Capturing and Processing Slaking Images With a Multi-Well Plate
Claire Phillips, Robert E Meadows III, Joaquin Casanova, Bryan Emmett
Abstract
This describes a process to measure soil wet aggregate stability through slaking, or rapid immersion it water. It uses a multi-well plate to process many aggregates at one time. Air dry, pea-sized aggregates (3-10 mm) are submerged in water and time lapse images are collected with a web cam to measure their dispersion (slaking) over 10 minutes. Image-J software is used to measure the projected area of the aggregates over time. Python code is also provided to automate image analysis. Slaking index is calculated from the change in projected area of the aggregates. This protocol accompanies a manuscript Phillips et al. submitted to Soil Science Society of America Journal (July 2023)
Before start
Obtain a soil sample containing at least 20 pea-sized aggregates and air dry it.
Steps
Sample preparation
Start with air-dried soil that was not previously sieved. It is recommended to measure at least 20 pea-sized (3 to 10 mm) aggregates per soil sample. More replication is useful for unstable soils. See Phillips et al. (submitted 2023) for more information on necessary replication.
If the soil sample is consolidated and does not have enough pea-sized aggregates, it can be sieved through a 6 mm mesh sieve to break it up.
Establish how you will count the wells (across or down) and create a spreadsheet identifying the sample in each well.
Example Document: SiteName P16_P17_P20_Time4_Samples.csv
Make sure the tray is clean and dry, and the mesh is secured to the bottom (no tears or gaps).
Use webcam to collect images
In Microsoft Windows, type camera into search bar to find Camera app
Adjust the brightness as needed to ensure strong contrast between the aggregates and the background.
Carefully remove the multi-well tray from the soaking dish. Fill the dish with at least 2 cm water.
Let the time lapse run at a 5 second interval for the first minute of recording, then toggle the time lapse to the 10 second interval without pausing or stopping the time lapse.
Within the file directory where project data will be saved create a folder representing the sample that was tested. E.g. C:\Users\REM\Documents\SLAKES\SampleNameTime1_Depth1
Within the Samples folder create a folder for the images collected from the sample. E.g. C:\Users\REM\Documents\SLAKES\SampleNameTime1_Depth1\Images
Move the captured images to the Images folder created in step 12.1 . The default save location for the Camera app is the Camera Roll subfolder in the active user's Pictures folder. E.g. C:\Users\REM\Pictures\Camera Roll
Ensure the Camera Roll folder is empty before starting a different sample.
Process The images in ImageJ
Set the Scale of the Image with the Straight Line Tool in ImageJ.
Use the oval or rectangle tool to select the first well.
In the ROI Manager click Add or use the T as a shortcut to add the selection to the ROI manager tool.
Make the image stack a set of Binary images.
Images in the stack should generate a red layer over the soil aggregates. If any shadows are picked up during this stage they will also appear in red. If this happens cancel the Make Binary action and adjust the Contrast through the Menu by selecting Image then Adjust and Brightness/Contrast until shadows no longer appear in the selected ROIs.
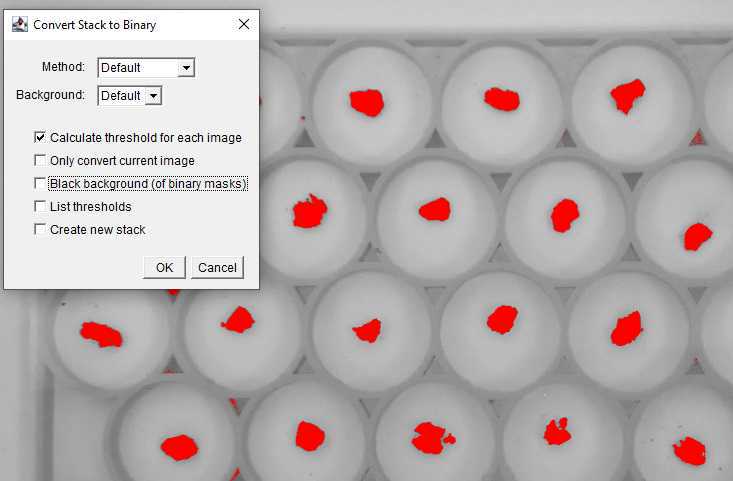
Uncheck the "Black background" box.
In the Multi Measure menu check the boxes for 'Measure all # Slices' and One row per slice.
Save the Multi Measure results in the sample folder created in step 12 . The filename should match the format being used with '_Results' appended at the end.
E.g. SampleNameTime1_Depth1_Results.csv
Computations
Several versions of slaking index calculations have been suggested. The slaking index originally recommended by Fajardo et al. (2016) involved fitting a rise-to-threshold (Goempertz function) model to timeseries of aggregate area, and computing the function's limit as the slaking index (SI).
Flynn et al. (2020) recommended instead computing the observed change in aggregate area over time:
_SI600_ = ( _A600 - A0_ <sub>600</sub> - A<sub>0</sub>)/ _A0_ <sub>0</sub>
where A0 0 is the initial projected area of the dry aggregate and A600 600 is the projected
area after 600 seconds of slaking. Note that higher SI600 value indicates lower aggregate stability, as a less stable ped will spread out more over time.
By contrast, Rieke et al. (2022) reported a slaking score as 1/(1 + SI600), such that higher values indicate greater aggregate stability, which follows the same direction as other aggregate stability measures.
This R script provides example code for using the Image-J output to plot area timeseries and compute slaking index following Flynn et al. (2020). ComputeSlakingIndex.R
Slaking index tends to follow a log-normal distribution with a long right tail. Because arithmetic means are influenced by high outliers, it is recommended to use a geometric mean (and geometric standard deviation) to summarize the aggregates for each soil sample.
The geometric mean is equivalent to computing the arithmetic mean of log-transformed
data, and back-transforming the mean:






















