CODEX® Multiplexed Imaging | Tissue Sectioning
Diane Saunders, Marcela Brissova, Alvin C. Powers, Conrad Reihsmann, Alexander Hopkirk, Regina Jenkins
Disclaimer
This protocol is adapted from the CODEX User Manual, revision C (Akoya Biosciences, Dec. 2020). See also: Tissue Processing - Best Practices.
Abstract
This protocol describes the tissue preparation processes for the CODEX® (now PhenoCycler™) system by Akoya Biosciences. For the comprehensive multiplexed imaging workflow currently in use at the Vanderbilt Diabetes Research Center, please see CODEX® Multiplexed Imaging | Modality overview .
Steps
Coverslip Preparation
Gather reagents:
Gently place coverslips at the bottom of a 500-mL glass beaker and swirl to spread the coverslips out. Add approximately 20mL poly-L-lysine solution to the beaker, ensuring coverslips are fully submerged. Rotate the beaker at a 45° angle for 0h 1m 0s to mix.
Cover beaker with parafilm and incubate for minimum of 12h 0m 0s at Room temperature. Coverslips can sit in the poly-L-lysine solution for up to one week.
Slowly pipet off the poly-L-lysine solution and dispose.
Fill the beaker containing the coverslips to half volume with Milli-Q® water. Swirl gently, let sit for 0h 0m 30s, and then slowly pour off water into the sink.
and repeat for a total of 5-7 washes.
Spread WypAll® towels on the benchtop. In small batches, remove coverslips from the beaker, separate from one another, and place individual coverslips side by side on towels. Layer another towel on top and gently dab to remove excess liquid. Leave on benchtop until completely dry.
ⓘ Coated coverslips can be stored in a petri dish for up to 2 months.
Cryosectioning
Set the cryostat temperature to -20°C. Remove a fresh blade, clean with ethanol and a lint free wipe as needed, and insert blade.
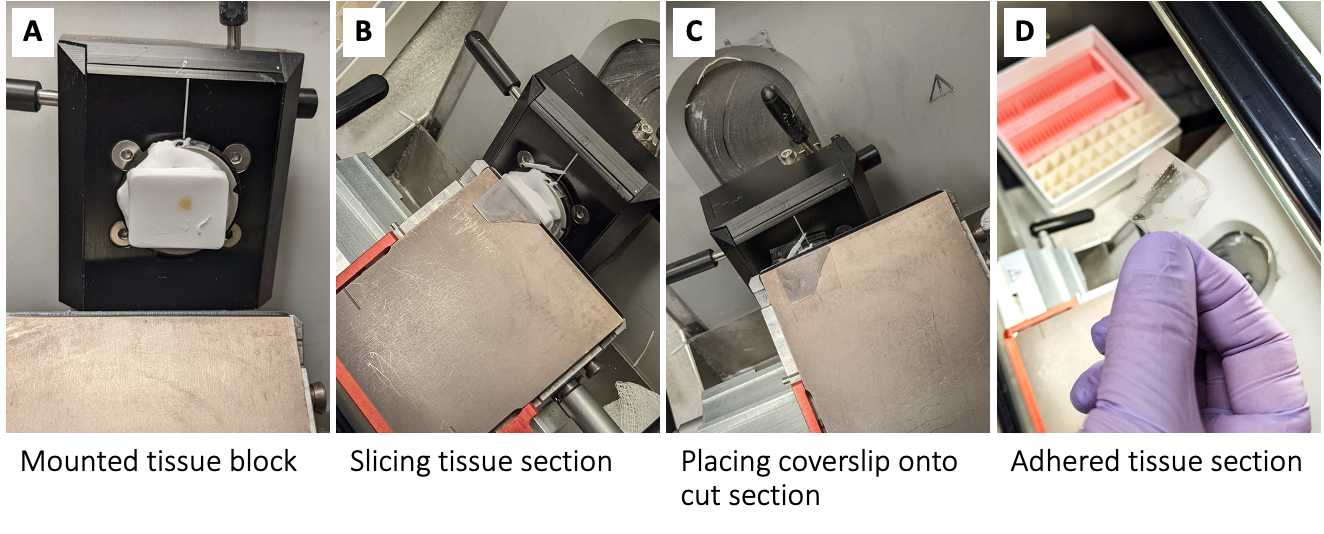
Obtain 6 µm to 10 µm sections, using a fine-tipped paintbrush to guide the section onto base and place coverslip onto section ( Figure 1b-1d ).
Using forceps, slot coverslips with mounted tissue into a plastic coverslip holder under the appropriate donor/block ID label(s).
As necessary, mount adjacent sections onto Gold Plus slides (for traditional immunohistochemistry) and/or ITO slides (for mass spectrometry). Record all sections cut, the thickness, and any additional block shaved off in the process.
Coverslip Storage
Store coverslips at -80°C for up to 6 months.