A Sensitive and Quantitative Assay to Enumerate and Measure Mycobacterium leprae Viability in Clinical and Experimental Specimens
Jaymes H. Collins, Jaymes H. Collins, Shannon M. Lenz, Shannon M. Lenz, Nashone A. Ray, Nashone A. Ray, Marivic F. Balagon, Marivic F. Balagon, Deanna A. Hagge, Deanna A. Hagge, Ramanuj Lahiri, Ramanuj Lahiri, Linda B. Adams, Linda B. Adams
Abstract
Mycobacterium leprae , the etiologic agent of leprosy, cannot be cultured on artificial media. This characteristic, coupled with its long generation time, presents a number of unique challenges to studying this pathogen. One of the difficulties facing both researchers and clinicians is the absence of a rapid test to measure the viability of M. leprae in clinical or experimental specimens. The lack of such a tool limits the understanding of M. leprae immunopathogenesis and makes determining the efficacy of drug treatments difficult. With this in mind, we developed a robust two-step molecular viability assay (MVA) that first enumerates the M. leprae in the tissue; then, this data is used to normalize bacterial RNA quantities for the second step, in which the expression of M. leprae esxA and hsp18 are measured. This assay is specific and sensitive enough to be used on most clinical samples. This protocol describes the steps required to extract DNA and RNA from M. leprae- infected tissue, enumerate M. leprae , and measure M. leprae viability based on the normalized expression of two M. leprae -specific genes (hsp18 and esxA). This protocol also outlines an optimal laboratory design and workflow for performing this assay. © 2022 The Leprosy Mission Nepal. Current Protocols published by Wiley Periodicals LLC. This article has been contributed to by US Government employees and their work is in the public domain in the USA.
This article was corrected on 19 February 2022. See the end of the full text for details.
Basic Protocol 1 : DNA and RNA P purification from M. leprae -infected tissue
Basic Protocol 2 : Enumeration of M. leprae by RLEP qPCR on the DNA fraction
Basic Protocol 3 : Calculation of M. leprae per tissue and normalization of RNA
Basic Protocol 4 : Reverse-transcription of normalized RNA to generate cDNA
Basic Protocol 5 : Determination of M. leprae viability using HSP18 and ESXA qPCR on the cDNA
Support Protocol 1 : M. leprae qPCR primer/probe stock preparation
Support Protocol 2 : Preparation of plasmid stocks and standard curves
INTRODUCTION
Leprosy, also known as Hansen's Disease, is caused by the acid-fast bacterium, Mycobacterium leprae. The disease manifests as an immunohistopathological spectrum that depends on the immune capabilities of the host and affects the skin, mucous membranes of the upper respiratory tract, and peripheral nerves. At one end of the spectrum is multibacillary lepromatous disease, in which enormous numbers of M. leprae are found in the lesions and there is poorly developed cell-mediated immunity (CMI) but a strong antibody response. At the other end of the spectrum is tuberculoid disease, in which few bacilli are present and there is a strong CMI to M. leprae antigens. Between these two extremes are varying levels of CMI and bacterial growth. Confounding this complicated disease presentation are reactional episodes that can arise from an apparent change in the host's immune response and frequently manifest during or following anti-leprosy treatment (Scollard et al., 2006).
Leprosy is diagnosed primarily by clinical examination. Diagnostic confirmation is via histopathological observation of acid-fast bacilli inside the peripheral nerves of a leprosy lesion. Recently, an M. leprae -specific quantitative polymerase chain reaction (qPCR) assay has become standard-of-care for leprosy diagnostics at the National Hansen's Disease Programs (NHDP) (Sharma et al., 2020). However, this diagnostic qPCR test detects M. leprae DNA but does not distinguish between live and dead bacilli. In fact, the assessment of M. leprae viability has been the bane of both clinical and research studies (Lahiri & Adams, 2016) due to three characteristics of M. leprae. First, this organism has yet to be cultivated on artificial medium and must be grown in animal models, such as the mouse footpad or the nine-banded armadillo. Second, M. leprae has an extremely long division time of 12-14 days; therefore, cultivation experiments can take up to 1 year to complete. Third, due to its dense lipid cell wall, it can take months to years for the immune system to clear dead bacilli from tissues, so the presence of acid-fast bacteria in tissue does not necessarily indicate viable organisms. Thus, the difficulty in determining whether an immunological manifestation of leprosy is induced or exacerbated by metabolites generated by live bacilli or by cellular remnants released by dead bacteria cannot be overstated. The capacity to assess the viability of M. leprae in a tissue sample in a sensitive, specific, easy, and relatively short-term manner would be beneficial.
The NHDP maintains multiple clinical strains of M. leprae in continuous programmed passage in athymic mouse footpads. This colony provides a regular, consistent, and reproducible supply of highly viable M. leprae suspensions and infected tissues for use as research reagents (Adams, 2021; Lenz, Collins, Lahiri, & Adams, 2020; Truman & Krahenbuhl, 2001). This resource was instrumental in developing, refining, and validating this two-step molecular viability assay (MVA) for M. leprae (Davis et al., 2013). In the first step, M. leprae is quantified in the tissue sample, and this data is used to normalize bacterial RNA quantities for the second step, which measures the expression of M. leprae esxA and hsp18. This article details how to perform this MVA. DNA and RNA purification from M. leprae -infected host tissues, including human biopsies and mouse footpads, is presented in Basic Protocol 1. Enumeration of the number of M. leprae per tissue by qPCR is described in Basic Protocol 2. A key step in the MVA protocol is the normalization of the M. leprae RNA based on the M. leprae count. This procedure is explained in Basic Protocol 3. The normalized RNA is then reverse transcribed to cDNA (Basic Protocol 4), and M. leprae viability is measured by quantifying esxA and hsp18 transcripts (Basic Protocol 5). Preparation of the M. leprae -specific primers and probe for the qPCR assays are detailed in Support Protocol 1, and preparation of the standard curves are presented in Support Protocol 2.
SAFETY PRECAUTIONS
- M. leprae is a BSL-2 pathogen in the United States. However, once the infected tissue is fixed in 70% ethanol for at least 24 hr, it is no longer infectious. Therefore, all molecular work can be performed in a BSL-1 laboratory following basic biosafety protocols. However, laboratories should consult their institutional biosafety guidelines before starting work.
- Disposable gloves are to be worn at all times throughout the procedure. Powdered latex gloves are not recommended.
- All work should be performed in a still air hood dedicated for either DNA or RNA extraction/handling, as well as a separate hood for loading 96-well plates for qPCR. Use of a chemical fume hood is recommended when handling organic reagents.
- After work is completed, all surfaces and pipettors should be disinfected with DNA AWAY and RNase AWAY or a similar product.
STRATEGIC PLANNING
- It is important to follow a unidirectional workflow to prevent contamination. Separate areas or rooms, complete with their own equipment, should be dedicated to each part of the workflow. Primers/probes and other qPCR reagents should be stored and handled in a Reagent Area separate from where specimens are handled (Sample Prep Area). qPCR should be performed in a Product Area, and qPCR products should never be taken into the Reagent or Sample Prep Areas. Pipettors should be designated for either DNA or RNA work and not used interchangeably. Refer to the Clinical and Laboratory Standards Institute's publication MM19-A for comprehensive guidance and recommendations on molecular biology laboratory design.
- If there are multiple tissue samples to evaluate, we have found that a group of 12 samples is a reasonable number to process at one time.
- This assay takes multiple days to complete, so it is important to plan and allocate sufficient time for the entire procedure. Figure 1 gives a brief overview of the MVA.
Basic Protocol 1: DNA AND RNA PURIFICATION FROM M. LEPRAE-INFECTED TISSUE
This protocol describes purifying DNA and RNA from M. leprae -infected tissues and using DNase to isolate the RNA. Infected tissue is first homogenized, and then DNA and RNA are extracted and purified. Purified RNA is DNase treated using Turbo DNase.
Materials
-
Specimens fixed in 70% ethanol, e.g., 4-5 mm human leprosy lesion biopsy or M. leprae -infected mouse footpad tissue
-
Nuclease-free water (Fisher ThermoScientific, cat. no. AM9930)
-
Trizol (Fisher Thermo Scientific, cat. no. 15596-018)
-
Chloroform isoamyl mixture (Sigma, cat. no. 25666)
-
GlycoBlue co-precipitant (Fisher ThermoScientific, cat. no. AM9516)
-
Ammonium acetate (5M) (Fisher Thermo Scientific, cat. no. AM9070G)
-
Molecular grade isopropanol (Fisher Scientific, cat. no. BP2618500) 500 ml bottles
-
Molecular grade ethanol 200 proof (Fisher Scientific, cat. no. BP2818) 500 ml bottles
-
1× TE buffer (Sigma, cat. no. T9285, 100× Tris EDTA buffer solution)
-
Turbo DNAse Kit (Thermo Fisher Scientific, cat. no. AM1907)
-
Ice and ice bucket
-
10, 20, 200, and 1000 μl pipettors
-
Filtered pipette tips
-
Refrigerated microcentrifuge
-
Petri dishes, 35 mm × 10 mm (Fisher Scientific, cat. no. FB0875711YZ)
-
Scalpels, No. 10 BP (Fisher Scientific, cat. no. 14-823-45)
-
Micro dissecting scissors: curved; sharp points; 24 mm blade length; 4.5 in. overall length (Fisher Scientific, cat. no. RS5913)
-
Blue cap FastPrep tubes containing MP Biomedicals Lysing Matrix B, 2.0 ml tubes, RNase/DNase free (Fisher Scientific, cat. no. 6911-500)
-
FastPrep FP-24 instrument (MP Bio, SKU:116005500)
-
1.5-ml microcentrifuge tubes (Fisher Scientific, cat. no. 50809238)
-
Heat block
-
PCR reaction tubes (Thermo Fisher Scientific cat. no. N8010540)
-
PCR tube adapters (Thomas Scientific, cat. no. 2508Z68)
This protocol is optimized for human leprosy lesion tissue (4-5 mm punch biopsy) or mouse footpad tissue (∼20 to 40 mg) placed immediately upon collection into 1 ml of 70% ethanol and fixed for at least 24 hr. Tissues in 70% ethanol can be kept at room temperature (RT; 20°C to 22°C) during transport to the laboratory. However, we recommend using cold packs if tissues must be shipped to another site. Long-term storage should be at –20°C.
Tissue Homogenization
1.Retrieve tubes containing tissue samples in 70% ethanol from –20°C and place them on ice.
2.Centrifuge tubes at 10,000 × g for 10 min at 4°C if the sample is in multiple small pieces.
3.Pipette to remove ethanol.
4.Add 1 ml nuclease-free water to each tube to rehydrate samples.
5.Incubate at RT for 1 hr for human biopsy tissue or 15 min for mouse footpad tissue.
6.Spin tubes at 10,000 × g for 10 min at 4°C.
7.Pipette to remove the water and place each tissue sample in the lid of a 35 mm × 10 mm Petri dish. Use a pipette tip to aid in removing the tissue sample from the tube.
8.Pipette 100 μl water onto the tissue and mince in with No. 10 scalpels for 1 min.
9.Continue to mince with scissors for 3 min until fine. Tissue pieces should be <1 mm in size after mincing.
10.Transfer each tissue homogenate to a blue cap FastPrep tube using the scissors.
11.Add 900 µl TRIzol to each tube for a total volume of 1 ml.
12.Place tubes in the FastPrep FP-24 instrument and homogenize the tissue using a speed setting of 6.5 m/s for 45 s.
13.Allow the blue cap FastPrep tubes containing the homogenates to sit in the FastPrep FP-24 instrument for 5 min.
14.Repeat step 12.
15.Chill tubes on ice for 5 min.
DNA and RNA Extraction
16.Transfer the blue cap FastPrep tubes to a tube rack and add 200 µl chloroform/isoamyl alcohol (CIA) to each tube. Vortex for 10 s.
17.Let tubes sit at RT for 3 min.
18.Centrifuge the tubes at 700 × g for 5 min at 4°C. Remove the tubes from the centrifuge and place them on a rack.
19.Transfer the contents of each tube (both the pink and clear layers) to a new 1.5-ml microcentrifuge tube.
20.Centrifuge the tubes at 14,000 × g for 10 min at 4°C. Remove the tubes from the centrifuge and place them on a rack.
21.Transfer only 400 µl of the top colorless aqueous phase (RNA) to new 1.5-ml microcentrifuge tubes.
22.Place the original tube, which contains the pink phenol-chloroform lower phase and solid interphase, on ice.
RNA Precipitation
23.Add 400 µl of CIA to the 400 µl aqueous phase (tubes from step 21) and vortex for 5 s to mix.
24.Let tubes sit at RT for 3 min.
25.Centrifuge tubes at 14,000 × g for 2 min at 4°C.
26.Transfer 300 µl of each top aqueous phase to a new 1.5-ml microcentrifuge tube.
27.Add 2.0 µl GlycoBlue to each tube.
28.Add 30 µl of 5M ammonium acetate to each tube.
29.Precipitate the RNA by adding 332 µl of cold 100% Isopropanol to each tube. Vortex for 5 s to mix.
30.Incubate tubes at RT for 5 min.
31.Incubate the tubes at –70°C overnight or until ready to complete precipitation.
DNA Precipitation
32.Transfer the contents of each tube saved at step 22 (containing the interphase and phenol-chloroform phases) to a new 1.5-ml microcentrifuge tube.
33.Add 100 µl of 1× TE to each tube.
34.Add 150 µl CIA to each tube.
35.Place tubes in the FastPrep FP 24 instrument.
36.Homogenize twice at a speed setting of 6.5 m/s for 45 s with 5 min rest in between each cycle.
37.Centrifuge tubes at 14,000 × g for 10 min at 4°C.
38.Transfer the aqueous phase (∼200 µl) to a new 1.5-ml tube.
39.Add 1.5 µl GlycoBlue to each tube.
40.Precipitate the DNA by adding 20 µl of 5M ammonium acetate followed by 400 µl of cold 100% ethanol to each tube. Vortex for 5 s to mix.
41.Store tubes at –70°C overnight or until ready to complete.
DNA Purification
42.Thaw the tubes from step 41 on ice.
43.Centrifuge at 10,000 × g for 10 min at 4°C.
44.Remove the supernatant and wash the DNA pellet by resuspending in 1 ml cold 70% ethanol. Vortex for 5 s to mix.
45.Centrifuge at 10,000 × g for 10 min at 4°C.
46.Remove the supernatant, then air dry the pellet by inverting the tube on sterile gauze at RT for 30 min.
47.Resuspend the DNA in 30 μl of sterile 1× TE by vortexing for 5 s.
48.Spin at 10,000 × g for 30 s.
49.Store purified DNA at –70°C until use.
RNA Purification
50.Thaw tubes from step 31 on ice.
51.Centrifuge tubes at 14,000 × g for 30 min at 4°C.
52.Pipette off the supernatant from each tube without disturbing the pellet.
53.Wash the pellets by adding 1 ml cold 75% ethanol. Vortex each tube briefly for 2 s.
54.Centrifuge tubes at 12,000 × g for 5 min at 4°C.
55.Let the tubes sit at RT for 5 min.
56.Centrifuge again at 12,000 × g for 5 min at 4°C.
57.Carefully remove the supernatant from each tube without disturbing the pellet.
58.Repeat steps 53-57 once more.
59.Invert the tubes on sterile gauze and let air dry for 10 min ONLY.
60.Resuspend each RNA pellet in 25 µl of nuclease-free water at RT. Vortex for 5 s.
61.Spin at 10,000 × g for 30 s.
62.Place tubes in a 55°C heat block for 15 min.
63.Remove tubes from heat block and place on ice.
DNase treatment
64.Add 17 µl nuclease-free water from the Turbo DNase Kit to a PCR reaction tube, one for each RNA sample.
65.Add 5 µl Turbo DNase 10× buffer to each tube.
66.Add 25 µl purified RNA from step 63 to each tube.
67.Add 1.5 µl Turbo DNase to each tube. Vortex for 5 s and spin for 30 s at 10,000 × g.
68.Incubate for 30 min at 37°C in a thermocycler.
69.Add 1.5 µl Turbo DNase to each tube. Vortex for 5 s and spin for 30 s at 10,000 × g.
70.Incubate for 30 min at 37°C in a thermocycler.
DNase Inactivation
71.Remove the PCR tubes from the thermocycler.
72.Add 10 µl DNase Inactivation Reagent to each tube.
73.Set a timer for 5 min. Vortex each tube individually for 10 s. Once each tube has been vortexed for 10 s, start back at the first tube and repeat the 10 s vortex for each tube until the 5 min is over.
74.Centrifuge the tubes at 10,000 × g for 2 min at RT.
75.Transfer 45 µl of each supernatant to a sterile microcentrifuge tube.
76.Keep the DNase-treated RNA on ice or store at 4°C while setting up the reverse transcription reaction.
Basic Protocol 2: ENUMERATION OF M. LEPRAE BY RLEP qPCR OF THE DNA FRACTION
This protocol describes how to set up and run RLEP qPCR to quantify the number of M. leprae in each sample. Purified DNA from the previous protocol is used as a template to target and amplify the M. leprae- specific repetitive element, RLEP, using qPCR. RLEP is unique to M. leprae , having been tested against 17 mycobacterial species, including the closely related M. lepromatosis (Sharma et al., 2020), and RLEP qPCR correlates with microscopic counting of acid-fast bacilli for the enumeration of M. leprae in tissues (Davis et al., 2013; Truman et al., 2008).
Materials
- DNA samples from Basic Protocol 1, step 49
- 1× TE buffer (Sigma, cat. no. T9285; 100× Tris EDTA buffer solution)
- TaqMan Universal Master Mix II, with UNG (Fisher ThermoScientific, cat. no. 4440045)
This cat. no. is for a 5-ml five-pack.
-
Nuclease-free water (Fisher ThermoScientific, cat. no. AM9930)
-
RLEP primers (1× concentration, see Support Protocol 1)
-
RLEP probe (1× concentration, see Support Protocol 1)
-
Corning 96-well clear PVC assay microplate (Sigma, cat. no. CLS2797)
-
Reagent reservoir (Vistalab Technologies, cat. no. 3054-1004)
-
Applied Biosystems MicroAmp Fast Optical 96-Well Reaction Plate with Barcode, 0.1 ml (Fisher Thermo Scientific, cat. no. 4346906)
-
PCR reaction tubes (Thermo Fisher Scientific cat. no. N8010540)
-
10, 20, 200, and 1000 μl pipettors
-
200 μl multichannel pipette
-
Pipette tips
-
Vortex
-
Mini centrifuge
-
Real-time PCR machine
-
Microsoft Excel
Dilution of DNA
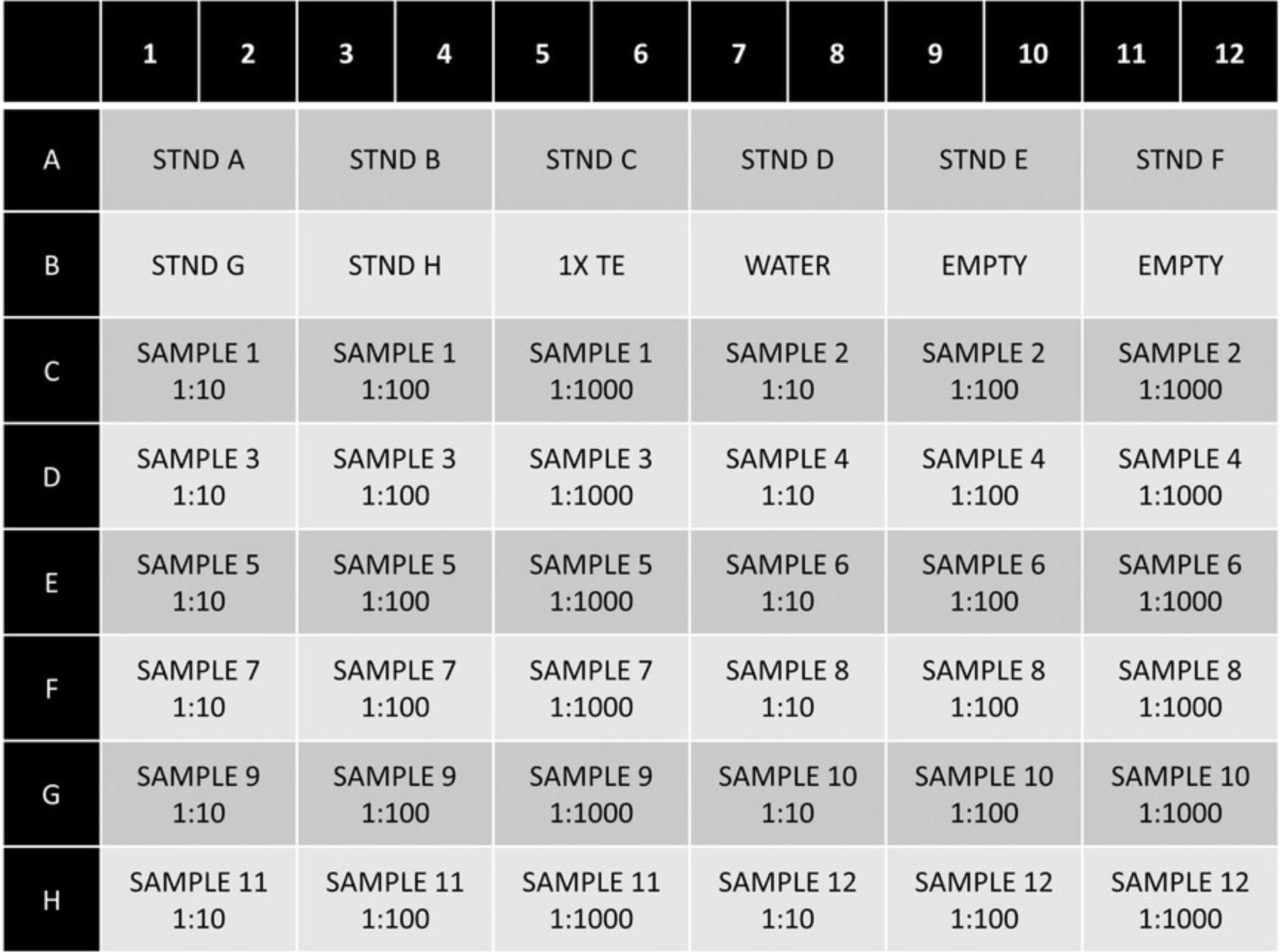
1.Prepare serial 10-fold dilutions of 1:10, 1:100, and 1:1000 of each extracted DNA sample.
2.Pipette 45 µl of 1× TE into each tube. For the first dilution (1:10), add 5 µl of the undiluted extracted DNA into the first tube. Briefly vortex the tube and centrifuge at 10,000 × g for 30 s to mix. For the second dilution (1:100), add 5 µl of the 1:10 DNA dilution to the tube. Briefly vortex the tube and centrifuge at 10,000 × g for 30 s to mix. For the final dilution (1:1000), add 5 µl of the 1:100 dilution to the tube. Briefly vortex the tube and centrifuge at 10,000 × g for 30 s to mix.
3.DNA dilutions should be stored at –20°C until ready to use.
Master mix preparation
4.Prepare the qPCR master mix as shown in Table 1 using the RLEP primers and probe.
| Reagent | Volume per reaction (μl) | Total for 96-well plate (μl) |
|---|---|---|
| Universal Master Mix | 12.5 | 1350 |
| Sterile water | 7 | 756 |
| Forward primer | 1.25 | 135 |
| Reverse primer | 1.25 | 135 |
| 1× Probe | 0.5 | 54 |
| Total | 22.5 | 2430 |
RLEP qPCR
5.Distribute 22.5 μl master mix per well in a 96-well plate using a multichannel pipette.
6.Add 2.5 μl of each DNA dilution to duplicate wells.
7.Add 2.5 μl of each standard curve dilution to duplicate wells.
8.Add 2.5 μl of 1× TE and nuclease-free water to duplicate wells as negative controls.
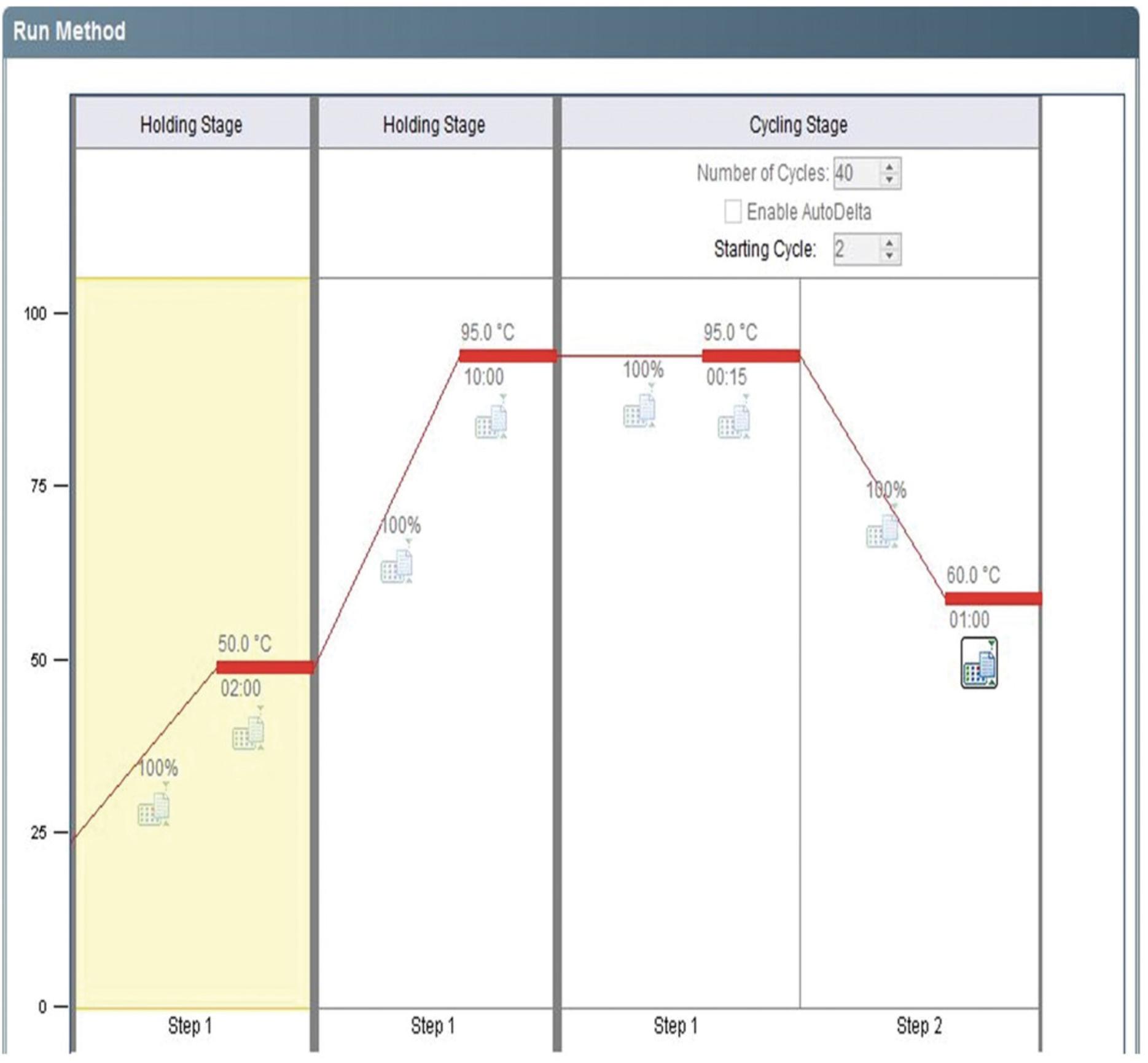
9.Run qPCR on a real-time PCR machine using the following thermal profile:
- Step 1: 50°C for 2 min (Holding Stage)
- Step 2: 95°C for 10 min (Holding Stage)
- Step 3: 95°C for 15 s (denaturation)
- Step 4: 60°C for 1 min (annealing/extension)
- Repeat steps 3 and 4 for 40 cycles.
This thermal profile is shown in Figure 4.
10.Export the data into Excel.
Basic Protocol 3: CALCULATION OF M. LEPRAE PER TISSUE AND NORMALIZATION OF RNA
This protocol explains how to use the NHDP MVA calculator to calculate the number of M. leprae per sample and determine the dilutions necessary to normalize the RNA for the reverse transcription reaction. The number of dilutions required will be dependent on the number of M. leprae in the sample, not on the weight of the tissue or the amount of total RNA recovered (see Critical Parameters).
Materials
-
DNase-treated RNA samples from Basic Protocol 1, step 76
-
Nuclease-free water (Fisher ThermoScientific, cat. no. AM9930)
-
RLEP qPCR Data from Basic Protocol 2 step 10.
-
NHDP MVA Calculator (supplemental Excel File)
-
PCR reaction tubes (Thermo Fisher Scientific cat. no. N8010540)
-
10, 20, 200, and 1000 μl pipettors
-
Pipette tips
-
Vortex
-
Mini centrifuge
Calculation of the number of M. leprae per tissue sample
1.Refer to the RLEP qPCR data from Basic Protocol 2, step 10, and compare the linearity of the Mean Quantity values to the 10-fold DNA dilutions for each sample. Choose the Mean Quantity value of the dilution closest to the “C” dilution of the standard curve.
2.Enter the Mean Quantity value of your chosen dilution into the “Mean Quantity” column (Column B) of the NHDP MVA Calculator.
3.Enter the DNA dilution for that chosen sample, for example, 10, 100, or 1000 into the “Dilution” column (Column C).
4.The NHDP MVA Calculator will calculate the number of M. leprae per tissue sample (Column E).
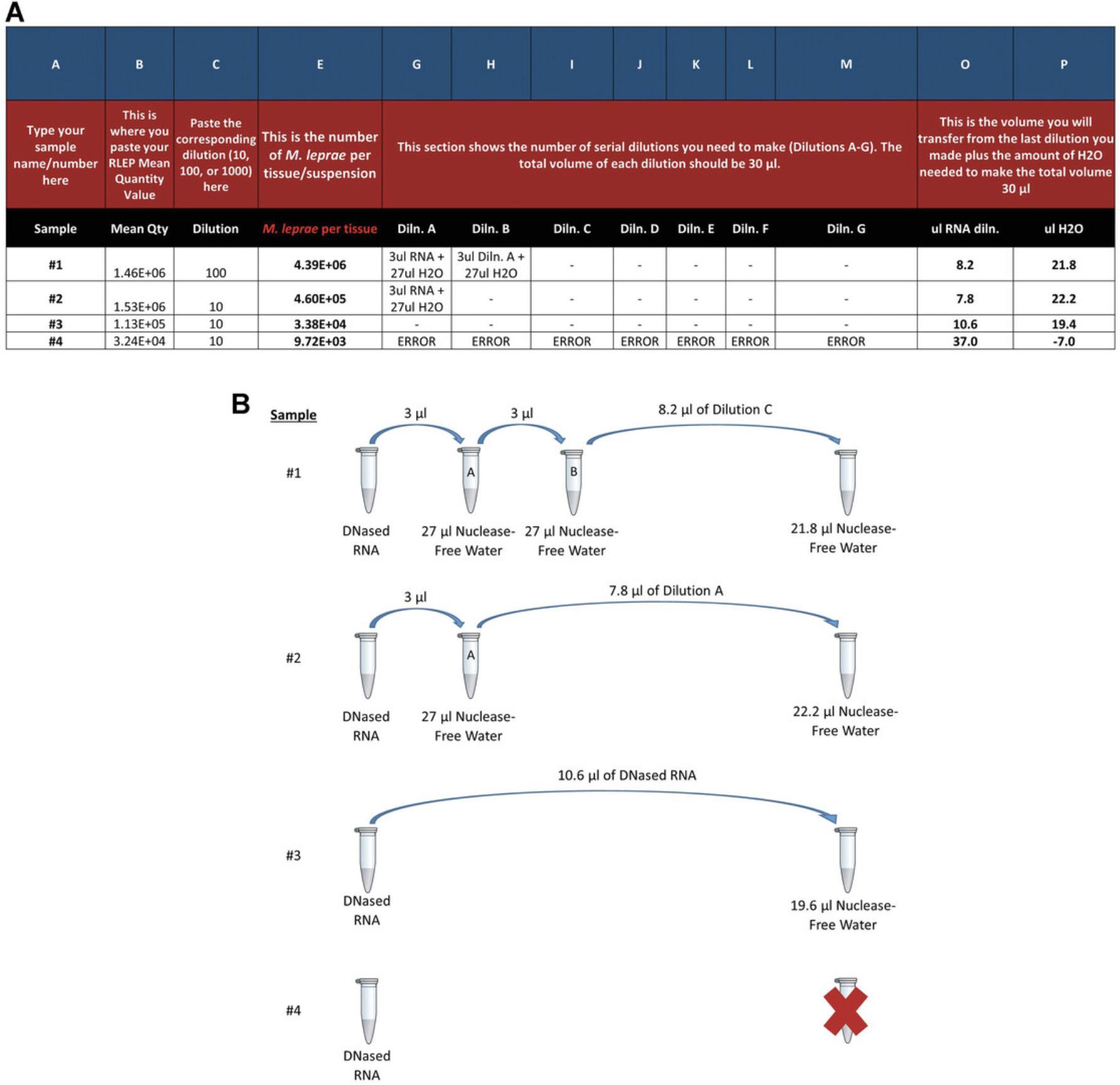
Calculation of dilutions to normalize each M. leprae RNA sample
5.The NHDP MVA Calculator will also calculate the RNA dilutions needed to normalize each RNA sample to 3000 M. leprae equivalents per reverse transcription reaction.
6.Prepare the required number of serial 10-fold dilutions (Columns G-M) by transferring 3 µl of the DNase-treated RNA into 27 μl of nuclease-free water. Vortex and quick-spin after each dilution.
7.Transfer the calculated volume of RNA (Column O) from the last dilution into the calculated volume of nuclease-free water (Column P) for a total volume of 30 μl.
8.If an ERROR message appears in the Dilution columns (Columns G-M), there are not enough M. leprae (<1.9 × 104) present in the sample to normalize the RNA. Disregard any values in columns O and P for samples that give an ERROR. The viability assay cannot be performed on that sample.
9.Store the normalized RNA samples on ice.
Basic Protocol 4: REVERSE-TRANSCRIPTION OF NORMALIZED M. LEPRAE RNA TO GENERATE cDNA FOR THE VIABILITY qPCR
This protocol explains the steps required to reverse-transcribe the normalized RNA from step 7 in Basic Protocol 3 to generate cDNA using the Advantage RT-for-PCR Kit from Clontech. The cDNA generated in this protocol will be used as the qPCR template in Basic Protocol 5 to measure the viability of M. leprae in the sample.
Materials
-
Normalized RNA samples from Basic Protocol 3, step 9
-
Nuclease-free water (Fisher ThermoScientific, cat. no. AM9930)
-
Advantage RT-for-PCR Kit (Clontech, cat. no. 639506)
-
PCR reaction tubes (Thermo Fisher Scientific cat. no. N8010540)
-
10, 20, 200, and 1000 μl pipettors
-
Pipette tips
-
Vortex
-
Mini centrifuge
-
Thermocycler
1.Prepare the reverse transcription master mix using the reagents provided in the Advantage RT-for-PCR kit, as shown in Table 2.Store the master mix on ice until it is ready to add to the PCR tube.
| Reagent | Volume per reaction (μl) | Volume for 12 samples (μl) |
|---|---|---|
| 5× Reaction buffer | 4 | 56 |
| dNTP mix (10 mM each) | 1 | 14 |
| Recombinant RNase inhibitor | 0.5 | 7 |
| MMLV reverse transcriptase | 1 | 14 |
| Total | 6.5 | 91 |
2.Prepare a second, Mock master mix by substituting nuclease-free water for the MMLV Reverse Transcriptase (Table 3). Do not use the DEPC-treated water supplied in the kit for this step. Store the master mix on ice until it is ready to add to the PCR tube.
| Reagent | Volume per reaction (μl) | Volume for 12 samples (μl) |
|---|---|---|
| 5× Reaction buffer | 4 | 56 |
| dNTP mix (10 mM each) | 1 | 14 |
| Recombinant RNase inhibitor | 0.5 | 7 |
| Nuclease-free water | 1 | 14 |
| Total | 6.5 | 91 |
3.Label a set of “RT” tubes, one per sample.
4.Label a set of “Mock” tubes, one per sample.
5.Add 1.0 µl of Random Hexamer to each tube and close tubes.
6.Add 12.5 µl of each normalized RNA sample to an “RT” tube.
7.Add 12.5 µl of each normalized RNA sample to its corresponding “Mock” tube.
8.Mix the PCR tube contents by vortexing for 5 s followed by a quick spin in a microcentrifuge.
9.Transfer the PCR tubes to a thermocycler.
10.Heat the RNA at 70°C for 2 min.
11.Remove the PCR tubes from the thermocycler.
12.Add 6.5 µl of RT Master Mix to each RT sample tube. Vortex and quick-spin tubes.
13.Add 6.5 µl of Mock Master Mix to each Mock sample tube. Vortex and quick-spin tubes.
14.Place the RT and Mock tubes in a thermocycler and run the program:
- • 42°C for 1 hr
- • 94°C for 5 min
- • 4°C for 5 min
- • 4°C Hold
15.Remove the tubes from the thermocycler.
16.Add 30 µl of nuclease-free water to each tube for a final volume of 50 µl, vortex for 5 s, and quick spin.
17.Transfer each cDNA and Mock into an appropriate long-term storage tube and store at –70°C.
Basic Protocol 5: DETERMINATION OF M. LEPRAE VIABILITY USING hsp18 AND esxA qPCR ON THE cDNA
This protocol describes how to set up and run a qPCR assay to determine the viability of M. leprae in a sample by measuring the expression of hsp18 , which encodes the 18 kD heat shock protein, and esxA , which encodes the ESAT-6 protein transcripts. These genes were chosen because of their high expression levels by M. leprae actively growing in athymic nude mouse footpads and for their ability to discriminate loss of viability after bacterial killing by antimicrobial drugs or immunologically mediated methods (Davis et al., 2013; Williams, Torrero, Wheeler, Truman, & Yoder, 2004).
Materials
-
cDNA and Mock samples from Basic Protocol 4, step 17
-
1× TE buffer (Sigma, cat. no. T9285; 100× Tris EDTA buffer solution)
-
TaqMan Universal Master Mix II, with UNG (Fisher ThermoScientific, cat. no. 4440045) This cat. no. is for a 5 ml five-pack.
-
Nuclease-free water (Fisher ThermoScientific, cat. no. AM9930)
-
hsp18 primers (1× concentration, see Support Protocol 1)
-
hsp18 probe (1× concentration, see Support Protocol 1)
-
esxA primers (1× concentration, see Support Protocol 1)
-
esxA probe (1× concentration, see Support Protocol 1)
-
16S primers (1× concentration, see Support Protocol 1)
-
16S probe (1× concentration, see Support Protocol 1)
-
Corning 96-well clear PVC assay microplate (Sigma, cat. no. CLS2797)
-
Reagent reservoir (Vistalab Technologies, cat. no. 3054-1004)
-
Applied Biosystems MicroAmp Fast Optical 96-Well Reaction Plate with Barcode, 0.1 ml (Fisher Thermo Scientific, cat. no. 4346906)
-
PCR reaction tubes (Thermo Fisher Scientific cat. no. N8010540)
-
10, 20, 200, and 1000 μl Pipettors
-
200 μl Multichannel pipette
-
Pipette tips
-
Vortex
-
Mini centrifuge
-
Real-time PCR machine
1.Prepare 3 sets of qPCR master mix (refer to Table 1):
- • 1 set using hsp18 primers and probe
- • 1 set using esxA primers and probe
- • 1 set using 16S primers and probe
16S qPCR is used as an internal RNA extraction control. It is NOT a viability indicator.
2.Distribute 22.5 μl of each qPCR master mix per well in a 96 well plate using a multichannel pipette.
3.Add 2.5 μl of each cDNA sample, in duplicate, to each master mix set.
4.Add 2.5 μl of each Mock sample, in duplicate, to each master mix set.
5.Add 2.5 μl of each standard curve dilution, in duplicate, to each master mix set.
6.Add 2.5 μl of 1× TE and nuclease-free water, in duplicate, to each master mix set. These serve as negative controls.
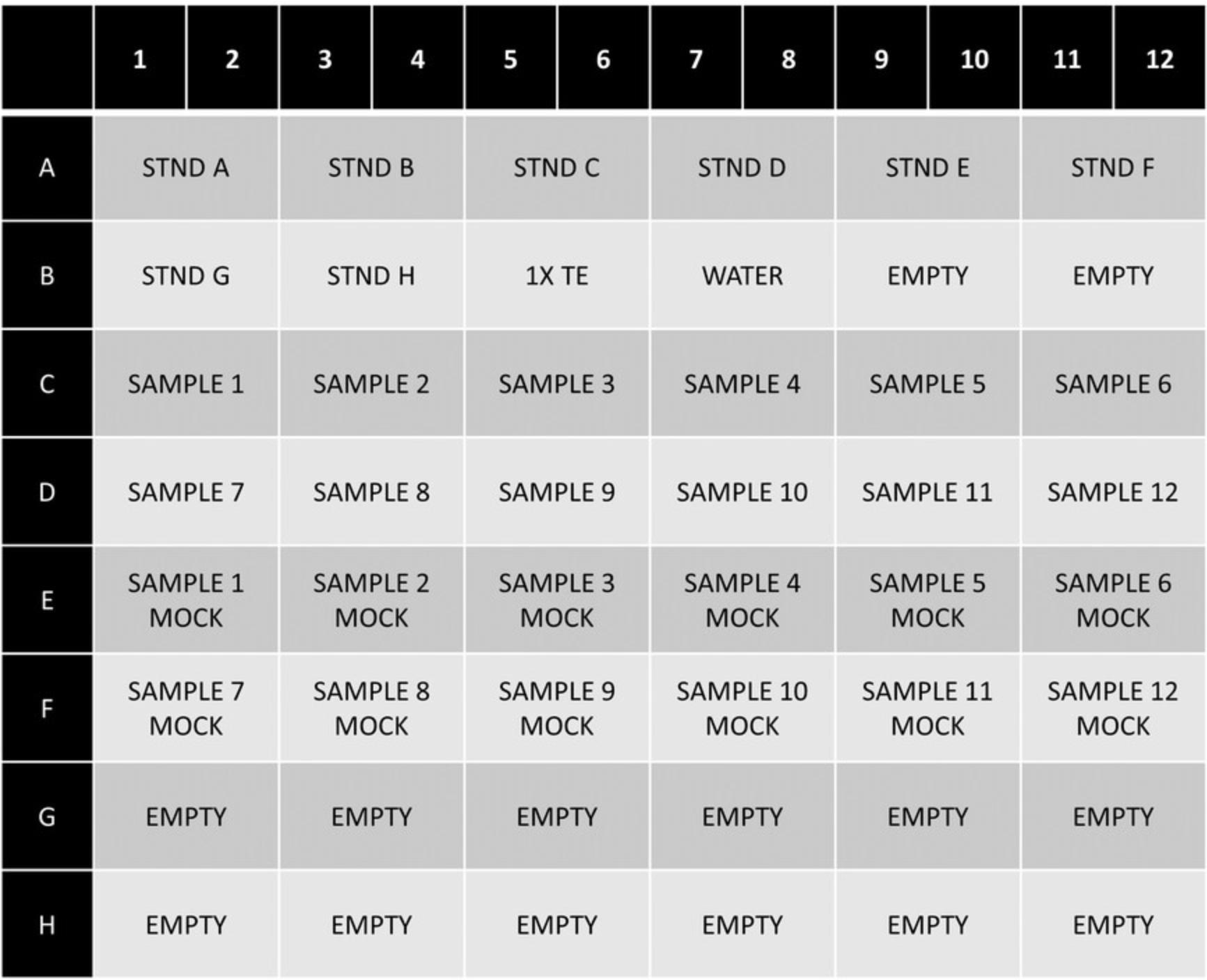
7.Run qPCR reactions on a real-time PCR machine using the following thermal profile:
- Step 1: 50°C for 2 min (Holding Stage)
- Step 2: 95°C for 10 min (Holding Stage)
- Step 3: 95°C for 15 s (denaturation)
- Step 4: 60°C for 1 min (annealing/extension)
- Repeat steps 3 and 4 for 40 cycles.
The thermal profile for each qPCR is shown in Figure 4.
8.Export the data into Excel.
Support Protocol 1: M. leprae qPCR PRIMER/PROBE STOCK PREPARATION
This protocol describes how to rehydrate and prepare the qPCR primers and probes used in the MVA to quantify M. leprae and measure viability. The primer and probe sequences are shown in Table 4.All primers and probes used in this assay have been extensively tested to ensure no expression of the targets in non-infected mouse and human tissue.
| M. leprae target | Primer and probe sequence |
|---|---|
| RLEP | FORWARD: 5′ GCAGCAGTATCGTGTTAGTGAA 3′ |
| REVERSE: 5′ CGCTAGAAGGTTGCCGTAT 3′ | |
| PROBE: 5′ CGCCGACGGCCGGATCATCGA 3′ | |
| hsp18 | FORWARD: 5′ CGATCGGGAAATGCTTGC 3′ |
| REVERSE: 5′ CGAGAACCAGCTGACGATTG 3′ | |
| PROBE: 5′ ACACCGCGTGGCCGCTCG 3′ | |
| esxA | FORWARD: 5′ CCGAGGGAATAAACCATGCA 3′ |
| REVERSE: 5′ CGTTTCAGCCGAGTGATTGA 3′ | |
| PROBE: 5′ TGCTTGCACCAGGTCGCCCA 3′ | |
| 16S | FORWARD: 5′ GCATGTCTTGTGGTGGAAAGC 3′ |
| REVERSE: 5′ CACCCCACCAACAAGCTGAT 3′ | |
| PROBE: 5′ CATCCTGCACCGCA 3′ |
Materials
-
TaqMan Universal Master Mix II, with UNG (Fisher ThermoScientific, cat. no. 4440045)
-
This catalog number is for a 5 ml five-pack.
-
Lyophilized RLEP forward and reverse primers
-
RLEP probe (liquid)
-
Lyophilized hsp18 forward and reverse primers
-
hsp18 probe (liquid)
-
Lyophilized esxA forward and reverse primers
-
esxA probe (liquid)
-
Lyophilized 16s forward and reverse primers
-
16S probe (liquid)
-
Nuclease-free water (Fisher ThermoScientific, cat. no. AM9930)
-
1× TE (Sigma, cat. no. T9285) 100 × Tris EDTA buffer solution. Use at 1 ×.
-
Corning 96-well clear PVC assay microplate (Sigma, cat. no. CLS2797)
-
Reagent reservoir (Vistalab Technologies, cat. no. 3054-1004)
-
Applied Biosystems MicroAmp Fast Optical 96-Well Reaction Plate with Barcode, 0.1 ml (Fisher Thermo Scientific, cat. no. 4346906)
-
1.5-ml microcentrifuge tubes (Fisher Scientific, cat. no. 50809238)
-
Mini centrifuge
-
Vortex mixer
-
10, 20, 200, and 1000 μl Pipettors and tips
M. leprae Primers
1.Centrifuge tubes at 10,000 × g for 1 min to ensure lyophilized primers are at the bottom of the tube. Primers are ordered at a concentration of 80,000 pmol and come lyophilized.
2.Add 400 μl nuclease-free water to each tube containing the lyophilized primer.
3.Vortex for 10 s and incubate at RT for at least 30 min.
4.Vortex each tube for 10 s and quick-spin in the mini centrifuge.
5.Prepare a 1:20 dilution by adding 25 μl of the 20× RLEP primer to 475 μl of nuclease-free water to make a 1× working stock of RLEP primer. Store at -20°C.
6.Prepare a 1:10 dilution by adding 50 μl of the hsp18, esxA , or 16S primers to 450 μl nuclease-free water to make a 1× working stock of hsp18, esxA , or 16S primers.
7.Store all primers at -20°C.
M. leprae Probes
8.Perform a 1:10 dilution to make a 1× working stock for all probes (RLEP, hsp18 , esxA , and 16s). Probes are ordered as a liquid at 100 μM.
9.Store all probes at –20°C.
Support Protocol 2: PREPARATION OF PLASMID STOCKS AND STANDARD CURVES
This protocol describes how to rehydrate and prepare stock solutions of plasmids and dilute plasmids to create a standard curve used for qPCR. Custom-designed plasmids are used to prepare the standard curves for qPCR. One plasmid contains the RLEP sequence, another includes the genes for both esxA and hsp18 , and the third includes 16S, all of which are specific to M. leprae. The sequence for RLEP (supplemental Excel file) was cloned into a pUCIDT (Amp) plasmid using custom gene synthesis by Integrated DNA Technologies (IDT). Both sequences for hsp18 and esxA (supplemental Excel file) were cloned into a separate pUCIDT (Amp) plasmid using the same custom gene synthesis from IDT. These genes can be cloned separately if desired. The sequence for 16S was also cloned into a separate pUCIDT (Amp) plasmid using the same custom gene synthesis from IDT.
Materials
-
Lyophilized plasmid from IDT
-
Nuclease-free water (Fisher ThermoScientific, cat. no. AM9930)
-
1× TE (Sigma, cat. no. T9285) 100× Tris EDTA buffer solution. Use at 1×.
-
1.5-ml microcentrifuge tubes (Fisher Scientific, cat. no. 50809238)
-
Mini centrifuge
-
Vortex Mixer
-
10, 20, 200, and 1000 μl Pipettors
-
Pipette Tips
Preparation of plasmid stocks
1.Spin the tube for 30 s at 10,000 × g before opening.
2.Resuspend the plasmid DNA in 20 µl TE by pipetting.
3.Vortex for 20 s.
4.Incubate the tube at RT for 30 min.
5.Centrifuge for 1 min at 10,000 × g.
6.Store at –20°C. The plasmid is now at a concentration of 200 ng/µl.
Preparation of “A” dilution for plasmid standard curve
7.Make a 1:100 dilution of the plasmid by adding 2 µl plasmid DNA to 198 µl TE in a 1.5-ml tube. Mix by gently pipetting for 30-45 s. Let tube sit at RT for 2 min. Label this tube “Stock (insert name) Plasmid DNA.” Store at –20°C.
8.Perform a series of five 1:4 serial dilutions for RLEP, starting with 25 μl of “Stock RLEP Plasmid DNA” and 75 μl TE. Mix each dilution by gently pipetting for 30-45 s. Let tube sit at room temperature for 2 min before making the next dilution. The tube you stop at will be the “A” dilution of the standard curve.
9.Perform a series of six 1:4 serial dilutions for esxA, hsp18 , and 16S, starting with 25 μl of “Stock (esxA, hsp18 , or 16S) Plasmid DNA” and 75 μl TE. Mix each dilution by gently pipetting for 30-45 s. Let each tube sit at RT for 2 min before making the next dilution. The final tube will be the “A” dilution of the standard curve.
Preparation of plasmid standard curves
10.Perform a series of seven 1:4 serial dilutions from the “A” dilution, starting with 25 μl of the “A” dilution and 75 μl of TE to make dilutions for the standard curve. Mix each dilution by gently pipetting for 30-45 s. Let each tube sit at RT for 2 min before making the next dilution. These are dilutions “A” through “H” for making the standard curves. Store at 4°C.
| Standard curve dilution | Number of M. leprae per ml |
|---|---|
| A | 2 × 107 |
| B | 5 × 106 |
| C | 1.25 × 106 |
| D | 3.13 × 105 |
| E | 7.81 × 104 |
| F | 1.95 × 104 |
| G | 4.88 × 103 |
| H | 1.22 × 103 |
COMMENTARY
Background Information
Since M. leprae cannot be grown on axenic media, traditional methods of determining viability, such as calculating colony-forming units, are impossible. The gold standard for measuring M. leprae viability is the mouse footpad assay (Levy & Ji, 2006; Shepard, 1962). In this assay, M. leprae bacilli are recovered from the clinical or experimental tissue sample and counted by microscopic examination of acid-fast stained smears. This count is total bacteria recovered, both live and dead. The presence of viable M. leprae is assessed by adjusting the suspension to an inoculum of 5000 bacilli and injected into the footpads of passage mice. After 1 year, M. leprae is harvested from the footpads of the passage mice and counted to determine if there was bacterial growth. Theoretically, if there is one viable M. leprae in the inoculum, it will multiply to a countable level of ∼105 in 1 year. No growth after 1 year means that the bacteria in the original sample were dead. While this technique is effective, the amount of time required to obtain results and the costs associated with purchasing and maintaining scores of mice make it impractical for most laboratories.
Several studies have made significant progress toward developing alternative M. leprae viability assays that are simpler and less time-consuming than the mouse footpad assay (Lahiri & Adams, 2016). The most widely used are viability stains (Lahiri, Randhawa, & Krahenbuhl, 2005) and metabolic assays (Franzblau SG, 1988), although both of these techniques require large numbers (106 to 107) of freshly harvested M. leprae from unfixed host tissues. Furthermore, most metabolic assays utilize radioactive compounds such that appropriate radioactive material user licenses, safety protocols, and disposal methods must be maintained. Due to these limitations, both techniques are primarily used in research laboratories rather than clinical settings. qPCR-based assays have so far shown the greatest promise for developing a simple and robust viability assay that is specific, sensitive, rapid, field-friendly, and reliable.
The advantages of the MVA are that it is (1) highly specific for M. leprae , so any co-infection or contamination in the host tissue will not influence the assay results; (2) effective with as few as 1.9 × 104 M. leprae per tissue sample, a sensitivity that makes it available for most human biopsy samples; (3) field-friendly and can be used on any appropriately fixed host tissue, making it clinically useful; (4) relatively rapid as results can be obtained as little as 4 days; and (5) reliable and has been extensively validated by comparison with other proven viability assays and optimized using M. leprae preparations with known high and low viability. In addition, the user-friendly NHDP MVA Calculator makes assay quality control, normalization, data calculation, and result interpretation straightforward. There are, however, numerous steps in the protocol, and rather sophisticated equipment is required; consequently, this assay is most likely suited for clinical reference laboratories or research laboratories.
We have gone to great lengths to seek the most accurate viability indicators for the MVA, choosing genes that are abundantly expressed by viable M. leprae but are rapidly degraded once the bacilli become non-viable (Borah et al., 2020; Ojo, Williams, Adams, & Lahiri, 2022). Most molecular assays measure the presence or absence of the 16S rRNA transcript because it is produced in copious amounts and easily detected. M. leprae 16S rRNA, however, is extremely stable. Like M. leprae DNA, 16S rRNA can be detected in dead bacilli that have remained in tissues for years (manuscript in preparation). For this reason, 16S rRNA is not a valid indicator of M. leprae viability. Conversely, 16S rRNA expression is an excellent internal control for M. leprae RNA extraction. For example, if a sample is “Undetermined” for esxA and hsp18 expression, indicating dead M. leprae in the sample, one can verify that the RNA extraction was successful by running the 16S rRNA qPCR.
The MVA has been utilized to evaluate new anti-leprosy drugs (Lahiri, Adams, Thomas & Pethe, 2021), study experimental models of the immunological spectrum of leprosy (Hagge et al., 2014), and search for potential transmission vectors (Tongluan et al., 2021). The MVA is also proving useful in clinical settings to determine whether a patient has viable M. leprae following completion of anti-leprosy regimens or when relapse or re-infection is suspected. The MVA would also be of great benefit in clinical trials of new drugs for leprosy, as programs currently must rely on the mouse footpad assay for confirmation of anti-mycobacterial efficacy.
Critical Parameters
The most crucial aspect of the MVA is the normalization of M. leprae RNA at the reverse transcription step based on the number of M. leprae in the sample. Most reverse transcription-PCR assays normalize the RNA on the basis of total RNA quantity, e.g., 500 ng RNA per reverse transcription reaction. However, in M. leprae -infected tissues, whether human biopsy or mouse footpad, the majority of the RNA recovered will be host derived, and it is impossible to distinguish between host and M. leprae RNA through normal RNA quantification methods. Furthermore, depending on a patient's position in the leprosy spectrum, the number of M. leprae per biopsy specimen will vary greatly. Therefore, since we are measuring expression of M. leprae transcripts, normalization of RNA based on the number of M. leprae in the sample is imperative for the data to have any meaning.
It is also important to emphasize that a minimum of 1.9×104 M. leprae must be present in the sample in order to normalize the RNA to 3000 bacterial equivalents per reverse transcription reaction. The NHDP MVA Calculator will show an ERROR for samples that do not meet the threshold for normalizing RNA. If the threshold is not met, the MVA cannot be performed on that sample.
While care should be taken in every step of any molecular-based assay, our experience has taught us that certain steps in the MVA protocol are especially sensitive to user manipulation. Do not over-dry the RNA (Basic Protocol 1: Step 59); any moisture seen on the pellets after 10 min of drying is residual water, not ethanol. Second, be sure to frequently agitate the tubes during DNase inactivation (Basic Protocol 1: Step 73). Third, allow a minimum of 2 min incubation at each step when diluting the plasmids (Support Protocol 2). Following these steps precisely will promote a successful outcome. We also recommend making small aliquots of the primers and probes to reduce freezing and thawing. When working with the light-sensitive probes, one should cover the tubes with some form of light-blocking material, such as aluminum foil.
The MVA has been optimized for human leprosy lesion biopsy tissues (4-5 mm punch biopsy) or mouse footpad tissues fixed in 70% ethanol for at least 24 hours. Ethanol fixation kills the bacilli (Lahiri et al., 2005) and preserves the M. leprae nucleic acids (Fiallo, Williams, Chan, & Gillis, 1992). Human biopsies are best collected from the leading edge of an active leprosy lesion and should be placed in 70% ethanol immediately upon collection. We do not recommend using RNA-later as it makes the tissue rubbery, and the fixed tissue does not homogenize well.
Troubleshooting
| Problem | Possible cause | Solution |
|---|---|---|
| No DNA/RNA pellet after precipitation |
Improper fixation of tissue Inefficient precipitation Extraction failure Contamination |
Add 1.5 μl GlycoBlue during the precipitation step Obtain fresh sample and re-extract |
| No amplification of standard curve or amplification of negative controls |
Master mix prepared incorrectly Contamination |
Prepare new master mix correctly Decontaminate working area |
| Amplification of cDNA that is similar to DNA amplification | DNase failure | Perform the DNase protocol from the beginning with fresh RNA |
Understanding Results
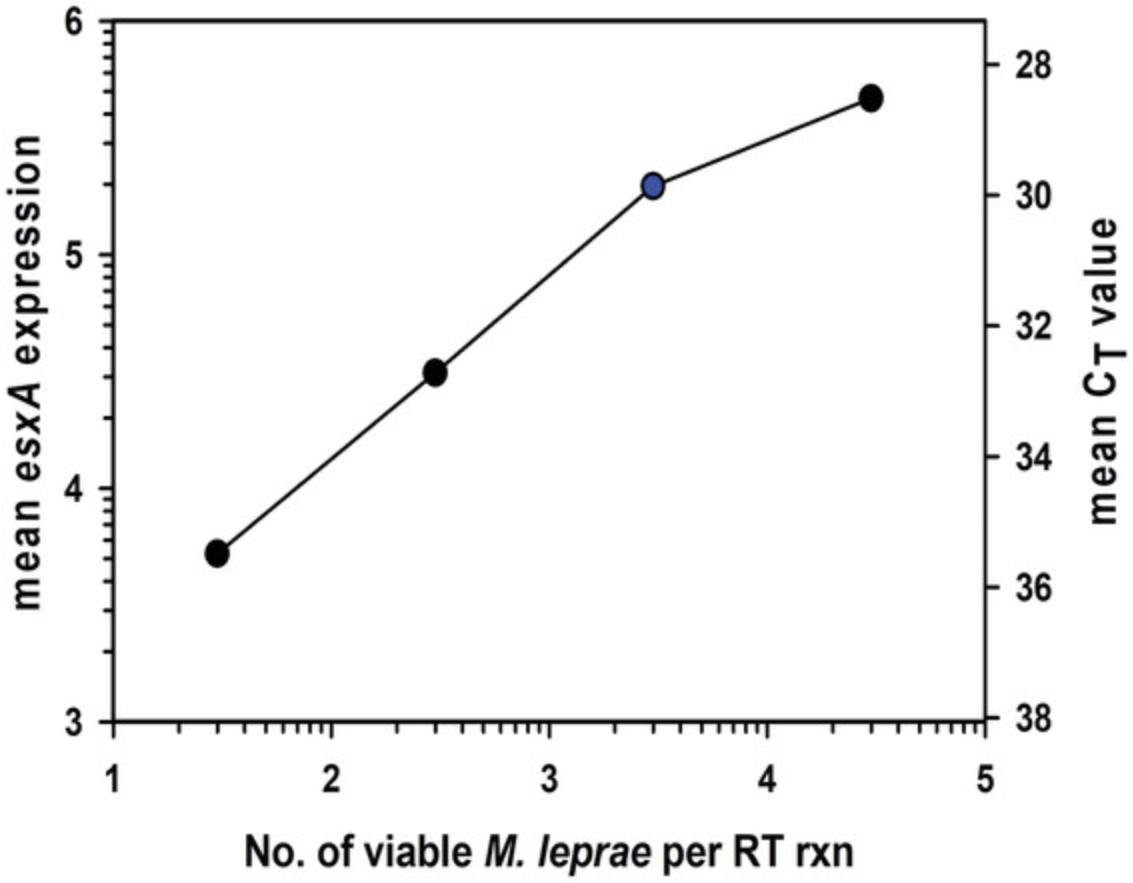
Immunocompetent BALB/c mice are quite resistant to M. leprae. If viable M. leprae are inoculated with a low dose of 5000 bacilli as in Shepard's mouse footpad assay explained above, the organisms will multiply until they reach a plateau of ∼105 to 106 per footpad. Growth ceases at this point due to immune recognition (Welch et al., 1980). If BALB/c mice are inoculated with ≥106 M. leprae per footpad, the bacilli will not multiply as the mice will be immunized. We used this high-dose mouse infection model to validate the MVA and evaluate human leprosy lesion biopsies in which viability is unknown. As shown in Figure 9, footpad tissues from BALB/c mice infected with a high dose of viable M. leprae were harvested on Day 1 (MFP-D1) and at 3 months (MFP-3m) post-infection. Human leprosy lesion biopsies were collected from untreated lepromatous leprosy patients. All tissues were fixed in 70% ethanol and stored at –20°C. DNA and RNA were harvested from the infected tissues and processed in the MVA.
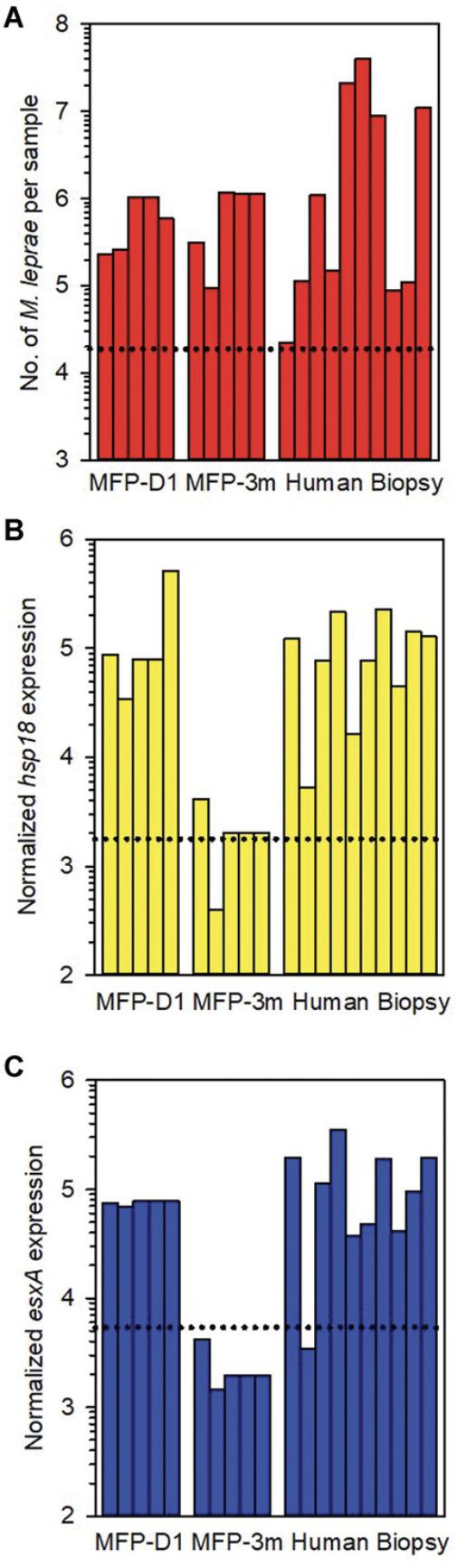
M. leprae were first enumerated on the DNA fraction by RLEP qPCR (Fig. 9A). Approximately 105 M. leprae were recovered from all mouse footpad tissues. There was much greater variability in the number of M. leprae in the human biopsy tissues. All tissues, however, had at least 1.9 × 104 M. leprae per sample (dotted line).
Each DNase-treated RNA was normalized to 3000 M. leprae equivalents based on these counts and reverse transcribed. Viability was determined by hsp18 and esxA qPCR. As shown in Figures 9B and C, M. leprae from footpads harvested on Day 1 showed high viability, whereas M. leprae from footpads harvested at 3 months had low viability. M. leprae from human biopsies had varying levels of viability.
Normalized RNA from an M. leprae population with high percent viability will yield approximately 105 hsp18 and esxA transcripts. This expression will decrease to about 103 for normalized RNA from a population of M. leprae that is only ∼1% viable. Upon examination of >300 M. leprae -infected tissues, a receiver operating characteristic (ROC) curve analysis of the results yielded a viability cut-off value of 1.785 × 103 for hsp18 expression and 5.395 × 103 for esxA expression (manuscript in preparation). These levels are depicted in Figures 9B and C, respectively. Therefore, each sample can be independently assessed for viability.
It is also important to note that there is no correlation between M. leprae counts and viability. While there was little difference in the number of bacilli at Day 1 and 3 months, the M. leprae from the former population had high viability, whereas those from the latter had very low viability. This lack of correlation also held for the human biopsy samples, further confirming the importance of the normalization step in the MVA.
Time Considerations
It is important to plan properly before starting this assay to allow enough time to reach the appropriate safe stopping points. The MVA can be completed in as few as 4 days. Working stocks of the primers and probes (Support Protocol 1) and a properly diluted standard curve (Support Protocol 2) can be made prior to beginning Basic Protocols 1-5. Basic Protocol 1 takes 2 full days to complete. Basic Protocols 3 and 4 can be completed on the third day and Basic Protocol 5 on the fourth day. It is recommended to begin this protocol at the beginning of the week to allow enough time to finish all the steps as presented.
Ethics Statement
Human leprosy lesion biopsies were collected after signed informed consent under country approvals (CLTRFI/LWM-IEC 2015-004; NHRC 108/2015) that conform to the standards indicated by the Declaration of Helsinki. Mouse footpad tissues were obtained in accordance with the United States Public Health Service Policy on the Humane Care and Use of Laboratory Animals. The NHDP Institutional Animal Care and Use Committee (Assurance #D16-00019 [A3032-01]) reviewed and approved the protocol, and experiments were conducted in accordance with The Guide to Care and Use of Laboratory Animals, Eighth Edition.
Acknowledgments
This work was supported by the Leprosy Research Initiative (LRI) and the Turing Foundation under LRI Grant number 703.15.43, and by the National Institutes of Health, National Institute of Allergy and Infectious Diseases through an interagency agreement (number AAI21005) with the Health Resources and Services Administration National Hansen's Disease Program.
Author Contributions
Jaymes H. Collins : Conceptualization, Data curation, Formal analysis, Methodology, Software, Validation, Manuscript draft, review, and editing; Shannon M. Lenz : Conceptualization, Data curation, Formal analysis, Methodology, Validation, Manuscript draft; Nashone A. Ray : Data curation, Methodology, Validation, Manuscript draft; Marivic F. Balagon : Funding acquisition, Resources, Validation; Deanna A. Hagge : Funding acquisition, Resources, Validation; Ramanuj Lahiri : Conceptualization, Formal analysis, Funding acquisition, Validation, Manuscript draft, review, and editing; Linda B. Adams : Conceptualization, Formal analysis, Funding acquisition, Project administration, Supervision, Validation, Manuscript draft, review, and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data are available on request from the authors.
Supporting Information
| Filename | Description |
|---|---|
| cpz1359-sup-0001-SuppMat1.xlsx10.9 KB | Supplementary material |
| cpz1359-sup-0002-SuppMat2.xlsx247.2 KB | Supplementary material |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Adams, L. B. (2021). Susceptibility and resistance in leprosy: Studies in the mouse model. Immunological Reviews , 301, 157–174. doi: 10.1111/imr.12960.
- Borah, K., Kearney, J. L., Banerjee, R., Vats, P., Wu, H., Dahale, S., … McFadden, J. (2020). GSMN-ML—a genome-scale metabolic network reconstruction of the obligate human pathogen Mycobacterium leprae. PLoS Neglected Tropical Diseases , 14(7), e0007871. doi: 10.1371/journal.pntd.0007871.
- Davis, G. L., Ray, N. A., Lahiri, R., Gillis, T. P., Krahenbuhl, J. L., Williams, D. L., & Adams, L. B. (2013). Molecular assays for determining Mycobacterium leprae viability in tissues of experimentally infected mice. PLoS Neglected Tropical Diseases , 7, e2404. doi: 10.1371/journal.pntd.0002404.
- Fiallo, P., Williams, D. L., Chan, G. P., & Gillis, T. P. (1992). Effects of fixation on polymerase chain reaction detection of Mycobacterium leprae , Journal of Clinical Microbiology , 30, 3095–3098. doi: 10.1128/jcm.30.12.3095-3098.
- Franzblau, S. G. (1988). Oxidation of palmitic acid by Mycobacterium leprae in an axenic medium. Journal of Clinical Microbiology , 26, 18–21. doi: 10.1128/jcm.26.1.18-21.1988.
- Hagge, D. A., Scollard, D. M., Ray, N. A., Marks, V. T., Deming, A. T., Spencer, J. S., & Adams, L. B. (2014). IL-10 and NOS2 modulate antigen-specific reactivity and nerve infiltration by T cells in experimental leprosy. PLoS Neglected Tropical Diseases , 8, e3149. doi: 10.1371/journal.pntd.0003149.
- Lahiri, R., & Adams, L. B. (2016). Cultivation and viability determination of Mycobacterium leprae. In International Textbook of Leprosy. D. M. Scollard & T. P. Gillis (Eds.). Retrieved from https://www.internationaltextbookofleprosy.org/chapter/assays-determining-viability-mycobacterium-leprae.
- Lahiri, R., Randhawa, B., & Krahenbuhl, J. (2005). Application of a viability-staining method for Mycobacterium leprae derived from the athymic (nu/nu) mouse footpad. Journal of Medical Microbiology , 54, 235–242. doi: 10.1099/jmm.0.45700-0.
- Lahiri, R, Adams, L. B., Thomas, S. S., & Penthe, K. (2021). Mycobacterium leprae is exquisitely sensitive to Telacebec. Emerging Infectious Diseases , Accepted 2021.
- Levy, L., & Ji, B. (2006). The mouse footpad technique for cultivation of Mycobacterium leprae. Leprosy Review , 77, 5–24. doi: 10.47276/lr.77.1.5.
- Lenz, S. M., Collins, J. H., Lahiri, R., & Adams, L. B. (2020). Rodent models in leprosy research. In International Textbook of Leprosy. D. M. Scollard & T. P. Gillis (Eds.). Retrieved from https://www.internationaltextbookofleprosy.org/chapter/rodents.
- Ojo, O., Williams, D. L., Adams, L. B., & Lahiri R. (2022). Mycobacterium leprae transcriptome during in vivo growth and ex vivo stationary phases. Frontiers in Cellular and Infection Microbiology. 11, 817221. doi: 10.3389/fcimb.2021.817221.
- Scollard, D. M., Adams, L. B., Gillis, T. P., Krahenbuhl, J. L., Truman, R. W., & Williams, D. L. (2006). The continuing challenges of leprosy. Clinical Microbiology Reviews , 19, 338–381. doi: 10.1128/CMR.19.2.338-381.2006.
- Sharma, R., Singh, P., McCoy, R. C., Lenz, S. M., Donovan, K., Ochoa, M. T., … Adams, L. B. (2020). Isolation of Mycobacterium lepromatosis and development of molecular diagnostic assays to distinguish M. leprae and M. lepromatosis. Clinical Infectious Diseases , 71, e262–e269. doi: 10.1093/cid/ciz1121.
- Shepard, C. C. (1962). Multiplication of Mycobacterium leprae in the footpad of the mouse, International Journal of Leprosy , 30, 291–306.
- Tongluan, N., Shelton, L. T., Collins, J. H., Ingraffia, P., McCormick, G., Peña, M., … Macaluso, K. R. (2021). Mycobacterium leprae infection in ticks and tick-derived cells. Frontiers in Microbiology , 12, 761420. doi: 10.3389/fmicb.2021.761420.
- Truman, R. W., & Krahenbuhl, J. L. (2001). Viable M. leprae as a research reagent. International Journal of Leprosy and Other Mycobacterial Diseases , 69, 1–12.
- Truman, R. W., Andrews, P. K., Robbins, N. Y., Adams, L. B., Krahenbuhl, J. L., & Gillis, T. P. (2008). Enumeration of Mycobacterium leprae using real-time PCR. PLoS Neglected Tropical Diseases , 2, e328. doi: 10.1371/journal.pntd.0000328.
- Welch, T. M., Gelber, R. H., Murray, L. P., Ng, H., O'Neill, S. M., & Levy, L. (1980). Viability of Mycobacterium leprae after multiplication in mice, Infection and Immunity , 30, 325–328. doi: 10.1128/iai.30.2.325-328.1980.
- Williams, D. L., Torrero, M., Wheeler, P. R., Truman, R. W., Yoder, M., Morrison, N, … Gillis, T. P. (2004). Biological implications of Mycobacterium leprae gene expression during infection. Journal of Molecular Microbiology and Biotechnology , 8, 58–72. doi: 10.1159/000082081.
Corrections
In this publication, the primer and probe sequences for M. leprae listed in Table 4 were updated and corrected.
The current version online now includes this correction and may be considered the authoritative version of record.
Citing Literature
Number of times cited according to CrossRef: 4
- Abigail Marie Klever, Kathleen A. Alexander, Deepak Almeida, Matthew Z. Anderson, Ray L. Ball, Gillian Beamer, Paola Boggiatto, Jane E. Buikstra, Bruce Chandler, Tiffany A. Claeys, Aislinn E. Concha, Paul J. Converse, Keith M. Derbyshire, Karen M. Dobos, Kathryn M. Dupnik, Janice J. Endsley, Mark A. Endsley, Kevin Fennelly, Carlos Franco-Paredes, Deanna A. Hagge, Luanne Hall-Stoodley, Don Hayes, Katherine Hirschfeld, Courtney A. Hofman, Jennifer R. Honda, Natalie M. Hull, Igor Kramnik, Karen Lacourciere, Ramanuj Lahiri, Elise A. Lamont, Michelle H. Larsen, Timothy Lemaire, Sandrine Lesellier, Naomi R. Lee, Christopher A. Lowry, Najmus S. Mahfooz, Temet M. McMichael, Marlena R. Merling, Michele A. Miller, Jyothi F. Nagajyothi, Elizabeth Nelson, Eric L. Nuermberger, Maria T. Pena, Claudia Perea, Brendan K. Podell, Charlie J. Pyle, Fred D. Quinn, Murugesan V.S. Rajaram, Oscar Rosas Mejia, Michelle Rothoff, Saydie A. Sago, Liliana C.M. Salvador, Andrew W. Simonson, John S. Spencer, Srinand Sreevatsan, Selvakumar Subbian, James Sunstrum, David M. Tobin, K.K.Vidya Vijayan, Caelan T.O. Wright, Richard T. Robinson, The Many Hosts of Mycobacteria 9 (MHM9): A conference report, Tuberculosis, 10.1016/j.tube.2023.102377, 142 , (102377), (2023).
- Sofie Marijke Braet, Patrícia Sammarco Rosa, John Stewart Spencer, Charlotte Avanzi, Leprosy Agents and Principal Methods of Detection, Identification, and Characterization of the Leprosy Agents, Hansen’s Disease, 10.1007/978-3-031-30893-2_5, (45-57), (2023).
- Shannon M. Lenz, Nashone A. Ray, Tsehaynesh Lema, J. Hunter Collins, Ruby Thapa, Selfu Girma, Marivic Balagon, Kidist Bobosha, Deanna A. Hagge, Diana L. Williams, David M. Scollard, Ramanuj Lahiri, Linda B. Adams, Utility of a Mycobacterium leprae molecular viability assay for clinical leprosy: An analysis of cases from the Philippines, Ethiopia, and Nepal, Frontiers in Tropical Diseases, 10.3389/fitd.2022.967351, 3 , (2022).
- Rubikah Vimonish, Janaina Capelli-Peixoto, Wendell C. Johnson, Hala E. Hussein, Naomi S. Taus, Kelly A. Brayton, Ulrike G. Munderloh, Susan M. Noh, Massaro W. Ueti, Anaplasma marginale Infection of Dermacentor andersoni Primary Midgut Cell Culture Is Dependent on Fucosylated Glycans, Frontiers in Cellular and Infection Microbiology, 10.3389/fcimb.2022.877525, 12 , (2022).

