Cathepsin D assay to verify the retention of lysosomal content
Dario R. Alessi, Rotimi Y. Fasimoye
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
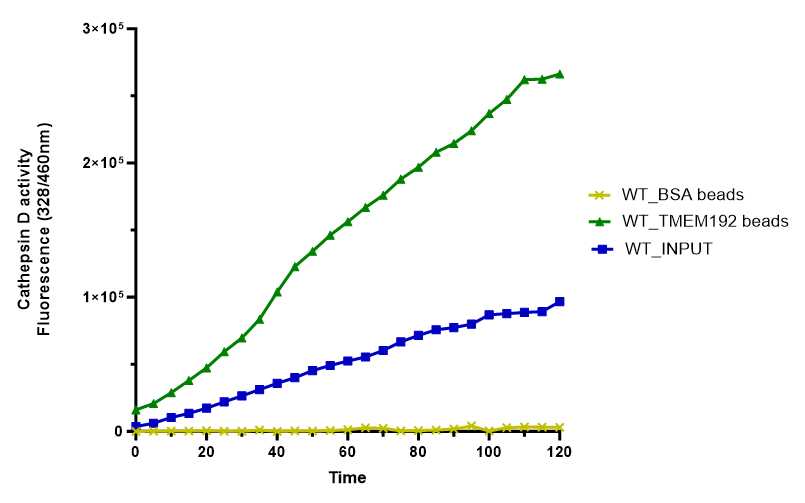
Cathepsin D assay is a fluorescence-based assay that leverage on the activity of cathepsin D, a lysosomal enzyme, to monitor the intactness of lysosome in the cell. Here, we describe a method where we used the measurement of cathepsin D activity to verify the intactness of lysosomes that were isolated from HEK293 cells based on anti-TMEM192 Lyso-IP. Our data showed an increase in the cathepsin D activity of lysosomal fraction when compared with whole cell fraction and Mock-IP fraction, an indication that the lysosomes are intact and viable.
Attachments
Steps
Seeding cells and performing Lyso-IP with anti-TMEM192 beads
Seed HEK293 cells in 15cm plates and allow to reach 80-90% confluency.
Perform Lyso-IP (using anti-TMEM192 beads) and Mock-IP (using BSA coated beads) as previously described in dx.doi.org/10.17504/protocols.io.x54v9yp51g3e/v1
Preparing sample for Cathepsin D assay
Add 2µg of protein from Lyso-IP and whole cell lysate into the wells FLUOTAC flat bottom black 96-well plate. This should be done in duplicate.
Top up to 50µLwith lysis buffer provided in the kit.
Prepare Blank sample in duplicate. This should contain only lysis buffer.
Prepare a reaction master mix for 9 wells:
450µLreaction buffer (from the kit) per well.18µLsubstrate (from the kit)
Add 52µL of master mix into each well. Gently mix but avoid bubbles. Cover plate with foil to avoid light exposure.
Plate reading and analysis.
Start the ClarioStar plate reading machine and initiate the software.
Set temperature to 37°C.
Set reading time to 0h 5m 0s for 24 cycles. This is total reading time of 2h 0m 0s.
Set reading wavelength to Ex/Em =328 / 460 .
Highlight and name the virtual wells, ensuring they correspond with the orientation of the plate.
Set direction of plate reading.
Open the plate holder and insert plate. Remember to remove foil covering before inserting the plate into the equipment.
Close plate holder and run the program.
After the completion of the run, export data in excel format and analyse it using GraphPad Prism.