Optimizing PCR for Mouse Genotyping: Recommendations for Reliable, Rapid, Cost Effective, Robust and Adaptable to High-Throughput Genotyping Protocol for Any Type of Mutation
Sylvie Jacquot, Sylvie Jacquot, Nathalie Chartoire, Nathalie Chartoire, Françoise Piguet, Françoise Piguet, Yann Hérault, Yann Hérault, Guillaume Pavlovic, Guillaume Pavlovic
3Rs
replacement
reduction
refinement
automation
genotyping
molecular biology
mouse model
PCR
reproducibility
Abstract
Genotyping consists of searching for a DNA sequence variation localized at a well-defined locus in the genome. It is an essential step in animal research because it allows the identification of animals that will be bred to generate and maintain a colony, euthanized to control the available space in the animal facility, or used in experiment protocols. Here we describe polymerase chain reaction (PCR) genotyping protocols for fast, sensitive, easy, and cost-effective characterization of mouse genotype. We discuss optimization of parameters to improve the reliability of each assay and propose recommendations for enhancing reproducibility and reducing the occurrence of inconclusive genotyping. All steps required for efficient genotyping are presented: tissue collection; sample verification and direct DNA lysis; establishment of a robust genotyping strategy with reliable, rapid, and cost-effective assays; and finally, transition to high-throughput automatized PCR, including mix miniaturization and automation. © 2019 The Authors.
Basic Protocol 1 : Tissue sampling methods and procedure
Basic Protocol 2 : Sample verification and DNA lysis
Basic Protocol 3 : Design of a genotyping strategy
Basic Protocol 4 : Moving to high-throughput genotyping
INTRODUCTION
Many rodent genotyping protocols are based on polymerase chain reaction (PCR) amplification of genes or genetic markers, as PCR is easy, fast, sensitive, and cost effective. Wrongly, PCR genotyping is often considered as a straightforward and easy step; in reality, however, providing robust, accurate, and fast results is frequently more challenging that it seems. Because of its high sensitivity, PCR is subject to frequent false positive results (i.e., amplification of a contaminant). Conversely, because of its low tolerance of inhibitors, frequent false negative results are also encountered (i.e., no amplification; Bacich, Sobek, Cummings, Atwood, & O'Keefe, 2011; Schrader, Schielke, Ellerbroek, & Johne, 2012). PCR can also fail to amplify certain templates, such as GC-rich sequences or secondary structures. Although it is a routine technique in mouse research laboratories, the establishment of reliable, rapid, and cost-effective genotyping protocols for every mutation is generally low on the list of priorities for scientific researchers. Researchers are very attentive to the reliability of data from their experimental protocols, but the genotype of the animals used is not always verified.
Analysis of our genotyping over a 5-year period indicated that ∼6% of the animals that were sampled and genotyped twice had different genotype outcomes (Table 1). Additionally, when phenotyping cohorts (groups consisting of six to eight animals per genotype and sex) were sampled and genotyped twice, before and after phenotyping, 30% were found to include at least one animal with a discordant genotype (data not shown). These data are consistent with the finding that over 15% of lines deposited to public repositories, such as the Mutant Mouse Resource & Research Centers (MMRRCs) and the Jackson Laboratory (JAX), do not carry the mutation specified by the depositor (Lloyd, Franklin, Lutz, & Magnuson, 2015). Inconclusive genotyping is one factor that can impact preclinical studies and basic research reproducibility, contributing to the “reproducibility crisis” (Cinelli, Rettich, Seifert, Bürki, & Arras, 2007; Picazo & García-Olmo, 2015). Moreover, genotyping errors can lead to genetic contamination of stocks and even to the extinction of a genetically unique mouse line (Lloyd et al., 2015). Cryopreservation of mutant sperm or embryos and PCR quality control of the preserved stocks are therefore essential (Scavizzi et al., 2015). Finally, the development of CRISPR/Cas9 technology has enhanced the possibility of achieving mutations in mice (Birling, Herault, & Pavlovic, 2017; Birling, Schaeffer, et al., 2017) but also required further genotyping of the resulting mouse models (Birling, Schaeffer, et al., 2017; Mianné et al., 2017). The consequences of genotyping errors and animal misidentification should not be underestimated and must be controlled through efficient and robust PCR genotyping (Bonaparte et al., 2013).
| Comparison between the two results | |||||
|---|---|---|---|---|---|
| Year | Total animals analyzed | Animals genotyped with two independent biopsiesb | Confirmed genotypes | Distinct genotypes | Inconclusive genotypes (%) |
| 2013 | 56,331 | 4140 | 3867 | 273 | 6.6 |
| 2014 | 51,309 | 4588 | 4328 | 260 | 5.7 |
| 2015 | 58,250 | 7166 | 6673 | 493 | 6.9 |
| 2016 | 50,267 | 4411 | 4152 | 259 | 5.9 |
| 2017 | 51,272 | 6900 | 6576 | 324 | 4.7 |
- a We analyzed the genotype outcomes of animals that were sampled and genotyped at least twice, for various different reasons: e.g., genotype verification before or after phenotyping experiments or before mutant line freezing or shipment.
- b The analysis included only the animals for which the results of both genotypes were interpretable (no PCR inhibition or PCR contamination observed).
Based on our expertise in standardized and high-throughput genotyping (60,000 animals per year for hundreds of different genetic markers or gene mutations), we describe here some recommendations and protocols for reproducible PCR genotyping. The establishment of a robust genotyping strategy begins with the choice of tissue to be sampled, the verification of samples before direct DNA lysis, and finally reliable, rapid, and cost-effective testing. The last part of this article details how to translate a procedure to high-throughput PCR, including recommendations for reaction mix miniaturization and automation. The four protocols presented in this paper are optimized for the most common samples used for genotyping transgenic mice: DNA in crude extracted from tail, ear, or toe tissue, with specific authorizations according to your local ethical regulations.
Basic Protocol 1: TISSUE SAMPLING METHODS AND PROCEDURE
There are several ways to obtain DNA for mouse genotyping: tail biopsy, ear or toe clipping, hair, blood, or fecal or oral samples. The method selected depends upon several parameters, including the established practice in your laboratory or institute and the quantity of DNA required for the assay. Table 2 describes the different tissues typically used as samples for genotyping and the key considerations for choosing the most appropriate sampling method.
| Choice of biopsy method depending on age of animal | |||||||
|---|---|---|---|---|---|---|---|
| Sampling method | <2 wk | 3-4 wk | >4 wk | Recommended sample size | Invasiveness of sampling | Possible repetition of sampling | Specific remarks |
| Tail biopsy | Yes | Yes | Yes | 0.3-0.5 cm | Amputation | Yes1 | 1At 14-17 days after birth, mouse tails are incompletely ossified; at >4 wk, anesthesia is mandatory to make the procedure less painful for the animal. |
| Ear punch | No | No | Yes | 0.2-cm hole | Amputation | Yes2 |
|
| Toe clipping | Yes | No | No | Distal phalanx | Amputation | No | In young mice, the ossification process is not yet complete. |
| Fecal pellet | No3 | Yes | Yes | 10-50 mg4 | Noninvasive | Yes |
|
| Blood sample | No | Yes | Yes | 20-50 µl5 | Minor | Yes6 |
|
| Hair roots | No7 | Yes | Yes | One tuft of hair8 | Noninvasive | Yes |
|
| Oral swab | Yes | Yes | Yes | 6-8 mm | Noninvasive9 | Yes |
|
Material
-
70% (v/v) ethanol
-
0.75-ml screw-cap tubes (e.g., Matrix tubes, blank; cat. no. 446015, Dutscher) and caps (e.g., Sepra Seal Cap Mats for Matrix tubes; cat. no. 446045, Dutscher)
-
Sterile compresses
-
Animals for tissue collection
-
Class II microbiological safety cabinet or changing station
-
Personal protection equipment: lab coat, gloves
-
Ear puncher for mice (e.g., cat. no. AT7020, Agnthos) or sharp surgical scissors (e.g., cat. no. 14106-09, Fine Science Tools)
-
Clean cages
Preparation of materials
1.Five minutes before starting the procedure, turn on the safety cabinet or changing station.
2.Clean the work surface with 70% ethanol.
3.Place the following equipment on the work surface: rack of screw-cap tubes, clean cage, compress soaked in 70% diluted ethanol, and scissors or ear punch.
4.Place the cage containing the animals to be sampled under the safety cabinet or changing station and open it.
5.Transfer the parents (if present) to the clean cage. Animals to be genotyped will be placed individually in the cage after each biopsy.
Preparation of sterile tools for biopsy procedure
Use of sterile tools for biopsy procedure is crucial (see Critical Parameters). The ear puncher or scissors must be sanitized using an appropriate method (e.g., with 70% ethanol).
6.Disinfect the scissors or ear puncher with the compress soaked in 70% ethanol.
7.Clean the scissors or ear puncher between mice to avoid sample contamination.
Sampling procedure
8.Check that the tube in which the sample will be placed is clean and correctly labeled.
9.Manually restrain the mouse between thumb and forefinger.
10.Using the sanitized sharp scissors or ear puncher, precisely excise a piece of tissue, of homogeneous size relative to other samples (appropriate sizes: tail biopsy, 5 mm; ear punch, 2-mm hole; toe clipping, one distal phalanx; see Critical Parameters).
11.Place the sampled pup in the clean cage with its parents.
12.For each mouse, place the sample into the corresponding tube.
13.Close the tube and check that the biopsy is at the bottom of the tube.
14.Once all mice have been sampled, the tubes can be stored at −20°C for later genotyping.
Basic Protocol 2: SAMPLE VERIFICATION AND DNA LYSIS
There are many alternative protocols that can be used to prepare samples for PCR. They extend from the use of a raw lysate to a variety of purification protocols (e.g., organic extraction with phenol/chloroform or silica column) designed to remove contaminants and inhibitors. Purification protocols suffer the disadvantages that they are expensive, may have low performance (Miller, Bryant, Madsen, & Ghiorse, 1999), and are quite difficult to implement on a workstation. In contrast, direct lysis methods are quick, easy, and inexpensive, but do not remove inhibitors, and therefore cannot be used for all applications. Here, we will describe the use of DirectPCR Lysis Reagents with the addition of proteinase K for routine DNA isolation. In our hands, this approach provides a low level of PCR failure for standard PCR genotyping (Table 3, percentage of failed genotype) and good DNA yields (Table 3, average quantity of DNA obtained).
| Sampling methoda | Extraction method | Average quantity of DNA obtainedb | Percentage of failed genotypingc | Easily adaptable to high throughput |
|---|---|---|---|---|
| Tail biopsies | Direct PCR lysis | 26 ± 4 ng/μl | 2.6 | Yes |
| Ear punching | Direct PCR lysis | 34 ± 5 ng/μl | 2.8 | Yes |
| Toe clipping | Direct PCR lysis | 20 ± 2 ng/μl | 3.3 | Yes |
- a
These three kind of tissue biopsies give enough DNA for PCR genotyping. Their performance in PCR is quite similar as the percentage of failed genotyping is very similar, indicating that similar levels of inhibitors are present in the crude extracts. Data were collected from 27,070 tail, 8470 ear, and 2900 toe biopsies to estimate genotyping failure.
- b
Amount of isolated DNA was determined by spectrophotometry with a NanoDrop ND-1000 system (N = 5).
- c
Percentage of biopsies that could not be genotyped (2018 data) because results were not interpretable. Causes are multifactorial and include uncalibrated biopsy, inefficient lysis, and presence of inhibitors in the crude extract.
Materials
-
Tubes containing mouse tissue samples (Basic Protocol 1)
-
DirectPCR Lysis Reagent (Mouse Tail; 102-T, Viagen)
-
10 mg/ml proteinase K (dissolve 1 g lyophilized proteinase K powder [cat. no. P6556, Merck] in 100 ml water to obtain a clear solution; if desired, store aliquots in 1.5-ml tubes (e.g., cat. no. 72.690001, Sarstedt) at −20°C)
-
Manual pipets
-
Centrifuge (e.g., Allegra 25R centrifuge Beckmann Coulter)
-
Heated water bath (e.g., GLF 1083), 85°C
-
Heating oven (e.g., Memmert), 55°C
-
Personal protection equipment: lab coat, gloves
Visually verify each tube to be processed
1.Check that only one sample is present in the tube.
2.Visually check that the size of the tissues corresponds to the recommended size (see Table 2 for recommended sizes). If not, mark the tube for later adjustment of the lysis buffer volume.
Prepare buffers
3.Prepare a premix lysis buffer: Add 200 μl DirectPCR Lysis Reagent (Mouse Tail) and 6 μl 10 mg/ml proteinase K solution (10 mg/ml) per reaction.
Incubate and lyse sample
4.To each sample, add 200 μl premix lysis buffer for a 0.5-cm tail biopsy or 100 μl for a 0.2-cm ear punch or toe biopsy.
5.Hermetically seal the tubes.
6.Centrifuge tubes 2 min at 4000 × g , room temperature.
7.Check that the biopsies are at the bottom of the tube and covered by the solution.
8.Incubate overnight at 55°C in a heating oven.
9.Remove tubes and check that they are still hermetically sealed.
10.Shake vigorously by turning over.
Deactivate proteinase K
11.Incubate tubes at 85°C in a heated water bath for 45 min to 1 hr to inactivate the proteinase K.
12.Centrifuge tubes 2 min at 4000 × g , room temperature.
13.Store tubes at 4°C.
Basic Protocol 3: DESIGN OF A GENOTYPING STRATEGY
A genotyping assay must provide a rapid and cost-effective method of identifying animals. It also needs to be reliable and robust, as it will be used to select mutant animals for experiments and for ensuring the integrity of a unique genetic resource (i.e., animals for future production, for cryopreservation, or to be received by a collaborator or resource).
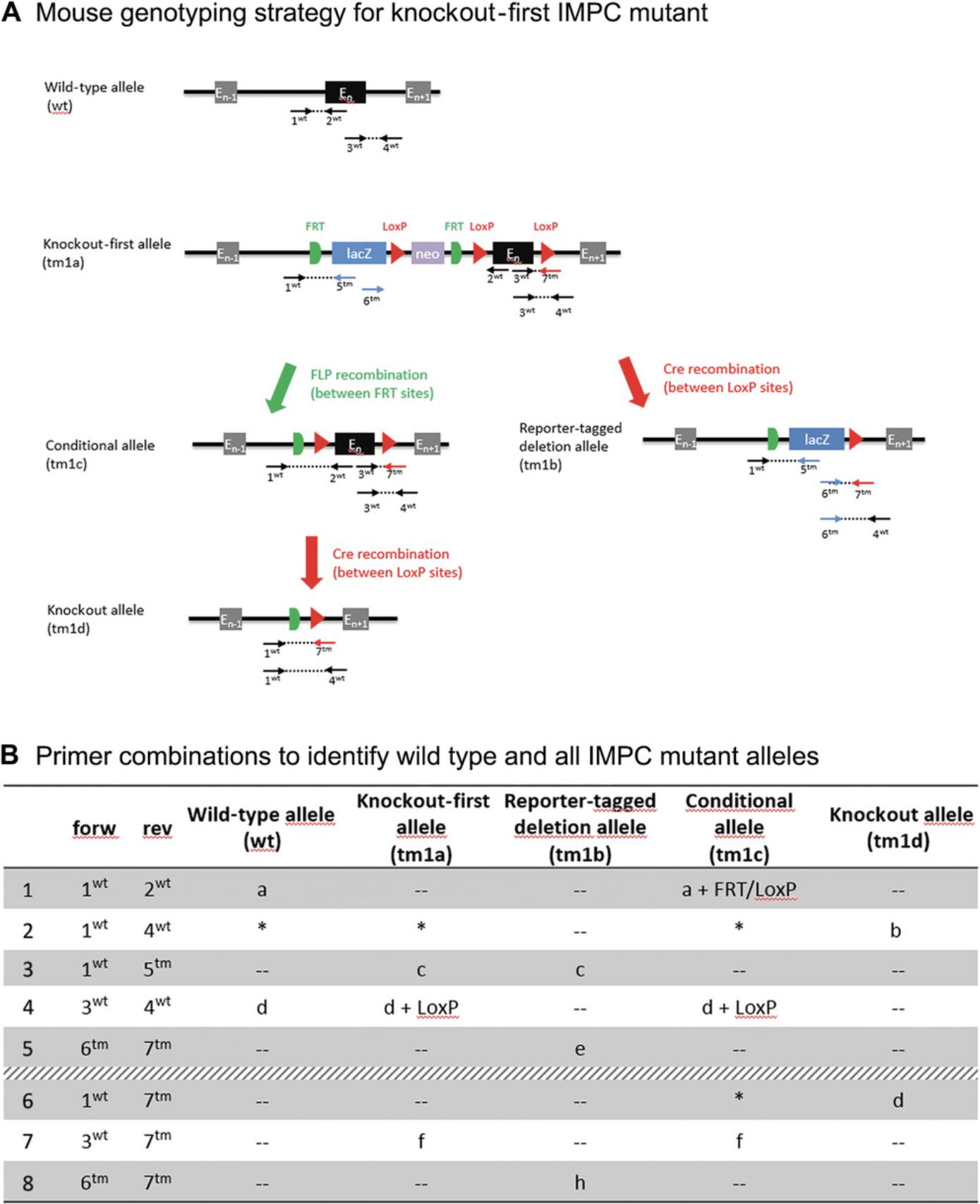
Designing genotyping assays around markers that are found in many transgenic lines, such as the neomycin selection marker, GFP, or Cre, is discouraged as it does not verify that you are using the anticipated mutant line and is not suitable for genotyping double-transgenic strains that contain common transgenic markers. Instead, it is recommended that the PCR assay specifically identify each line and each possible allele derived from the stock mutant line. For example, the International Knockout Mouse Consortium is creating a catalog of mammalian gene function (Meehan et al., 2017) and reports the generation of over 5000 new mouse mutant strains all harboring a lacZ reporter cassette and providing conditional inactivation potential. An optimized protocol allows researchers to evaluate all possible genotypes and the integrity of the targeting event (Fig. 2A).
Defining PCR design rules that can be applied to all protocols (see Troubleshooting) is recommended. This simplifies genotyping because specific conditions (reaction mix, thermocycling program, agarose gel analysis) are not required for any project, and it thus allows multiple mutant lines to be assayed in the same experiment. This strategy is especially recommended when large volumes and/or high throughput are involved.
Materials
-
Biological material: aliquot from crude sample lysate, to be used as template for PCR typing
-
Primers for mouse-specific target of interest
-
Master mix: e.g., FastStart PCR Master (50 ml; cat. no. 4710452001, Merck)
-
20× speed buffer (SB), prepared as described by Zhang, Wang, & Wang (2011) using boric acid (cat no. 5935, Euromedex), sodium hydroxide (cat. no. 06203, Merck), and ethidium bromide (cat. no. EU0170, Euromedex)
-
Water, PCR grade (for primer dilution and reaction fill-up)
-
Adjuvants (optional): 5% (v/v) dimethyl sulfoxide (DMSO; e.g., cat. no. D8418-100ML, Merck), 5% (v/v) glycerol (e.g., cat. no., 15524-1L-R, Merck), or 0.5 µg/µl bovine serum albumin (BSA; e.g., cat. no. B9001S, New England Biolabs) in final reaction volume
-
Agarose, DNA grade (e.g., cat. no. D5, Euromedex)
-
Homemade loading dye stock (prepared by dissolving 3 ml glycerol and 8 mg bromophenol blue in 7 ml H2O)
-
DNA molecular weight marker: e.g., GeneRuler 50-bp DNA Ladder (cat. no. SM0372, Thermo Fisher Scientific)
-
Appropriate restriction enzymes with buffers recommended by suppliers
-
0.75-ml screw-cap tubes (e.g., Matrix tubes, blank; cat. no. 446015, Dutscher) and caps (e.g., Sepra Seal Cap Mats; cat. no. 446045, Dutscher)
-
1.5-ml Microtubes (e.g., cat. no. 72.690001, Sarstedt)
-
Computer with software for sequence analyses and tools for primer design, either commercial (e.g., Vector NTI, SnapGene, Geneious) or free (e.g., U-GENE, BioEdit, SeaView)
-
4titude FrameStar 96-well plates (cat. no. 44760, Dutscher) or 0.2-ml PCR tubes
-
Centrifuge for microcentrifuge tubes (cat. no. 016000, Dutscher), optional
-
PCR machine (e.g., Eppendorf thermal cycler)
-
Agarose gel electrophoresis equipment: Tank for electrophoresis, support where the gel is poured, combs for forming wells (where samples are deposited)
-
Acquisition imager using UV light as excitation source (e.g., U:Genius, Syngene)
-
Personal protection equipment: lab coat, gloves, safety glasses
-
Manual pipets
Search for targeted and wild-type sequence maps
1.Obtain from mouse provider the sequences of the wild-type and mutant alleles.
2.Verify that the wild-type sequence you have matches the latest release of the Mus musculus C57BL/6J reference genome assembly in NCBI (https://www.ncbi.nlm.nih.gov/genome?term=mus%20musculus) or Ensembl (https://www.ensembl.org/Mus_musculus/). If it does not, you will need to carefully check the wild-type sequence provided.
3.Use genomic sequence analysis software (such as commercial Vector NTI, SnapGene, Geneious or free U-GENE, BioEdit, or SeaView software) to construct all the sequence maps for each of the alleles you need to genotype.
Design of genotyping primers
Primers should be designed to fit the targeted sequences. It is possible to design primers using a variety of tools (genomic sequence analysis software), or even by eye following the rules below.
4a. Primer structure:
1.20-24 nucleotides in length. 2.G or C at 3′ end.
The bases G or C at the 3′ end serve as the starting binding site for the DNA polymerase.
3.40%-60% GC composition. 4. Comparable melting temperatures (T m) for both primers.
Comparable Tm (within 5°C of each other) will determine the stability of the hybrids once the match between primers and matrix is achieved.
5.Specific to the appropriate genomic DNA sequence.
Check the specificity of each primer against genomic DNA with NCBI Nucleotide blastn on the Mus musculus genome only (https://blast.ncbi.nlm.nih.gov/Blast.cgi). You do not need to modify any algorithm parameters or program selection (i.e., use megablast).
The specificity of a primer set is related to whether the primers will bind only to the sequence that we want to detect or also to additional sequences.
CAUTION: It is not necessary that there be 100% homology between primers and a genomic sequence for nonspecific PCR amplification to occur. Primers with a few mismatches or with a nonspecific 5′ end can give rise to nonspecific amplifications.
6.No internal secondary or primer-primer annealing structures.
Internal secondary structure can be checked by using primer design software (e.g., OligoArchitect from Sigma-Aldrich) to analyze duplex formation.
Primer pairs should lack significant internal secondary structure to avoid internal folding. Primer-primer annealing caused by homology within the primer pair creates primer dimers and disrupts the amplification process and is thus to be avoided.
4b. Amplicon structure: 100-500 bp amplicon size.
5.Select the best primer position on the wild-type or mutant(s) allele(s).
6.Optional : Detect mutant alleles with single-nucleotide polymorphism (SNP) mutations.
7.Verify that each PCR amplicon produces a size that can be readily separated on 2% agarose gel.
8.Order primers from any provider.
9.Dilute primers to 100 µM in PCR-grade H2O and store at −20°C.
Setup of polymerase chain reaction
The PCR strategy described below should be tested on three to five samples. Ideally, a biopsy from mutant animals is used for the test PCR. If no biopsy is available, embryonic stem cell clone DNA diluted in crude extract, targeting vector diluted in crude extract, or chimera biopsy can be used. A tissue biopsy from a wild-type animal should always be included as a control. Likewise, PCR reactions with no template act as a negative (or water) control to confirm the absence of PCR contamination.
10.Centrifuge tubes containing biological samples for 2 min at 4000 × g , room temperature, to sediment debris.
11.Prepare the PCR reactions as described below, either in a sterile 1.5-ml tube for a few samples or in a 96-well plate for larger sample numbers.
- Add 14 µl master mix (FastStart PCR Master, Roche) per tube or well.
Using a premade mixture of the enzyme, dNTPs, and reagents, such as FastStart PCR master, minimizes errors and contamination risk and reduce the time for PCR preparation.
-
Add 0.2 µl each of 100 µM forward and reverse primers (from step 9) per tube or well.
-
Add 7.6 µl sterile water per tube or well.
12.To each 0.2-ml PCR tube or each well of a 96-well plate, add 3 µl crude extract and then 22 µl PCR reaction mix.
13.Mix thoroughly by gently pumping the plunger of a micropipet up and down two or three times.
14.Prepare a negative control: Add all reagents with the exception of the DNA template (increase the water to compensate for the missing volume).
15.Seal the tubes or plate.
16.Centrifuge the reaction mixture briefly so that it falls to the bottom of the tube or plate.
17.Insert PCR tubes or plates into the thermal cycler and begin PCR program following the parameters described in Table 4.
| Temperature | Time | Number of cycles |
|---|---|---|
| 95°C | 4 min | 1 |
| 94°C | 30 s | |
| 62°C | 30 s | 34 |
| 72°C | 1 min | |
| 72°C | 7 min | 1 |
| 20°C | 5 min | 1 |
- a
These cycling conditions work well with our protocols (Basic Protocols 3 and 4) but may require modification if other conditions (e.g., other Taq polymerase) are used.
Image acquisition and analyses
PCR amplicons are separated using a 2% agarose gel and the results are visualized using a digital camera.
18.Dilute 20× concentrated SB stock to 1× to be used to make and run the gel.
19.Prepare a 2% (w/v) gel agarose containing 1× SB. Before polymerization, wearing gloves and safety glasses, add 15 µl 10 mg/ml ethidium bromide stock per 600 ml of agarose gel in the solution. Shake slowly without making bubbles.
20.To prepare PCR samples for migration, add 5 µl homemade loading dye to 15 µl of PCR reaction.
21.Carefully load samples into the wells of the gel.
22.Run the gel at 280 V (400 mA, 100 W) until the dye line is ∼80% of the way down the gel.
23.Use a digital camera (U:Genius) to visualize the DNA fragments.
Assay validation
24.Check that each amplicon band corresponds to the expected size for each assay. Verify absence of additional bands.
If yes, the PCR setting is validated, and the primer pairs should be selected for genotyping.
If not, new primer sets should be tested.
Additional steps for SNP mutation detection via diagnostic digest
If the wild-type and mutant PCR amplicon are of the same size (see step 6), PCR product cleavage with restriction enzyme can be performed to differentiate the two amplicons.
25.Verify the presence of DNA amplicons after PCR by electrophoresis as detailed in steps 18-23 but using only 5 µl of the PCR reaction in step 20.
26.Prepare the digestion reaction mix, either in sterile 1.5-ml Microtubes for few samples or in a 96-well plate.
- Add components in the following order: 2.5 µl of the 10× buffer supplied with the enzyme, 1 µl restriction enzyme, and 11.5 µl water.
The amount of restriction enzyme you use for a given digestion will depend on the amount of DNA you want to cut. By definition, one unit of enzyme will cut 1 µg DNA in a 50-µl reaction in 1 hr. Reactions are often performed with 0.5-1 µl enzyme.
-
Add 10 µl of PCR reaction.
27.Incubate tubes at digestion temperature (usually 37°C) for 1 hr.
28.Visualize the digested PCR products by migration on an agarose gel as described in steps 18-23.
Basic Protocol 4: MOVING TO HIGH-THROUGHPUT GENOTYPING
All classical genotyping methods have relatively low throughput. Automating genotyping through the use of a workstation allows parallel genotyping of a large number of genetic modifications, at many genetic loci, in many individuals. In addition to improving throughput, automation reduces the potential for contamination and error by limiting pipetting steps and preventing tube switching. The conventional endpoint PCR method is easily adapted for automation and is indeed an effective, proven, and affordable method for high-throughput screening. Moreover, PCR assays can be miniaturized in 384-well plates on a workstation, reducing the cost of animal genotyping.
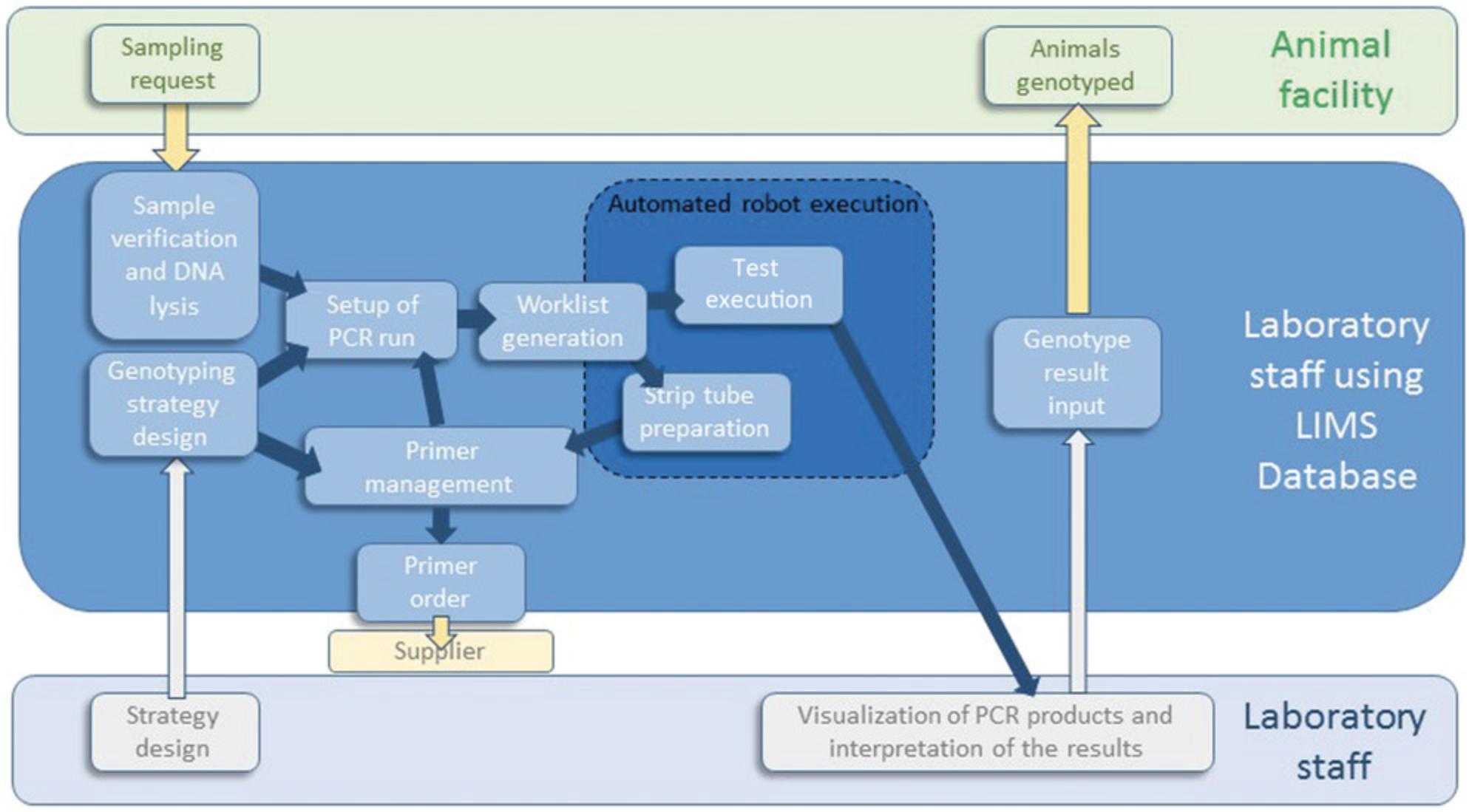
Developing a Laboratory Information Management System (LIMS) to effectively manage workstations, samples, and associated data is also essential (Fig. 3; Critical Parameters).
Materials
-
Biological material: Crude sample lysates in 96-tubes racks
-
Primers for mouse-specific target of interest
-
Water, PCR grade (for primer dilution and reaction fill-up)
-
FastStart PCR Master (50 ml; cat. no. 4710452001, Merck)
-
Adjuvant: 5% (v/v) DMSO (cat. no. D8418-100ML, Merck), 5% (v/v) glycerol (cat. no. 15524-1L-R, Merck), or 0.5 µg/µl BSA (cat. no. B9001S, New England Biolabs)
-
Workstation or liquid handler (e.g., Freedom EVO200, Tecan, or STARplus, Hamilton Microlab)
-
4titude FrameStar 384-well PCR plates (cat. no. 384 44751, 4titude)
-
8-Strip PCR tubes with caps (e.g., cat. no. 016000, Dutscher)
-
15-ml conical tube (e.g., cat. no. 352097, Corning)
-
Integrated centrifuge (e.g., Sias)
-
Integrated heat sealer (e.g., PlateLoc, Agilent
-
Sealable film clear seal (cat. no. 4Ti-0542, 4titude)
-
PCR thermocycler with motorized heated lid (e.g., T-robot, Biometra)
-
Manual pipets
Design of genotyping strategy and assay validation
1.Follow steps 1-9 of Basic Protocol 3 to design a high-throughput genotyping strategy.
2.For assay validation, follow steps 10-24 of Basic Protocol 3.Use conditions described in Table 5 for 384-well plates.
| Reagent | Volume for 384-well plate | Volume for 96-well plate |
|---|---|---|
| FastStart PCR Master (Roche) | 7.5 µl | 14 µl |
| Crude extract | 1.5 µl | 3 µl |
| 5′ primer (100 mM) | 0.06 µl | 0.2 µl |
| 3′ primer (100 mM) | 0.06 µl | 0.2 µl |
| Sterile H2O | Up to 15 µl | Up to 25 µl |
Prepare the automatized PCR run
We present here the protocol used on our two workstations (the Tecan Freedom EVO 200/8 and the Hamilton Microlab STARplus). This protocol must be adapted to each installation according to your particular workstation specifications.
3.Turn on the workstation, the associated computer, and all integrated instruments.
4.Open the software controlling the workstation (e.g., Freedom EVOware for Freedom EVO 200/8 Tecan or VENUS software for STARplus Hamilton).
5.Initialize the instrument.
6.Flush the instrument.
7.Load or generate the worklist.
-
If you have developed a LIMS to manage your sample (see Critical Parameters), load your worklist using the software controlling the workstation.
-
If not, generate the worklist as indicated by the workstation provider.
Load reagents, tubes, and plates onto the robot worktable
A visualization of the worktable (in worktable windows) is generated by the software controlling the workstation following step 7.This windows represents the working surface (deck) of the instrument. Follow the indications on the screen to place all the reagents, tubes, and plates.
8.Place all 96-tube racks containing crude extract on the workstation as indicated by the worktable windows.
9.Position the primer sets as described in the worktable windows.
-
Prepare 12-24 tube strips per mutant line and store them at −20°C.Add 6.5 µl each of 100 mM forward and reverse primers per microtube.Supplement with 187 µl sterile water per microtube.Repeat this step for all PCR sets used for genotyping the mutant lines (up to eight PCRs per mouse line).
-
Place all 8-tube strips needed for the PCR run on the workstation as indicated by the worktable windows.
10.Prepare the reaction mix.
-
Add 3.3 ml FastStart PCR Master and 1.7 ml sterile water in a 15-ml conical tube for each 384-well PCR plate to be generated during the run. An extra 15% of reaction mix is included to accommodate pipetting loss.
-
Place the 15-ml tube as indicated on the worktable windows.
The reaction mix volume is based on the total number of samples, number of PCR amplifications to be conducted per sample, and required reagent dead volumes.
11.Place 384-well PCR plates in the indicated locations.
Set up a PCR run on an automated workstation
12.Start the appropriate pipetting script (e.g., method for PCR with 384-well plates and sealing).
13.Visually check that the pipetting process has started correctly before leaving the workstation.
In our configuration, the liquid handler will distribute to each well of the plate, in order:
-
11.5 µl of reaction mix;
-
2 µl of the relevant primer set;
-
1.5 µl of the relevant sample crude extract.
This pipetting order (reaction mix then primers and sample crude extracts) is optimized to reduce pipetting steps and duration, decontamination steps (bleaching), flushing, and risk of contaminations.
Post-run process
14.Each 384-well plate is automatically sealed, then centrifuged (2 min at 4000 × g) and placed in a thermocycler by the gripper arm. The liquid handler software controls the thermal cycler and starts the thermal cycling program (program parameters are described in Table 4).
15.At the end of a run, it is possible to perform a visual inspection to check that all the wells of the plate have the same and expected volume.
Image acquisition and analyses
16.Follow steps 18-23 of Basic Protocol 3.
Decontamination
We recommend thoroughly decontaminating the instrument at the end of each week either by pipetting a 10% diluted bleach solution or by using a UV lamp.
COMMENTARY
Critical Parameters
Age of animals for biopsy
Table 2 shows the age(s) at which each tissue sampling method can be used. Sampling should, where possible, be done on young animals for the reasons listed below:
- DNA from tissues of young mice is more optimal for genotyping than that from older animals (Picazo & García-Olmo, 2015).
- In a newborn mouse (particularly before 12 days of age), discomfort due to the sampling is reduced because the sampled tissue is not fully ossified and because the nociceptive stimulus may not result in the conscious perception of pain due to the lack of a competent pain pathway at this age (Hankenson, Garzel, Fischer, Nolan, & Hankenson, 2008; Silverman & Hendricks, 2014; Wever, Geessink, Brouwer, Tillema, & Ritskes-Hoitinga, 2017).
- If genotyping is completed before weaning, extra animals or those of nondesired genotype can be sacrificed before separation of young animals into different cages.
Tail biopsy is ideally performed between ∼10 and 21 days of age. At this age, the distal tail is not fully ossified in most mouse strains, making the procedure less painful and reducing the likelihood of complications (Hankenson et al., 2008). After 4 weeks of age, anesthesia is mandatory for this procedure. Removal of more than 5 mm of tail must be avoided, as the bone is thicker in more proximal parts of the tail and this increases the likelihood of causing tissue trauma and suffering.
Ear clipping (also ear notching or ear punching) should not be carried out on mice <2 weeks of age because the ear is not yet fully developed and the removal of even a small piece of tissue can represent a significant proportion of the pinna. Ear clipping is the preferred sampling method for animals >4 weeks of age because it produces little discomfort in older mice (Picazo & García-Olmo, 2015) and therefore does not require anesthesia. During sampling, care should be taken not to accidentally drop or lose the very small ear tissue sample.
Toe clipping consists of the removal of the distal phalanx of a neonatal animal as a means of identification. This can be used as a source of sample for genotyping. Toe clipping of very young mice (ideally up to postnatal day 7) is an acceptable method as the ossification process is not yet completed and the peripheral nervous system is not yet fully myelinated (and thus the nerve conduction of pain is dramatically reduced; Dorit et al., 2001; Lee et al., 2012).
Sample calibration
Sample calibration is verified by visual inspection of each tube (see Basic Protocol 2, step 2).
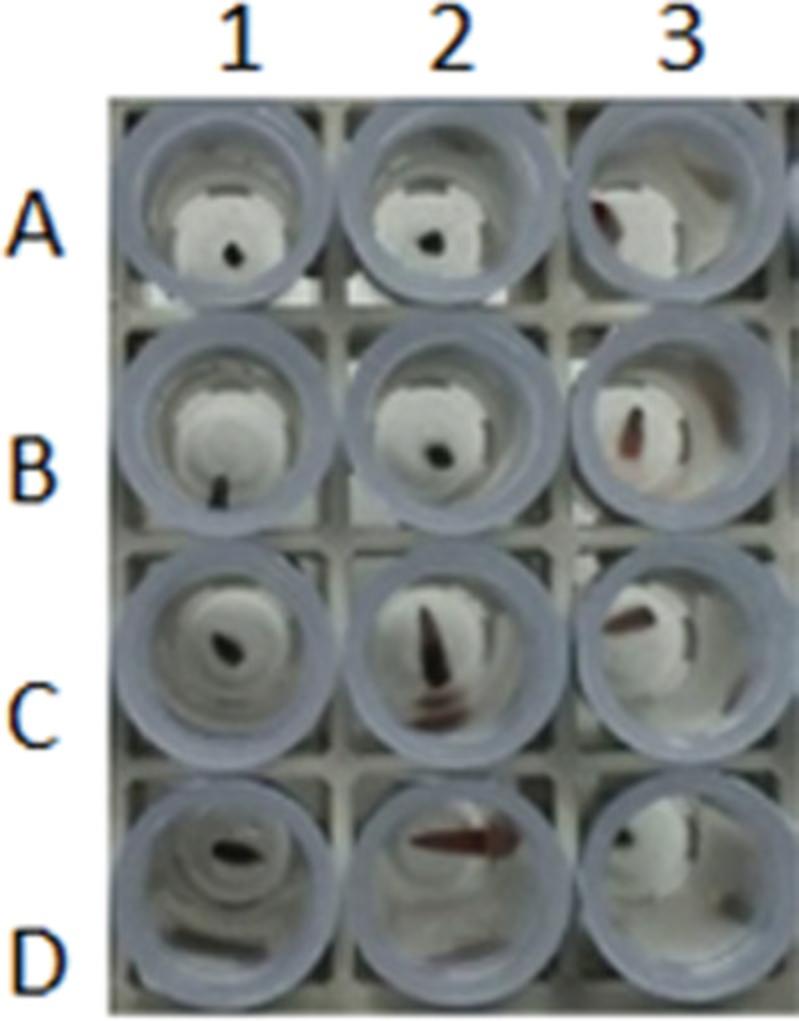
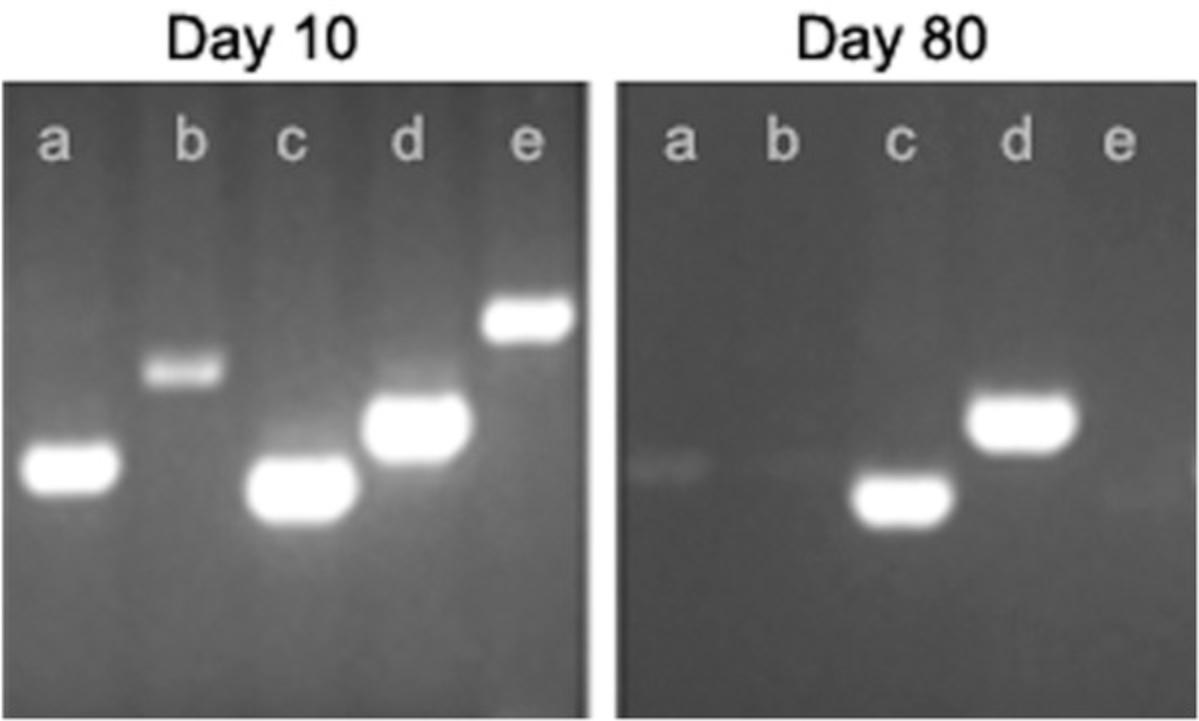
Ear punching (0.9% total oversized samples; 2018 data corresponding to more than 40,000 samples visually checked) and toe clipping (1.0% oversized samples) provide more calibrated samples than tail biopsies (8.7% oversized samples), as shown in Figure 1. Respecting recommended and calibrated sample size reduces the risk of false negative results due to either increased inhibitors or insufficient template concentration.
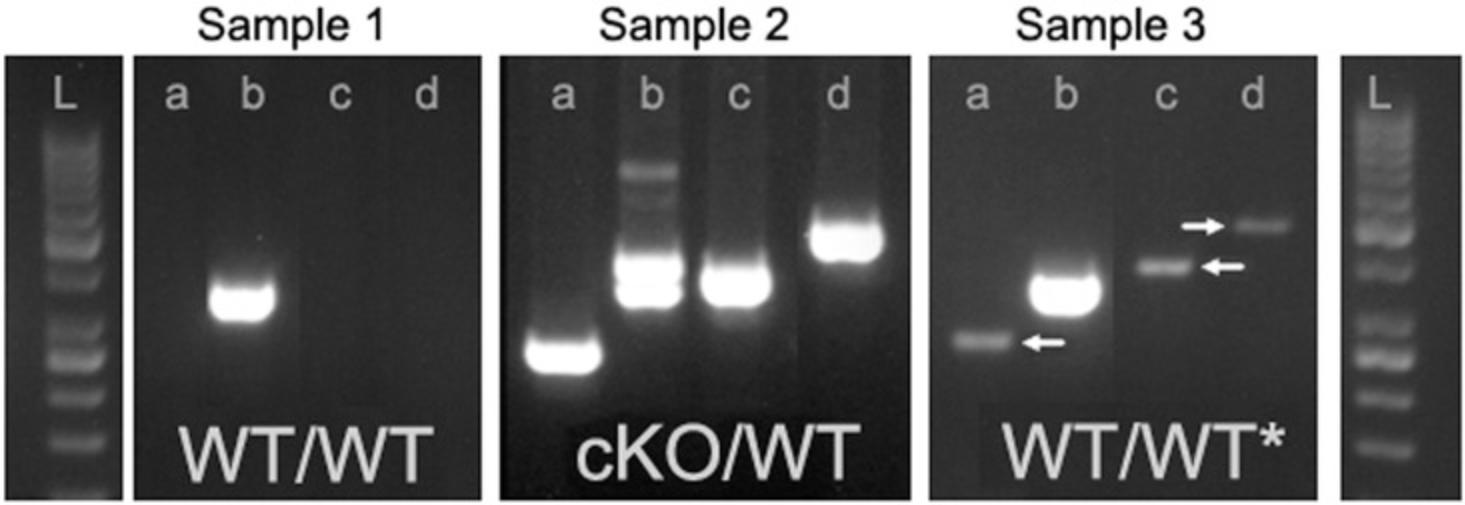
Impact of improperly cleaned instruments
When collecting tissue samples, the instruments must be cleaned between individual animals to avoid cross-contamination of genetic material. As shown in Figure 4, false positive PCR amplification can result from sample cross-contamination.
Sample storage
Tail, ear, or toe biopsies contain DNase that will slowly reduce the quantity and integrity of genomic DNA after tissue collection (Al-Griw et al., 2017). To slow this process, samples are usually stored at −20°C. However, native high-molecular-weight DNA is not required to amplify a target sequence by PCR: as only the target sequences are required to be intact, partially degraded or denatured DNA could be successfully used for PCR applications (Wever et al., 2017). From our experience, tissues stored at room temperature can be used if storage is for <24 hr and temperature does not exceed 20°C (for example, avoid shipment of samples at room temperature in summer).
Quality check of each tube before lysis
Before starting lysis, we recommend each tube be checked for absence of (or presence of additional) biopsy or oversized samples (see guide in Table 6). This quick quality check will reduce inconclusive genotyping (see section on error rate, below) and allow you to adapt the lysis buffer volume if needed.
| Observation | Possible cause | Solution |
|---|---|---|
| More than one sample in tube | Toe biopsy cut for identification of the animal remained stuck to the scissors and fell into the tube with the sample to be genotyped | Remove the incorrect sample (if can be distinguished: e.g., toe biopsy in a tail sample) or ask for a new biopsy. |
| An earlier sample was mistakenly placed in the tube |
|
|
| No sample in the tube | Sample tissue was accidentally dropped or lost | Discard the tube or even the whole genotyping sample set, if there is any doubt about tube switching, and ask for a new biopsy. |
| Type of biopsies in the tube different from what is mentioned by the animal facility | Writing error occurred | Modify the type of biopsy indicated on the genotyping request and add the corresponding premix lysis buffer. |
| Uncalibrated biopsy: Sample too large | Part of the ear was torn off | Cut the sample to obtain a calibrated size or adjust the premix lysis buffer. |
| Tail biopsy is too big (Fig. 1) | ||
| Uncalibrated biopsy: Sample too small | Tail biopsy is too small | Adjust the premix lysis buffer. |
| Tail biopsy is from embryonic day 8.5 mouse |
Error rate
Inconclusive or incorrect genotyping can occur for a variety of reasons, including errors during the genotyping procedure itself and the misidentification of samples or animals. Genotyping errors can result in irreproducible results and genetic contamination or loss of a mouse line. They are sometimes detected because of inconsistent Mendelian patterns in pedigrees (i.e., the observed genotypes are not consistent with the transmission pattern). In these cases the parents are biopsied again to confirm their genotypes.
Genotype verification is advised for all key animals: that is, before or after phenotyping experiments or freezing or shipment of mutant lines (Fig. 5). Without verification of the genotype during a phenotyping study, in our experience 30% of the cohorts will have at least one animal whose genotype does not correspond to the one expected (data not shown). Likewise, >15% of lines deposited to public repositories do not carry the mutation specified by the depositor (Lloyd et al., 2015).
How to design a genotyping strategy with minimal sequence information
To ensure accurate assay design, it is crucial to know the precise sequences of the wild-type and mutant alleles. The sequence of the wild-type allele can easily be retrieved from the Ensembl or NCBI databases. However, the sequence of the mutant allele is not systematically provided when you receive a new mutant model. Where possible, insist on obtaining this key information from the researcher who generated the mouse.
In cases where you do not know the mutant allele sequence, you may have received a genotyping protocol. You can create a hypothetical mutant map by aligning the primers from this protocol onto the wild-type sequence, which will allow you to determine the position of modified sequences (such as loxP sites or selection cassettes). Additionally, the PCR products can be sequenced to obtain a partial mutant sequence. If necessary, a new assay can be designed once this information is available (see Basic Protocol 3 for recommended design strategy). Sequencing the mutant allele PCR product can also provide quality control of the mutant model (presence of an SNP mutation or loxP site).
If the primers provided do not give you accurate information on the genomic structure of the targeted allele, you will only be able to design your strategy to target marker sequences, such as the neomycin selection marker, GFP, or Cre. These sequences are very frequent in many genetically modified mouse strains, meaning that you will not be able check reliably for animal misidentification (critical parameters) or genotype double-transgenic models.
Genotyping assay optimization
The genotyping protocol presented here is very robust in most cases. Of >4000 different PCR primer sets tested in our lab, only 9% were not validated (data not shown). Instead of changing PCR parameters when a PCR primer pair does not work, we advise first testing a different primer pair.
It is often possible to retrieve the genotyping protocol from the researcher who generated the mouse. This protocol can of course be used for genotyping. As it will be necessary to optimize it to adapt this protocol to your laboratory conditions, however, we recommend instead designing a new genotyping protocol according to the standardized parameters we described here. This will allow you to verify the line you received with different PCRs. The PCR design rules we describe here can also be applied to any mutant line. If you systematically follow them, all steps from DNA preparation to gel agarose analysis will be standardized and can thus be performed in parallel for all your mutant models, saving time and effort.
The ramping conditions of your PCR thermocycler is a critical parameter. The ramping conditions used here are a heating rate of 4°C/s and a cooling rate of 3°C/s. Differences in ramping conditions between two thermal cyclers can explain why the same PCR protocol may work or fail in different laboratories.
Optimizing PCR for complex template (GC or AT rich template, repeated sequence, secondary structure)
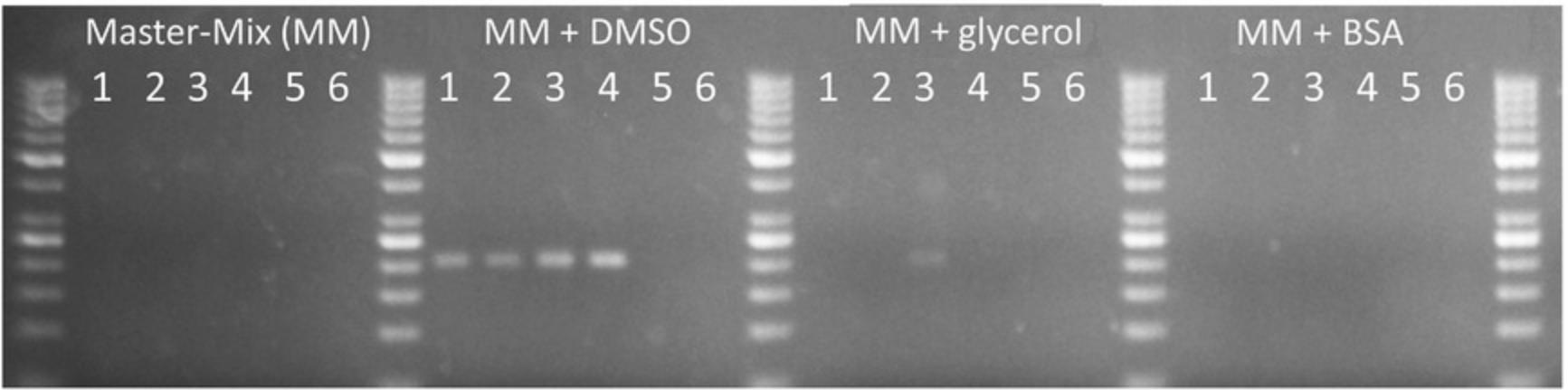
There are many parameters to optimize in a PCR, such as magnesium, dNTP, and Taq concentrations, as well as cycling conditions. Optimization of such parameters is described, for example, by Lorenz (2012). Our approach is to keep the PCR protocol as standard as possible. We therefore propose, as a first step, merely trying different additives, such as DMSO, glycerol, or BSA. Such adjuvants improve PCR amplification efficiency and specificity. Figure 6 illustrates an instance in which adding 5% (v/v) DMSO substantially improved the amplification of a GC-rich region. Multiple additives usually need to be tested for a complex template, as the most efficient additive will depend of the sequence to be amplified and cannot be anticipated.
Presence of inhibitors in reaction or template too concentrated (oversized biopsy)
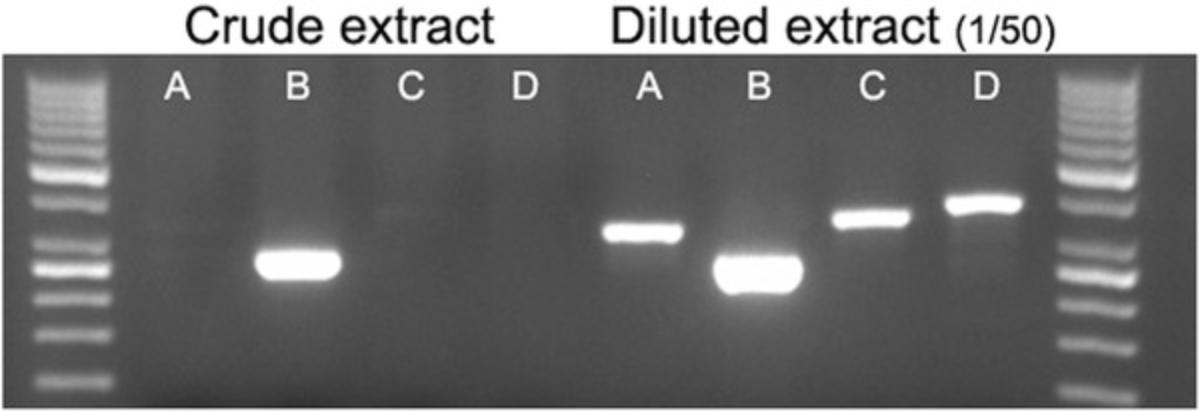
For samples that produce inconclusive results, a second trial can be performed with a sample diluted 1/50 (Fig. 7). Dilution of the crude extract is an easy solution to reduce inhibitor or template concentration.
Purchasing a liquid handling workstation
Ideally, the workstation should accommodate all steps from sample lysis to gel electrophoresis. Among these, automation of sample lysis is far from essential, as this is a very quick step. Automation of gel electrophoresis (or other PCR reading methods) is very complex and thus not advised. We therefore recommend purchasing a workstation that can prepare PCR mix and perform thermal cycling. This will be composed of a liquid handler with a robotic gripper arm (for instance, a Tecan or Hamilton workstation), an automated thermal cycler, and an automated heat sealer. In addition, including a centrifuge is highly recommended (see section on centrifugation before PCR amplification, below). The volume of samples that the multipurpose liquid-handling automated workstation can manage is one feature to consider when making a purchase.
The programming of sampling, mixing, and combining of liquid samples automatically on the workstation, called the worklist, is usually done by the liquid handler supplier, but as you may have to perform protocol optimization, the ease of use of the provided software interface is another important feature to consider.
Use of disposable tips versus fixed-steel washable needles
Workstation suppliers advise the use of disposable tips to reduce risk of cross-contamination between biological samples. From our experience with more than 600,000 samples, fixed-steel washable needles produce very reproducible results for PCR genotyping and do not induce contamination of PCR assays. When using these needles, the workstation needs to be programmed to include a decontamination step (parameters are specific for each workstation): aspiration of bleach followed by flushing of the system with water (the water flow will efficiently clean the needles) after each sample or primer dispensing.
Specific plasticware used with workstations
Not all plasticware can be adapted for use on a workstation. For example, some 384-well plates are not suitable for handling by robotic gripper arms: the plates must be rigid enough for the arm to grasp them. Plates must also be thermosealable: i.e., resistant to the temperature used for the sealing step (165°C).
For tissue sampling, using individual tubes is easier for animal caretakers. Choosing 0.75-ml microtubes with an independent cap that can be adapted to use in a 96-tubes rack (for instance Matrix tubes; cat. no. 446015, Dutscher) simplifies the processing of a very large number of biopsies.
Any changes to consumables or reagents must be tested before being used for high-throughput genotyping.
Laboratory Information Management System
Workstation suppliers will develop scripts for their automate piloting software according to your specifications. This will manage the sampling, mixing, and combining of liquid samples. We also recommend developing an in-house LIMS that manages sample traceability (Fig. 3). This reduces genotyping errors as it avoids transposition of data between samples. When a high number of different mutant lines are to be genotyped, a LIMS will also allow easy communication to the workstation, via worklists.
Important recommendations for designing genotyping strategy for high-throughput workflow
Reduce the number of primers used per mutant line : Primers are used in multiple PCR combinations in order to detect all possible alleles and minimize the number of primers ordered and stored. Figure 2 illustrates this genotyping strategy.
Use as many common primers as possible : In a high-throughput workflow, it is important to use as many common primers as possible. Most mutant models contain selection markers, tags, and reporter genes. These sequences can be used to design primers that will be suitable on multiple mutants. However, as stated in the Basic Protocol 3 introduction, designing genotyping primers only for markers that can be found in many transgenic lines is discouraged. Using common primers means using one common primer in combination with a mutation-specific primer for each PCR assay (see Fig. 2 for an example).
Define a standard annealing temperature (Tm) and thermal cycling conditions for all mutant lines : See Table 4 for recommended conditions. This is especially important when multiple mutant lines are genotyped. Without standardization of this step, PCR cannot be automated.
Reducing PCR failure in high-throughput genotyping
In a high-throughput workflow, making a duplicate or even a triplicate for each PCR point is common and ensures reliable and robust results. We do not advise making a duplicate of the same PCR design, but rather using two independent PCR designs for each allele being genotyped, to further increase genotyping reliability and robustness.
Reducing cost by multiplex PCRs
Another way to optimize a high-throughput workflow is to set up multiplex PCRs: The different PCR sets are combined into one tube to detect all the relevant alleles for a mutant line. Optimization is usually required for PCR multiplexing.
For multiplex PCRs, we recommend checking the parameters below.
- Sequence structures : All primers must not contain complementary regions of more than 3 nucleotides in 3′.
- Amplicon lengths : The choice of primer must be made so that the sizes of the sequence to be amplified are sufficiently distinct from each other to be identified by gel electrophoresis.
Setting up a multiplex PCR is done in two steps: First, validate each PCR set by simplex PCR, and then multiplex the PCR sets.
If the intensity of each amplicon is not similar on an agarose gel, weak PCR signals can be improved by increasing the concentrations of the primers that produce weak signals and/or decreasing the concentrations of the primers that give strong signals.
Managing a large number of primer sets
By analyzing our genotyping dataset (∼1500 mutant lines genotyped using ∼10,000 primers), we observed that thawing/freezing cycles lead to variable and unpredictable degradation of primers. We thus recommend the use of a master primer bank (that is used as backup) and a working primer bank (smaller single-use aliquots). This will reduce primer degradation issues by decreasing the number of thawing cycles to only two.
Using 8-tube strips (discarded after each thawing) to optimize the working primer bank improves large-volume primer management. Each tube of a strip contains the primer pair that is used to detect a specific allele, so that up to eight different PCRs can be done for a mutant line genotyping. Using 8-tube strips reduces the time needed to prepare the workstation for PCR genotyping and the number of pipetting steps on the automated workstation (as PCR pairs are already mixed together), and avoids repeated primer thawing-freezing cycles and the risk of primer contamination.
Automation using 384- plate versus 1536-well plates
384-well plates allow a volume of 10 to 130 µl per well, whereas 1536-well plate are adapted to volume of 3 to 10 µl. Thus, using 1536-well plate further reduces the volume of the reaction mix used and increases the throughput. However, 1536-well plates are not adapted for PCR genotyping if multiple PCR sets are to be used. If you need not only to genotype many individuals for one mutation but to analyze numerous genetic modifications and/or genetic loci, the pipetting step will last more than 1 hr and some samples will evaporate in 1536-well plates before the start of the PCR thermal cycling.
Centrifugation before PCR amplification
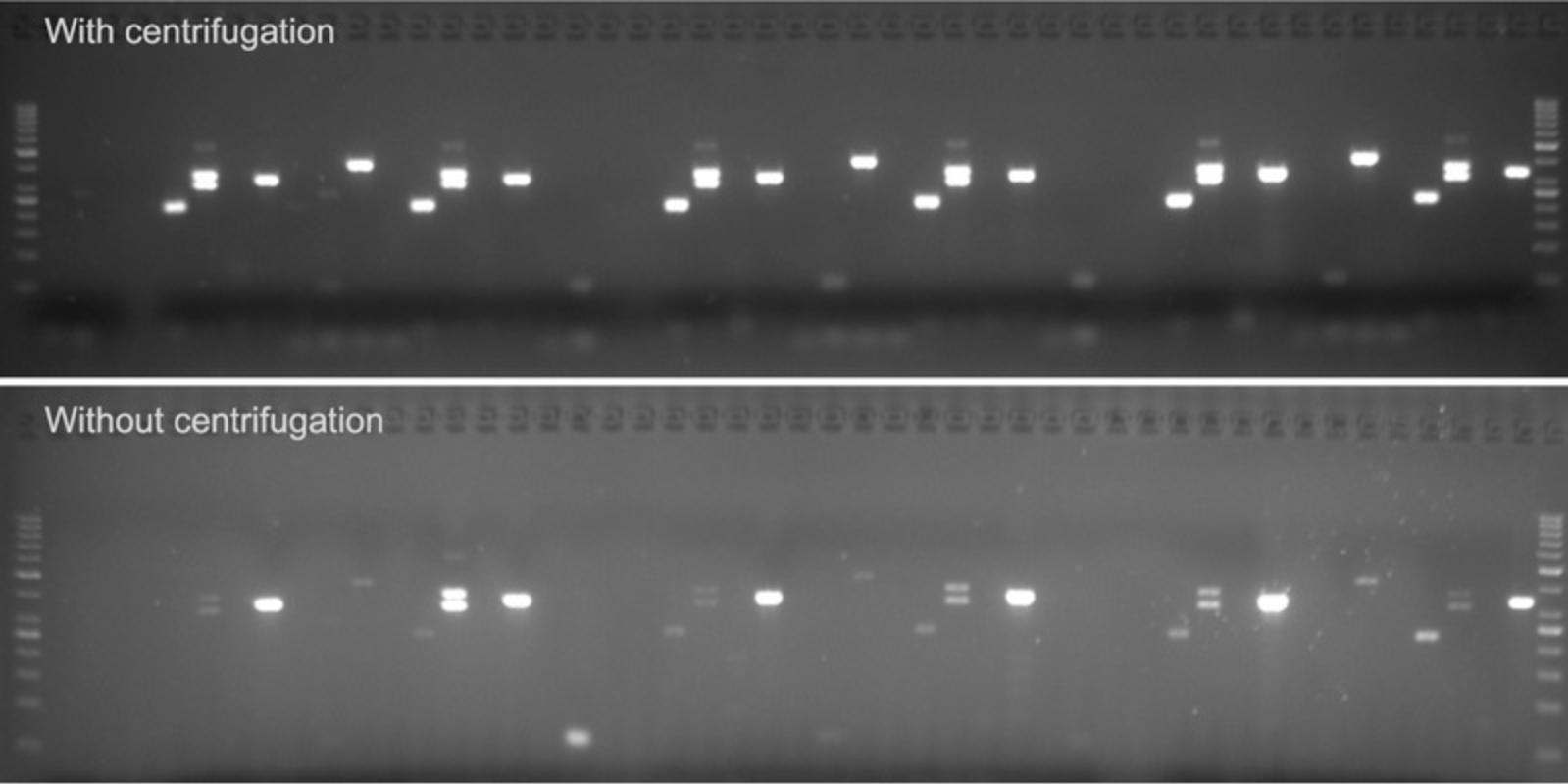
After dispensing of all PCR components into the 384-well plates and sealing, centrifugation of each 384-well plate will strongly reduce failed or weak PCR by improving homogenization of the PCR mix (Fig. 8).
Avoiding sample cross-contamination in high-throughput design
The large number of samples analyzed, and particularly the resulting PCR amplicons, logically increases the risk of contamination for high-throughput platforms. It is therefore crucial to use negative controls and follow good laboratory practices (wearing gloves, using filter tips) in all experiments. Daily sterilization of the workstation using UV light (to break contaminating DNA molecules) is also recommended.
No PCR products should be opened or otherwise handled near workstations. It is very important that a room be dedicated to the migration of PCR products on agarose gel. A PCR tube that has been opened and then closed again is very contaminating and should not be thrown into a refuse bin near workstations.
Troubleshooting
A troubleshooting guide for PCR assay design (Basic Protocol 3) is provided in Table 7. A troubleshooting guide for the use of the automated workstation (Basic Protocol 4) is provided in Table 8.
| Observation | Most probable cause(s) | Solution(s) |
|---|---|---|
| No product obtained with most of the PCR sets | Missing reaction component | Check the experimental plan and repeat reaction setup. |
| Degraded reaction component (due to multiple freeze-thaw cycles or bad storage) | Repeat the PCR with reagents from another provider, or change batch number. | |
| Low quality of reaction component batch sent by the provider, or issue during shipment | Repeat the PCR with reagents from another provider or batch number. | |
| Thermal cycler malfunction | Validate performance of your thermal cycler (Kim, Yang, Bae, & Park, 2008). | |
| Problem with agarose gel |
|
|
| Presence of inhibitors in plasticware | Check the compatibility of plasticware for PCR reaction. | |
| No product obtained with more than one PCR set | High levels of inhibitor in the template (oversized biopsy) | Dilute the sample (see Fig. 7). |
| Poor biopsy lysis | Check if the biopsy is dislocated, add proteinase K, or start from a new biopsy. | |
| Poor template quality (DNA too degraded or too old) | Start from a new biopsy. | |
| Poor genotyping design | Check the experimental plan and repeat reaction setup. | |
| No product obtained with one PCR set | Poor primer design | Design and order new primer. |
| Inefficient PCR amplicon or primer pair | Design and order new primer pair. | |
| Poor quality of primer synthesis | Order new primers. | |
| Complex template (GC- or AT-rich template, repeated sequence, or secondary structure) | Try an additive like DMSO or betaine (see Fig. 6), or even a specific GC-rich DNA polymerase. | |
| Missing primer or missing template | Check the experimental plan and repeat reaction setup. | |
| Multiple or nonspecific band amplification | Problem with primer specificity | Run a blast search on the NCBI website to check the target specificity of the primers. |
| Amplification of related pseudogenes or homologs | Run a blast search on the NCBI website to check the target specificity of the primers. | |
| Complex template (GC- or AT-rich template, repeated sequence, or secondary structure) | Try an additive like DMSO (see Fig. 6) or even a specific GC-rich DNA polymerase. | |
| Contamination of reagent, pipets, or working area with other PCR products | Use new reagents; clean the pipets and working area. Always use filter tips. | |
| Mutant allele sequence that is inaccurate | Sequence the mutant allele by an appropriate method. | |
| Sample contamination by another biopsy during tissue sampling (see Fig. 4) | Start from a new biopsy. | |
| Poor template quality (DNA too degraded or too old) | Start from a new biopsy. | |
| Nonspecific band amplification in blank | Contamination of reagent, pipet, or working area with other PCR products | Use new reagents; clean the pipets and working area. Always use filter tips. |
| Weak target amplification | Poor genotyping design | Check the experimental plan and repeat reaction setup. |
| Complex template (GC- or AT-rich template, repeated sequence, or secondary structure) | Try an additive like DMSO or betaine (see Fig. 6), or even a specific GC-rich DNA polymerase. | |
| Template insufficiently concentrated (for example biopsy too small) | Increase volume of crude extract per PCR. | |
| Template too concentrated (biopsy oversized) | Dilute the sample 1/50 in water (see Fig. 7). | |
| Presence of inhibitors in reaction | Dilute the sample 1/50 in water (see Fig. 7). | |
| Poor biopsy lysis | Check if the biopsy is dislocated, add proteinase K, or start from a new biopsy. | |
| Poor template quality (DNA too degraded or too old) | Start from a new biopsy. | |
| Nonreproducibility of genotyping protocol or smearing of amplification | Use of a different thermal cycler for reaction | Use the original cycler or optimize PCR program. |
| Reaction component degradation (due to multiple freeze-thaw cycles or incorrect storage) | Repeat the PCR with reagents from another batch number. | |
| Low quality of reaction component batch sent by the provider, or issue during shipment | Repeat the PCR with reagents from another batch number. |
| Observation | Possible cause(s) | Solution(s) |
|---|---|---|
| Automated workstation stops during the run | Gripper stopped and plate fell | Check if plasticware (plates) is adapted for automation. Verify errors in worklist program. |
| Arm collision occurred |
|
|
| Loss of connectivity with peripheral devices occurred | Possible power or network failure: add a power inverter. | |
| Plate was incorrectly positioned on the workstation | Verify that each plate and tube is in a good position on the workstation. | |
| Pipetting or dispensing error occurred | Check for problems such as clogged or loose tips, faulty O-rings, or poorly optimized pipetting settings. | |
| Wrong volumes in PCR plate | Evaporation of PCR mix occurred | Check if cooler is broken or was not turn on. Verify that sealer is well configured. |
| Level of PCR mix in the tube was too low | Verify that enough reaction mix and primers are added to perform all reactions. | |
| Pipetting or dispensing errors occurred | Check problems such as clogged or loose tips, faulty O-rings or pipetting setting poorly optimized. Check for errors in worklist program. | |
| No PCR band | Fixed-washable needle is clogged | Run a washing program and reboot the robot. |
| Fixed-washable needle is broken | Replace needle. | |
| Inversion of plates on the workstation: reagents such as primers and crude extract 96-well plates are incorrectly positioned on the workstation | Check position of each plate, and if wrong, change and restart the run. | |
| Incorrect worklist was loaded | Check which worklist was used; restart with the appropriate worklist. | |
| No centrifugation step occurred before thermocycling | Check that the centrifuge is working properly. Check for errors in worklist program. | |
| Level of PCR mix in the tube was too low | Verify that enough reaction mix and primers were added to perform all reactions. | |
| Reagents were incorrectly dispensed into PCR plates | Check for errors in worklist program. | |
| Contamination (PCR amplicon in the negative control) | No decontamination step was included in worklist | Program into the worklist a decontamination step that include bleach aspiration followed by flushing the system with water. |
| During run, decontamination between dispensing did not occur | Verify that enough bleach and water are available to perform all decontamination steps between each sample and primer. | |
| Drop is visible on pipet tip when dispensing samples | Check liquid class, dispenser seals, and dispensing position in the well. |
Time Considerations
Tissue sampling methods and procedure
Sampling (including preparation of the materials, instrument sterilization, restraint of the animal, and tissue biopsy) lasts around 2 to 3 min per animal.
Sample verification and DNA lysis
The duration of the protocol does not increase linearly with the number of treated samples. In Table 9, we detail experiment duration per 96-sample plate. Note that the incubation in lysis buffer is done overnight.
| Steps | Time (min) |
|---|---|
| Biopsy quality check | 30 |
| Premix preparation and dispensing into tubes | 15 |
| Incubation of the lysis buffer | Overnight |
| Proteinase K inactivation and storage | 50 |
Design of a genotyping strategy
Table 10 shows the experiment duration for 96 samples.
| Steps | Time (min) |
|---|---|
| Primer design (sequences validation, design, primer order) | 60 |
| Samples preparation | 30 |
| PCR cycling (cycler and target size dependent) | 90 |
| Gel electrophoresis (loading of samples and migration) | 60 |
| Image acquisition with a digital camera | 5 |
| Interpretation of results | 30 |
High-throughput genotyping
The deployment of a high-throughput platform requires months from workstation design to efficient automation. Table 11 describes the duration of a run that generates six 384-well PCR plates.
| Steps | Time (min) |
|---|---|
| Run preparation in the database | 15 |
| Workstation preparation | 15 |
| Reagent preparation | 30 |
| Run timing (automates pipetting, sealing, centrifugation, PCR) | Overnighta |
| Gel electrophoresis (loading of samples and migration) | 85 |
| Image acquisition with a digital camera | 30 |
| Interpretation of results | 180 |
- a A run of six 384-well plates requires about 17 hr with our configuration.
Acknowledgements
We thank Dr. Marie Wattenhofer-Donze for critically reading the manuscript, and Valerie Rousseau, Amélie Jeanblanc, and Laurence Luppi from the PHENOMIN-ICS genotyping service for their contribution to generating the data presented in this manuscript. This work was supported by the National Centre for Scientific Research (CNRS), the French National Institute of Health and Medical Research (INSERM), the University of Strasbourg (UDS), and the Centre Européen de Recherche en Biologie et en Médecine. This study also received support from French state funds through the Agence Nationale de la Recherche awarded under the frame program Investissements d'Avenir, grants ANR-10-IDEX-0002-02 and ANR-10-INBS-07 PHENOMIN to Y.H.
Literature Cited
- Al-Griw, H. H., Zraba, Z. A., Al-Muntaser, S. K., Draid, M. M., Zaidi, A. M., Tabagh, R. M., & Al-Griw, M. A. (2017). Effects of storage temperature on the quantity and integrity of genomic DNA extracted from mice tissues: A comparison of recovery methods. Open Veterinary Journal , 7, 239–243. doi: 10.4314/ovj.v7i3.7.
- Bacich, D. J., Sobek, K. M., Cummings, J. L., Atwood, A. A., & O'Keefe, D. S. (2011). False negative results from using common PCR reagents. BMC Research Notes , 4, 457. doi: 10.1186/1756-0500-4-457.
- Birling, M.-C., Herault, Y., & Pavlovic, G. (2017). Modeling human disease in rodents by CRISPR/Cas9 genome editing. Mammalian Genome , 28(7–8), 291–301. doi: 10.1007/s00335-017-9703-x.
- Birling, M.-C., Schaeffer, L., André, P., Lindner, L., Maréchal, D., Ayadi, A., … Hérault, Y. (2017). Efficient and rapid generation of large genomic variants in rats and mice using CRISMERE. Scientific Reports , 7, 43331. doi: 10.1038/srep43331.
- Bonaparte, D., Cinelli, P., Douni, E., Hérault, Y., Maas, M., & Pakarinen, P. … Federation of European Laboratory Animal Science Associations Working Group. (2013). FELASA guidelines for the refinement of methods for genotyping genetically-modified rodents: A report of the Federation of European Laboratory Animal Science Associations Working Group. Laboratory Animals , 47, 134–145. doi: 10.1177/0023677212473918.
- Cinelli, P., Rettich, A., Seifert, B., Bürki, K., & Arras, M. (2007). Comparative analysis and physiological impact of different tissue biopsy methodologies used for the genotyping of laboratory mice. Laboratory Animals , 41, 174–184. doi: 10.1258/002367707780378113.
- Dorit, R. L., Ohara, O., Hwang, C. B., Kim, J. B., & Blackshaw, S. (2001). Direct DNA sequencing of PCR products. Current Protocols in Molecular Biology , 56(1), 15.2.1–15.2.13. doi: 10.1002/0471142727.mb1502s56.
- Hankenson, F. C., Garzel, L. M., Fischer, D. D., Nolan, B., & Hankenson, K. D. (2008). Evaluation of tail biopsy collection in laboratory mice (Mus musculus): Vertebral ossification, DNA quantity, and acute behavioral responses. Journal of the American Association for Laboratory Animal Science , 47, 10–18.
- Kim, Y. H., Yang, I., Bae, Y.-S., & Park, S.-R. (2008). Performance evaluation of thermal cyclers for PCR in a rapid cycling condition. Biotechniques , 44, 495–496. doi: 10.2144/000112705.
- Lee, P. Y., Costumbrado, J., Hsu, C.-Y., & Kim, Y. H. (2012). Agarose gel electrophoresis for the separation of DNA fragments. Journal of Visualized Experiments , (62), 3923. doi: 10.3791/3923.
- Lloyd, K., Franklin, C., Lutz, C., & Magnuson, T. (2015). Reproducibility: Use mouse biobanks or lose them. Nature , 522, 151–153. doi: 10.1038/522151a.
- Lorenz, T. C. (2012). Polymerase chain reaction: Basic Protocol plus troubleshooting and optimization strategies. Journal of Visualized Experiments , (63), e3998. doi: 10.3791/3998.
- Meehan, T. F., Conte, N., West, D. B., Jacobsen, J. O., Mason, J., Warren, J., … Smedley, D. (2017). Disease model discovery from 3,328 gene knockouts by The International Mouse Phenotyping Consortium. Nature Genetics , 49(8), 1231–1238. doi: 10.1038/ng.3901.
- Mianné, J., Codner, G. F., Caulder, A., Fell, R., Hutchison, M., King, R., … Teboul, L. (2017). Analysing the outcome of CRISPR-aided genome editing in embryos: Screening, genotyping and quality control. Methods , 121–122, 68–76. doi: 10.1016/j.ymeth.2017.03.016.
- Miller, D. N., Bryant, J. E., Madsen, E. L., & Ghiorse, W. C. (1999). Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Applied and Environmental Microbiology , 65, 4715–4724.
- Picazo, M. G., & García-Olmo, D. C. (2015). DNA from tissues of young mice is optimal for genotyping. Electronic Journal of Biotechnology , 18, 83–87. doi: 10.1016/j.ejbt.2014.12.002.
- Scavizzi, F., Ryder, E., Newman, S., Raspa, M., Gleeson, D., Wardle-Jones, H., … Doe, B. (2015). Blastocyst genotyping for quality control of mouse mutant archives: An ethical and economical approach. Transgenic Research , 24, 921–927. doi: 10.1007/s11248-015-9897-1.
- Schrader, C., Schielke, A., Ellerbroek, L., & Johne, R. (2012). PCR inhibitors — occurrence, properties and removal. Journal of Applied Microbiology , 113, 1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x.
- Silverman, J., & Hendricks, G. (2014). Sensory neuron development in mouse coccygeal vertebrae and its relationship to tail biopsies for genotyping. PLoS One , 9, e88158. doi: 10.1371/journal.pone.0088158.
- Wever, K. E., Geessink, F. J., Brouwer, M. A. E., Tillema, A., & Ritskes-Hoitinga, M. (2017). A systematic review of discomfort due to toe or ear clipping in laboratory rodents. Laboratory Animals , 51, 583–600. doi: 10.1177/0023677217705912.
- Zhang, J.-H., Wang, F., & Wang, T.-Y. (2011). A simple and effective SuperBuffer for DNA agarose electrophoresis. Gene , 487, 72–74. doi: 10.1016/j.gene.2011.05.018.
Citing Literature
Number of times cited according to CrossRef: 24
- Olivia C. Ihedioha, Haley Q. Marcarian, Anutr Sivakoses, Stephen M. Beverley, Diane McMahon-Pratt, Alfred L. M. Bothwell, Leishmania major surface components and DKK1 signalling via LRP6 promote migration and longevity of neutrophils in the infection site, Frontiers in Immunology, 10.3389/fimmu.2024.1473133, 15 , (2024).
- Audrey E Kaye, Jacob W Proctor-Bonbright, Jai Y Yu, Rectal swab DNA collection protocol for PCR genotyping in rats, BioTechniques, 10.2144/btn-2024-0023, 76 , 6, (275-283), (2024).
- Deng-ying Fan, Hao-yan Zhai, Yuan Zhao, Xing Qiao, De-chao Zhu, Hui-Juan Liu, Chunyan Liu, The role of cannabinoid receptor 2 in bone remodeling during orthodontic tooth movement, BMC Oral Health, 10.1186/s12903-023-03810-5, 24 , 1, (2024).
- Albert Blanch-Asensio, Catarina Grandela, Christine L. Mummery, Richard P. Davis, STRAIGHT-IN: a platform for rapidly generating panels of genetically modified human pluripotent stem cell lines, Nature Protocols, 10.1038/s41596-024-01039-2, (2024).
- Liu Liu, Kimberley El, Diptadip Dattaroy, Luiz F. Barella, Yinghong Cui, Sarah M. Gray, Carla Guedikian, Min Chen, Lee S. Weinstein, Emily Knuth, Erli Jin, Matthew J. Merrins, Jeffrey Roman, Klaus H. Kaestner, Nicolai Doliba, Jonathan E. Campbell, Jürgen Wess, Intra-islet α-cell Gs signaling promotes glucagon release, Nature Communications, 10.1038/s41467-024-49537-x, 15 , 1, (2024).
- Lydia Teboul, James Amos-Landgraf, Fernando J. Benavides, Marie-Christine Birling, Steve D. M. Brown, Elizabeth Bryda, Rosie Bunton-Stasyshyn, Hsian-Jean Chin, Martina Crispo, Fabien Delerue, Michael Dobbie, Craig L. Franklin, Ernst-Martin Fuchtbauer, Xiang Gao, Christelle Golzio, Rebecca Haffner, Yann Hérault, Martin Hrabe de Angelis, Kevin C. Kent Lloyd, Terry R. Magnuson, Lluis Montoliu, Stephen A. Murray, Ki-Hoan Nam, Lauryl M. J. Nutter, Eric Pailhoux, Fernando Pardo Manuel de Villena, Kevin Peterson, Laura Reinholdt, Radislav Sedlacek, Je Kyung Seong, Toshihiko Shiroishi, Cynthia Smith, Toru Takeo, Louise Tinsley, Jean-Luc Vilotte, Søren Warming, Sara Wells, C. Bruce Whitelaw, Atsushi Yoshiki, Atsushi Yoshiki, Chi-Kuang Wang, Jacqueline Marvel, Ana Zarubica, Sara Wells, Jason Heaney, Sara Wells, Ian F. Korf, Cathleen Lutz, Andrew J. Kueh, Paul Q. Thomas, Ruth M. Arkell, Graham J. Mann, Guillaume Pavlovic, Improving laboratory animal genetic reporting: LAG-R guidelines, Nature Communications, 10.1038/s41467-024-49439-y, 15 , 1, (2024).
- Claire E. Ryan, Thomas R. Salvetti, Ilana R. Baum, Brandon A. Figueroa, Brittany E. LeBere, Michael O. Alberti, Single-tube Ptprc SNP genotyping of JAXBoy (CD45.1) and C57BL/6J (CD45.2) mice by endpoint PCR and gel electrophoresis, Molecular and Cellular Probes, 10.1016/j.mcp.2024.101962, 75 , (101962), (2024).
- Emmanuelle Totain, Loïc Lindner, Nicolas Martin, Yolande Misseri, Alexandra Iché, Marie-Christine Birling, Tania Sorg, Yann Herault, Alain Bousquet-Melou, Pascale Bouillé, Christine Duthoit, Guillaume Pavlovic, Severine Boullier, Development of HPV16 mouse and dog models for more accurate prediction of human vaccine efficacy, Laboratory Animal Research, 10.1186/s42826-023-00166-3, 39 , 1, (2023).
- Xinyun Xu, Xinge Hu, Guodong Ma, Tiannan Wang, Jayne Wu, Xiaojuan Zhu, Guoxun Chen, Ling Zhao, Jiangang Chen, Detecting fa leptin receptor mutation in Zucker rats with tetra-primer amplification-refractory mutation system (ARMS)-PCR, Heliyon, 10.1016/j.heliyon.2023.e20159, 9 , 9, (e20159), (2023).
- Julien M. D. Legrand, Robin M. Hobbs, Defining Gene Function in Spermatogonial Stem Cells Through Conditional Knockout Approaches, Spermatogonial Stem Cells, 10.1007/978-1-0716-3139-3_15, (261-307), (2023).
- Laurence Schaeffer, Loic Lindner, Guillaume Pavlovic, Yann Hérault, Marie-Christine Birling, CRISMERE Chromosome Engineering in Mouse and Rat, Transgenesis, 10.1007/978-1-0716-2990-1_12, (277-297), (2023).
- Dmitry P. Karabanov, Eugeniya I. Bekker, Dmitry D. Pavlov, Elena A. Borovikova, Yulia V. Kodukhova, Alexey A. Kotov, New Sets of Primers for DNA Identification of Non-Indigenous Fish Species in the Volga-Kama Basin (European Russia), Water, 10.3390/w14030437, 14 , 3, (437), (2022).
- Sara Carpi, Ambra Del Grosso, Miriam De Sarlo, Laura Colagiorgio, Luca Scaccini, Ilaria Tonazzini, Gabriele Parlanti, Marco Cecchini, Reliable and Fast Genotyping Protocol for Galactosylceramidase (Galc) in the Twitcher (Twi) Mouse, Biomedicines, 10.3390/biomedicines10123146, 10 , 12, (3146), (2022).
- Jiangang Chen, Xinyun Xu, Paul Dalhaimer, Ling Zhao, Tetra-Primer Amplification-Refractory Mutation System (ARMS)—PCR for Genotyping Mouse Leptin Gene Mutation, Animals, 10.3390/ani12192680, 12 , 19, (2680), (2022).
- Richard E. Brown, Genetically modified mice for research on human diseases: A triumph for Biotechnology or a work in progress?, The EuroBiotech Journal, 10.2478/ebtj-2022-0008, 6 , 2, (61-88), (2022).
- Shutaro Kobayashi, Kazunori O’Hashi, Keisuke Kaneko, Satomi Kobayashi, Shouhei Ogisawa, Morio Tonogi, Satoshi Fujita, Masayuki Kobayashi, A new phenotype identification method with the fluorescent expression in cross-sectioned tails in Thy1 -GCaMP6s transgenic mice, Journal of Oral Science, 10.2334/josnusd.21-0528, 64 , 2, (156-160), (2022).
- Kelli A. McCord, Matthew S. Macauley, Transgenic mouse models to study the physiological and pathophysiological roles of human Siglecs, Biochemical Society Transactions, 10.1042/BST20211203, 50 , 2, (935-950), (2022).
- Sharda P. Singh, Jihyun Lee, Chhanda Bose, Hongzhi Li, Yate-Ching Yuan, Ashly Hindle, Sharad S. Singhal, Jonathan Kopel, Philip T. Palade, Catherine Jones, Rakhshanda L. Rahman, Sanjay Awasthi, Haploinsufficiency Interactions between RALBP1 and p53 in ERBB2 and PyVT Models of Mouse Mammary Carcinogenesis, Cancers, 10.3390/cancers13133329, 13 , 13, (3329), (2021).
- Véronique Brault, Thu Lan Nguyen, Javier Flores-Gutiérrez, Giovanni Iacono, Marie-Christine Birling, Valérie Lalanne, Hamid Meziane, Antigoni Manousopoulou, Guillaume Pavlovic, Loïc Lindner, Mohammed Selloum, Tania Sorg, Eugene Yu, Spiros D. Garbis, Yann Hérault, Dyrk1a gene dosage in glutamatergic neurons has key effects in cognitive deficits observed in mouse models of MRD7 and Down syndrome, PLOS Genetics, 10.1371/journal.pgen.1009777, 17 , 9, (e1009777), (2021).
- Loic Lindner, Pauline Cayrou, Thomas W. Rosahl, Heather H. Zhou, Marie-Christine Birling, Yann Herault, Guillaume Pavlovic, Droplet digital PCR or quantitative PCR for in-depth genomic and functional validation of genetically altered rodents, Methods, 10.1016/j.ymeth.2021.04.001, 191 , (107-119), (2021).
- Diane Gleeson, Debarati Sethi, Radka Platte, Jonathan Burvill, Daniel Barrett, Shaheen Akhtar, Michaela Bruntraeger, Joanna Bottomley, Sanger Mouse Genetics Project, James Bussell, Edward Ryder, High-throughput genotyping of high-homology mutant mouse strains by next-generation sequencing, Methods, 10.1016/j.ymeth.2020.10.011, 191 , (78-86), (2021).
- M.-C. Birling, M. D. Fray, P. Kasparek, J. Kopkanova, M. Massimi, R. Matteoni, L. Montoliu, L. M. J. Nutter, M. Raspa, J. Rozman, E. J. Ryder, F. Scavizzi, V. Voikar, S. Wells, G. Pavlovic, L. Teboul, Importing genetically altered animals: ensuring quality, Mammalian Genome, 10.1007/s00335-021-09908-x, 33 , 1, (100-107), (2021).
- Louhanna Pinheiro Rodrigues Teixeira, Francisco Eder de Moura Lopes, André Saraiva Leão Marcelo Antunes, Matheus Soares Alves, André Marrocos Miranda, Saul Gaudencio Neto, Leonardo Tondello Martins, Ana Cristina de Oliveira Monteiro Moreira, Kaio Cesar Simiano Tavares, Application of a cost-effective DNA extraction protocol for screening transgenic and CRISPR-edited primary goat cells, PLOS ONE, 10.1371/journal.pone.0239435, 15 , 9, (e0239435), (2020).
- Romain Joubert, Virginie Mariot, Julie Dumonceaux, One‐hour universal protocol for mouse genotyping, Muscle & Nerve, 10.1002/mus.26841, 61 , 6, (801-807), (2020).

