Quantifying the Probing and Selection of Microenvironmental Pores by Motile Immune Cells
Janina Kroll, Janina Kroll, Mauricio J. A. Ruiz-Fernandez, Mauricio J. A. Ruiz-Fernandez, Malte B. Braun, Malte B. Braun, Jack Merrin, Jack Merrin, Jörg Renkawitz, Jörg Renkawitz
Abstract
Immune cells are constantly on the move through multicellular organisms to explore and respond to pathogens and other harmful insults. While moving, immune cells efficiently traverse microenvironments composed of tissue cells and extracellular fibers, which together form complex environments of various porosity, stiffness, topography, and chemical composition. In this protocol we describe experimental procedures to investigate immune cell migration through microenvironments of heterogeneous porosity. In particular, we describe micro-channels, micro-pillars, and collagen networks as cell migration paths with alternative pore size choices. Employing micro-channels or micro-pillars that divide at junctions into alternative paths with initially differentially sized pores allows us to precisely (1) measure the cellular translocation time through these porous path junctions, (2) quantify the cellular preference for individual pore sizes, and (3) image cellular components like the nucleus and the cytoskeleton. This reductionistic experimental setup thus can elucidate how immune cells perform decisions in complex microenvironments of various porosity like the interstitium. The setup further allows investigation of the underlying forces of cellular squeezing and the consequences of cellular deformation on the integrity of the cell and its organelles. As a complementary approach that does not require any micro-engineering expertise, we describe the usage of three-dimensional collagen networks with different pore sizes. Whereas we here focus on dendritic cells as a model for motile immune cells, the described protocols are versatile as they are also applicable for other immune cell types like neutrophils and non-immune cell types such as mesenchymal and cancer cells. In summary, we here describe protocols to identify the mechanisms and principles of cellular probing, decision making, and squeezing during cellular movement through microenvironments of heterogeneous porosity. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Immune cell migration in micro-channels and micro-pillars with defined pore sizes
Support Protocol 1 : Epoxy replica of generated and/or published micro-structures
Support Protocol 2 : Dendritic cell differentiation
Basic Protocol 2 : Immune cell migration in 3D collagen networks of variable pore sizes
INTRODUCTION
Migrating cells face microenvironments of various mechano-chemical compositions, for example with varying microenvironmental porosity (Yamada & Sixt, 2019). Particularly immune cells have to cross large distances to fulfill their homeostatic functions, to patrol the organism, and to initiate and execute immune responses, such as the transport of antigens by dendritic cells (DCs) for presentation in lymph nodes (Kameritsch & Renkawitz, 2020). During their trafficking, these cells constantly encounter pores within the microenvironment that are variable in size and frequently smaller than the nucleus (Starborg, Lu, Kadler, & Holmes, 2008; Stoitzner, Pfaller, Stössel, & Romani, 2002; Weigelin, Bakker, & Friedl, 2012; Wolf et al., 2009). This raises the question of how immune cells repeatedly make directional decisions for alternative narrow paths, how they generate the necessary cellular forces for squeezing, and how they withstand the considerable deformation of their cell body and their intracellular organelles.
Recent advances in imaging immune cell motility within the native microenvironment of living animals beautifully illustrated the complexity of the mechano-chemical composition of the microenvironment, including differences in collagen density and porosity (Ilina et al., 2020; Lerchenberger et al., 2013). Here, we describe in vitro micro-channel and micro-pillar assays with alternatively sized cellular paths as precisely controllable and tunable microenvironments (Basic Protocol 1). These assays allow us to investigate (1) whether the cellular model of interest migrates along larger pore sizes and thus the path of least resistance (Renkawitz et al., 2019), (2) how cells probe (Leithner et al., 2016; Sitarska et al., 2021) and select (Renkawitz et al., 2019) differently sized environmental pores, and (3) how cells coordinate their protrusions when encountering multiple pores simultaneously (Kopf et al., 2020). Further, micro-channels with defined pore sizes are also suitable to study the consequences of nuclear squeezing, such as nuclear envelope ruptures and DNA damage (Cho et al., 2019; Denais et al., 2016; Raab et al., 2016; Thiam et al., 2016; Wallis et al., 2021). As a complementary approach to the micro-channel and micro-pillar protocol, we describe the usage of three-dimensional collagen networks with different pore sizes (Basic Protocol 2). This protocol has the advantage that it does not require any micro-engineering expertise.
Whereas we here describe detailed protocols to investigate the probing and selection of microenvironmental pores by immune cells, these assays can also be performed with other cell types like cancer cells (Denais et al., 2016; Sabeh, Shimizu-Hirota, & Weiss, 2009; Wolf et al., 2013). Moreover, these assays can be modified to symmetric path junctions composed of micro-channels of the same diameter, enabling the investigation of directional decisions independent of pore decisions (Hadjitheodorou et al., 2022; Hadjitheodorou et al., 2021; Tweedy et al., 2020; Wang, Hossain, Bogoslowski, Kubes, & Irimia, 2020).
STRATEGIC PLANNING
Before starting, we highly recommend planning the experimental procedure according to the research question and cellular model of interest (Fig. 1A). For instance, if the research question aims to investigate how cells navigate through microenvironments of heterogeneous porosity, we recommend careful selection of the most suitable experimental approach for the individual cellular model (Fig. 1B): Fast and directionally migrating cells such as dendritic cells (DCs), T cells, or neutrophilic granulocytes (neutrophils) can be investigated in micro-channels that divide at junctions into different paths with initially differentially sized pores (Fig. 1B, part a; Basic Protocol 1). These “pore-decision micro-channels” composed of the transparent and gas-diffusible polymer polydimethylsiloxane (PDMS) offer a highly reductionistic environment, which simplifies the analysis of cellular pore size preference and the cellular coordination within these junctions. Yet, when investigating motile cells with slower velocities or a frequent turning behavior, such as mesenchymal cells and cancer cells, these micro-channels are often less practicable due to their single direction of interest and their (on a cellular scale) relatively long initial straight channel part. For these rather slowly migrating cells, we recommend using PDMS-based micro-pillars that are arranged in varying distances to form an environment of heterogeneous porosity (Fig. 1B, part b; Basic Protocol 1). Using these “pore-decision micro-pillar” assays allow investigation of the pore size preference of slowly and non-directionally migrating cells directly upon their entrance from the loading area into the pillar area. As a comparison for pore-decision micro-channels and pore-decision micro-pillar assays, we suggest using micro-channels with single pores inside straight channels (Fig. 1B, part c) to measure the squeezing capability and translocation time through individual pores by the cellular model of interest. This approach will also reveal the smallest pore size that the cellular model can cross, and thus helps to design the most suitable pore sizes in the pore-decision micro-channels and pore-decision micro-pillars. When generating new porous micro-structures, we strongly recommend that standard PDMS-based architectural requirements are followed, such as the aspect ratio (height-to-width ratio) of micro-channels and micro-pillars (Velve-Casquillas, Berre, Piel, & Tran, 2010), and smallest possible features (e.g., pores <2 micrometers are difficult to generate).
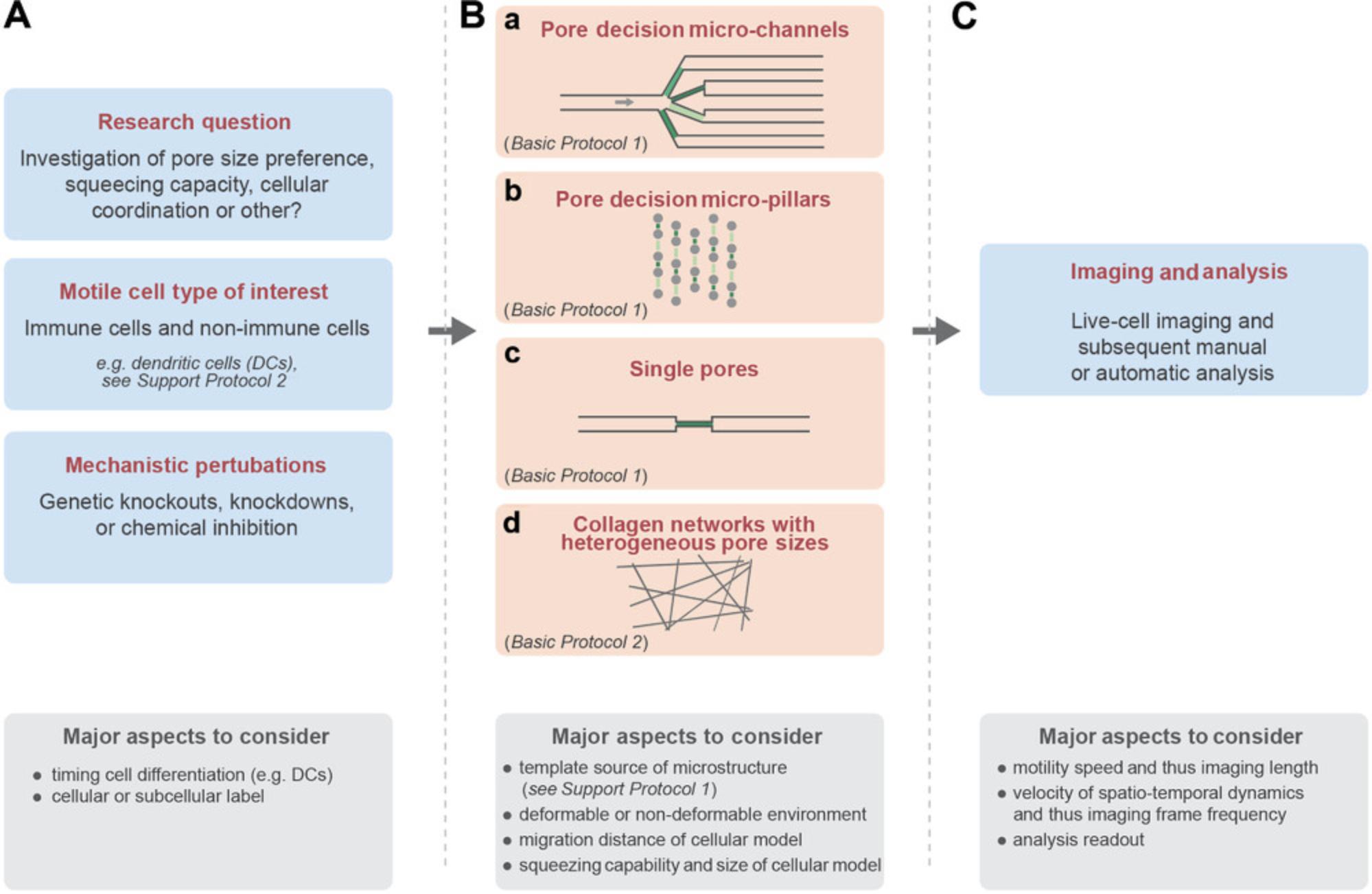
As a complementary approach, collagen networks with heterogeneous pore sizes (Fig. 1B, part d; Basic Protocol 2) offer a microenvironment that is close to physiological conditions, given their deformability and proteolytic cleavability. Moreover, these collagen networks do not require specific laboratory equipment and allow simultaneous imaging of many cells for in-depth statistical analysis, yet it is more difficult to precisely measure the pore sizes within the collagen network.
Before starting one of these assays, we highly recommend considering the imaging parameters and the subsequent analysis readout (Fig. 1C). For instance, fast migrating cells require a rather fast imaging frame frequency to capture the underlying spatiotemporal dynamics. However, too frequent imaging might result in phototoxicity. Further, imaging over long time periods with high temporal sampling leads to large datasets, which may be challenging in subsequent processing and analysis of the microscopy data. Similarly, fluorescent labeling of entire cells or subcellular structures (e.g., the actin cytoskeleton or the nucleus) simplifies the later analysis (e.g., for cellular segmentation followed by automatic tracking), yet it should be carefully controlled so that the fluorescent labeling does not alter the cellular phenotype.
Basic Protocol 1: IMMUNE CELL MIGRATION IN MICRO-CHANNELS AND MICRO-PILLARS WITH DEFINED PORE SIZES
This protocol uses PDMS-based micro-channels and micro-pillars to mimic the complex 3D microenvironment of motile cells in vitro , while giving the user the advantage of precisely controlling the experimental parameters like pore sizes, chemokine gradients, the number of possible cellular paths, and the number of path decisions. Specifically, we describe PDMS-based micro-channels and micro-pillars to quantify cell migration along paths of differentially sized pores (Kopf et al., 2020; Renkawitz et al., 2019; Renkawitz, Reversat, Leithner, Merrin, & Sixt, 2018).
Whereas the micro-structures are different, the usage of both micro-channels and micro-pillars follows the same protocol: First, a mold with a template of the desired micro-structures is needed to prepare micro-structures with defined width, height, length, and constriction/pore sizes. These templates can either be a custom-made wafer produced by photolithography, a commercially acquired wafer, or an “epoxy replicate” of an already existing micro-structure shared by another research group (the latter option is a simple option for research groups without micro-engineering expertise; see Support Protocol 1).
Once the micro-fabricated mold is available, the transparent and gas-diffusible polymer PDMS can be used to generate a replica of the micro-structure. For this purpose, PDMS is mixed with a curing agent to start a polymerization reaction. This still liquid mixture is then added to the mold (Fig. 2A-C), such as a wafer or an epoxy replicate. After solidification of the PDMS, the device is diced (Fig. 2E-H), loading holes are punched (Fig. 2I), and the PDMS device is irreversibly attached to a glass surface using a plasma cleaner. This generates a microenvironment in which the channel/pillar walls and the ceiling are composed of PDMS, whereas the bottom is made of a glass coverslip (Fig. 2J, 2K) enabling high-resolution microscopy (Fig. 2L-O).
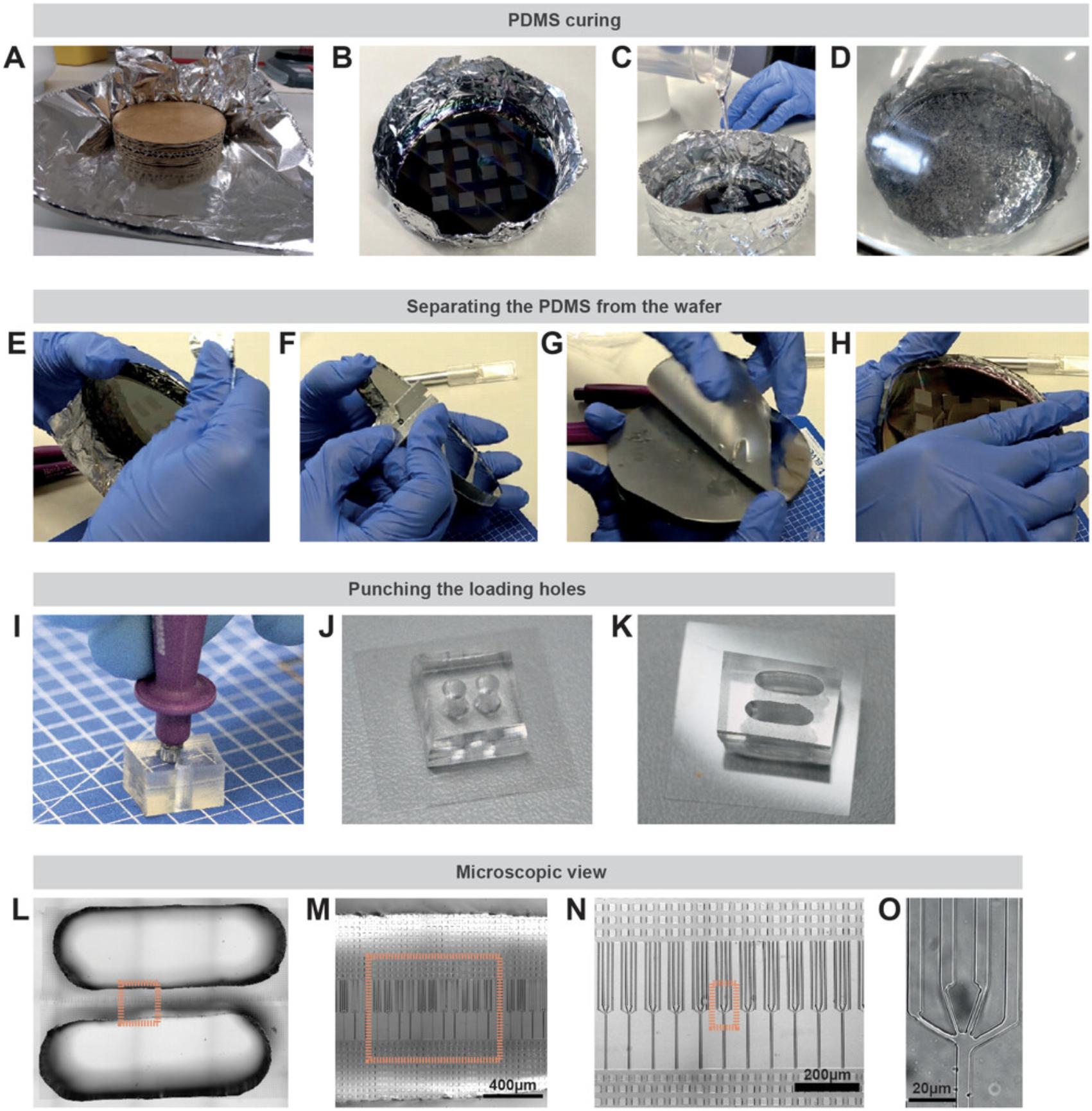
In the next step, the PDMS device has to be prepared for loading with cells by filling the micro-channels/micro-pillars with cell culture medium. The devices are placed into a plasma cleaner followed by addition of the cell culture medium to the PDMS device. Thereby, the surface of the device is converted from hydrophobic to hydrophilic, enabling subsequent flooding of the whole device and the micro-structures. A vacuum chamber can be used to remove residual air bubbles inside the micro-structures.
The assay can be combined with a chemokine gradient to guide migrating cells into the micro-structures. After addition of the respective chemoattractant (for the cellular model of interest; e.g., CCL19 for DCs, fMLP for human neutrophils) into one loading hole, the cells are added into a second loading hole. Subsequently, the cells can be imaged inside the pore-decision micro-channels and micro-structures by live cell microscopy (Fig. 3).
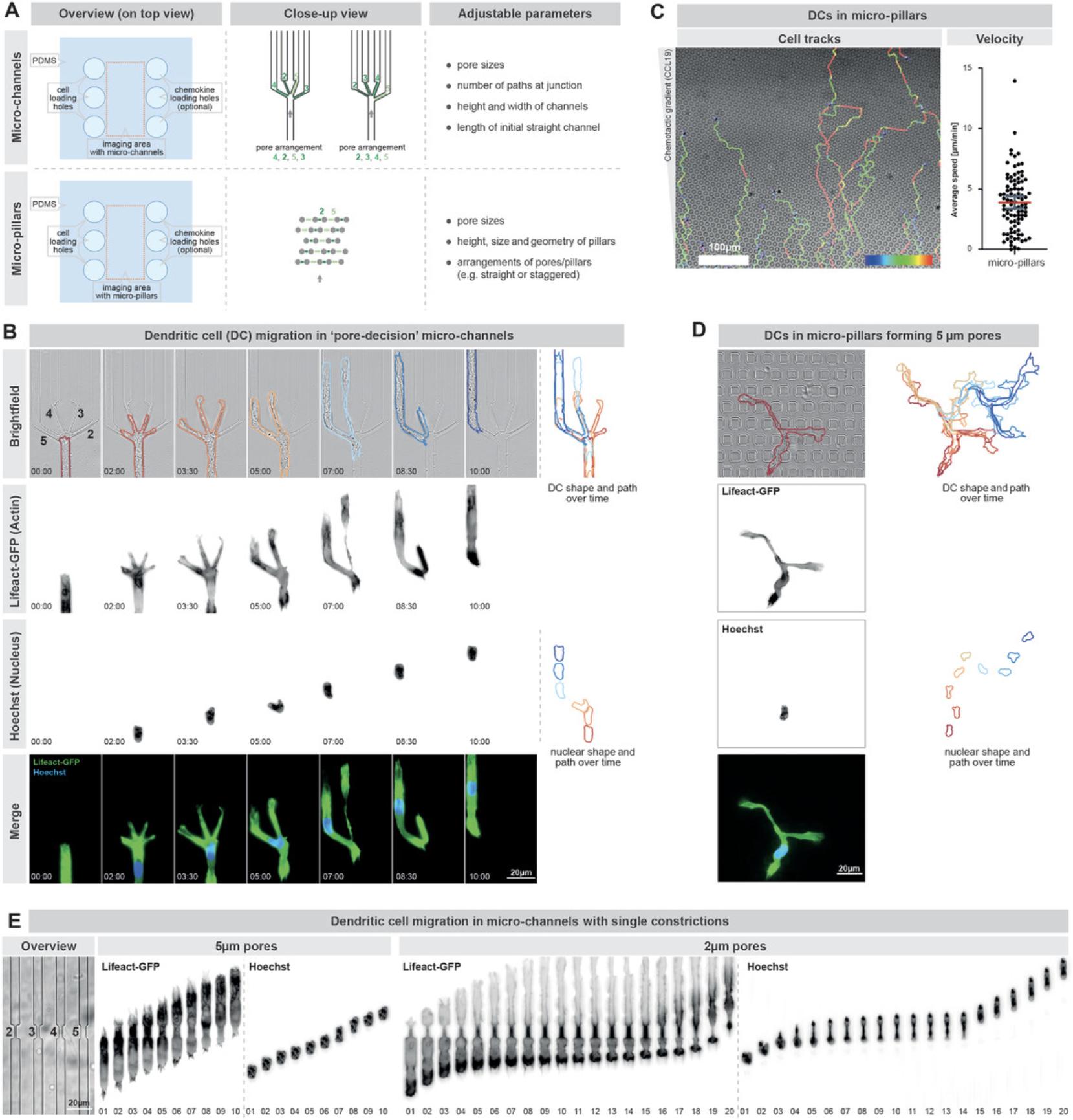
We have used these PDMS-based migration micro-structures already with a wide variety of different cell types. For instance, DCs (see Support Protocol 2), neutrophils, and T cells efficiently migrate in the described pore-decision micro-channels as well as in the pore-decision micro-pillars (Fig. 3; Leithner et al., 2016; Renkawitz et al., 2019; Renkawitz et al., 2018). Further, we successfully employed macrophages (MCs), HT1080 fibrosarcoma cells, and 3T3 fibroblasts inside pore-decision micro-pillars (Renkawitz et al., 2019). These pore-decision micro-pillars are particularly advantageous when investigating slowly and/or nondirectionally migrating cells, as they enable imaging directly upon exit from the loading hole.
Imaging cell migration inside micro-structures with alternative pore sizes allows the ability to receive information about general migration parameters like the speed of the cell but also more specific information like (1) the time a cell requires to cross pores of a specific size, (2) the mechanisms underlying cellular decision making, and (3) cellular preference for specific pore sizes. It is also possible to perform these experiments with genetic and non-genetic fluorescent labels to visualize specific cellular compartments or proteins during migration. For example, it is possible to use cells labeled with fluorescent Lifeact (Riedl et al., 2008) to visualize F-actin dynamics during migration through narrow pores and decision making, or to label the cellular nucleus with DNA binding dyes like Hoechst 33342 to visualize nuclear positioning and squeezing during migration. The described assay can also be coupled with the CRISPR/Cas9 technology to investigate the function of specific proteins or protein subdomains on cellular migration (Hammerschmidt et al., 2018; Leithner et al., 2018; Orosz, Walzog, & Mócsai, 2020; Redecke et al., 2013; Xu et al., 2021).
Materials
-
Polydimethylsiloxane (PDMS) SYLGARD™ 184 Silicone Elastomer Kit (Biesterfeld)
-
Curing agent: SYLGARD™ 184 Silicone Elastomer Kit (Biesterfeld)
-
Wafer (custom-made) or epoxy replica (see Support Protocol 1)
-
Isopropanol, Certified AR for Analysis (Thermo Fisher Scientific, 10366430)
-
Ethanol, Absolute 99.8+ %, Certified AR for Analysis (Thermo Fisher Scientific, 10437341)
-
Imaging medium (see recipe)
-
Chemokine: Recombinant Mouse CCL19/MIP-3 beta Protein (Bio-Techne, 440-M3-025)
-
Dendritic cells (DCs; see Support Protocol 2)
-
DNA stain: Invitrogen NucBlue™ Live ReadyProbes™ (Hoechst 33342; Thermo Fisher Scientific, R37605)
-
ThinkyMixer Cups (Thinky-250AD-200DSP-Disposable Cups; C3 Prozess- und Analysentechnik)
-
Scotch tape (Scotch® RemovableTM M8111933)
-
Razor blade (Thermo Fisher Scientific, 11904325)
-
Puncher (Harris Uni-Core™, 3 mm)
-
Petri dish: CELLVIEW cell culture dish (Greiner Bio-One, 627860)
-
Beaker (SCHOTT DURAN® Beaker, low form, with spout, 211063604)
-
Pipet tips (starlab):
- 10 µl graduated TipOne® Tip (S1111-3700-C)
- 1,000 µl graduated TipOne® Tip (S1111-6701-C)
- 200 µl UltraPoint® graduated TipOne® Tip (S1113-1706-C)
-
Scale (Ohaus CL501 or equivalent)
-
ThinkyMixer (Thinky ARE-250)
-
Vacuum desiccator (Kartell filtration and vacuum desiccators, Art. 230 or equivalent)
-
Vacuum pump (Dekker RVR003H-OT or equivalent)
-
Oven (mini oven STZ 5.4 720.2070.04 or equivalent)
-
Ultrasonication bath (Elma Elmasonic S or equivalent)
-
Pressured air (from house gas supply)
-
Air blower (Bahco BP218 or equivalent)
-
Forceps
-
Plasma cleaner: Expanded Plasma Cleaner (Harrick Plasma, PDC-002-CE)
-
Pipet (Eppendorf Research plus or equivalent)
-
Cell culture microscope (Leica or equivalent)
-
Cell culture incubator (Eppendorf Galaxy 170R or equivalent)
-
Microscope with automated stage, incubation chamber, temperature control unit, and CO2 control unit (Leica DMI 8, Incubator i8, Pecon® TempController 2000, Pecon® CO2-Controller 2000 or equivalent)
Templates of micro-channels and micro-pillars
This protocol is based on molds with templates for the respective micro-channels and micro-pillars. As a template one can either use (1) a custom-made wafer generated by photolithography, (2) a commercially generated wafer, or (3) an epoxy replicate of an existing wafer. The generation of custom-made wafers by high-resolution photomasks to pattern photoresists has been described earlier (Merrin, 2019; Renkawitz et al., 2018). The generation of epoxy replicates of existing wafers represents a suitable possibility to replicate and thus share already existing micro-structures (see Support Protocol 1). Once the template with the respective micro-channels and micro-pillars is available, our protocol starts with pouring PDMS onto the template (the wafer or epoxy replicate) to generate the microstructure of interest as a PDMS block (steps 1-6).
Generating a PDMS block bearing the respective micro-channels or micro-pillars
1.Weigh a 10:1 (w/w) mixture of PDMS and PDMS curing agent in a Thinky mixer cup.
2.Mix PDMS and PDMS curing agent in the Thinky ARE-250: 2,000 rpm for 2 min in mixing mode and 2,200 rpm for 2 min in defoaming mode.
3.Pour PDMS mix into the aluminum mold with the wafer (Fig. 2C).
4.Place wafer in a vacuum desiccator to remove remaining air bubbles (Fig. 2D).
5.Degas in the vacuum desiccator until no air bubbles are left.
6.Place wafer in an oven set to 80°C overnight (at least 4 hr).
Plasma bonding of the PDMS block to a glass surface
7.Remove PDMS from the wafer.
8.Cut each PDMS piece in size.
9.Punch holes for cell and chemokine administration into the PDMS piece with a puncher (Fig. 2I).
10.Cover the surface with Scotch tape for storage to avoid dust particles on the micro-structures.
11.Clean glass-bottom petri dishes with isopropanol to provide a glass surface suitable for imaging with an inverted microscope (see Materials section) by placing them into a beaker. Sonicate for 3 min.
12.Transfer imaging dish into a new beaker with pure ethanol and repeat sonication step.
13.Remove remaining ethanol from the imaging dish with compressed air.
14.Place PDMS pieces from step 10 with micro-structures facing upwards and the cleaned imaging dish from step 13 into a plasma cleaner.
15.Plasma clean PDMS pieces and imaging dishes at high power for 2 min.
16.Turn off plasma cleaner and carefully press PDMS pieces (features now facing downwards) onto the imaging dish.
17.Place bonded PDMS devices in an oven set to 80°C overnight.
Filling the micro-channels with medium
18.Prepare imaging medium (see recipe section).
19.Activate device for 2 min on high power in the plasma cleaner.
20.Flush PDMS device by pipetting 20 µl imaging medium into each entry hole.
21.If the plasma cleaning is not sufficient for flooding the entire micro-structure with imaging medium (e.g., narrow constrictions inside micro-channels might take longer to be filled with medium), place the dish in a vacuum desiccator for 5 min.
22.Overlay the entire PDMS device with medium (if not done during step 20).
23.Incubate for at least 1 hr in a humidified incubator at 37°C and 5% CO2.
Generating a chemokine gradient and cell administration
24.Remove most of the medium from the petri dish and entirely from all entry holes.
25.Add 20 µl chemoattractant solution (0.625 µg/µl CCL19 in imaging medium, 40× stock: 25 µg/µl CCL19 in 0.1% BSA in PBS) to each entry hole on one side of the micro-channels.
26.Place device in an incubator for 30 min to let the gradient be established.
27.Add cell suspension to each of the entry holes on the other side of the micro-channels.
28.Place device in an incubator and wait until the cells migrate to the structure of interest.
Imaging
29.Use an inverted microscope with an incubation chamber heated to 37°C and supplied with humidified 5% CO2. Set up the microscope according to your experimental setup and research question.
30.Optional: In addition to brightfield and epifluorescence microscopy, this assay can also be used with a wide range of other microscopic techniques. Total internal reflection fluorescence (TIRF) microscopy can be used to visualize structures in close proximity to the glass surface of the imaging dish (e.g., actin, integrins). Fluorescence lifetime imaging microscopy (FLIM) can be used to measure membrane tension in migrating cells, while a spinning disk confocal microscope allows the possibility of observing fast and dynamic processes within the migrating cells such as microtubule polymerization.
Support Protocol 1: EPOXY REPLICA OF GENERATED AND/OR PUBLISHED MICRO-STRUCTURES
Micro-structures can be replicated with epoxide resin (Kamande, Wang, & Taylor, 2015). As an alternative to wafers, epoxide replicas are recommended when access to wafers is limited. Epoxide replicas can be used to avoid the risk of breaking the fragile wafers accidentally during repeated production of PDMS devices. Manufacturing new wafers may also not always be feasible because the generation of wafers requires specialized laboratory equipment. Additionally, epoxide replicas can be used to share existing micro-structure designs with other research laboratories. To produce an epoxide replica, the micro-structures on the PDMS block (described in Basic Protocol 1) are imprinted into the resin, resulting in a replica of structures of wafer. The epoxide resin is mixed with a hardener to start a polymerization reaction, then the PDMS block is placed with the micro-structures pointing downwards into the still liquid epoxide. Upon solidification of the epoxide resin and removal of the PDMS, a negative replica of the PDMS is generated, which features the same micro-structures as the wafer and hence can be repeatedly used as a template to produce further PDMS blocks. However, it should be noted that minimal size alterations of the micro-structures (e.g., height, width) can occur during the replication procedure.
Materials
-
Epoxy Resin: Epoxy Resin L + Hardener S kit (R&G Faserverbundwerkstoffe)
-
Hardener: Epoxy Resin L + Hardener S kit (R&G Faserverbundwerkstoffe)
-
Petri dish: Cell culture dish, 150 mm, TC-treated (VWR, 734-2322)
-
Serological pipets: Advantage™ Serological Pipets (Corning, FALC 356543)
-
Vacuum desiccator (Kartel filtration and vacuum desiccators Art. 230 or equivalent)
-
Vacuum pump (Dekker RVR003H-OT or equivalent)
1.Cure PDMS as indicated in Basic Protocol 1 to generate structures for replication.
2.Mix epoxy components as indicated by the manufacturer on the bottle.
3.Mix carefully by stirring and transfer into a clean petri dish.
4.Wait for 15 min.
5.Remove the majority of air bubbles by putting the dish into a vacuum desiccator.
6.Add PDMS piece(s) on top of the epoxy, features facing downwards.
7.Put petri dish under vacuum for 1 hr.
8.Let epoxy harden at room temperature overnight.
9.Remove PDMS from the epoxy mold carefully.
Support Protocol 2: DENDRITIC CELL DIFFERENTIATION
Dendritic cells (DCs) are an important cell type linking the innate and adaptive immune system (Worbs, Hammerschmidt, & Förster, 2017). Immature DCs take up antigens in the tissue for MHC-II presentation. After antigen uptake, the DCs mature and migrate towards the lymph nodes where they present their captured antigens via MHC II to T cells and B cells. Their high migratory capacity in combination with the ability to migrate directly towards chemokines such as CCL21 and CCL19 makes DCs a versatile tool to investigate directed migration mechanisms in three dimensional (3D) environments.
In this support protocol, we describe the generation of immature and mature DCs using bone marrow derived cells (Lutz et al., 1999) or the immortalized progenitor cell line Hoxb8-FL (Leithner et al., 2018; Redecke et al., 2013). To initiate differentiation, the precursor cells are stimulated with granulocyte-macrophage colony-stimulating factor (GM-CSF). This activates the differentiation of monocytes and other progenitor cells into immature DCs and proinflammatory M1 macrophages. The macrophages are “sticky” (adhesive) and adhere to surfaces while the immature DCs stay in suspension or just weakly adhere to surfaces, allowing removal of the macrophages by collecting only “floating” (non-adhesive) cells (Sixt & Lämmermann 2011). Stimulation of immature DCs with bacterial derived lipopolysaccharides (LPS) leads to differentiation into mature DCs (Lutz et al., 1999). Mature DCs are chemotactic to CCL21 and CCL19 and can be identified by surface expression of Cd11c and MHC-II, while immature DCs do not express MHC II.
CAUTION : DC maturation can be triggered by freezing, thus immature DCs can only be used directly after differentiation without freezing.
Materials
-
Hoxb8-FL cells (kindly provided by Prof. Michael Sixt's lab, ISTA)
-
Bone marrow cells (isolated from male C57Bl6/J mice aged 2 to 3 months, bred in the Core Facility - Animal Models at Biomedical Center Munich)
-
R10 medium supplemented with 10% GM-CSF supernatant (see recipe)
-
R10 medium supplemented with 20% GM-CSF supernatant (see recipe)
-
Lipopolysaccharide (LPS) from Escherichia coli O26:B6 (MilliporeSigma, L2654-1MG)
-
Freezing medium (see recipe)
-
100-mm petri dish (PS, 94/16; Greiner Bio-One, MM 632180)
-
Serological pipets: Advantage™ Serological Pipets (Corning, FALC 356543, FALC 356551)
-
Pipet tips (starlab):
- 10 µl graduated TipOne® Tip (S1111-3700-C)
- 1,000 µl graduated TipOne® Tip (S1111-6701-C)
- 200 µl UltraPoint® graduated TipOne® Tip (S1113-1706-C)
-
Cryotubes: Nunc™ Biobanking and Cell Culture Cryogenic Tubes (Thermo Fisher Scientific™, 368632) or equivalent
-
Cell culture treated petri dish (Eppendorf® Cell Culture Dishes, 0030702115)
-
Cell counting chamber (Marienfeld Superior™ Counting Chamber or equivalent)
-
PIPETBOY (ClearLine® pipet controller or equivalent)
-
Pipet (Eppendorf Research plus or equivalent)
-
Cell culture incubator (Eppendorf Galaxy 170R or equivalent)
-
Cell culture centrifuge (Hettich Rotina 35R or equivalent)
-
Styrofoam box
-
-80°C freezer (Thermo Scientific HERAfreeze™ HFU T Series or equivalent)
-
Liquid nitrogen tank (Thermo Scientific™ CryoPlus™ Storage Systems or equivalent)
Day 0
DCs can be differentiated from either murine bone marrow or an immortalized progenitor Hoxb8-FL cell line. For detailed information see Lutz et al., 1999; Redecke et al., 2013; and Leithner et al., 2018.
1.Count cells and seed 2 × 105 cells for Hoxb8-FL derived DCs or 2.5 × 106 cells for bone marrow derived DCs per 100-mm petri dish containing 10 ml R10 medium supplemented with 10% GM-CSF.
2.Incubate in a humidified incubator at 37°C and 5 % CO2.
Day 3
3.Add 10 ml R10 medium supplemented with 20% GM-CSF.
4.Incubate in a humidified incubator at 37°C and 5% CO2.
Day 6
5.Remove 10 ml medium.
6.Add 10 ml R10 medium supplemented with 20% GM-CSF.
7.Incubate in a humidified incubator at 37°C and 5% CO2.
Day 8
Cells can be used directly as immature DCs or stimulated with LPS to generate mature DCs. Cells can also be frozen for storage and stimulated after thawing at a later time point.
8.Collect nonadherent and loosely adherent cells by taking up the medium and flushing the plate multiple times.
9.Centrifuge at 300 × g for 5 min.
10.Optional: Resuspend cells in freezing medium at a density of 5 × 106 cells/ml and place into a Styrofoam box at −80°C.
11.Count cells, resuspend in R10 medium supplemented with 10% GM-CSF, and seed cells on a 100-mm tissue culture treated petri dish.
12.Add 200 ng/ml LPS (5,000× stock in PBS).
13.Incubate in a humidified incubator at 37°C and 5% CO2 overnight.
Basic Protocol 2: IMMUNE CELL MIGRATION IN 3D COLLAGEN NETWORKS OF VARIABLE PORE SIZES
The described generation of PDMS-based “pore-choice” micro-channels and micro-pillars (Basic Protocol 1) requires equipment, which may not be readily available, such as physical templates of the desired micro-structures and a plasma cleaner. In contrast, 3D collagen networks are defined microenvironments that can be generated with equipment available in most laboratories.
In the following protocol, we generate 3D in vitro networks of collagen type I which represents a major constituent of most interstitial tissues (Wolf et al., 2009). As an experimental setup, we use 6-well plates or 30-mm petri dishes in which we drill holes of 17-mm diameter. Closing these holes with glass slides on both sides generates chambers with defined dimensions and suitable optical properties for imaging (Fig. 4A). These chambers are filled with a suspension of collagen and cells. It is important to note that the formed collagen network is heterogeneous in its porosity, meaning that different pore sizes are next to each other. Notably, the pore sizes and complexity of these collagen networks can be adjusted by changing the amount of collagen added to the mixture. For this purpose, we describe the usage of three different bovine collagen type I preparations: PureCol® which contains 3 mg/ml collagen, Nutragen® containing 6 mg/ml collagen, and FibriCol® containing 10 mg/ml collagen. Accordingly, collagen networks generated with PureCol® are the least dense collagen networks with the largest average pore sizes while the collagen networks generated with FibriCol® have the highest collagen density with the smallest average pore sizes (Renkawitz et al., 2019).
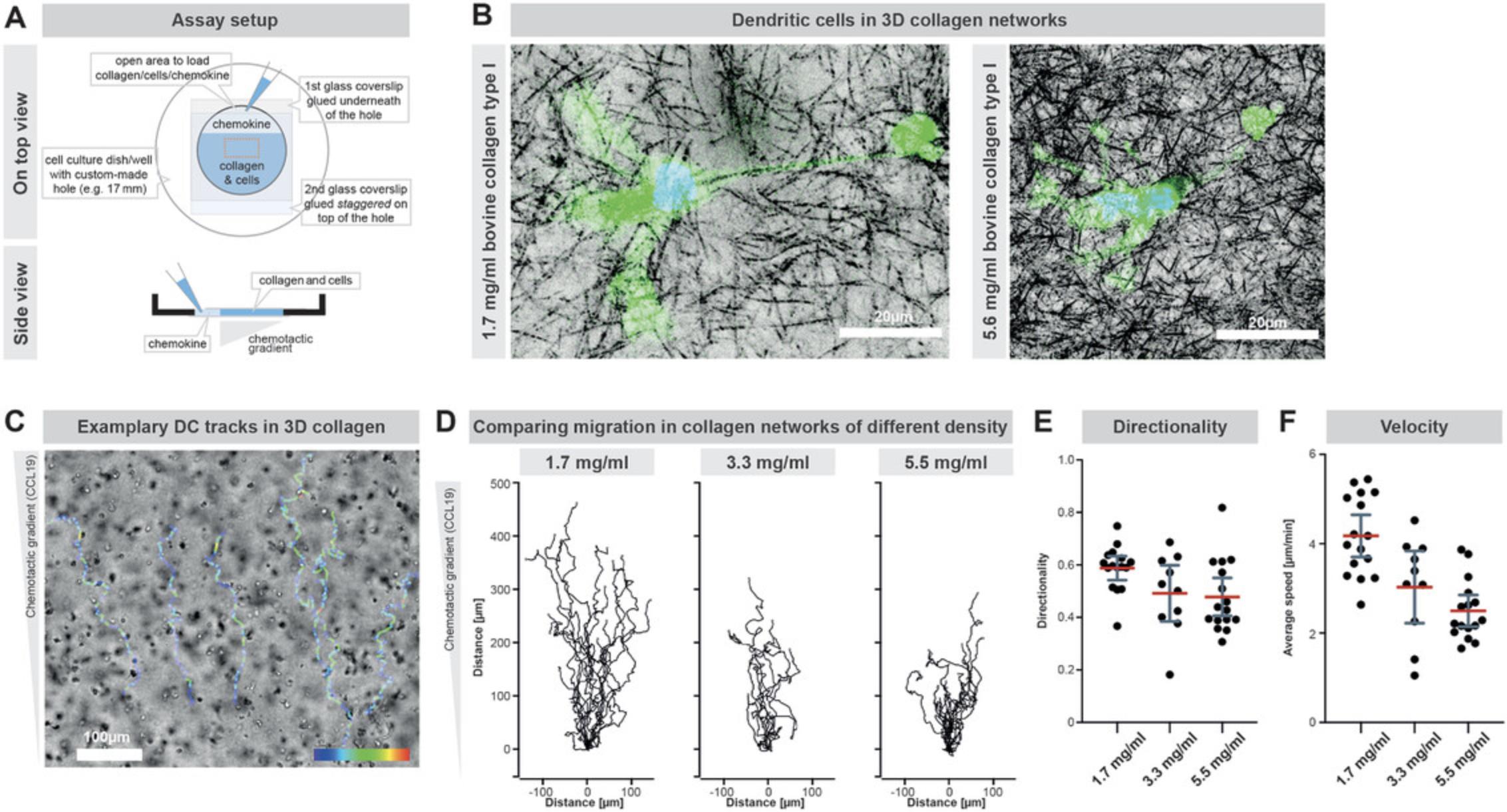
Similar to the PDMS-based micro-channels, a chemokine gradient can be used to guide directed cell migration. For this purpose, the chemokine is diluted in cell culture medium and added on one side of the solidified collagen gel. A chemokine gradient is formed upon diffusion into the collagen network, which attracts the cells towards the respective side of the collagen gel.
We here describe the usage of collagen networks as porous environments for DC migration (Lämmermann et al., 2008; Sixt & Lämmermann 2011), yet collagen networks are versatile as they have been already used with neutrophils (François et al., 2021; Salvermoser et al., 2018; Wolf et al., 2013), T cells (Wolf et al., 2013), fibroblasts, and cancer cells (Doyle, Sykora, Pacheco, Kutys, & Yamada, 2021; Ilina et al., 2020; Infante et al., 2018; Sabeh et al., 2009; Wolf et al., 2013). In summary, collagen networks give insights into cell migration in physiologically close porous environments with different complexity and pore sizes. Thereby, collagen networks can be used to investigate migration mechanisms like cellular squeezing through individual pores and cell motility through less or more dense microenvironments. From a technical perspective, this assay further allows the imaging of large numbers of motile cells simultaneously. Yet, quantifying individual pore sizes is not trivial.
Materials
-
Paraffin wax (MilliporeSigma, 76242)
-
Dendritic cells (DCs; see Support Protocol 2)
-
R10 medium (see recipe)
-
Gibco™ MEM, 10×, no glutamine (Thermo Fisher Scientific, 11-430-030)
-
7.5% sodium bicarbonate (NaHCO3) solution (MilliporeSigma, S8761)
-
Collagen (CellSystems):
- Bovine collagen solution Nutragen® (5010-50ML)
- PureCol® (5005-100ML)
- FibriCol™ (5133-20 ml)
-
Chemokine: Recombinant Mouse CCL19/MIP-3 beta Protein (Bio-Techne, 440-M3-025)
-
60-mm petri dish with 17-mm hole (Falcon™ Easy-Grip Tissue Culture Dishes, 60 mm; Corning, 353004; holes need to be added manually)
-
Paintbrush
-
22- × 22-mm coverslip: Karl Hecht™ Microscope Cover Slips (Thermo Fisher Scientific, 41014515)
-
20- × 20-mm cover slip: Karl Hecht™ Microscope Cover Slips (Thermo Fisher Scientific, 41014512)
-
Cell counting chamber (Marienfeld Superior™ Counting Chamber or equivalent)
-
Pipet tips (starlab):
- 10 µl graduated TipOne® Tip (S1111-3700-C)
- 1,000 µl graduated TipOne® Tip (S1111-6701-C)
- 200 µl UltraPoint® graduated TipOne® Tip (S1113-1706-C)
-
Heating plate (Ikamag RH or equivalent)
-
Pipet (Eppendorf Research plus or equivalent)
-
Cell culture incubator (Eppendorf Galaxy 170R or equivalent)
-
Microscope with incubation chamber and temperature control unit (Leica DMI 8, Incubator i8, Pecon® TempController 2000 or equivalent)
Preparing collagen chamber
1.Make use of a petri dish with a hole (diameter 17 mm; Fig. 4A).
2.Make a circle of liquid paraffin around the hole on the bottom of the plate using a paintbrush.
3.Carefully press a coverslip (22 × 22 mm) on the hole and heat paraffin by holding the plate over a hot plate. Wait until the paraffin has melted and thereby tightly sealed the coverslip to the plate.
4.Seal coverslip from the bottom by painting around the coverslip with paraffin.
5.Place a coverslip (20 × 20 mm) on the inside of the dish leaving a small opening on the top.
6.Fix coverslip with paraffin.
Prepare cells and collagen mixture
7.Adjust the number of mature dendritic cells to 3–5 × 105 cells in 100 µl R10 medium (per well).
8.Prepare collagen mixture in the order given in Table 1 (indicated volumes are needed for one well).
| Component | Volume |
|---|---|
| MEM, 10× | 30 µl |
| NaHCO3 | 15 µl |
| Collagen | 225 µl |
| Problem | Possible cause | Solution |
|---|---|---|
| Collagen does not polymerize | The ratio of components used is not correct | Collagen solidifies at neutral pH, NaHCO3 neutralizes the HCl from the collagen solution. The ratio of the two components has to be accurate. |
| NaHCO3 is too old. The color should turn purple when adding NaHCO3 to 10× MEM and orange-red when adding the collagen | Make sure the pipetting is accurate and test with fresh NaHCO3 | |
| The collagen needs more time to fully polymerize | The gel should solidify at 37°C and 5% CO2 after 45 min for high collagen concentrations (5.6 mg/ml). However, lower collagen concentrations may need up to 75 min. | |
| A drift of the gel is observed | Drift can be caused by the microscope stage or by gel distorting when not fully polymerized. For the analysis, discard the first 30 min of recording. | |
| Collagen does not polymerize homogeneously (streaks are visible) | Collagen solidifies partially while mixing or before adding it into the chamber | Especially for high collagen concentrations, pipet each well individually to reduce the time between mixing and adding collagen mix in the chamber |
| Components are not mixed properly | Make sure to properly mix collagen mixture and cells. If necessary, prepare larger volumes to be able to pipet without introducing bubbles. A combination of swirling the pipet and pipetting up and down can be used to improve mixing. | |
| Collagen does not build a flat line at the top | Highly concentrated collagen may not easily flow to the bottom of the chamber | Strongly knock the dish against the metal surface of the incubator repeatedly until the cell-collagen suspension fills the chamber and creates a flat surface line. This process also can eliminate larger bubbles. |
| Cells are dying in the collagen (e.g., during imaging) | 37°C, 5% CO2 or humidity is not maintained, which leads to high pH or evaporation | The chamber can be sealed directly after adding the chemokine solution but if the seal is not air tight the gel will turn purple, indicating a basic pH and cells will die quickly. Make sure to seal chamber properly or use 5% CO2 + high humidity during imaging. |
| High laser power might lead to phototoxicity | Make sure laser intensity and exposure time is not too high. Especially during setting autofocus under constant illumination, cells might die quickly. | |
| Staining bleaches quickly | Collagen scatters light, leading to lower fluorescence signals. Compensation with higher light intensity can lead to bleaching. For discussion of bleaching see above. | Scattering can be limited by use of longer wavelengths (using respective fluorophores), improving the signal and necessary light intensity. Also, by imaging relatively close to the glass surface facing the objective. Note that collagen polymerization might be different close to the glass compared to the center of the gel. Typically, 100-200 µm distance to the glass is appropriate for fluorescence imaging. |
| Cells are not migrating and stay spherical | Insufficient chemokine is present for the cell type | Check the appropriate concentration of chemokine. Different chemokines may require other concentrations. |
| Chemokine needs more time to diffuse to cells or chemokine gradient is too shallow for cells to detect it, e.g., if imaging started too late | Avoid air bubbles in the gel and chemokine solution, as they prevent local diffusion of chemokine. Depending on the hydrodynamic radius of the chemokine, the time to diffusion to the cells may vary. Optimize distance of the imaging area to the chemokine or incubate it longer before imaging. To this end, regularly check if the cells start migrating. With CCL19, the chemokine gradient establishes ∼30-60 min after adding chemokine, depending on the distance to the chemokine solution. After 3-5 hr the gradient will get shallower and cells will no longer migrate in a directional manner. | |
| Gel does not solidify well. In particular, if the gel does not stick to the glass surface, which is the case when the gel can slide around in the chamber. | Make sure collagen is completely solidified. If the gel can “turn” in the chamber (e.g., when you turn the dish and it slides around), the chemokine will diffuse at the glass wall and diffuse into the gel from the top and bottom. |
9.Add 200 µl collagen mix to the cell suspension and gently pipet up and down to avoid air bubbles.
10.Pipet 280 µl of the mixture into the prepared chamber.
11.Incubate chamber for 45-75 min (see Table 2 for troubleshooting suggestions) at 37°C and 5% CO2 to allow the polymerization of the gel.
Gradient formation and imaging
12.Dilute chemokine to a final concentration of 0.625 µg/µl in R10 medium supplemented with 10% GM-CSF (40× stock with 0.1% BSA in PBS).
13.Fill chamber on top of the gel with chemokine solution.
14.Seal the small opening of the chamber with pre-warmed liquid paraffin.
Imaging
15.Use an inverted microscope with an incubation chamber heated to 37°C. Set up the microscope according to your experimental setup and research question.
16.Optional: Normally, we record the collagen assay at a relatively low resolution (e.g., 4× or 10×; Fig. 4C) for a time period of 5 hr with an image recorded every minute. Here, cells should start migrating towards the chemotactic gradient (Fig. 4C-F), which diffuses to the cells after ∼10-30 min at the gel side facing the chemokine. This allows the possibility of recording and quantifying (Fig. 4D-F; see also the section Understanding Results) large cell numbers and different conditions in one assay (using an automatic microscope stage). In addition to brightfield microscopy, this assay can also be used for a wide range of other microscopic techniques. Dark-field microscopy can be used to remove background light in the image (cells appear white on a dark background) for automatic tracking. Furthermore, the collagen migration assay is compatible with confocal and epifluorescence microscopy (Fig. 4B). For this purpose, we recommend using lower collagen concentrations as collagen fibers scatter light which can interfere with the fluorescence signal (see Table 2 for troubleshooting suggestions). Instead of live-cell imaging, the collagen gel can be used for immunofluorescence imaging. To this end, the collagen gel with the cells is carefully removed from the plate after a desired migration period and fixed in 4% paraformaldehyde (PFA), followed by subsequent staining steps and confocal imaging.
REAGENTS AND SOLUTIONS
Freezing medium
- 45 ml Gibco™ FBS, qualified, Brazil (Thermo Fisher Scientific, 11573397)
- 5 ml methyl sulfoxide (DMSO), 99.8+ %, for molecular biology (Thermo Fisher Scientific, 10397841)
Store for up to 1 year at −20°C.
Imaging medium
- 500 ml RPMI 1640 medium phenol free (Gibco™ RPMI 1640 medium, without phenol red)
- 50 ml Gibco™ FBS, qualified, Brazil (Thermo Fisher Scientific, 11573397)
- 5 ml Gibco™ Penicillin-Streptomycin, 10,000 U/ml (MilliporeSigma, P0781-100 ml)
- 550 µl Gibco™ 2-mercaptoethanol, 50 mM (Thermo Fisher Scientific, 31350010)
- 50 μM L-ascorbic acid (MilliporeSigma, A92902-100G)
Store for up to 4 weeks at 4°C.
R10 medium
- 500 ml RPMI 1640 medium (Thermo Fisher Scientific, 21875059)
- 50 ml Gibco™ FBS, qualified, Brazil (Thermo Fisher Scientific, 11573397)
- 5 ml Gibco™ Penicillin-Streptomycin, 10,000 U/ml (MilliporeSigma, P0781-100 ml)
- 550 µl Gibco™ 2-mercaptoethanol, 50 mM (Thermo Fisher Scientific, 31350010)
Store for up to 4 weeks at 4°C.
R10 medium supplemented with 10% GM-CSF supernatant
- 500 ml RPMI 1640 medium (Thermo Fisher Scientific, 21875059)
- 50 ml Gibco™ FBS, qualified, Brazil (Thermo Fisher Scientific, 11573397)
- 5 ml Gibco™ Penicillin-Streptomycin, 10,000 U/ml (MilliporeSigma, P0781-100 ml)
- 550 µl Gibco™ 2-mercaptoethanol, 50 mM (Thermo Fisher Scientific, 31350010)
- 61 ml GM-CSF supernatant (harvested from cell line expressing recombinant GM-CSF, cell line kindly provided by Prof. Michael Sixt's lab, ISTA)
Store for up to 4 weeks at 4°C.
R10 medium supplemented with 20% GM-CSF supernatant
- 500 ml RPMI 1640 medium (Thermo Fisher Scientific, 21875059)
- 50 ml Gibco™ FBS, qualified, Brazil (Thermo Fisher Scientific, 11573397)
- 5 ml Gibco™ Penicillin-Streptomycin, 10,000 U/ml (MilliporeSigma, P0781-100 ml)
- 550 µl Gibco™ 2-mercaptoethanol, 50 mM (Thermo Fisher Scientific, 31350010)
- 122 ml GM-CSF supernatant (harvested from cell line expressing recombinant GM-CSF, cell line kindly provided by Prof. Michael Sixt's lab, ISTA)
Store for up to 4 weeks at 4°C.
COMMENTARY
Background Information
Immune cells migrate highly efficiently through the diverse microenvironments of multicellular organisms. Our understanding of the underlying principles is supported by experimentation in vitro (Garcia-Arcos et al., 2019) upon pioneering research to reconstruct 3D microenvironments for immune cell migration (Faure-André et al., 2008; Heuzé, Collin, Terriac, Lennon-Duménil, & Piel, 2011; Lämmermann et al., 2008; Lautenschläger & Piel, 2013; Vargas et al., 2016). These 3D and 3D-like microenvironments enable the reductionistic investigation of individual cellular and microenvironmental parameters, such as the regulation of immune cell migration by environmental confinement (Lomakin et al., 2020), topography (Reversat et al., 2020), hydraulic resistance (Moreau et al., 2019), chemokine gradients (Schwarz et al., 2017), fluid flow (Pruenster et al., 2015), and porosity (Renkawitz et al., 2019; Thiam et al., 2016; Wolf et al., 2013).
In this protocol, we focus on experimental assays to investigate cell motility through microenvironments of heterogeneous porosity. Specifically, we describe the usage of (1) micro-channels with path junctions of different pore sizes, (2) micro-pillars arranged in various distances to form different pore sizes, and (3) collagen networks with different pore sizes.
Upon pioneering migration experiments with dendritic cells in linear micro-channels (Faure-André et al., 2008), micro-channels have become a highly valuable experimental tool to elucidate the mechanisms of immune cell motility (Kameritsch & Renkawitz, 2020; Moreau, Piel, Voituriez, & Lennon-Duménil, 2018), such as actomyosin dynamics (Chabaud et al., 2015; Wilson et al., 2013), nuclear deformation (Estabrook, Thiam, Piel, & Hawkins, 2021; Thiam et al., 2016), pore decisions (Renkawitz et al., 2019), directional decisions (Ambravaneswaran, Wong, Aranyosi, Toner, & Irimia, 2010; Wang et al., 2020), protrusion coordination (Hadjitheodorou et al., 2021; Kopf et al., 2020), and signaling responses such as calcium signaling and responses to extracellular ATP (Sáez et al., 2017; Solanes et al., 2015). Further, micro-channels have also been successfully employed with non-immune cells like the single cell amoeba Dictyostelium (Tweedy et al., 2020) and cancer cells (Bergert et al., 2015; Tweedy et al., 2020).
When investigating either slowly or nondirectionally moving cells, micro-pillar assays have the advantage of enabling cellular analysis directly upon cellular entrance from the loading hole to the micro-pillar area. Micro-pillars that cover one surface have been widely applied as cellular adhesion sites for adhesive cells (Brand et al., 2017; Rossier et al., 2010), nuclear deformation structures (Hanson et al., 2015), and as a readout for cellular traction forces (Shiu, Aires, Lin, & Vogel, 2018). Yet, when micro-pillars are connecting two surfaces, they form a barrier for cell migration and thus a heterogenous microenvironment with pore sizes according to the distance between individual micro-pillars (Denais et al., 2016; Renkawitz et al., 2018). In this protocol we describe the usage of such micro-pillars connecting a PDMS surface and a glass surface. Notably, adjusting the distance, size, and arrangement of the micro-pillars enables designing of the experimental setup according to the research model and cell type of interest (Denais et al., 2016; Kopf et al., 2020; Renkawitz et al., 2019).
We further describe collagen networks as a third in vitro assay to investigate cell migration in porous microenvironments. Advantages of collagen networks are their closer physiological properties, such as deformability and proteolytic cleavability (Sabeh et al., 2009; Wolf et al., 2013). However, measuring the size of individual pores during cell migration requires fast and high-resolution microscopy and subsequent computational analysis (Renkawitz et al., 2019) based on collagen visualization by confocal reflection microscopy or fluorescent collagen labeling (Sixt & Lämmermann 2011). Whereas we here describe the usage of non-crosslinked bovine collagen type I networks that have been pioneered earlier for migration studies with immune cells and non-immune cells (Lämmermann et al., 2008; Wolf et al., 2013; Wolf et al., 2009), alternative protocols using rat tail collagen preparations (Wolf et al., 2013), or cross-linked collagen networks (Sabeh et al., 2009) have been described. Collagen networks from rat tail even enable the generation of different sized pores by polymerization of the same collagen concentration at different temperatures (Wolf et al., 2013). Thus, the physical and molecular properties depend on the collagen source and preparation (Wolf et al., 2009), including its porosity and bundle thickness.
Critical Parameters
Micro-channels and micro-pillars
Critical parameters of these assays are optimal cell numbers, the proper formation of a stable chemokine gradient, and accurate usage of inhibitors (see also Table 3). The optimal cell number has to be determined depending on the experiment, pattern design, and cell type. For cells migrating without a chemokine gradient, sufficient cell numbers may be needed to allow cells to directly touch the edge of the PDMS device after sedimentation. For a loading hole with a 3-mm diameter, we recommend using 10,000 to 20,000 cells when using DCs as a cellular model (Fig. 2J). With a custom-made puncher or by punching several overlapping holes next to each other, a larger hole can be produced with a loading area of ∼3 × 9 mm (Fig. 2K, L). For this size, we recommend using 50,000 to 100,000 DCs. When too many cells enter the micro-structures, cells can touch each other, influencing experimental results or leading to a traffic jam of cells that block each other's paths. If a traffic jam is observed (many cells touching each other and arrested migration), cell numbers should be reduced in the next experiment. Patterns with unidirectional paths are less susceptible to traffic jams compared to patterns where cell paths can regularly cross and migrate in different directions. However, high cell densities can still lead to blockages at the entry sites of the loading hole into the micro-structures. To avoid this, the design of micro-channels can be adjusted to include a region of micro-pillars with large distances in front of the channels, where cells can distribute before reaching the entrances to the micro-channels (see Fig. 2M, N). For cells with reduced migration capabilities (e.g., genetic knockouts or cells treated with inhibitors) higher cell numbers (e.g., two times compared to wild type and/or untreated) might have to be used in order to observe sufficient numbers of migrating cells in the micro-structures.
| Problem | Possible cause | Solution |
|---|---|---|
| Cells do not migrate into the chamber | Cells need more time to migrate into the micro-structures | Start imaging when cells start to migrate into the micro-structures by checking with a microscope when the first cells start to enter the channels. The start of the imaging strongly depends on the cell number used and the distance between the two holes (see below). |
| Too few or too many cells: Cells are not close enough to the entrance of the micro-structures or the cells block each other at the entrance or in the channels | Observe the cell-loaded hole at the microscope: Cell density after sedimentation can be used to estimate adequate cell numbers, even when changing the size of the loading hole. If cells seem too dense in the hole or block each other in the micro-structures, reduce cell number per hole. | |
| The distance between the two loading holes is too large so that the chemokine gradient does not establish in time | Reduce distance between holes, if possible. If your pattern does not allow it, incubate cells longer before imaging or incubate device longer before adding cells (cell loading hole can be left empty during this step). | |
| Chemokine solution was swapped into the cell loading hole (on top of the PDMS block) | Make sure chemokine solution does not get into the loading hole of the cells. To this end, the area outside of the PDMS should not be filled too high (to prevent swapping of medium over the PDMS) and there should be some space between the liquid surface in the hole and the surface of the PDMS (∼1 mm). Make sure to remove all liquid that is on top of the PDMS (in particular on top between the loading holes) before adding cells and chemokine. | |
| Fluid flow between holes due to different fill heights, preventing formation of chemokine gradients | Make sure medium in both holes is filled to the same height to prevent hydrodynamic pressure. If the holes do not have the same area, the volume added to each should be adjusted. Flow can be observed if small, e.g., dust particles flow through the channels. | |
| Cells get stuck in a traffic jam inside the micro-structures | Too many cells are in the loading hole and enter the micro-structures at the same time | Try reducing cell number per hole. If possible, adjust the design of the micro-channels to have micro-pillars with large distances in front of the channel entrances to allow distribution of cells on their way to the channel entrances, thus ensuring that cells do not arrive at the entrances at the same time. |
| Cells at the front stop migrating and block the other cells | Make sure you have a stable chemokine gradient (e.g., the distance between the holes should not be too large, allow sufficient incubation time after adding chemokine) | |
| Cells die in the channel and block other cells | Make sure to use healthy cells. This can be difficult when using harmful inhibitors or drugs. In this case try to optimize cell number or incubation time, or the concentration of the drug. See also below (37°C and 5% CO2). | |
| Cells are dying during imaging | Humidity, 37°C, or 5% CO2 is not maintained, which can lead to high pH or evaporation and osmotic shock | Do not unnecessarily leave the device outside of the incubator. Make sure CO2 and heating (37°C and 5% CO2) works properly in the microscope. We strongly recommend preheating the microscope chamber for 2 hr. Make sure humidity is very high in the incubator; if needed preheat water through which the CO2-air mix flows. Add water in reservoirs of the imaging chamber where possible (e.g., into the empty space between the wells of a 6-well plate). |
| High laser power leads to phototoxicity | Make sure laser intensity and exposure time is not too high. We recommend not using an autofocus system based on fluorescence, as setting the autofocus under constant illumination might quickly result in cell death, especially if the laser power is high. | |
| Cell viability is too low | Use healthy cells (see row above) | |
| Staining bleaches quickly | Bleaching is typically caused by high light dosage or intensity, sensitive fluorophores, or inadequate imaging medium | Improve imaging settings (e.g., lower laser intensity and exposure time). The addition of ascorbic acid (vitamin C) in the imaging medium should limit photo bleaching. Fluorophores with longer wavelengths are typically more photostable. |
High background signals (e.g., due to autofluorescence) may require the use of high laser power to reach a sufficient signal-to-noise ratio |
Background signal due to autofluorescence of cellular components can also be reduced by choosing fluorophores with longer wavelengths. Also, use medium without phenol red. |
Two parameters are critical for the formation of chemokine gradients in micro-channels and micro-pillars: (1) The distance between the chemokine hole and the cell hole can substantially influence the timing of chemokine gradient formation, as larger distances will lead to more shallow gradients that can be sensed by the cells only after a longer time period. Note that the design of the micro-structures determines the minimal required distance, e.g., for the here-described four-way junctions we recommend using a distance of 1 to 1.5 mm. Shorter distances can be used with micro-pillar designs, for which we recommend using 1 mm. (2) Fluid flow from one loading hole to the other loading hole prevents the formation of a stable chemokine gradient. This fluid flow can occur when the two holes are not filled to the same height: Fluid flows through the micro-structures to equalize the pressure. This flow can result in leakage of chemokine into the loading hole of the cells (disrupting a gradient) or fluid flow towards the chemokine loading hole prevents chemokine from diffusing towards the loading hole of the cells. To prevent this fluid flow, we recommend punching both loading holes with the same puncher, followed by adding the same volume to both holes precisely. When generating loading holes that consist of several overlapping holes (see paragraph above), we recommend carefully adjusting the volume in the different holes to reach the same fill height. Flow can occasionally be observed in a microscope as particles (dust or cell debris) flow through the micro-structures. In this case, small volumes (in steps of 1 to 2 µl) of imaging medium can be added to the hole that the particles are flowing towards.
When using inhibitors or other chemical drugs in micro-channel and micro-pillar assays, we recommend the following steps to ensure accurate distribution and concentration of the inhibitor in the micro-channels. It is crucial to add inhibitors to the medium used for initial filling as well as the medium of the cells and chemokine. Inhibitors should always be freshly dissolved in medium from (frozen) stock. Also note that inhibitor stock solutions with DMSO or ethanol should not be used at lower dilutions than 1:1,000 and 1:10,000, respectively, as they might influence cellular properties. For the use of heat or light sensitive inhibitors, the PDMS device should be equilibrated for only 1 hr in the incubator before adding the chemokine or cells or lights should be turned off during the pipetting step, respectively.
When investigating intracellular dynamics (e.g., cytoskeletal dynamics), the glass coverslip of the PDMS device offers ideal imaging qualities and can be used with a high numerical aperture (NA) and high magnification objective (63× to 100×) to record a single cell at high resolution. Here, cells have to be imaged individually to record sufficient numbers of cells. However, the large area of the micro-patterns in the PDMS device also allows the recording of many at once. Here, we recommend using 20× to 40× objectives and recording multiple tiles in parallel, when a motorized microscope stage is available. Depending on the required temporal resolution and the speed of the microscope, the maximum number of parallel areas recorded may be limited. We typically record approximately ten to fifteen tiles and three channels per minute in parallel.
DC differentiation
It is critical for proper DC differentiation that the cells do not grow too dense. Hoxb8-FL cells tend to still divide during the first few days of differentiation. Depending on the replication rate of the Hoxb8-FL cells, in particular for different genetically modified Hoxb8-FL cell lines, initial cell numbers should be adjusted accordingly (higher cell numbers for slower replication cell lines). A measure of whether cells grew too dense is the color of the medium on day 8, where the medium should be orange-red and not bright yellow. It is normal that during the differentiation adherent cells (macrophage-like cells) appear as well. It is critical to use untreated petri dishes for DC differentiation. Tissue culture treated dishes should be used for LPS stimulation to remove any remaining adherent cells while collecting the suspension cells the next day. Further, we noted differences in the efficiency of DC differentiation depending on the FBS batch.
Collagen
Collagen polymerization occurs at neutral pH and 37°C. To prevent cell damage, the components of the collagen mix should be prewarmed to room temperature (prewarming to 37°C might induce polymerization already during pipetting; see also Table 2). Lower concentration collagen gels will take longer for complete polymerization. We recommend incubating the collagen for 75 min in the incubator. The incubation time may be reduced to 45 min for highly concentrated collagen gels, if necessary. Note that there might be significant lateral deformation of the gel during the first 10 to 30 min of recording if the gel is not completely polymerized, impeding subsequent analysis. Furthermore, it is critical to avoid the generation of any bubbles in the collagen gel (e.g., by pipetting larger volumes). Bubbles limit the area that can be imaged but also can locally disturb the formation of a chemokine gradient. For comparable results, images should always be recorded at the same distance to the chemokine-gel interface. Also, bubbles in the chemokine solution, which can form when the chamber is not completely filled with chemokine solution, should be avoided. If such a bubble ends up touching the gel, chemokine diffusion might be hindered locally. Cell migration velocity towards the chemokine typically increases until the full chemokine gradient is established (after 30 to 60 min, depending on the distance to the chemokine solution) and subsequently slows down as the gradient gets shallower. Migration analysis should take this into account and a specific time span (e.g., 30 to 170 min after adding chemokine) that is equal in all conditions should be analyzed.
Troubleshooting
See Tables 2 and 3 for common problems encountered when performing these protocols and suggested solutions.
Statistical Analysis
Migrating cells can behave considerably inhomogeneously, resulting in a large variance of quantified properties (such as migration speed) between cells and over time for each single cell. Depending on the parameter measured, data points may not be normally distributed, requiring appropriate statistical tests to compare different conditions. Depending on the variance in the sample, sufficient cell numbers have to be measured and the experimental setup has to be adjusted accordingly (more frames recorded, more repetitions). At least three biologically independent experiments (DCs differentiated from different mice or DCs differentiated from Hoxb8-FL cells independently, i.e., not at the same time) should be conducted.
In comparison to micro-channels, collagen gels allow the analysis of even more cells simultaneously (∼600 × 600 µm area per frame possible), allowing for robust quantification of migration speeds. However, if manual quantification (e.g., tracking) is required, an appropriate compromise between manual work time and statistical results has to be established. Based on our experience (Kopf et al., 2020; Renkawitz et al., 2019), at least ten cells per replicate have to be measured to capture a meaningful range of the quantified property, whereas a higher cell number may be necessary for parameters with large variances. Great care has to be taken not to introduce a bias, for instance by nonrandomly selecting cells for analysis. Thus, defined and reproducible parameters on how to select cells have to be determined before starting the quantification, e.g., dead or round nonmoving cells should not be quantified.
Understanding Results
In order to efficiently navigate through complex microenvironments, immune cells have to find suitable paths among alternative microenvironmental pores, while maintaining cellular integrity during intense cellular deformation. The here described assays replicate many of the challenges migrating cells face in vivo and can therefore be used to investigate the cellular properties required for efficient migration in complex environments.
The pore-decision micro-channels (Fig. 3B) offer a highly reductionist environment that simplifies quantification of individual migration parameters of cells migrating through porous microenvironments. First, counting the cellular passages through individual pore sizes allows the investigation of a cellular pore size preference and thus the investigation of migration along the path of least resistance. Second, the passage time through the path junction can be measured (e.g., from the moment when the cell touches the junction until it completely leaves it) to quantify the pace of cellular decision making for a specific path. Third, the assay enables the precise quantification of the number of explored pores by cellular protrusions. Thereby, one can draw conclusions on the microenvironment exploration activity of the cell type of interest. Further, this allows us to precisely investigate the protrusion and retraction dynamics of individual cell fronts during cellular path selection and thus represents a measure of intracellular coordination. Likewise, the assay enables the investigation of intracellular components and organelles, such as the measurement of nuclear deformation and probing as a mechanical gauge into the individual pores. Lastly, when the cellular coordination is impaired (e.g., upon microtubule inhibition), migrating cells can self-fragment during path decisions (Kopf et al., 2020), which can be reliably quantified in these highly reductionistic microenvironments. Comparing these results with the cellular passage through single pores (Fig. 3E) distinguishes cellular squeezing defects from pore-decision and navigation defects.
The described micro-pillars represent an additional reductionist microenvironment with precisely defined pore sizes, yet they offer a complementary set of results. Whereas the cell shape and thus the protrusion-retraction dynamics are more difficult to be quantified in a standardized manner (Fig. 3D), micro-pillars enable the investigation of consecutive pore decisions along longer cellular paths. This allows the usage of cell tracking and visualization tools (such as ImageJ Manual Tracking plugin or Imaris-based tracking of fluorescently labeled nuclei) to visualize and quantify motility parameters like the cellular velocity (Fig. 3C) next to the quantification of the pore size preference.
Collagen networks represent even more complex environments with various pore sizes. Notably, the pore sizes can be adjusted by using different collagen concentrations, where higher collagen concentrations result in smaller pore sizes and an altered cellular shape (Fig. 4B). Cell tracking (e.g., using ibidi's chemotaxis and migration tool; Fig. 4D-F) quantifies the cellular directionality and velocity during local navigation through the collagen network (Fig. 4C), and establishes faster and more directed motility in networks of lower collagen density and pore size (Fig. 4D-F).
In summary, using these complementary assays enables the investigation of different cellular models and genetically or pharmacologically perturbed cells to elucidate the migration mechanisms through microenvironments of heterogeneous porosity.
Time Considerations
Basic Protocol 1
Starting from a microfabricated mold, the protocol is relatively simple, inexpensive, and quick as it can be performed in 3 days.
Day 1: Setting up to mix and degas the PDMS takes ∼10 to 15 min. After a 60 min incubation time in a vacuum desiccator, the PDMS is left to harden overnight.
Day 2: On the next day it is possible to directly proceed with dicing and bonding of the PDMS, which takes at least 30 min depending on the number of devices generated (alternatively, the generated PDMS blocks can be stored for weeks to months). Proper bonding is achieved by leaving the device in an oven overnight.
Day 3: This day can be used for the migration experiment or the devices can be stored until needed. Instead of flushing the device on the day of the experiment, it is also possible to flush the devices with medium 1 day before the experiment and leave them overnight in a humidified incubator. Chemokine gradient formation and cell administration take ∼1 hr. The time needed for setting up the microscope strongly depends on the experimental question and the microscope itself. For epifluorescence imaging at least 20 min should be calculated. For this setup we normally image the cells for a time period of 5 hr. In summary, it takes 3 days in total from preparation to imaging.
Support Protocol 1
The preparation of the epoxy takes ∼1 hr followed by an incubation step overnight to let the epoxy harden.
Support Protocol 2
The protocol for DC differentiation takes 10 days in total. Seeding of the cells on day 0 takes ∼1 hr. However, the time periods for this specific protocol always depend on the number of cells and cell culture dishes used for differentiation. The addition of 10 ml medium on the third day takes ∼10 min. For day 6, ∼20 to 30 min have to be considered. If cells are frozen on day 8, ∼1 hr has to be calculated. Thawing or stimulating the cells 1 day before the experiment requires 15 min.
Basic Protocol 2
The first step of preparing the collagen chamber becomes much faster with experience but takes roughly between 20 and 30 min per plate. Forty-five minutes is needed for preparing the cells and loading the collagen mixture into the plate. After 75 min incubation, adding the chemokine and sealing the chamber with paraffin takes 15 min. Again, the time needed for setting up the microscope and the imaging time strongly depend on the experimental question and the microscope type. For brightfield imaging you should calculate at least 15 min.
Acknowledgments
We thank Kasia Stefanowski for excellent technical assistance, and the Core Facility Bioimaging of the Biomedical Center (BMC) of the Ludwig-Maximilian University for excellent support. We gratefully acknowledge financial support from the Peter Hans Hofschneider Professorship of the Stiftung Experimentelle Biomedizin (to J.R), from the DFG (Collaborative Research Center SFB914, project A12; and Priority Programme SPP2332, project 492014049; both to J.R) and from the LMU Institutional Strategy LMU-Excellent within the framework of the German Excellence Initiative (to J.R).
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Janina Kroll : Data curation, investigation, methodology, visualization, writing original draft; Mauricio J. A. Ruiz-Fernandez : Data curation, investigation, methodology, visualization, writing original draft; Malte B. Braun : Data curation, investigation, methodology, visualization, writing original draft; Jack Merrin : Methodology, writing review and editing; Jörg Renkawitz : Conceptualization, funding acquisition, investigation, supervision, writing original draft.
Conflict of Interest
The authors declare no competing interests.
Open Research
Data Availability Statement
The data, tools and material (or their source) that support the protocol are available from the corresponding author upon reasonable request.
Literature Cited
- Ambravaneswaran, V., Wong, I. Y., Aranyosi, A. J., Toner, M., & Irimia, D. (2010). Directional decisions during neutrophil chemotaxis inside bifurcating channels. Integrative Biology , 2(11–12), 639–647. doi: 10.1039/c0ib00011f.
- Bergert, M., Erzberger, A., Desai, R. A., Aspalter, I. M., Oates, A. C., Charras, G., … Paluch, E. K. (2015). Force transmission during adhesion-independent migration. Nature Cell Biology , 17(4), 524–529. doi: 10.1038/ncb3134.
- Brand, C. A., Linke, M., Weißenbruch, K., Richter, B., Bastmeyer, M., & Schwarz, U. S. (2017). Tension and elasticity contribute to fibroblast cell shape in three dimensions. Biophysical Journal , 113(4), 770–774. doi: 10.1016/j.bpj.2017.06.058.
- Chabaud, M., Heuzé, M. L., Bretou, M., Vargas, P., Maiuri, P., Solanes, P., … Lennon-Duménil, A.-M. (2015). Cell migration and antigen capture are antagonistic processes coupled by myosin II in dendritic cells. Nature Communications , 6, 7526. doi: 10.1038/ncomms8526.
- Cho, S., Vashisth, M., Abbas, A., Majkut, S., Vogel, K., Xia, Y., … Discher, D. E. (2019). Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell-cycle arrest. Developmental Cell , 49(6), 920–935.e5. doi: 10.1016/j.devcel.2019.04.020.
- Denais, C. M., Gilbert, R. M., Isermann, P., McGregor, A. L., te Lindert, M., Weigelin, B., … Lammerding, J. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science , 352(6283), 353–358. doi: 10.1126/science.aad7297.
- Doyle, A. D., Sykora, D. J., Pacheco, G. G., Kutys, M. L., & Yamada, K. M. (2021). 3D mesenchymal cell migration is driven by anterior cellular contraction that generates an extracellular matrix prestrain. Developmental Cell , 56(6), 826-841. doi: 10.1016/j.devcel.2021.02.017.
- Estabrook, I. D., Thiam, H. R., Piel, M., & Hawkins, R. J. (2021). Calculation of the force field required for nucleus deformation during cell migration through constrictions. PLoS Computational Biology , 17(5), e1008592. doi: 10.1371/journal.pcbi.1008592.
- Faure-André, G., Vargas, P., Yuseff, M.-I., Heuzé, M., Diaz, J., Lankar, D., … Lennon-Duménil, A.-M. (2008). Regulation of Dendritic Cell Migration by CD74, the MHC Class II-Associated Invariant Chain. Science , 322(5908), 1705–1710. doi: 10.1126/science.1159894.
- François, J., Kandasamy, A., Yeh, Y.-T., Schwartz, A., Ayala, C., Meili, R., … del Álamo, J. C. (2021). The interplay between matrix deformation and the coordination of turning events governs directed neutrophil migration in 3D matrices. Science Advances , 7(29), eabf3882. doi: 10.1126/sciadv.abf3882.
- Frick, C., Dettinger, P., Renkawitz, J., Jauch, A., Berger, C. T., Recher, M., … Mehling, M. (2018). Nano-scale microfluidics to study 3D chemotaxis at the single cell level. PLoS One , 13(6), e0198330. doi: 10.1371/journal.pone.0198330.
- Garcia-Arcos, J. M., Chabrier, R., Deygas, M., Nader, G., Barbier, L., Sáez, P. J., … Piel, M. (2019). Reconstitution of cell migration at a glance. Journal of Cell Science , 132(4), jcs225565. doi: 10.1242/jcs.225565.
- Hadjitheodorou, A., Bell, G. R. R., Ellett, F., Shastry, S., Irimia, D., Collins, S. R., & Theriot, J. A. (2021). Directional reorientation of migrating neutrophils is limited by suppression of receptor input signaling at the cell rear through myosin II activity. Nature Communications , 12(1), 6619. doi: 10.1038/s41467-021-26622-z.
- Hadjitheodorou, A., Bell, G. R. R., Ellett, F., Irimia, D., Tibshirani, R., Collins, S. R., & Theriot, J. A. (2022). Mechanical competition promotes selective listening to receptor inputs to resolve directional dilemmas in neutrophil migration. BioRxiv , 2022.02.21.481331. doi: 10.1101/2022.02.21.481331.
- Hammerschmidt, S. I., Werth, K., Rothe, M., Galla, M., Permanyer, M., Patzer, G. E., … Förster, R. (2018). CRISPR/Cas9 immunoengineering of Hoxb8-immortalized progenitor cells for revealing CCR7-mediated dendritic cell signaling and migration mechanisms in vivo. Frontiers in Immunology , 9, 1949. doi: 10.3389/fimmu.2018.01949.
- Hanson, L., Zhao, W., Lou, H.-Y., Lin, Z. C., Lee, S. W., Chowdary, P., … Cui, B. (2015). Vertical nanopillars for in situ probing of nuclear mechanics in adherent cells. Nature Nanotechnology , 10, 554–562. doi: 10.1038/nnano.2015.88 for.
- Heuzé, M. L., Collin, O., Terriac, E., Lennon-Duménil, A.-M., & Piel, M. (2011). Cell migration in confinement: A micro-channel-based assay (Vol. 769, pp. 415–434). Totowa, New Jersey: Humana Press. doi: 10.1007/978-1-61779-207-6_28.
- Ilina, O., Gritsenko, P. G., Syga, S., Lippoldt, J., Porta, C. A. M. L., Chepizhko, O., … Friedl, P. (2020). Cell–cell adhesion and 3D matrix confinement determine jamming transitions in breast cancer invasion. Nature Cell Biology , 22(9), 1103–1115. doi: 10.1038/s41556-020-0552-6.
- Infante, E., Castagnino, A., Ferrari, R., Monteiro, P., Agüera-González, S., Paul-Gilloteaux, P., … Chavrier, P. (2018). LINC complex-Lis1 interplay controls MT1-MMP matrix digest-on-demand response for confined tumor cell migration. Nature Communications , 9(1), 2443. doi: 10.1038/s41467-018-04865-7.
- Kamande, J. W., Wang, Y., & Taylor, A. M. (2015). Cloning SU8 silicon masters using epoxy resins to increase feature replicability and production for cell culture devices. Biomicrofluidics , 9(3), 036502. doi: 10.1063/1.4922962.
- Kameritsch, P., & Renkawitz, J. (2020). Principles of leukocyte migration strategies. Trends in Cell Biology , 30(10), 818–832. doi: 10.1016/j.tcb.2020.06.007.
- Kopf, A., Renkawitz, J., Hauschild, R., Girkontaite, I., Tedford, K., Merrin, J., … Sixt, M. (2020). Microtubules control cellular shape and coherence in amoeboid migrating cells. The Journal of Cell Biology , 219(6), e201907154. doi: 10.1083/jcb.201907154.
- Lämmermann, T., Bader, B. L., Monkley, S. J., Worbs, T., Wedlich-Söldner, R., Hirsch, K., … Sixt, M. (2008). Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature , 453(7191), 51–55. doi: 10.1038/nature06887.
- Lautenschläger, F., & Piel, M. (2013). Microfabricated devices for cell biology: All for one and one for all. Current Opinion in Cell Biology , 25(1), 116–124. doi: 10.1016/j.ceb.2012.10.017.
- Leithner, A., Renkawitz, J., de Vries, I., Hauschild, R., Häcker, H., & Sixt, M. (2018). Fast and efficient genetic engineering of hematopoietic precursor cells for the study of dendritic cell migration. European Journal of Immunology , 48(6), 1074–1077. doi: 10.1002/eji.201747358.
- Leithner, A., Eichner, A., Müller, J., Reversat, A., Brown, M., Schwarz, J., … Sixt, M. (2016). Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nature Cell Biology , 18(11), 1253–1259. doi: 10.1038/ncb3426.
- Lerchenberger, M., Uhl, B., Stark, K., Zuchtriegel, G., Eckart, A., Miller, M., … Reichel, C. A. (2013). Matrix metalloproteinases modulate ameboid-like migration of neutrophils through inflamed interstitial tissue. Blood , 122(5), 770–780. doi: 10.1182/blood-2012-12-472944.
- Lomakin, A. J., Cattin, C. J., Cuvelier, D., Alraies, Z., Molina, M., Nader, G. P. F., … Piel, M. (2020). The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science , 370(6514), eaba2894. doi: 10.1126/science.aba2894.
- Lutz, M. B., Kukutsch, N., Ogilvie, A. L., Rössner, S., Koch, F., Romani, N., & Schuler, G. (1999). An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. Journal of Immunological Methods , 223(1), 77–92. doi: 10.1016/S0022-1759(98)00204-X.
- Merrin, J. (2019). Frontiers in microfluidics, a teaching resource review. Bioengineering , 6(4), 109. doi: 10.3390/bioengineering6040109.
- Moreau, H. D., Blanch-Mercader, C., Attia, R., Maurin, M., Alraies, Z., Sanséau, D., … Lennon-Duménil, A.-M. (2019). Macropinocytosis overcomes directional bias in dendritic cells due to hydraulic resistance and facilitates space exploration. Developmental Cell , 49(2), 171–188.e5. doi: 10.1016/j.devcel.2019.03.024.
- Moreau, H. D., Piel, M., Voituriez, R., & Lennon-Duménil, A.-M. (2018). Integrating physical and molecular insights on immune cell migration. Trends in Immunology , 39(8), 632–643. doi: 10.1016/j.it.2018.04.007.
- Orosz, A., Walzog, B., & Mócsai, A. (2020). In vivo functions of mouse neutrophils derived from HoxB8-transduced conditionally immortalized myeloid progenitors. The Journal of Immunology , 206(2), 432–445. doi: 10.4049/jimmunol.2000807.
- Pruenster, M., Kurz, A. R. M., Chung, K.-J., Cao-Ehlker, X., Bieber, S., Nussbaum, C. F., … Sperandio, M. (2015). Extracellular MRP8/14 is a regulator of β2 integrin-dependent neutrophil slow rolling and adhesion. Nature Communications , 6, 6915. doi: 10.1038/ncomms7915.
- Raab, M., Gentili, M., de Belly, H., Thiam, H. R., Vargas, P., Jimenez, A. J., … Piel, M. (2016). ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science , 352(6283), 359–362. doi: 10.1126/science.aad7611.
- Redecke, V., Wu, R., Zhou, J., Finkelstein, D., Chaturvedi, V., High, A. A., & Häcker, H. (2013). Hematopoietic progenitor cell lines with myeloid and lymphoid potential. Nature Methods , 10(8), 795–803. doi: 10.1038/nmeth.2510.
- Renkawitz, J., Kopf, A., Stopp, J., de Vries, I., Driscoll, M. K., Merrin, J., … Sixt, M. (2019). Nuclear positioning facilitates amoeboid migration along the path of least resistance. Nature , 568(7753), 546–550. doi: 10.1038/s41586-019-1087-5.
- Renkawitz, J., Reversat, A., Leithner, A., Merrin, J., & Sixt, M. (2018). Micro-engineered “pillar forests” to study cell migration in complex but controlled 3D environments. Methods in Cell Biology , 147, 79–91. doi: 10.1016/bs.mcb.2018.07.004.
- Reversat, A., Gaertner, F., Merrin, J., Stopp, J., Tasciyan, S., Aguilera, J., … Sixt, M. (2020). Cellular locomotion using environmental topography. Nature , 582(7813), 582–585. doi: 10.1038/s41586-020-2283-z.
- Riedl, J., Crevenna, A. H., Kessenbrock, K., Yu, J. H., Neukirchen, D., Bista, M., … Wedlich-Soldner, R. (2008). Lifeact: A versatile marker to visualize F-actin. Nature Methods , 5(7), 605–607. doi: 10.1038/nmeth.1220.
- Rossier, O. M., Rossier, O. M., Gauthier, N., Gauthier, N., Biais, N., Biais, N., … Sheetz, M. P. (2010). Force generated by actomyosin contraction builds bridges between adhesive contacts. The EMBO Journal , 29(6), 1055–1068. doi: 10.1038/emboj.2010.2.
- Sabeh, F., Shimizu-Hirota, R., & Weiss, S. J. (2009). Protease-dependent versus -independent cancer cell invasion programs: Three-dimensional amoeboid movement revisited. Journal of Cell Biology , 185(1), 11–19. doi: 10.1083/jcb.200807195.
- Sáez, P. J., Vargas, P., Shoji, K. F., Harcha, P. A., Lennon-Duménil, A.-M., & Sáez, J. C. (2017). ATP promotes the fast migration of dendritic cells through the activity of pannexin 1 channels and P2X7 receptors. Science Signaling , 10(506), eaah7107. doi: 10.1126/scisignal.aah7107.
- Salvermoser, M., Pick, R., Weckbach, L. T., Zehrer, A., Löhr, P., Drechsler, M., … Walzog, B. (2018). Myosin 1f is specifically required for neutrophil migration in 3D environments during acute inflammation. Blood , 131(17), 1887–1898. doi: 10.1182/blood-2017-10-811851.
- Schneider, C. A., Rasband, W. S., & Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods , 9(7), 671–675. doi: 10.1038/nmeth.2089.
- Schwarz, J., Bierbaum, V., Vaahtomeri, K., Hauschild, R., Brown, M., de Vries, I., … Sixt, M. (2017). Dendritic cells interpret haptotactic chemokine gradients in a manner governed by signal-to-noise ratio and dependent on GRK6. Current Biology , 27(9), 1314–1325. doi: 10.1016/j.cub.2017.04.004.
- Shiu, J.-Y., Aires, L., Lin, Z., & Vogel, V. (2018). Nanopillar force measurements reveal actin-cap-mediated YAP mechanotransduction. Nature Cell Biology , 7, 1. doi: 10.1038/s41556-017-0030-y.
- Sitarska, E., Almeida, S. D., Beckwith, M., Stopp, J., Schwab, Y., Sixt, M., … Diz-Muñoz, A. (2021). Curvature sensing steers actin-driven cell migration. BioRxiv , 2021.03.26.437199. doi: 10.1101/2021.03.26.437199.
- Sixt, M., & Lämmermann, T. (2011). In vitro analysis of chemotactic leukocyte migration in 3D environments. Methods in Molecular Biology , 769, 149–165. doi: 10.1007/978-1-61779-207-6_11.
- Solanes, P., Heuzé, M. L., Maurin, M., Bretou, M., Lautenschlaeger, F., Maiuri, P., … Lennon-Duménil, A.-M. (2015). Space exploration by dendritic cells requires maintenance of myosin II activity by IP3 receptor 1. The EMBO Journal , 34(6), 798–810. doi: 10.15252/embj.201489056.
- Starborg, T., Lu, Y., Kadler, K. E., & Holmes, D. F. (2008). Electron microscopy of collagen fibril structure in vitro and in vivo including three-dimensional reconstruction. Methods in Cell Biology , 88, 319–345. doi: 10.1016/s0091-679×(08)00417-2.
- Stoitzner, P., Pfaller, K., Stössel, H., & Romani, N. (2002). A close-up view of migrating Langerhans cells in the skin. The Journal of Investigative Dermatology , 118(1), 117–125. doi: 10.1046/j.0022-202×.2001.01631.x.
- Thiam, H.-R., Vargas, P., Carpi, N., Crespo, C. L., Raab, M., Terriac, E., … Piel, M. (2016). Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nature Communications , 7, 10997. doi: 10.1038/ncomms10997.
- Tweedy, L., Thomason, P. A., Paschke, P. I., Martin, K., Machesky, L. M., Zagnoni, M., & Insall, R. H. (2020). Seeing around corners: Cells solve mazes and respond at a distance using attractant breakdown. Science , 369(6507), eaay9792. doi: 10.1126/science.aay9792.
- Vargas, P., Chabaud, M., Thiam, H.-R., Lankar, D., Piel, M., & Lennon-Duménil, A.-M. (2016). Study of dendritic cell migration using micro-fabrication. Journal of Immunological Methods , 432, 30–34. doi: 10.1016/j.jim.2015.12.005.
- Velve-Casquillas, G., Berre, M. L., Piel, M., & Tran, P. T. (2010). Microfluidic tools for cell biological research. Nano Today , 5(1), 28–47. doi: 10.1016/j.nantod.2009.12.001.
- Wallis, S. S., Ventimiglia, L. N., Otigbah, E., Infante, E., Cuesta-Geijo, M. A., Kidiyoor, G. R., … Agromayor, M. (2021). The ESCRT machinery counteracts Nesprin-2G-mediated mechanical forces during nuclear envelope repair. Developmental Cell , 56(23), 3192-3202. doi: 10.1016/j.devcel.2021.10.022.
- Wang, X., Hossain, M., Bogoslowski, A., Kubes, P., & Irimia, D. (2020). Chemotaxing neutrophils enter alternate branches at capillary bifurcations. Nature Communications , 11(1), 1–12. doi: 10.1038/s41467-020-15476-6.
- Weigelin, B., Bakker, G.-J., & Friedl, P. (2012). Intravital third harmonic generation microscopy of collective melanoma cell invasion. IntraVital , 1(1), 32–43. doi: 10.4161/intv.21223.
- Wilson, K., Lewalle, A., Fritzsche, M., Thorogate, R., Duke, T., & Charras, G. (2013). Mechanisms of leading edge protrusion in interstitial migration. Nature Communications , 4(1), 2896. doi: 10.1038/ncomms3896.
- Wolf, K., Alexander, S., Alexander, S., Schacht, V., Schacht, V., Coussens, L. M., … Friedl, P. (2009). Collagen-based cell migration models in vitro and in vivo. Seminars in Cell & Developmental Biology, 20(8), 931–941. doi: 10.1016/j.semcdb.2009.08.005.
- Wolf, K., te Lindert, M., Krause, M., Krause, M., Alexander, S., te Riet, J., … Friedl, P. (2013). Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. The Journal of Cell Biology , 201(7), 1069–1084. doi: 10.1083/jcb.201210152.
- Worbs, T., Hammerschmidt, S. I., & Förster, R. (2017). Dendritic cell migration in health and disease. Nature Reviews Immunology , 17(1), 30–48. doi: 10.1038/nri.2016.116.
- Xu, H., Look, T., Prithiviraj, S., Lennartz, D., Cáceres, M. D., Götz, K., … Zenke, M. (2021). CRISPR/Cas9 editing in conditionally immortalized HoxB8 cells for studying gene regulation in mouse dendritic cells. European Journal of Immunology , doi: 10.1002/eji.202149482.
- Yamada, K. M., & Sixt, M. (2019). Mechanisms of 3D cell migration. Nature Reviews Molecular Cell Biology , 20(12), 738–752. doi: 10.1038/s41580-019-0172-9.
Key References
- Denais et al. (2016). See above.
This key reference employs micro-pillars and discovered, together with Raab et al. (2016), transient nuclear breakages and their repair during cell migration through tight pores.
- Faure-André et al. (2008). See above.
This key reference pioneers the investigation of dendritic cell migration in micro-channels.
- Lautenschläger and Piel (2013). See above.
This key reference provides a pioneering overview on micro-engineered devices to investigate immune cell motility and cell biology in general.
- Lämmermann et al. (2008). See above.
This key reference employed in vitro collagen networks with tunable density and thus pore sizes to investigate myosin-mediated cellular squeezing during integrin-independent immune cell migration.
- Raab et al. (2016). See above.
This key reference discovered, together with Denais et al., 2016, transient nuclear breakages and their repair during immune cell migration through tight pores.
- Renkawitz et al. (2019). See above.
This key reference identified that immune cells are pore size selective by using their nucleus as a mechanical gauge.
- Thiam et al. (2016). See above.
This key reference pioneered the use of micro-channels with single constrictions, identifying the cytoskeletal forces moving the nucleus through tight microenvironmental pores.
- Wolf et al. (2013). See above.
This key reference pioneered the knowledge of nuclear translocation through narrow pores by using collagen networks of different porosity.
Internet Resources
Chemotaxis and Migration Tool (ibidi): This software enables the analysis of manual tracks of motile cells, the measurement of the cellular velocity and directionality, and the ability to plot the respective cellular tracks (see e.g., Fig. 4D-F). For more detail we recommend reading the manual that explains many analysis methods such as directionality and rose blots in more detail.
Imaris (Bitplane): Imaris microscopy image analysis software enables automatic tracking of cells and visualization of tracks colored by quantified parameters (e.g., cell speed, see Fig. 3C and Fig. 4C). It can also be used to visualize and quantify 3D microscopy data.
The software ImageJ (Schneider, Rasband, & Eliceiri, 2012) with the Manual Tracking plugin (https://imagej.nih.gov/ij/plugins/manual-tracking.html) can be used to manually generate tracks of migrating cells. Further, ImageJ is useful for general browsing through and editing of microscopy data and in particular to generate z-stacks of 3D microscopy data (e.g., see Fig. 4A).
Citing Literature
Number of times cited according to CrossRef: 3
- Janina Kroll, Robert Hauschild, Artur Kuznetcov, Kasia Stefanowski, Monika D Hermann, Jack Merrin, Lubuna Shafeek, Annette Müller‐Taubenberger, Jörg Renkawitz, Adaptive pathfinding by nucleokinesis during amoeboid migration, The EMBO Journal, 10.15252/embj.2023114557, 42 , 24, (2023).
- Hellen Ishikawa-Ankerhold, Janina Kroll, Dominic van den Heuvel, Jörg Renkawitz, Annette Müller-Taubenberger, Centrosome Positioning in Migrating Dictyostelium Cells, Cells, 10.3390/cells11111776, 11 , 11, (1776), (2022).
- Björn E. Clausen, Lukas Amon, Ronald A. Backer, Luciana Berod, Tobias Bopp, Anna Brand, Sven Burgdorf, Luxia Chen, Meihong Da, Ute Distler, Regine J. Dress, Diana Dudziak, Charles‐Antoine Dutertre, Christina Eich, Anna Gabele, Melanie Geiger, Florent Ginhoux, Lucila Giusiano, Gloria J. Godoy, Ahmed E.I. Hamouda, Lukas Hatscher, Lukas Heger, Gordon F. Heidkamp, Lola C. Hernandez, Lukas Jacobi, Tomasz Kaszubowski, Wan Ting Kong, Christian H. K. Lehmann, Tamara López‐López, Karsten Mahnke, Dominik Nitsche, Jörg Renkawitz, Rifat A. Reza, Pablo J. Sáez, Laura Schlautmann, Madeleine T. Schmitt, Anna Seichter, Malte Sielaff, Tim Sparwasser, Patrizia Stoitzner, Giorgi Tchitashvili, Stefan Tenzer, Nounagnon R. Tochoedo, Damir Vurnek, Fabian Zink, Thomas Hieronymus, Guidelines for mouse and human DC functional assays, European Journal of Immunology, 10.1002/eji.202249925, 53 , 12, (2022).

