Downregulation of Hypoxia Inducible Factor-1α in Primary Human Natural Killer Cells Using Small Interfering RNA Delivery with ExPERT ATx by MaxCyte
Sonia Y. Velásquez, Sonia Y. Velásquez, Gizem E. Baslar, Gizem E. Baslar, Jutta Schulte, Jutta Schulte, Tanja Fuderer, Tanja Fuderer, Holger A. Lindner, Holger A. Lindner, Anna Coulibaly, Anna Coulibaly
Abstract
Natural killer (NK) cells are innate cytokine-producing and cytolytic effector lymphocytes. Their function is responsive to environmental factors, e.g., hypoxia, a frequent feature of inflamed tissues. Such responses require that the NK cells up-regulate HIF-1α (hypoxia inducible factor-1α), the major mediator of cellular responses to hypoxia that affects cell survival as well as immune responses. Thus, a major approach to the study of NK cell effector function under hypoxic conditions involves the ability to regulate HIF-1α levels in primary human NK cells. One difficulty with this approach, however, is that NK cells are difficult-to-transfect cells and common transfection methods, including electroporation or lipofection, suffer from variable transfection efficiency and cell viability. Moreover, the detection of HIF-1α is technically challenging because of the rapid degradation of the protein under normoxic conditions. Here, using the commercially available ExPERT ATx by MaxCyte, we report a workflow for the reliable delivery of small interfering RNA (siRNA) for targeting HIF-1α expression in primary human NK cells. We further provide a protocol for the detection of HIF-1α by immunoblot analysis demonstrating its efficient downregulation by siRNA. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Isolation of natural killer cells from human peripheral blood mononuclear cells
Basic Protocol 2 : Delivery of non-coding small interfering RNA and HIF-1α targeting siRNA into natural killer cells using ExPERT ATx
Basic Protocol 3 : Assessing the downregulation of HIF-1α protein using immunoblot analysis
Support Protocol 1 : Exemplary assessment of transfection efficiency using fluorescently labeled non-targeting siRNA
Support Protocol 2 : Exemplary assessment of NK cell viability 20 hr post-transfection
Support Protocol 3 : Exemplary assessment of HIF-1α knockdown using immunoblot analysis
INTRODUCTION
Natural killer (NK) cells are cytokine-producing and cytolytic effector lymphocytes of the innate immune system. As such, they are a component of the first line of host defense against microbial infection and cancer. Sensing their environment, NK cells need to respond rapidly to cellular alterations caused by transformation and infection, often in areas with low oxygen tension. Responses to these conditions are largely regulated by hypoxia inducible factor (HIF)-1, a key transcription factor involved in the maintenance of oxygen homeostasis in mammalian cells and tissues. HIF-1 is a heterodimer consisting of a constitutively expressed beta subunit and an oxygen-regulated alpha subunit (HIF-1α) that is rapidly degraded under normoxic conditions via ubiquitination and subsequent proteasomal degradation (Semenza, 2011; Wang et al., 1995).
The role of HIF-1α for NK cell effector function remains poorly understood, in part because NK cells are difficult to transfect. Here, we overcome this problem for human NK cells isolated from peripheral blood mononuclear cells (PBMCs) with a technologically improved electroporation system. We report an efficient protocol for the siRNA-mediated downregulation of cytokine-induced accumulation of HIF-1α protein using a MaxCyte ExPERT ATx instrument to deliver commercially available siRNA targeting HIF-1α and a non-coding control into NK cells. Flow cytometry protocols for determination of transfection efficiencies and cell viabilities are also provided. Finally, a protocol for the detection of cytokine-induced HIF-1α accumulation under hypoxia by western blot analysis and for the confirmation of the reduced HIF-1α protein levels after siRNA transfection is described. Taken together, this protocol contains an electroporation procedure for the otherwise very challenging genetic manipulation of primary human NK cells as an alternative to chemical inhibition of HIF-1α that may have undesirable side effects.
NOTE : Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol 1: ISOLATION OF NATURAL KILLER CELLS FROM HUMAN PERIPHERAL BLOOD MONONUCLEAR CELLS
This protocol provides instructions for the isolation of NK cells from human PBMCs by negative selection, depleting non-NK cell populations through magnetic labeling of antigens not expressed by NK cells. Please note that several methods are available for the isolation of PBMCs. In this protocol for NK cell enrichment, we used freshly isolated PBMCs from leukocyte concentrates (“buffy coats”) obtained through density gradient centrifugation.
Materials
-
Human PBMCs obtained by density gradient centrifugation
-
Trypan blue solution (Thermo Fisher Scientific, cat. no. T10282)
-
MACS buffer (see recipe)
-
NK cell isolation kit, human (Miltenyi Biotec, cat. no. 130-092-657) containing:
- NK cell biotin-antibody cocktail
- NK cell MicroBead cocktail
-
50-ml conical tubes (Neolab, cat. no. C-8216)
-
Invitrogen Countess II automatic cell counter (Thermo Fisher Scientific, cat. no. AMQAX1000)
-
Countess cell counting chamber slides (Thermo Fisher Scientific, cat. no. C10283)
-
Centrifuge, 4°C
-
Refrigerator, 4°C
-
QuadroMACS separator (Miltenyi Biotec, cat. no. 130-090-976)
-
MACS MultiStand (Miltenyi Biotec, cat. no. 130-108-933)
-
LS columns (Miltenyi Biotec, cat. no. 130-042-401)
-
Chill 15 rack (Miltenyi Biotec, cat. no. 130-092-952)
-
15-ml conical tubes (Neolab, cat. no. C-8215)
-
Flow cytometer
NOTE : This protocol assumes familiarity with standard cell culture methods and the availability of a biosafety cabinet. The procedure of NK cell enrichment using the NK cell isolation kit is in accordance with the manufacturer's protocol for manual magnetic labeling.
NOTE : Keep NK cells and MACS buffer cold (4°C).
Preparation of PBMCs
1.Obtain PBMCs from leukocyte concentrates (“buffy coats”) and transfer them to 50-ml conical tubes.
2.Determine the number of viable PBMCs by trypan blue dye exclusion using a cell counter.
3.Centrifuge 10 min at 300 × g , 4°C.
4.Completely remove supernatant and resuspend cell pellet in 40 µl cold MACS buffer per 107 total cells.
Label PBMCs
5.Add 10 µl NK cell biotin-antibody cocktail per 10⁷ total cells and mix well.
6.Incubate 5 min in the refrigerator, 4°C.
7.Without centrifugation, add 30 µl MACS buffer per 10⁷ total cells.
8.Add 20 µl NK cell MicroBead cocktail per 10⁷ total cells.
9.Incubate 10 min at 4°C.
10.During incubation, prepare for the magnetic depletion of non-NK cell populations:
-
Place a QuadroMACS separator on the MACS MultiStand.
-
Insert LS column(s) into the QuadroMACS separator.
The maximal capacity of one LS MACS column is 1 × 108cells. For cell numbers >1 × 108, use two or more LS MACS columns. Resuspend up to 108total cells in 500 µl buffer. A minimum of 500 µl is required for magnetic separation. For higher cell numbers, scale up buffer volume accordingly.
-
Place a chill rack under the QuadroMACS separator with a MACS MultiStand and a 15-ml conical tube.
-
Rinse LS column with 3 ml cold MACS buffer. Retain the flow-through in the 15 ml tube.
Always wait until the column reservoir is empty before proceeding to the next step.
Deplete non-NK cells
11.Transfer magnetically labeled cell suspension to the column (from step 9).
12.Collect flow-through containing non-labeled NK cells.
13.Rinse column with 3 ml MACS buffer.
14.After rinsing is complete, remove 15-ml tube, and determine the cell number of obtained NK cells by trypan blue dye exclusion in a cell counter.
15.Remove 100 µl to determine the purity of NK cells by flow cytometry.
16.Proceed with Basic Protocol 2.
Basic Protocol 2: DELIVERY OF NON-CODING SMALL INTERFERING RNA AND HIF-1α TARGETING siRNA INTO NATURAL KILLER CELLS USING ExPERT ATx
This protocol provides information for the successful delivery of HIF-1α siRNA into primary human NK cells using the ExPERT ATx electroporation instrument. We describe how to prepare freshly isolated NK cells, obtained using Basic Protocol 1, for the electroporation procedure to avoid compromised cell viability and to ensure high knockdown efficiency.
Materials
-
HIF-1α silencer select siRNA with nuclease-free H2O (Thermo Fisher Scientific, cat. no. 4390826, assay ID: s6539)
-
Silencer Select Negative Control No. 1 siRNA with nuclease-free H2O (Thermo Fisher Scientific, cat. no. 4390844)
-
Enriched NK cells (see Basic Protocol 1)
-
Electroporation (EP) buffer (MaxCyte, cat. no. EPB-1)
-
Trypan blue solution (Thermo Fisher Scientific, cat. no. T10282)
-
RNase inhibitor, 20 U/µl (Thermo Fisher Scientific, cat. no. N8080119)
-
Culture medium (see recipe)
-
Ice in an ice container (e.g., Neolab, cat. no. 2-4994)
-
Centrifuge
-
Invitrogen Countess II automatic cell counter (Thermo Fisher Scientific, cat. no. AMQAX1000)
-
Countess cell counting chamber slides (Thermo Fisher Scientific, cat. no. C10283)
-
15-ml conical tubes (Neolab, cat. no. C-8215)
-
6-well plates (Sarstedt, cat. no. 83.3920500)
-
Tissue culture incubator, 37°C, 5% CO2, 95% humidity
-
ExPERT ATx instrument (MaxCyte, cat. no. E-ATx)
-
Processing assemblies, R-50 × 8 or R-50 × 3 (MaxCyte, cat. nos. ER050U8-03 and ER050U3-10, respectively)
-
0.5- or 1.5-ml reaction tubes, RNase-free (Eppendorf, cat. nos. 0030-121.023 and 0030-120.086, respectively), pre-cooled
NOTE : This protocol assumes familiarity with working in an RNase-free environment. As RNA oligonucleotides are susceptible to degradation by exogenous ribonucleases introduced during handling, it is essential that the users wear gloves, work on ice, use pre-cooled (ice-cold) tubes and execute the protocol without any delays. Make sure that all reagents and tubes are RNase-free. We recommend using barrier pipette tips.
NOTE : The electroporation procedure using the MaxCyte ExPERT ATx instrument is conducted in sterile, single use processing assemblies (PAs). The R-50 × 3 PA is a 3-well cuvette with 50 µl final working volume per well and the R-50 × 8 PA is an 8-well cuvette with 50 µl final working volume per well.
Preparation of siRNA
1.Thaw the nuclease-free water on ice (provided with siRNA).
2.Centrifuge the siRNA tubes 5 s 2000 × g , room temperature, to ensure that the dried siRNA is at the bottom of the tube.
3.Resuspend the siRNA in 400 µl nuclease-free water for a final concentration of 100 μM.
4.Aliquot siRNAs to limit the number of freeze-thaw cycles and store at or below –20°C.
5.Leave one aliquot on ice until NK cells are ready for the electroporation procedure.
Preparation of NK cells and a cell master mix for electroporation
After the isolation of NK cells from PBMCs, NK cells are resuspended in MACS buffer (see Basic Protocol 1, step 14).
6.Centrifuge 10 min at 300 × g , room temperature.
7.Completely remove supernatant and resuspend the cell pellet in 10 ml EP buffer.
8.Determine the number of NK cells by trypan blue dye exclusion in a cell counter.
9.Transfer cell suspension to a new 15-ml tube.
10.Centrifuge 10 min at 300 × g , room temperature.
11.During centrifugation, prepare working EP buffer for the electroporation procedure by adding RNase inhibitor to the ice-cold EP buffer:
-
For each sample to be transfected, a volume of 50 µl working EP buffer is required. Calculate the volume for all samples, plus one additional sample as an excess.
-
Add RNase inhibitor (1 U/µl).
12.During centrifugation, also prepare a 6-well plate with 4 ml culture medium per well according to the number of transfected samples and an additional untransfected control and place the 6-well plate into the tissue culture incubator.
13.During the last minutes of centrifugation, prepare the ExPERT ATx instrument for the electroporation procedure:
-
Log into the instrument, ideally before processing cells.
-
On the display, select “New Run”.
-
Under “Select Protocol” select “NK-4”.
-
Under “Select Processing Assembly” select the appropriate PA, “R-50 × 3” for three well (sample) transfection or “R-50 × 8” for eight well transfection.
-
Under “Select Assemblies” select the number of PAs to be electroporated.
-
Select “Next”.
You can add further information including a sample ID, cell type (NK), buffer lot, PA lot, volume of the sample size (50 µl), and notes.
-
Select “Next” and proceed.
14.Remove the 15-ml tube from the centrifuge containing NK cells and completely aspirate the supernatant.
15.Resuspend NK cell pellet in half of the calculated volume of working EP buffer, which was determined in step 11a.
16.Determine the volume of the cell suspension and fill up with working EP buffer to the calculated volume.
17.Prepare the PAs by appropriately labeling the wells. Prepare 0.5-ml tubes on ice and pipet the appropriate volume of working EP buffer (for untransfected cells), control siRNA, and HIF-1α siRNA.
18.Pipet 50 µl NK cell suspension (from step 16) in each of the appropriately labeled tubes with siRNA or working EP buffer (for the untransfected cells). Carefully pipet up and down to mix NK cell suspension with siRNA and transfer 50 µl of the mix to the appropriately labeled PA well.
19.Repeat for all samples to be transfected.
20.Close the lid of the PA.
21.Place the PA into the ExPERT ATx. Press “OK” and then “Start Processing”.
22.Directly transfer the PA and the tube for the “untransfected control” to the tissue culture incubator.
23.Incubate for 20 min at 37°C.
24.After 20 min, remove the PA, the untransfected control, and the prepared 6-well plate (with pre-warmed medium) from the incubator.
25.Carefully aspirate the electroporated NK cells from the PA and transfer 50 µl cell suspension into the appropriate well of the 6-well plate with the pre-warmed medium. Take 50 µl medium from the same well of the 6-well plate to “wash out” any possible remaining NK cells in the well of the PA.
26.Repeat for all wells within the PA.
27.Proceed with Basic Protocol 3.
Basic Protocol 3: ASSESSING THE DOWNREGULATION OF HIF-1α PROTEIN USING IMMUNOBLOT ANALYSIS
This protocol describes the procedure for the downregulation of the cytokine-induced accumulation of HIF-1α protein in NK cells under hypoxia. We have previously shown that the stimulation of NK cells with cytokines including IL-15, IL-12, and IL-18 (Coulibaly et al., 2021) induces the expression of HIF-1α protein in NK cells after 6 hr of cytokine-stimulation. In the following protocol, we use cytokine-stimulation of NK cells directly after the electroporation procedure (Basic Protocol 2) to demonstrate the efficiency of HIF-1α accumulation as well as the efficiency of the HIF-1α downregulation 20 hr post-transfection.
Materials
-
Electroporated NK cells (see Basic Protocol 2)
-
IL-15 (PeproTech, cat. no. 200-15)
-
IL-12 (PeproTech, cat. no. 200-12)
-
IL-18 (PeproTech, cat. no. B001-5)
-
Lysis buffer (see recipe)
-
4× NuPAGE LDS sample buffer (Thermo Fisher Scientific, cat. no. NP0007)
-
10× NuPAGE sample reducing agent (Thermo Fisher Scientific, cat. no. NP0009)
-
20× NuPAGE MOPS SDS running buffer (Thermo Fisher Scientific, cat. no. NP0001)
-
H2O, deionized
-
NuPAGE 10% Bis-Tris gels (e.g., Thermo Fisher Scientific, cat. no. NP0301BOX or NP0302BOX)
-
PageRuler Plus prestained protein ladder (Thermo Fisher Scientific, cat. no. 26619)
-
BLOTTO, non-fat dry milk (Santa Cruz, cat. no. sc-2324)
-
20× phosphate-buffered saline (PBS)-Tween (Thermo Fisher Scientific, cat. no. 28352)
-
20× Tris-buffered saline (TBS) (Thermo Fisher Scientific, cat. no. J60877.K2)
-
Bovine serum albumin (BSA) (e.g., Sigma-Aldrich, cat. no. A9418)
-
Recombinant anti-HIF-1α antibody [EPR16897], rabbit (Abcam, cat. no. ab179483)
-
β-Actin antibody 8H10D10, mouse (Cell Signaling, cat. no. 3700)
-
Secondary antibodies:
- Goat anti-rabbit IgG H&L, HRP (Abcam, cat. no. ab205718)
- Goat anti-mouse IgG H&L, HRP (Abcam, cat. no. ab205719)
-
1× PBS, pH 7.4 (Gibco, cat. no. 14190-094)
-
SuperSignal West Femto ultimate sensitivity ECL substrate (Thermo Fisher Scientific, cat. no. 34094)
-
Hypoxia incubator, 1% O2, 37°C, 5% CO2 (New Brunswick, cat. no. Galaxy 48R)
-
Ice in an ice container (e.g., Neolab, 2-4994)
-
2- and 1.5-ml tubes (Eppendorf, cat. nos. 0030-121.094 and 0030-120.086, respectively), pre-cooled
-
Centrifuge, pre-cooled to 4°C
-
ThermoMixer C (Eppendorf, cat. no. 5382000015)
-
XCell SureLock mini-cell gel tank (Thermo Fisher Scientific, cat. no. EI0001)
-
iBlot gel transfer stacks nitrocellulose (Thermo Fisher Scientific, cat. nos. IB301001 or IB301002), with blotting roller
-
iBlot dry blotting system (Thermo Fisher Scientific, cat. no. IB1001), or another blotting device
-
Western incubation trays (e.g., Carl Roth, cat. no. 8862.1)
-
Western blot and chemiluminescence imaging system (e.g., FUSION FX, Vilber)
CAUTION : Always wear protective gloves and safety glasses when handling polyacrylamide gels.
CAUTION : It is critical to wear gloves at all times during the entire blotting procedure. This prevents the contamination of gels and membranes. Do not touch the membrane with bare or gloved hands. This may interfere with further analysis. If you need to adjust the membrane, always use forceps.
NOTE : To avoid possible degradation of HIF-1α protein under normoxic conditions, execute the cell harvesting and lysis steps without any delays and perform all steps of harvesting and lysis on ice.
Cytokine stimulation and incubation under hypoxia to induce HIF-1α accumulation in NK cells
After the electroporation and the period of recovery in the PA for 20 min in the tissue culture incubator, NK cells are in the medium of a 6-well plate (4 × 106 NK cells/well) (see Basic Protocol 2, step 26).
1.Stimulate NK cells with IL-15 (45 ng/ml), IL-12 (10 ng/ml), and IL-18 (50 ng/ml).
2.Place the 6-well plate into hypoxia incubator.
3.Incubate overnight (20 h).
Cell harvesting and lysis
4.Place the 6-well plate from the hypoxia incubator directly on ice.
5.Resuspend NK cells in each well before transferring to a pre-cooled, appropriately labeled, 2-ml tube.
6.Place the tubes into the pre-cooled centrifuge, 10 min at 3500 × g , 4°C.
7.During centrifugation, prepare working lysis buffer by freshly adding NP-40 and PMSF to lysis buffer (see Reagents and Solutions).
8.Transfer the tubes with the pelleted NK cells directly on ice and remove the supernatant.
9.Resuspend NK cell pellet in 50 µl working lysis buffer.
10.Incubate 20 min on ice.
11.Centrifuge 20 min at 8000 × g , 4°C, to remove cell debris.
12.Transfer the lysates (supernatants) into new pre-cooled and labeled tubes and leave on ice.
Gel electrophoresis using NuPAGE Bis-Tris mini gels
13.Prepare new tubes.
14.For each sample to be loaded on a gel, prepare loading buffer. To do so, calculate 5 µl of 4× LDS sample buffer and 2 µl of 10× sample reducing agent for each sample, and include an excess for two additional samples.
15.Pipet 5 µl loading buffer into new tubes.
16.Add 15 µl NK cell lysate to the tubes containing loading buffer.
17.Resuspend and spin down if needed to collect the suspension at the bottom of the tube.
18.Heat samples 10 min at 70°C while shaking (400 rpm) in a ThermoMixer. Do not boil samples.
19.Add 50 ml of 20× NuPAGE MOPS SDS running buffer to 950 ml of deionized water to prepare 1× MOPS buffer.
20.Prepare a precast polyacrylamide gel:
-
Unpack the gel, remove the comb, and rinse the gel under deionized water.
-
Remove the white tape near the bottom of the gel cassette.
-
Place the gel in the mini gel tank.
Use a buffer dam instead of the second gel when you need to run only one gel.
-
Rinse the gel wells using 1× MOPS Buffer.
21.Fill the chambers of the mini gel tank with 1× MOPS.
22.Load 15 µl sample and 3 µl protein ladder on the gel in the appropriate wells.
23.Run the gel at 150 V constant.
Western blotting using the iBlot gel transfer device
24.Remove the gel from the gel cassette.
25.Rinse the gel in deionized water.
26.Assemble the iBlot gel transfer device with the iBlot gel transfer stacks and the gel.
27.Perform the transfer using the program “P3” for 8 min.
28.Disassemble the iBlot gel transfer device directly after the transfer.
29.Cut the membrane at 65 kDa to get two pieces of the membrane, with the upper part to detect HIF-1α at 100 kDa and the lower part to detect β-Actin at 40 kDa.
30.Place the membrane slides into clean incubation trays for further procedures.
31.Incubate the membrane slides with 10 ml blocking solution [5% (w/v) BLOTTO dissolved in 1× PBS-Tween] for 1 hr at room temperature under constant shaking.
32.Remove BLOTTO, rinse once with 5 to 10 ml 1× PBS-Tween, depending on the box size you use for incubation.
33.Incubate with 10 ml primary antibody overnight at 4°C under constant shaking:
-
To detect HIF-1α, dilute the antibody 1:1000 in 1× TBS containing 5% (w/v) BSA.
-
To detect β-Actin, dilute the antibody 1:10,000 in 1× TBS containing 5% (w/v) BSA.
34.Remove the antibodies and wash with 1× PBS-Tween for at least 5 to 10 min. Repeat for a total of three washes.
35.Incubate with the secondary antibody:
-
For HIF-1α: α-rabbit diluted 1:50,000 in 5% (w/v) BLOTTO dissolved in 1× PBS-Tween.
-
For β-Actin: α-mouse diluted at 1:10,000 in 5% (w/v) BLOTTO dissolved in 1× PBS-Tween.
36.Discard the antibodies and wash with 1× PBS-Tween for 5 to 10 min. Repeat for a total of three washes.
37.Wash twice with 1× PBS for 5 to 10 min.
38.Proceed with developing using an ECL substrate and your western blot imaging system.
Support Protocol 1: EXEMPLARY ASSESSMENT OF TRANSFECTION EFFICIENCY USING FLUORESCENTLY LABELED NON-TARGETING siRNA
This protocol describes the procedure to determine the transfection efficiency of primary human NK cells 4 hr after transfection. To demonstrate the efficiency of siRNA uptake, PBMCs were transfected with non-targeting control siRNAs, including a fluorescently labeled (FITC) siRNA. 4 hr post-transfection, the percentage of CD56+CD3– cells positive for the FITC fluorochrome was determined using flow cytometry.
Materials
-
Silencer Select Negative Control No. 1 siRNA with nuclease-free H2O (Thermo Fisher Scientific, cat. no. 4390844)
-
BLOCK-iT fluorescent oligo (Thermo Fisher Scientific, cat. no. 13750062)
-
Human PBMCs obtained by density gradient centrifugation, directly after isolation
-
Trypan blue solution (Thermo Fisher Scientific, cat. no. T10282)
-
1× PBS, pH 7.4 (Gibco, cat. no. 14190-094)
-
Electroporation (EP) buffer (MaxCyte, cat. no. EPB-1)
-
RNase inhibitor 20 U/µl (Thermo Fisher Scientific, cat. no. N8080119)
-
Culture medium (see recipe)
-
Cell wash (BD biosciences, 349524)
-
PE-conjugated anti-human CD56, NCAM16.2 (BD Biosciences, 345812)
-
APC-conjugated anti-human CD3, HIT3a (BD Biosciences, 555342)
-
Ice in an ice container (e.g., Neolab, cat. no. 2-4994)
-
Invitrogen Countess II automatic cell counter (Thermo Fisher Scientific, cat. no. AMQAX1000)
-
Countess cell counting chamber slides (Thermo Fisher Scientific, cat. no. C10283)
-
15-ml conical tubes (Neolab, C-8215)
-
Centrifuge
-
6-well plates (Sarstedt, cat. no. 83.3920500)
-
Tissue culture incubator, 37°C, 5% CO2, 95% humidity
-
ExPERT ATx instrument (MaxCyte, cat. no. E-ATx)
-
Processing assemblies, R-50 × 3 (MaxCyte, cat. no. ER050U3-10)
-
0.5-ml reaction tubes, RNase-free (Eppendorf, cat. no. 0030-121.023), ice-cold
-
5-ml round bottom polystyrene test tube (Corning, 352008)
-
Flow cytometer
NOTE : Obtain PBMCs by your method of choice. We used density gradient centrifugation for the isolation of human PBMCs from leukocyte concentrates (“buffy coats”).
1.Thaw Silencer Select Negative Control No. 1 siRNA and BLOCK-iT fluorescent oligo on ice.
2.Determine the number of viable PBMCs by trypan blue dye exclusion using a cell counter.
3.Transfer the cell suspension in a new 15-ml tube.
4.Centrifuge 10 min at 300 × g , room temperature.
5.Add 5 ml of 1× PBS to wash the cell pellet.
6.Centrifuge 10 min at 300 × g , room temperature.
7.Remove supernatant completely and resuspend the cell pellet in 5 ml EP buffer.
8.Determine the cell number by trypan blue dye exclusion in a cell counter.
9.Centrifuge 10 min at 300 × g , at room temperature.
10.During centrifugation prepare working EP buffer for the electroporation procedure by adding RNase inhibitor to ice- cold EP buffer:
-
For each sample, a volume of 50 µl working EP buffer is required. Calculate the volume for all samples, plus one additional sample as an excess.
-
Add RNase inhibitor (1 U/µl).
11.During centrifugation, prepare a 6-well plate with 4 ml culture medium per well according to the number of samples to be transfected plus a sample of untransfected cells. Place the 6-well plate into the tissue culture incubator to pre-warm the culture medium.
12.During the last minutes of centrifugation, prepare the ExPERT ATx for the electroporation procedure:
-
Log into the instrument.
-
On the display, select “New Run”.
-
Under “Select Protocol” select “NK-4”.
-
Under “Select Processing Assembly” select “R-50 × 3” for a three well (sample) transfection.
-
Under “Select Assemblies” select the number of PAs to be electroporated.
-
Select “Next”.
You can add further information including a sample ID, cell type (PBMCs), buffer lot, PA lot, volume of the sample size (50 µl), and notes.
-
Select “Next” and proceed with the cells.
13.Remove the 15-ml tube from the centrifuge containing PBMCs and completely aspirate the supernatant.
14.Resuspend the cell pellet in half of the calculated volume of working EP buffer, which was determined in step 10a.
15.Determine the volume of the cell suspension and fill with working EP buffer to the calculated volume.
16.Prepare the PAs by appropriately labeling the wells. Also prepare and label 0.5-ml tubes on ice and pipet the appropriate volume of working EP buffer (for untransfected cells and electroporated cells without siRNA), non-labeled control siRNA, and FITC-siRNA.
17.Pipet 50 µl PBMC suspension (from step 15) in each of the labeled tubes with siRNA or working EP buffer (for untransfected cells and cells electroporated without siRNA). Carefully pipet up and down to mix PBMC suspension with siRNA and transfer 50 µl of the mix to the appropriately labeled well of the PA.
18.Repeat for all samples to be transfected.
19.Close the lid of the PA.
20.Place the PA into the ExPERT ATx. Press “OK” and then “Start Processing”.
21.Directly transfer the PA and the tube for the “untransfected control” to the tissue culture incubator.
22.Incubate for 20 min at 37°C.
23.After 20 min, remove the PA, the untransfected control, and the prepared 6-well plate (with pre-warmed medium) from the tissue culture incubator.
24.Carefully aspirate the electroporated cells from the PA and transfer the 50 µl electroporated cell suspension into the appropriate well of the 6-well plate with the pre-warmed medium. Take 50 µl medium from the same well of the plate to “wash out” possible remaining cells in the PA-well.
25.Repeat for all wells within the PA.
26.Incubate for 4 hr.
27.After the incubation, transfer the cells into round-bottom polystyrene tubes.
28.Centrifuge 10 min at 300 × g , room temperature.
29.Wash with 2 ml cell wash.
30.Centrifuge 5 min at 300 × g , room temperature.
31.Resuspend cell pellet in 100 µl cell wash.
32.Add staining solution containing fluorochrome-conjugated antibodies for NK cell evaluation to the electroporated PBMCs.
33.Incubate 15 min at room temperature in the dark.
34.Wash with 2 ml cell wash.
35.Centrifuge 5 min at 300 × g , room temperature.
36.Resuspend in 250 µl cell wash.
37.Analyze the samples by flow cytometry.

Support Protocol 2: EXEMPLARY ASSESSMENT OF NK CELL VIABILITY 20 HR POST-TRANSFECTION
This protocol describes the procedure to determine the viability of primary human NK cells electroporated with 5 µM HIF-1α siRNA at 20 hr post-transfection using Zombie Violet fixable viability dye staining followed by flow cytometric analysis.
In this example, human NK cells were isolated from PBMCs (Basic Protocol 1) and electroporated using the MaxCyte ExPERT ATx (Basic Protocol 2). After electroporation, NK cells were stimulated with a combination of IL-15 (45 ng/ml), IL-12 (10 ng/ml), and IL-18 (50 ng/ml) and incubated under hypoxia overnight (20 h). NK cell viability was determined in untransfected NK cells and NK cells transfected with non-targeting control siRNA and HIF-1α siRNA 20 hr post-transfection.
Materials
-
Zombie Violet fixable viability kit (BioLegend, cat. no. 423113)
-
1× PBS, pH 7.4 (Gibco, cat. no. 14190-094)
-
Electroporated NK cells, stimulated with IL-15, IL-12, and IL-18 under hypoxia for 20 hr
-
Stain buffer (see recipe)
-
Cell wash (BD biosciences, cat. no. 349524)
-
5-ml round-bottom polystyrene test tubes (Corning, 352008)
-
Centrifuge
-
Flow cytometer
NOTE : A dilution of 1:2000 of Zombie Violet in 1× PBS has proven optimal to stain 1–10 × 106 NK cells.
1.Prepare staining solution containing viability dye by diluting Zombie Violet 1:100 in 1× PBS.
2.Transfer 1 × 106 NK cells in round-bottom polystyrene tubes.
3.Centrifuge NK cells 10 min at 300 × g , room temperature.
4.Wash with 2 ml of 1× PBS.
5.Centrifuge 5 min at 300 × g , room temperature.
6.Resuspend NK cells in 100 µl of 1× PBS and add 5 µl Zombie Violet.
7.Incubate 15 min at room temperature in the dark.
8.Wash with 2 ml stain buffer.
9.Centrifuge 5 min at 300 × g , room temperature.
10.Resuspend in 250 µl cell wash.
11.Analyze by flow cytometry.

Support Protocol 3: EXEMPLARY ASSESSMENT OF HIF-1α KNOCKDOWN USING IMMUNOBLOT ANALYSIS
This protocol describes the procedure to determine the knockdown of HIF-1α in primary human NK cells electroporated with 5 µM HIF-1α siRNA. In this example, human NK cells were isolated from PBMCs (Basic Protocol 1) and electroporated using the MaxCyte ExPERT ATx (Basic Protocol 2). Directly after seeding of electroporated NK cells, the cells were stimulated with a combination of IL-15 (45 ng/ml), IL-12 (10 ng/ml), and IL-18 (50 ng/ml) and incubated under hypoxia. HIF-1α protein was assessed using immunoblot analysis 20 hr post-transfection (Basic Protocol 3). After the incubation period, 1 × 106 of transfected NK cells can be used for flow cytometric analysis to evaluate NK cell viability (Support Protocol 2) or for other (flow cytometric) analyses. We determined 1−2 × 106 NK cells as an optimal NK cell count for HIF-1α detection by western blotting. The remaining electroporated NK cells can be used for further analyses.
Additional Materials (also see Basic Protocol 3)
- Electroporated NK cells, stimulated with IL-15, IL-12, and IL-18 under hypoxia for 20 hr
NOTE : The following steps are described for 2 × 106 NK cells/sample.
1.Harvest 2 × 106 NK cells in 2-ml polystyrene tubes.
2.Centrifuge NK cells 10 min at 3500 × g , 4°C.
3.Discard the supernatant and lyse NK cells in 50 µl working lysis buffer (see Basic Protocol 3, steps 9 to 12).
4.Perform SDS-PAGE as described in Basic Protocol 3, steps 13 to 23.
5.Perform immunoblot analysis as described in Basic Protocol, steps 24 to 38.
REAGENTS AND SOLUTIONS
Culture medium
- RPMI 1640 medium (Sigma-Aldrich, cat. no. R8758)
- 10% (v/v) fetal bovine serum (FBS) (Thermo Fisher, cat. no. A31604-02)
- Store up to 4 weeks at 4°C
Lysis buffer
- 50 mM Tris·HCl, pH 7.5 (e.g., Thermo Fisher, cat. no. 15567027)
- 120 mM NaCl (e.g., Sigma-Aldrich, cat. no. s9888)
- 20 mM NaF (e.g., Sigma-Aldrich, cat. no. 201154)
- 1 mM EDTA (e.g., Sigma-Aldrich, cat. no. 798681)
- 6 mM EGTA (e.g., Sigma-Aldrich, cat. no. E3889)
- 15 mM Na4P2O7 (e.g., Sigma-Aldrich, cat. no. P8010)
- Store up to 1 year at room temperature
- Add fresh:
- 0.1% (v/v) NP-40 (e.g., Thermo Fisher, cat. no. 85124)
- 1 mM PMSF (e.g., Sigma-Aldrich, cat. no. P7626)
Prepare “working lysis buffer” by freshly adding NP-40 and PMSF. Use it directly after preparation for cell lysis. Always use freshly prepared and pre-cooled working lysis buffer and do not store for longer periods.
MACS buffer
- 1× PBS, pH 7.4 (Gibco, cat. no. 14190-094)
- 2 mM EDTA (Sigma-Aldrich, E7889)
- 0.5% (v/v) BSA (Miltenyi Biotec, cat. no. 130-091-376)
- Prepare fresh
Stain buffer
- 1× PBS, pH 7.4 (Gibco, cat. no. 14190-094)
- 0.2% (v/v) BSA (Miltenyi Biotec, cat. no. 130-091-376)
- Store up to 2 weeks at 4°C
COMMENTARY
Background Information
Tissue hypoxia occurs when cellular oxygen demand exceeds supply. Infiltrating immune cells, including NK cells, respond to hypoxia through cellular stabilization and, thereby, activation of HIF-1α. As a major regulator of hypoxic response, HIF-1α induces cellular transcriptional programs to support cell survival and function. At the same time, HIF-1α is a critical regulator of both innate and adaptive immunity (Rius et al., 2008; Taylor et al., 2016). Mediators of inflammation, e.g., cytokines and pathogen- and damage-associated molecular patterns, can also induce the activation of HIF-1α in immune cells (Devraj et al., 2017). We recently have shown that pro-inflammatory cytokines, including IL-15, IL-12, and IL-18, synergize with hypoxia and chemical hypoxia (induced, for instance, by dimethyloxalylglycine) to increase HIF-1α protein levels in human peripheral NK cells (Coulibaly et al., 2021). However, the role of hypoxia, and in particular of HIF-1α in the regulation of human NK cell function, needs further investigation. Silencing gene expression by transfer of siRNA targeting HIF-1α mRNA represents a powerful tool to study the role of HIF-1α through loss of function. However, the transfection of NK cells is known to be technically challenging. Transfection protocols involving electroporation, viral transduction, and lipofection result in variable transfection efficiencies and compromised viability of NK cells (Carlsten & Childs, 2015). Here, we describe how to successfully downregulate cytokine-induced stabilization of HIF-1α protein in human NK cells under hypoxia using the ExPERT ATx instrument by MaxCyte. This protocol will facilitate investigations into HIF-1α´s importance for NK cell effector functions. Moreover, the same procedure can be optimized for the transfection of other siRNAs or mRNAs to study other genes of interest in NK cells.
Critical Parameters
To allow for optimal recovery of transfected primary human NK cells, before their transfer into culture plates it is critical to follow the described resting period of 20 min in the processing assembly directly after the electroporation procedure. After 20 min pre-warmed medium should be used for NK cell seeding to ensure high viability of NK cells after the electroporation procedure.
As a positive control, we recommend inducing the accumulation of HIF-1α protein in NK cells under hypoxia (Coulibaly et al., 2021) by stimulating the cells with a combination of IL-15, IL-12, and IL-18 directly after seeding the electroporated and recovered cells.
As HIF-1α protein is rapidly degraded under normoxic conditions, it is critical to execute the protocol without any delays as soon as the cells are removed from hypoxia. Perform all steps of cell harvesting and lysis on ice using pre-cooled centrifuges, tubes, and lysis buffer.
Troubleshooting
Maintaining the viability of NK cells after electroporation is essential for the successful assessment of HIF-1α knockdown. We therefore have emphasized a resting period before the seeding of electroporated NK cells. To assess whether cell viability changes as an on-target effect of the HIF-1α siRNA, we recommend including a sample with non-targeting control siRNA. We have provided detailed instructions to obtain NK cells with a viability >90% 20 hr post-transfection. For this, we titrated the concentrations of siRNA as well as the NK cells.
Resting NK cells cultured under normoxia do not show detectable levels of HIF-1α by immunoblotting, while small amounts of HIF-1α are readily detected in resting NK cells cultured under hypoxia for 4 hr. Cytokine (IL-15, IL-12, and IL-18) stimulation during the hypoxic culture clearly augments HIF-1α levels in NK cells, which are then maintained for up to 27 hr (Coulibaly et al., 2019; Coulibaly et al., 2021). For a positive control, we therefore recommend stimulating electroporated NK cells with cytokines directly after recovery and seeding of cells. To assess transfection efficiency, we recommend transfection with a fluorescently labeled non-targeting siRNA. We have provided a protocol to assess the efficiency of siRNA uptake using FITC-conjugated siRNA in electroporated PBMCs, gated on NK cells (CD56+CD3–) 4 hr post-transfection. The results reproducibly showed transfection efficiencies >90%.
HIF-1α is a highly degradable protein under normoxic conditions due to its oxygen dependent ubiquitination (Huang et al., 1996). To obtain reproducible HIF-1α signals using immunoblot analysis and to avoid HIF-1α degradation while harvesting cytokine-stimulated NK cells pre-cultured under hypoxia, place the culture plate directly on ice after completed hypoxic incubation. Also, make sure to use ice-cold tubes for cell harvesting as well as pre-cooled lysis buffer and centrifuges. We have provided a protocol to ensure the detection of a strong signal for HIF-1α protein in cytokine stimulated NK cells under hypoxia, which is used as a positive control for HIF-1α accumulation.
Understanding Results
Figure 1 displays the flow cytometric assessment of transfection efficiency in PBMCs electroporated with 5 µM FITC-siRNA 4 hr post-transfection (Support Protocol 1). Three control samples were included, a sample with cells that were not electroporated (untransfected), a sample of cells electroporated but without siRNA, and a sample electroporated with 5 µM non-targeting control siRNA. Figure 1 shows one representative example of flow cytometric evaluation of FITC after gating on NK cells (CD56+CD3–). The analysis confirmed an efficient uptake of siRNA into NK cells transfected with FITC-siRNA, reaching 91.4%.
Figure 2 demonstrates the flow cytometric assessment of NK cell viability 20 hr post-transfection using Zombie Violet fixable viability dye (Support Protocol 2). To confirm that the knockdown of HIF-1α did not result in cell death, the viability was determined in NK cells transfected with 5 µM non-targeting siRNA (control siRNA) and 5 µM siRNA targeting HIF-1α (HIF-1α siRNA). Untransfected cells were used as a control. No differences in viability dye staining were visible between cells transfected with non-targeting siRNA and HIF-1α siRNA. NK cell viability was >90% 20 hr post-transfection.
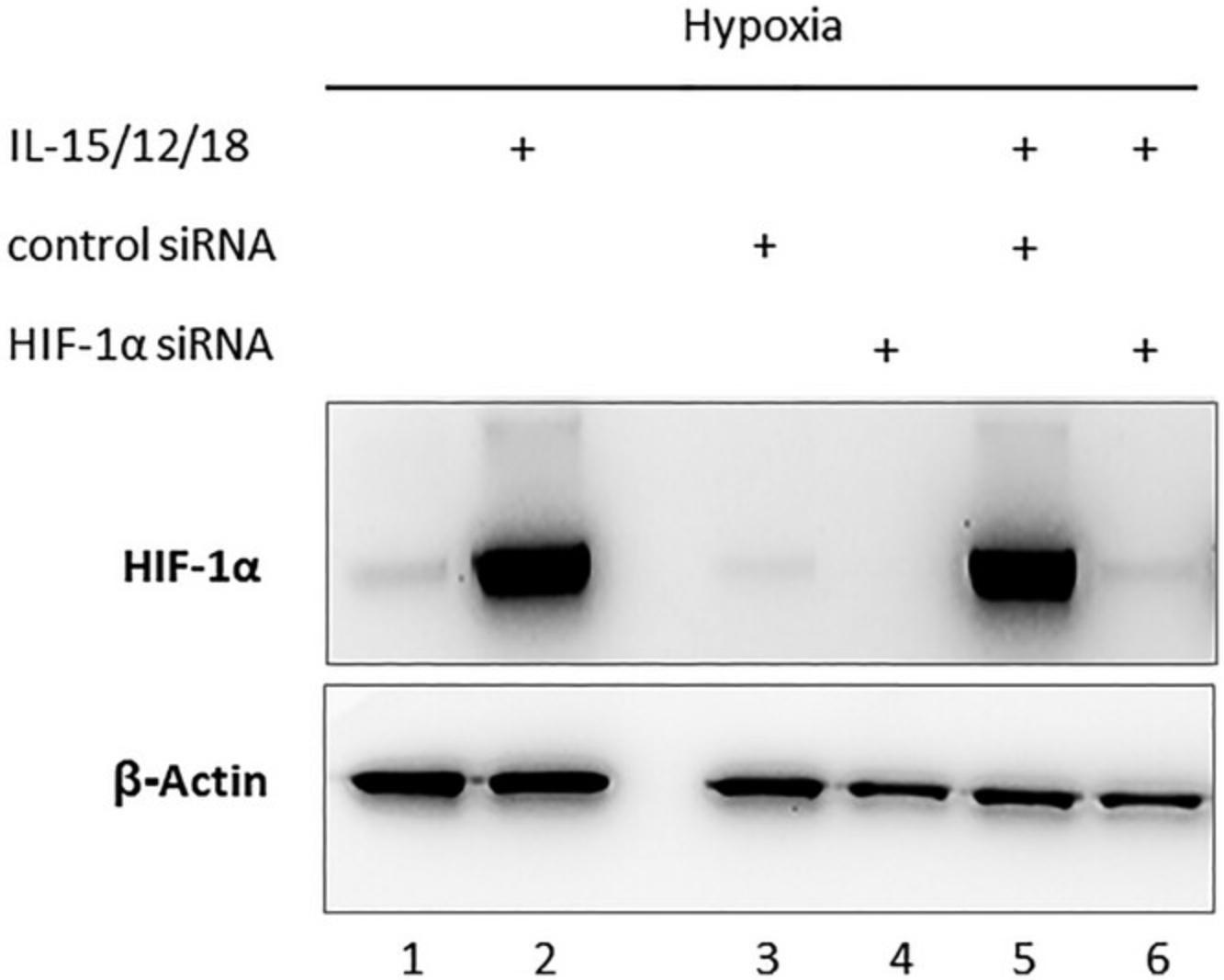
Figure 3 provides an example for the detection of HIF-1α protein using immunoblot analysis (Basic Protocol 3). β-Actin was used as a loading control. To further augment the expression of HIF-1α protein, we stimulated NK cells with a combination of IL-15, IL- 12, and IL-18 and cultured the cells for 20 hr under hypoxia. Untransfected cells that were either left unstimulated (lane 1) or cytokine-stimulated (lane 2) were used as positive controls for HIF-1α protein accumulation. Transfected cells included unstimulated NK cells (lanes 3 and 4) transfected with either non-coding siRNA (lane 3) or HIF-1α siRNA (lane 4), as well as cytokine-stimulated NK cells (lanes 5 and 6) transfected with control siRNA (lane 5) or HIF-1α siRNA (lane 6). These results confirm induction of HIF-1α protein expression by cytokines under hypoxia, which was not impaired by electroporation (lane 5). The transfection of siRNA targeting HIF-1α, however, efficiently blocked the accumulation of HIF-1α protein (lane 6), demonstrating high knockdown efficiency.
Time Considerations
The enrichment of NK cells from human PBMCs (Basic Protocol 1) takes ∼1 hr. The duration of the workflow of siRNA delivery including steps of NK cell preparation for electroporation, siRNA dilution and electroporation is ∼50 min. The subsequent period of NK cell recovery, seeding NK cells into culture plates, and the stimulation with cytokines can be completed in ∼30 min. For assessing knockdown efficiency, electroporated and cytokine-stimulated NK cells are incubated 20 hr under hypoxia. The detection of HIF-1α protein using immunoblot analysis is completed within 2 days. It is recommended to incubate the blot with HIF-1α-antibody overnight.
Acknowledgments
This work was funded by the German federal state Baden-Württemberg (reference 04HV.MED(21)/11/1, Clinical Cooperation Unit, Digital Data Spaces in Sepsis). G.E.B. was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) RTG2727–445549683.
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Sonia Y. Velásquez : Formal analysis; investigation; writing review and editing. Gizem E. Baslar : Formal analysis; investigation. Jutta Schulte : Formal analysis; investigation. Tanja Fuderer : Formal analysis; investigation. Holger A. Lindner : Funding acquisition; supervision; writing review and editing. Anna Coulibaly : Conceptualization; formal analysis; investigation; project administration; supervision; visualization; writing original draft; writing review and editing.
Conflict of Interest
The authors declare no conflicts of interest. The supporting source was not involved in the protocol design and in the collection, analysis, and interpretation of data.
Open Research
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
Literature Cited
- Carlsten, M., & Childs, R. W. (2015). Genetic manipulation of NK cells for cancer immunotherapy: Techniques and clinical implications. Frontiers in Immunology , 6, 266. https://doi.org/10.3389/fimmu.2015.00266
- Coulibaly, A., Bettendorf, A., Kostina, E., Figueiredo, A. S., Velásquez, S. Y., Bock, H. G., Thiel, M., Lindner, H. A., & Barbarossa, M. V. (2019). Interleukin- 15 signaling in HIF-1α regulation in natural killer cells, insights through mathematical models. Frontiers in Immunology , 10, 2401. https://doi.org/10.3389/fimmu.2019.02401
- Coulibaly, A., Velásquez, S. Y., Kassner, N., Schulte, J., Barbarossa, M. V., & Lindner, H. A. (2021). STAT3 governs the HIF-1α response in IL-15 primed human NK cells. Scientific Reports , 11(1), 7023. https://doi.org/10.1038/s41598-021-84916-0
- Devraj, G., Beerlage, C., Brüne, B., & Kempf, V. A. (2017). Hypoxia and HIF-1 activation in bacterial infections. Microbes and Infection , 19(3), 144–156. https://doi.org/10.1016/j.micinf.2016.11.003
- Huang, L. E., Arany, Z., Livingston, D. M., & Bunn, H. F. (1996). Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its α subunit. Journal of Biological Chemistry , 271(50), 32253–32259. https://doi.org/10.1074/jbc.271.50.32253
- Rius, J., Guma, M., Schachtrup, C., Akassoglou, K., Zinkernagel, A. S., Nizet, V., Johnson, R. S., Haddad, G. G., & Karin, M. (2008). NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature , 453(7196), 807–811. https://doi.org/10.1038/nature06905
- Semenza, G. L. (2011). Oxygen sensing, homeostasis, and disease. New England Journal of Medicine , 365(6), 537–547. https://doi.org/10.1056/NEJMra1011165
- Taylor, C. T., Doherty, G., Fallon, P. G., & Cummins, E. P. (2016). Hypoxia-dependent regulation of inflammatory pathways in immune cells. The Journal of Clinical Investigation , 126(10), 3716–3724. https://doi.org/10.1172/JCI84433
- Wang, G. L., Jiang, B. H., Rue, E. A., & Semenza, G. L. (1995). Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proceedings of the National Academy of Sciences , 92(12), 5510–5514. https://doi.org/10.1073/pnas.92.12.5510

