Canine Respiratory Pathogen Detection Assays
Come J Thieulent, Mariano Carossino, Laura Peak, Udeni B. R. Balasuriya
Disclaimer
Reference to any commercial materials, equipment, or processes do not in any way constitute approval, endorsement, or recommendation by the Food and Drug Administration.
Abstract
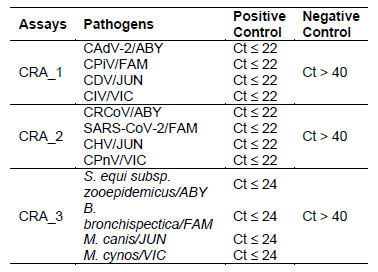
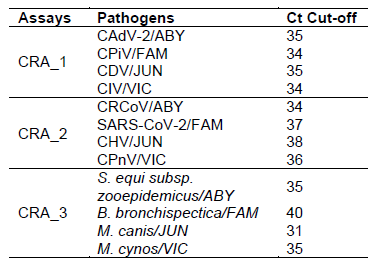
The Canine Respiratory Pathogen (CRP) Detection Assays is intended as an in vitro veterinary reagent set, based on Reverse Transcription quantitative PCR (RT-qPCR), for the detection of canine adenovirus-2 (CAdV-2), canine distemper (CDV), canine herpesvirus type 1 (CHV-1), canine influenza A virus (CIV), canine parainfluenza (CPiV), canine pneumovirus (CPnV), canine respiratory coronavirus (CRCoV), SARS-CoV-2, Bordetella bronchiseptica , Streptococcus equi subsp. zooepidemicus , Mycoplasma cynos and Mycoplasma canis in nasal and pharyngeal swab samples.
Steps
Reaction Setup
Thaw all reagents on ice.
Centrifuge all reagents on a benchtop centrifuge to ensure no liquid is in cap and keep on ice
Programming the Thermocycler
The following fluorescence channels should be selected: ABYTM, FAMTM, JUNTM, and VICTM.
ROXTM should be used as a passive reference dye.
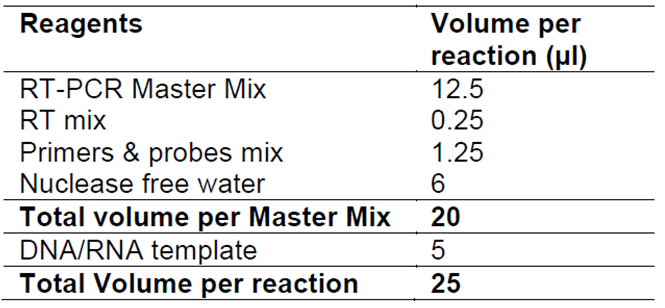
The standard mode should be selected. Setup cycling condition following table 2:
| A | B | C | D |
|---|---|---|---|
| Step | Number of cycles | Temp. (°C) | Time (min:sec) |
| Reverse transcriptase | 1 | 50 | 20:00 |
| Initial activation | 1 | 95 | 15:00 |
| Denaturation | 40 | 94 | 00:45 |
| Annealing/extension | 60 | 00:75 |
Table 2. Thermal profile. The data acquisition is performed during the annealing/extension step.
Results interpretation
Before results analysis, the threshold value of each fluorescent dye must be manually set in the region of exponential amplification, typically 0.1 × ΔRn value at the plateau phase.