neither of these techniques is as high throughput as spheroid models.
The 3D spheroid-based model is widely accessible due to its cost effectiveness. In addition, it is easy to reproduce, and long-term culture is possible. This has led to 3D spheroids being widely utilized in disease modeling, drug evaluation, and studies of cell migration and invasion (Friedrich et al., 2009; Mittler et al., 2017; Vinci et al., 2012, 2013; Zanoni et al., 2016). Spheroids can be produced by different methodologies, as highlighted in a recent review by Hoarau-Véchot, Rafii, Touboul, & Pasquier (2018). These include magnetic levitation (which is expensive and can be toxic to cells), suspension culture in spinner flasks (low throughput and hard to control size), hanging drop (technically challenging), and spontaneous spheroid formation in ULA plates (simple, with controllable spheroid size; hence, the method described here). Although many cancer cell lines have been optimized for this type of 3D spheroid growth (Vinci et al., 2012), there have been limited studies utilizing medulloblastoma cell lines in this model (Ivanov et al., 2014, 2015; Kumar et al., 2015; Neve, Kumar, Tripolitsioti, Grotzer, & Baumgartner, 2017; Santhana Kumar et al., 2018; Schönholzer et al., 2020). This may be because, in contrast to other cell types, medulloblastoma cells only proliferate in spheroids cultured in stem cell–enriching media, a finding that we and others have only recently described (Cheng et al., 2020; Roper et al., 2021). The methods described in this article therefore fulfill the need for an optimized and well-characterized 3D spheroid model of medulloblastoma suitable for a range of downstream applications.
Here, we describe how to produce actively growing spheroids using cells from different medulloblastoma subgroups in the same stem cell–enriching medium (Basic Protocol 1). Cell numbers required to generate spheroids of equivalent diameters and hence equivalent depth of drug penetration are also provided for routinely used cell lines (Table 1). These two features (same medium and diameter) allow direct comparison between lines. In Support Protocol 1, we provide a method for assessing assay uniformity, which we advise users to complete before proceeding with the remaining basic protocols. We also describe how to assay drug response and compare efficacy in spheroids in either 96-well plates (Basic Protocol 2) or 384-well plates (Support Protocol 2). Our findings regarding increased resistance to standard-of-care drugs in 3D relative to 2D culture and the fact that p53 modulation of drug response is only discernible in 3D spheroids have recently been published (Roper et al., 2021), exemplifying the utility of this approach. Target pathway activation or specific gene/protein pathway expression in spheroids can be verified by IHC analyses of sections (allowing signal localization; Basic Protocol 3) or by gene expression analyses with isolated RNA (Support Protocol 3). Finally, we show how measurement of cell migration from a spheroid across brain ECM–coated hydrogels can be used to mimic dissemination of cells from a tumor mass and to determine the influence of different ECM components (Basic Protocol 4). We selected a hyaluronan hydrogel because hyaluronan is a major constituent of the brain ECM (Rauch, 2004, 2007), making it a tissue-relevant matrix. We recently demonstrated, using RNA-Seq, that genes associated with fibronectin, another ECM component, were upregulated during spheroid migration across hyaluronan. In support of this, addition of fibronectin to hyaluronan hydrogels significantly enhanced migration of cells across the hydrogel (Roper et al., 2021). We have therefore described how the matrix can be tailored by addition of fibronectin and how to measure such changes in migration. Although we and others have also used more complex ECM-based models to study drug response and metastatic dissemination (Linke et al., 2021; Neve et al., 2017), these require more skill to set up, longer periods of time to run, and sophisticated imaging techniques and hence are better applied downstream in the drug screening process, facilitating studies of the mechanism of action of a smaller number of effective drugs in more detail.
In these protocols, we describe a manual seeding method using a multichannel pipet; however, the process can be made even easier and faster with automated liquid handling systems (e.g., MultiDrop Combi Reagent Dispenser, Thermo Fisher Scientific, 5840300). Likewise, manual imaging approaches are unsuitable for a higher-throughput drug screening study, but there are many automated imaging systems available that specialize in visualizing spheroids (e.g., IncuCyte S3, Celigo, EVOS). The simplicity of this model makes it easy to incorporate into routine laboratory studies for even a novice researcher, as well as offering the potential for the use of high-throughput robotics.
Critical Parameters
In standard 2D cultures, medulloblastoma cell lines grow in individually optimized media, which makes direct comparisons between cell lines difficult. The standard approach to spheroid growth is to grow cells in ULA plates in their individually optimized media. We have previously demonstrated, however, that medulloblastoma spheroids fail to grow in their standard 2D culture media (Roper et al., 2021). It is therefore essential to culture 3D-MB spheroids in the described stem cell–enriching medium (made fresh each week), containing growth factors and supplements typically used in neural stem cell maintenance. In 2D culture, medulloblastoma cells adopt different, subgroup-specific phenotypes, ranging from adherent (SHH subgroup) through semi-adherent (Group 3) to fully suspension (Group 4) growth. In 3D spheroids, different phenotypes are also observed, and not all medulloblastoma cell lines are highly suitable for 3D-MB spheroid culture. SHH cell lines produce tight 3D-MB spheroids with clearly defined edges, but additional modifications are recommended for Group 3 and 4 cell lines (described in Table 1). To allow direct comparison between lines in subsequent protocols (Basic Protocols 2 to 4), it is critical for cell seeding densities to be accurate during spheroid setup (Basic Protocol 1). Assay uniformity can be confirmed by CV analysis (Support Protocol 1). Optimized seeding densities (detailed in Table 1) should be used to produce 3D-MB spheroids that are 250 to 350 µm in diameter on day 4, at which point they are directly comparable in functional assays (Roper et al., 2021).
In Basic Protocols 2 to 4, spheroids need to be manipulated during medium changes or transferred from wells, and it is important to do this as gently as possible. In addition, as 3D culture medium should be made fresh each week, it is advised to calculate how much medium is required in order to avoid making it in excess, which will need to be discarded. As a guide, a minimum of 12 ml per 96-well plate is required (using a volume of 200 µl/well in 60 wells). To avoid edge effects, we recommend avoiding the use of the outer wells of the 96-well plate. In our experience, 3D-MB spheroids formed in these outer wells are consistently smaller, and therefore, any data generated could be unreliable. Instead, add 200 µl sterile HBSS or PBS to these wells to help maintain humidity across the plate.
In Basic Protocol 3, the size of the pores in the tissue processing cassette (Simport, M490) is critical; in our experience, using alternative cassettes can result in shrinkage. Please only use the recommended tissue processing cassette.
Basic Protocol 4 requires the use of a HyStem hydrogel kit, consisting of Glycosil (thiol-modified hyaluronic acid, or HA), Extralink-Lite (polyethylene glycol diacrylate, or PEGDA), and DG Water (degassed, deionized water). It is essential to work quickly when making HyStem hydrogels, as the matrix can gel at room temperature. Adjusting the Extralink-Lite concentration also affects gelation time; stiffer hydrogels (with a higher percentage of Extralink-Lite) solidify more quickly than softer gels. Addition of ECM factors can also speed up gelation time. It is recommended to reconstitute vials of Glycosil and Extralink-Lite in their entirety and to use them immediately. For Glycosil, do not uncap the vials because the material will crosslink in the presence of oxygen; instead, use a syringe and needle to add 1 ml DG Water and remove the product from the vials. For Extralink-Lite, we recommend reconstituting at a concentration of 2% (to achieve a matrix stiffness around 1.5 to 2 kPa) by adding 250 µl DG Water to the vial using a syringe and needle. Reconstituted Extralink-Lite can be stored at –20°C for 1 month.
Troubleshooting
See Table 2 for a list of common problems with the protocols, their causes, and potential solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Spheroids do not grow/stop growing | Medium composition | Check calculations and remake medium. Ensure medium is made fresh weekly. |
| Spheroids are smaller/larger than 250-350 µm at day 4 | Incorrect seeding density | Make sure cells are fully resuspended before counting. Use coefficient-of-variation analysis to assess assay uniformity and reproducibility (Support Protocol 1). |
| Spheroids lost during medium changes | Accidentally aspirating spheroids | Ensure spheroids remain undisturbed by placing the pipet at a 45° angle on the wall of the well. |
| Spheroid size is inconsistent within/across plates | Incorrect seeding density | Make sure cells are fully resuspended before counting. Check that pipets are calibrated. |
| HBSS/PBS not added to outer wells | Do not use outer wells for seeding. Instead, add 200 µl HBSS/PBS to these wells. | |
| No spheroids or fewer than expected in HistoGel block | Spheroids lost during transfer to HistoGel block | Allow spheroids to settle to the bottom of the tube and gently remove medium/PBS wash. |
| No staining present in spheroid sections following immunohistochemistry | Primary and secondary antibodies not compatible | Ensure isotypes of primary and secondary antibodies are compatible. |
| Not enough antibody bound to protein | Use higher concentration of primary antibody and/or incubate for longer. | |
| Protein of interest not present in the spheroid section | Check literature for a tissue type that expresses this protein and run that tissue as a positive control for the antibody. Include an additional control for a control protein such as tubulin. | |
| Hydrogel does not spread across surface of the well | Hydrogel is setting before it has spread evenly | Work quickly and ensure 50-µl volume is used. |
| Poor-quality RNA (from spheroid migration models) | Hydrogel is blocking RNA extraction spin column | Ensure tissue is fully lysed and homogenized before pelleting and loading onto a spin column. |
| Degraded RNA | Ensure use of RNase-free materials. |
Understanding Results
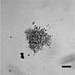
Table 1 lists details of the medulloblastoma cell lines that we have optimized so far in this spheroid model. Broadly speaking, 2D growth characteristics/molecular subgroup determine how readily spheroids are formed. SHH cell lines grow adherently in 2D culture and readily form spheroids. Group 3 cells grow semi-adherently in 2D and will form spheroids, although these tend to be less compact/looser than SHH spheroids. They also require the addition of methyl cellulose and gentle centrifugation. Group 4 cell lines typically grow as small clusters. Despite assay modifications, including the addition of methyl cellulose and a gentle centrifugation step, these cell lines fail to form large spheroids.
Figure 1 is a schematic overview of all of the protocols, demonstrating how they link together and with examples of the types of data that can be achieved.
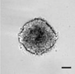
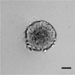
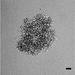
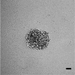
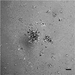
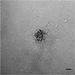
Figure 2 shows data from ONS76 spheroids obtained following Basic Protocol 1 and grown continuously over 21 days. In Figure 2A, a representative spheroid was microscopically imaged every 3 to 4 days. At day 1, the spheroid has clearly formed, although the outer edge is less well defined. By day 4, the edge is smooth and clearly defined, and a darker inner core that is characteristic of actively growing spheroids can be seen. The smooth edge remains as the spheroid continues to grow, although the edge of the inner darker core becomes less well defined. In order to assess which regions of the spheroid were actively growing or undergoing necrosis, spheroid sections produced in Basic Protocol 3 were stained for markers of proliferation (Ki67) and hypoxia (CA9). Figure 2B shows the results of staining 21-day-old ONS76 3D-MB spheroid sections for these markers of interest. Ki67 expression is visible throughout the spheroid, indicating that spheroids at this age are still actively proliferating. CA9 expression (absent at earlier stages) is restricted to the core, suggesting that this spheroid is hypoxic. Individual cells and nuclei can clearly be seen.
In Figure 3A, we show how ONS76 spheroids can be used to determine the IC50 of a drug, in this case the standard-of-care chemotherapeutic etoposide. Drug response curves and IC50 values depicting either ONS76 spheroid viability/luminescence (IC50 = 0.472 µM; Basic Protocol 2, steps 13a to 21a) or volume (IC50 = 0.483 µM; Basic Protocol 2, steps 13b to 16b) against concentration of etoposide are equivalent (p-value > 0.8). This indicates that these measurements can be used interchangeably in the context of Basic Protocol 2, and users can choose either method depending on resources or preference.
In Figure 3B, we demonstrate typical results that can be obtained from the spheroid migration model as described in Basic Protocol 4. Four-day old ONS76 spheroids were transferred onto either a hyaluronan (HA) or a hyaluronan and fibronectin (HA+FN) hydrogel-coated flat-bottomed 96-well plate. Cell migration from the 3D spheroid across the matrix was then imaged after 0, 24, 48, and 72 hr. Panel (i) shows representative images of 3D-MB spheroid migration across the surface of the hydrogels after 24 hr. Many more cells can be seen to have migrated in the presence of hyaluronan and fibronectin (HA+ FN) than on hyaluronan alone (HA). In (ii), spheroid outgrowth was calculated by measuring the area covered by migrating cells over a 72-hr period and normalizing to the initial spheroid size at 0 hr (following Basic Protocol 4, steps 16 to 23). The difference in migration was significant at all time points.
Time Considerations
The hours of hands-on time when culturing 3D-MB spheroids are not extensive. We have provided an estimation of timings within each protocol; however, these could be longer if processing multiple cell lines or plates in parallel. In addition, the time taken for some methods can be reduced by using multichannel pipets or automated liquid handling systems for seeding and maintenance, as well as automated imaging approaches.
It usually takes ∼2 hr to set up a 96-well plate of spheroids, followed by 4 days of incubation (Basic Protocol 1) until they reach the ideal size for downstream experiments. On day 4, we advise that users perform CV analysis to validate the model's reproducibility (Support Protocol 1) before moving on to downstream applications (Basic Protocols 2 to 4). This step takes <1 hr.
Setting up plates to assay drug response (Basic Protocol 2) takes up to 2 hr but can take longer if different drugs are being tested in parallel. Seeding 384-well plates (Support Protocol 2) is more time consuming but should only really be attempted with a multichannel pipet and then should again only take 2 hr.
The IHC analysis protocol (Basic Protocol 3) requires 3 hr to embed spheroids, followed by overnight processing (18 hr) and then 1 day for antibody staining.
Modeling migration (Basic Protocol 4) requires 2 hr to set up the hydrogels, followed by a further 2 hr to transfer spheroids onto the hydrogels. Downstream imaging typically takes 1 hr at each time point.
The RNA isolation protocol (Support Protocol 3) takes ∼1 hr to complete.
Acknowledgments
This research was supported by the Biotechnology and Biological Sciences Research Council Doctoral Training Partnership (S.J.R.; BB/J014508/1). The authors thank Dr. Franziska Linke for her contributions to optimizing hydrogel concentrations.
Author Contributions
Sophie J. Roper : Conceptualization, Data curation, Formal analysis, Writing-original draft, Writing-review and editing; Beth Coyle : Conceptualization, Formal analysis, Funding acquisition, Project administration, Supervision, Visualization, Writing-review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Literature Cited
- Cavalli, F. M. G., Remke, M., Rampasek, L., Peacock, J., Shih, D. J. H., Luu, B., … Taylor, M. D. (2017). Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell , 31, 737–754.e6. doi: 10.1016/j.ccell.2017.05.005.
- Cheng, Y., Franco-Barraza, J., Wang, Y., Zheng, C., Zhang, L., Qu, Y., … Yang, Z. (2020). Sustained hedgehog signaling in medulloblastoma tumoroids is attributed to stromal astrocytes and astrocyte-derived extracellular matrix. Laboratory Investigation , 100, 1208–1222. doi: 10.1038/s41374-020-0443-2.
- Edmondson, R., Broglie, J. J., Adcock, A. F., & Yang, L. (2014). Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay and Drug Development Technologies , 12, 207–218. doi: 10.1089/adt.2014.573.
- Friedrich, J., Seidel, C., Ebner, R., & Kunz-Schughart, L. A. (2009). Spheroid-based drug screen: Considerations and practical approach. Nature Protocols , 4, 309–324. doi: 10.1038/nprot.2008.226.
- Hoarau-Véchot, J., Rafii, A., Touboul, C., & Pasquier, J. (2018). Halfway between 2D and animal models: Are 3D cultures the ideal tool to study cancer-microenvironment interactions? International Journal of Molecular Sciences , 19, 181. doi: 10.3390/ijms19010181.
- Ivanov, D. P., Parker, T. L., Walker, D. A., Alexander, C., Ashford, M. B., Gellert, P. R., & Garnett, M. C. (2015). In vitro co-culture model of medulloblastoma and human neural stem cells for drug delivery assessment. Journal of Biotechnology , 205, 3–13. doi: 10.1016/j.jbiotec.2015.01.002.
- Ivanov, D. P., Parker, T. L., Walker, D. A., Alexander, C., Ashford, M. B., Gellert, P. R., & Garnett, M. C. (2014). Multiplexing spheroid volume, resazurin and acid phosphatase viability assays for high-throughput screening of tumour spheroids and stem cell neurospheres. PLOS ONE , 9, e103817. doi: 10.1371/journal.pone.0103817.
- Katt, M. E., Placone, A. L., Wong, A. D., Xu, Z. S., & Searson, P. C. (2016). In vitro tumor models: Advantages, disadvantages, variables, and selecting the right platform. Frontiers in Bioengineering and Biotechnology , 4, 12. doi: 10.3389/fbioe.2016.00012.
- Kool, M., Korshunov, A., Remke, M., Jones, D. T. W., Schlanstein, M., Northcott, P. A., … Pfister, S. M. (2012). Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathologica , 123, 473–484. doi: 10.1007/s00401-012-0958-8.
- Kumar, K. S., Pillong, M., Kunze, J., Burghardt, I., Weller, M., Grotzer, M. A., … Baumgartner, M. (2015). Computer-assisted quantification of motile and invasive capabilities of cancer cells. Scientific Reports , 5, 1–12. doi: 10.1038/srep15338.
- Linke, F., Aldighieri, M., Lourdusamy, A., Grabowska, A. M., Stolnik, S., Kerr, I. D., … Coyle, B. (2021). 3D hydrogels reveal medulloblastoma subgroup differences and identify extracellular matrix subtypes that predict patient outcome. Journal of Pathology , 253, 326–338. doi: 10.1002/path.5591.
- Mittler, F., Obeïd, P., Rulina, A. V., Haguet, V., Gidrol, X., & Balakirev, M. Y. (2017). High-content monitoring of drug effects in a 3D spheroid model. Frontiers in Oncology , 7, 293. doi: 10.3389/fonc.2017.00293.
- Neve, A., Kumar, K. S., Tripolitsioti, D., Grotzer, M. A., & Baumgartner, M. (2017). Investigation of brain tissue infiltration by medulloblastoma cells in an ex vivo model. Scientific Reports , 7, 1–12. doi: 10.1038/s41598-017-05573-w.
- Pui, C. H., Gajjar, A. J., Kane, J. R., Qaddoumi, I. A., & Pappo, A. S. (2011). Challenging issues in pediatric oncology. Nature Reviews Clinical Oncology , 8, 540–549. doi: 10.1038/nrclinonc.2011.95.
- Rauch, U. (2007). Brain matrix: Structure, turnover and necessity. Biochemical Society Transactions , 35, 656–660. doi: 10.1042/BST0350656.
- Rauch, U. (2004). Extracellular matrix components associated with remodeling processes in brain. Cellular and Molecular Life Sciences , 61, 2031–2045. doi: 10.1007/s00018-004-4043-x.
- Roper, S. J., Linke, F., Scotting, P. J., & Coyle, B. (2021). 3D spheroid models of paediatric SHH medulloblastoma mimic tumour biology, drug response and metastatic dissemination. Scientific Reports , 11, 1–17. doi: 10.1038/s41598-021-83809-6.
- Santhana Kumar, K., Neve, A., Guerreiro Stucklin, A. S., Kuzan-Fischer, C. M., Rushing, E. J., Taylor, M. D., … Baumgartner, M. (2018). TGF-β determines the pro-migratory potential of bFGF signaling in medulloblastoma. Cell Reports , 23, 3798–3812.e8. doi: 10.1016/j.celrep.2018.05.083.
- Schönholzer, M. T., Migliavacca, J., Alvarez, E., Santhana Kumar, K., Neve, A., Gries, A., … Baumgartner, M. (2020). Real-time sensing of MAPK signaling in medulloblastoma cells reveals cellular evasion mechanism counteracting dasatinib blockade of ERK activation during invasion. Neoplasia , 22, 470–483. doi: 10.1016/j.neo.2020.07.006.
- Sittampalam, G. S., Grossman, A., Brimacombe, K., Arkin, M., & Auld, D. (2004). HTS assay validation. In S. Markossian et al. (Eds.), Assay guidance manual (pp. 945–969). Eli Lilly & Company and the National Center for Advancing Translational Sciences.
- Vinci, M., Box, C., Zimmermann, M., & Eccles, S. A. (2013). Tumor spheroid-based migration assays for evaluation of therapeutic agents. Methods in Molecular Biology , 986, 253–266. doi: 10.1007/978-1-62703-311-4_16.
- Vinci, M., Gowan, S., Boxall, F., Patterson, L., Zimmermann, M., Court, W., … Eccles, S. A. (2012). Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biology , 10, 1–21. doi: 10.1186/1741-7007-10-29.
- Zanoni, M., Piccinini, F., Arienti, C., Zamagni, A., Santi, S., Polico, R., … Tesei, A. (2016). 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Scientific Reports , 6, 1–11. doi: 10.1038/srep19103.
- Zhang, S., Gong, Z., Oladimeji, P. O., Currier, D. G., Deng, Q., Liu, M., … Li, Y. (2019). A high-throughput screening identifies histone deacetylase inhibitors as therapeutic agents against medulloblastoma. Experimental Hematology and Oncology , 8, 1–10. doi: 10.1186/s40164-019-0153-x.
Citing Literature
Number of times cited according to CrossRef: 7
- Andrew S. Riching, Allyson Malloy, Emily M. Anderson, Jonathan Sheard, Piia Mikkonen, Anja Brabant Smith, Zaklina Strezoska, Josien Levenga, A Facile, Transfection‐Free Approach to siRNA Delivery in In Vitro 3D Spheroid Models, Current Protocols, 10.1002/cpz1.1121, 4 , 9, (2024).
- Xuechun Wang, Gautam Chauhan, Alison R. L. Tacderas, Aaron Muth, Vivek Gupta, Surface-Modified Inhaled Microparticle-Encapsulated Celastrol for Enhanced Efficacy in Malignant Pleural Mesothelioma, International Journal of Molecular Sciences, 10.3390/ijms24065204, 24 , 6, (5204), (2023).
- Elena M. Tosca, Davide Ronchi, Daniele Facciolo, Paolo Magni, Replacement, Reduction, and Refinement of Animal Experiments in Anticancer Drug Development: The Contribution of 3D In Vitro Cancer Models in the Drug Efficacy Assessment, Biomedicines, 10.3390/biomedicines11041058, 11 , 4, (1058), (2023).
- Florina-Nicoleta Grigore, Serena Johanna Yang, Clark C. Chen, Tomoyuki Koga, Pioneering models of pediatric brain tumors, Neoplasia, 10.1016/j.neo.2022.100859, 36 , (100859), (2023).
- Tanvir Ahmed, Biomaterial-based in vitro 3D modeling of glioblastoma multiforme, Cancer Pathogenesis and Therapy, 10.1016/j.cpt.2023.01.002, (2023).
- Huifen Ding, Kalyani Jambunathan, Guochun Jiang, David M. Margolis, Iris Leng, Michael Ihnat, Jian-Xing Ma, Jon Mirsalis, Yuanyuan Zhang, 3D Spheroids of Human Primary Urine-Derived Stem Cells in the Assessment of Drug-Induced Mitochondrial Toxicity, Pharmaceutics, 10.3390/pharmaceutics14051042, 14 , 5, (1042), (2022).
- Jafrin Jobayer Sonju, Achyut Dahal, Vivitri Dewi Prasasty, Prajesh Shrestha, Yong‐Yu Liu, Seetharama D. Jois, Assessment of Antitumor and Antiproliferative Efficacy and Detection of Protein‐Protein Interactions in Cancer Cells from 3D Tumor Spheroids, Current Protocols, 10.1002/cpz1.569, 2 , 10, (2022).