EPMotion - Normalization and Randomization
Oksana Polesskaya, Abraham Palmer, Khai-Minh H Nguyen, Katarina A Cohen
Abstract
This protocol normalizes and randomizes samples on the EPmotion for the Twist 96-Plex (Formerly riptide) library prep protocol. The procedure outlines how to complete normalization calculations and the randomization process for up to 96 samples, as well as how to set up the EPMotion robot to complete normalization on a 96-well plate.
Before start
This protocol assumes you have set up the EPmotion already. All required equipment and reagents are listed in the Materials section.
Steps
Excel Calculations
Enter sample information and values into the "Riptide Library Template" Excel worksheet:
Riptide Library Template for Low_Concentration Samples.xlsx
Riptide Library Template for High-Concentration Samples.xlsx
- Only use Low-Concentration Template when average concentration is lower than 90ng/ul.
- Enter all information under the "Sample_Information" tab (all information should be filled in from the Google Sheets Extraction Database you created in the "Sample Cutting/Processing Protocol."
- If missing a transponder ID, enter the sample's barcode ID.
- Note: This protocol is designed to input 100ng of DNA per sample.
Randomize
.csv files
The "Sample_Randomization" tab will be prepopulated with the values entered under "Sample_Information."
-
For samples with concentrations less than 35 ng/uL: Low-Concentration Samples -- set the sample volume to 4 uL and the water volume to 0 uL.
**High-Concentration Samples** -- set the sample volume to 30 uL and the water volume to 0 uL. -
"Sample (uL)" and "Water (uL)" cells will be highlighted in red if the volume is less than 1 uL. To rectify this, if the sample volume is below 1 uL, round up to 1 uL.
-
If the water volume is below 1 uL, adjust the sample volume so that the final water volume is either 0 uL or 1 uL.
Randomize the samples in the Sample_Randomization Tab.
- Drag and fill the "Rand" column to replace the random numbers.
- Select columns B-K
- Select "Sort & Filter" and "Custom Sort" the samples by "Rand"
Save and Rename the Library file.
- Our lab names the file as follows: Riptide## Library (DNA plate Code)
- EX . Riptide05 Library (John03)
Open the previously created Extraction Database on google sheets and enter the Riptide## you assigned to this library into column Q of the Extraction Database.
Create water and sample .csv file for the EPMotion robot using the following template:ep_normalization template.csv
For water , enter the following values in their respective columns:
-
Rack: 1
-
Source: 1
-
Rack: 1
-
Destination: A1-A12, B1-B12, etc.
-
Volume: Copy these values from Excel randomization, rounded to two decimal points - If the water volume for a well is 0 uL, delete the entire corresponding row.
-
Tool: '1' if volume is under 50 uL, '2' if it is over 50 uL
For samples , enter the following values in their respective columns:
- Rack: 1
- Source: Randomized list of the gDNA plate's well IDs from Excel
- Rack: 1
- Destination: A1-A12, B1-B12, etc.
- Volume: Copy these values from Excel randomization, rounded to two decimal points
- Tool: 1
Setting up the EPMotion Robot
Open epBlue Application.
In the epBlue Studio, open Application Editor.
Import the epBlue normalization protocol.
- High-concentration samples: Sample_Normalization.export7
- Low-concentration samples: Low_Concentration_Sample_Normalization.export7
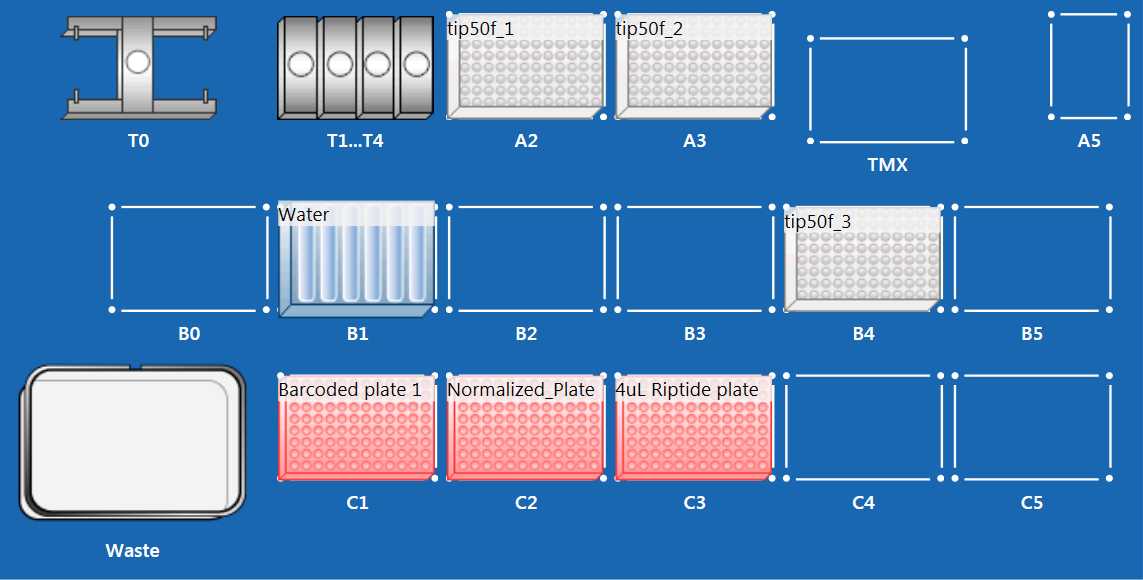
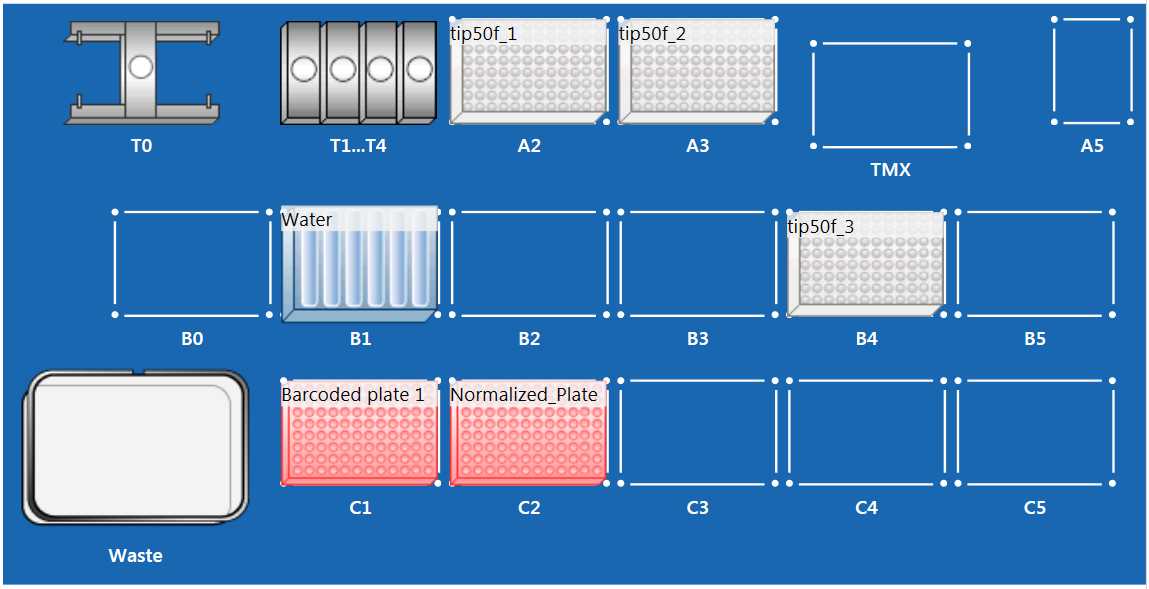
Open the imported protocol, and set up the EPMotion following the above worktable figure.
- Place 50 uL EPMotion tip racks in position A2, A3, and B4 without tip rack lid.
- Place EPMotion Reagent Reservoir Rack in position B1
- Place Eppendorf Thermoadapter PCR 96 in position C1 and C2
- Ensure tip waste basket is empty and liquid waste reservoir is empty
Label EPMotion 30 mL reservoirs.
- Label one reservoir "Water"
- Label one reservoir "Empty"
Place reservoir labelled "Empty" in position 7 of the EPMotion Reagent Reservoir Rack with its lid removed.
Add 30mL of
- Place "Water" in position 1 of the EPMotion Reagent Reservoir Rack with its lid removed.
Label a new ThermoScientific Thermo-fast 96 Detection Plate with Black Lettering (Catalog No. AB-1400-L) with the number of the Riptide library, user's initials, date of normalization, and sample volume.
_Ex. "Riptide 42 Normalized Plate K.C. 4/3/2021 30 uL"_
- Place labelled plate on the thermoadapter in position C2.
- Ensure that the plate is placed in the correct orientation. Letters and numbers should be right side up on the plate.
NOTE: For low-concentration plates, the samples are normalized to 4 uL. Because of this, only the "4-uL Plate" is required and is placed in position C2 (not a separate "Normalized Plate"). Skip Step 12, and proceed to Step 13.
Label a new ThermoScientific Thermo-fast 96 Detection Plate with Black Lettering (Catalog No. AB-1400-L) with the number of the Riptide library, user's initials, date of normalization, and sample volume.
_Ex. "Riptide 42 4-uL Plate K.C. 4/3/2021 4 uL"_
- Place labelled plate in position C3.
- Ensure that the plate is placed in the correct orientation. Letters and numbers should be right side up on the plate.
After thawing the gDNA plate, ensure the plate is properly sealed with a PCR adhesive seal. Briefly vortex and spin down the plate.
Remove seal from gDNA plate, and place it on the thermoadapter in position C1.
- NOTE: gDNA plate must be in MicroAmp™ Optical 96-Well Reaction Plate with Barcode Catalog No. 4306737
- The imported protocol is programmed for these specific plates.
Close EPMotion hood when all tips, plates, and reagents are in position.
Running the Normalization Protocol
Press the play button.
Select your EPMotion ID, and press "Next."
Select the "Input Volumes Manually" option under "Volume Settings," and press "Next."
Set volume for water at 30000 uL (30 mL), and press "Next."
Press "Run."
EQUIPMENT CHECK: The EPMotion report will show a warning that there is nothing located in space C3. Press "Ignore" to allow the protocol to continue
USER INTERVENTION: For high-concentration plates, after normalizing the first plate, the protocol will pause. Seal, shake, and spin down the normalized plate, and return it to C2. Remove the barcoded gDNA plate, and store it in a -20°C freezer. Move the Eppendorf Thermoadapter PCR 96 from position C1 to C3, and place the 4 uL plate in C3. Continue the protocol.
When the protocol ends, seal the plates. Store normalized plates and original gDNA plates in cardboard box.
- Box Label EX. Riptide 140/141, MM/DD/YYYY, initials, extracted gDNA
- 50mL falcon tube storage boxes with the dividers taken out can be used as storage. This will fit more than 4 plates. 2 libraries worth of plates can be stored in a single box
Store at plates -20°C
Enter the name of the storage box of the gDNA plates into the google sheets Extraction Database in column S.
Store 4uL plate at -20°C or continue with Twist 96-Plex (Riptide) protocol.