BridGE: a pathway-based analysis tool for detecting genetic interactions from GWAS
Chad L. Myers, Mehrad Hajiaghabozorgi, Mathew Fischbach, Michael Albrecht, Wen Wang































Supplementary information
Reporting Summary
Supplementary Data 1
The output of the original BridGE and BridGE 2.0 are compared for the two Parkinson’s disease GWAS cohorts, which were also analyzed in the original BridGE paper. The Excel files include summary tables and lists of BPM, WPM and PATH discovered at the FDR cutoff shown in the summary tables.
Supplementary Data 2
BridGE output files from the simulated 1000 Genomes GWAS data based on the combined disease model. The Excel files include BridGE results and interaction lists for BPMs and WPMs. The PDF file provides the visualization of pathway–pathway interactions.
Supplementary Data 3
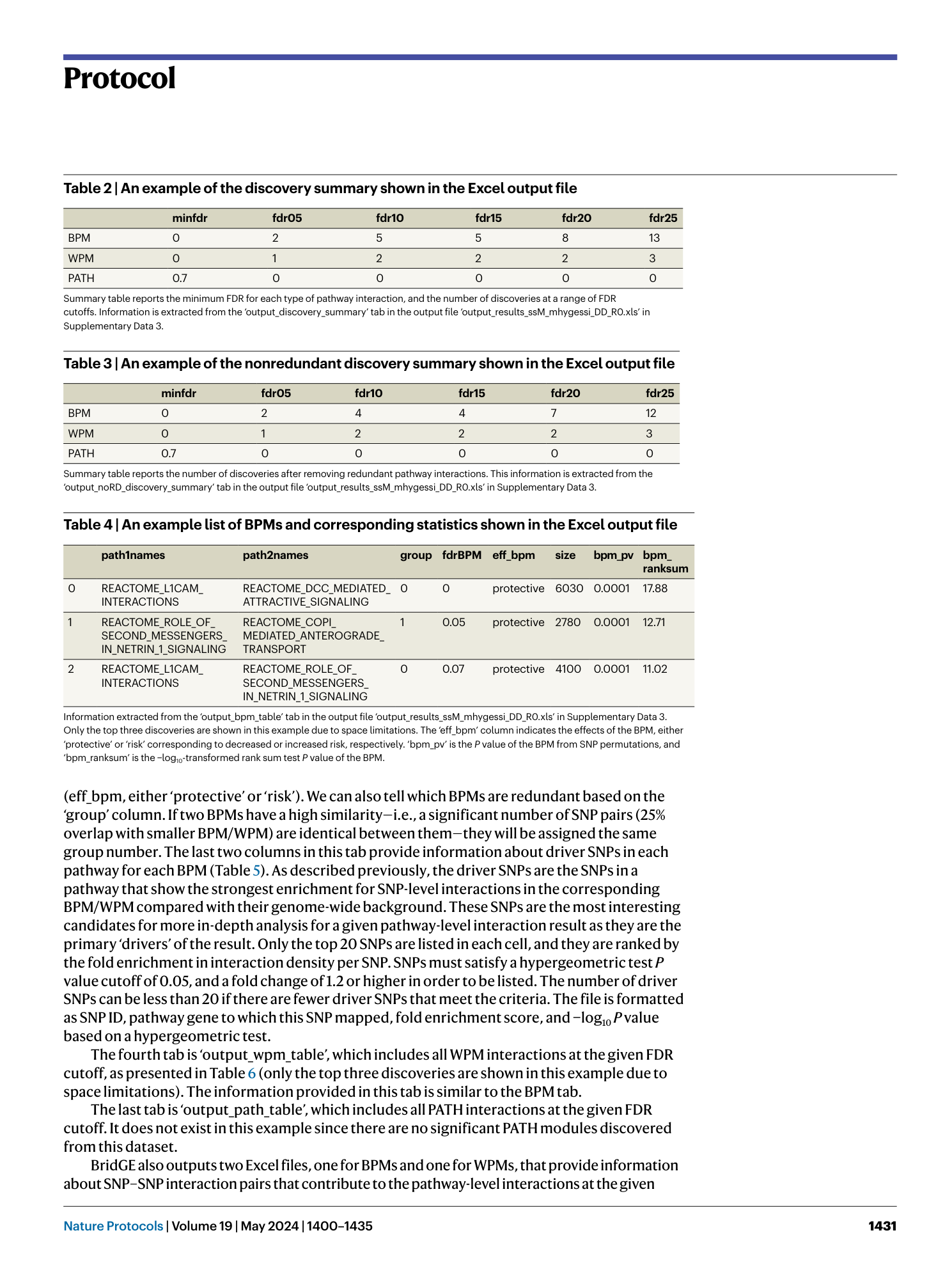
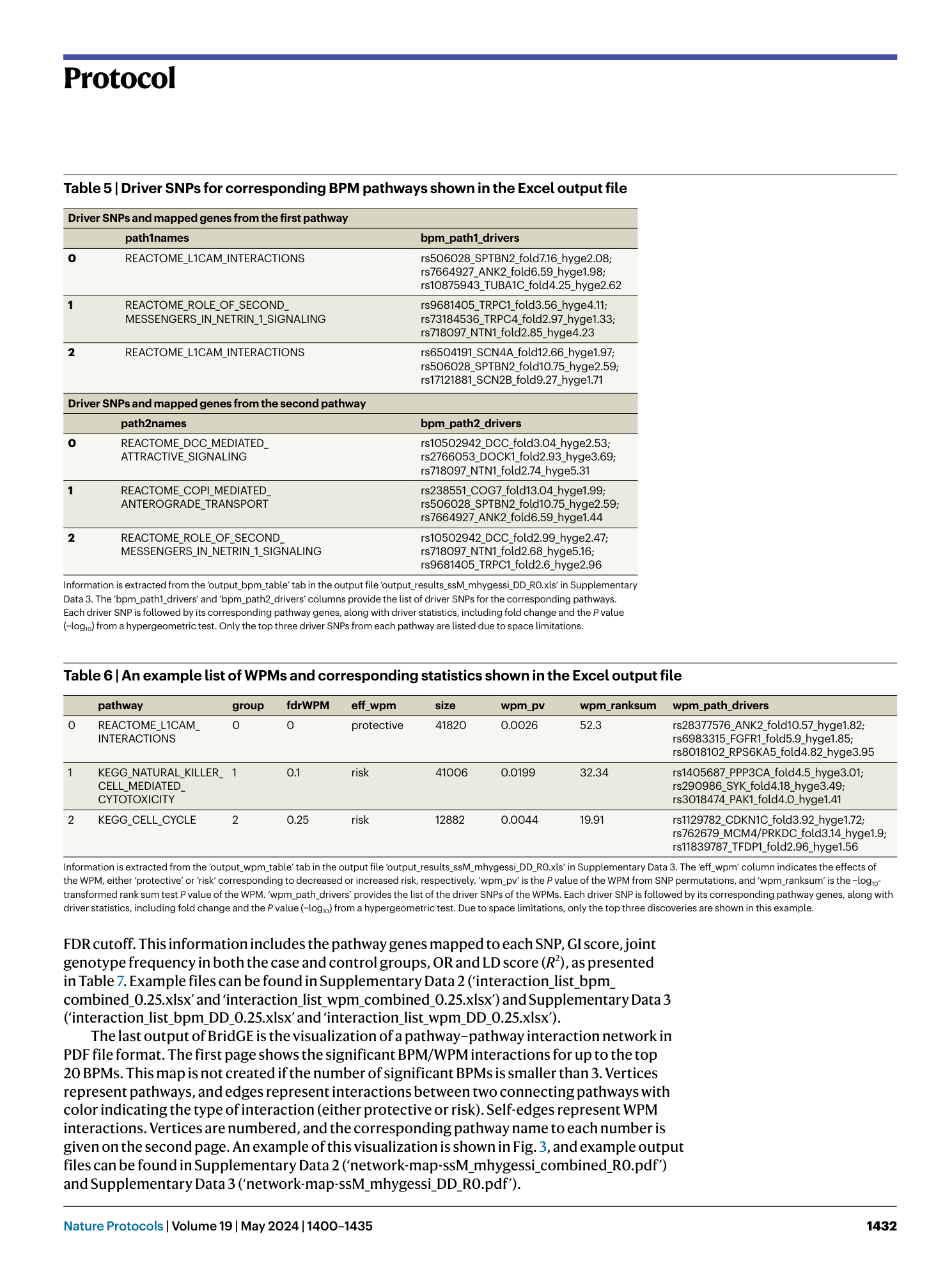
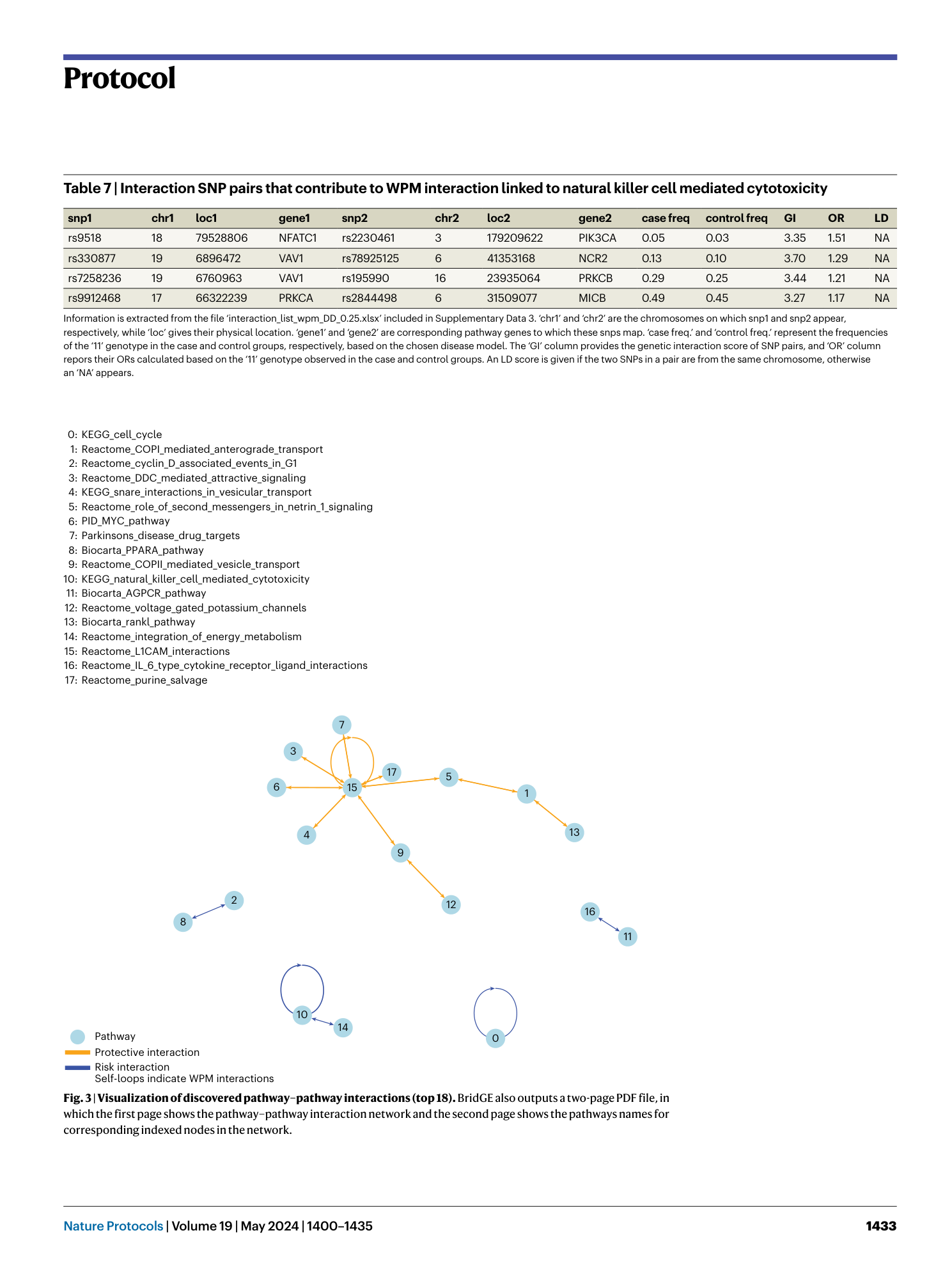
A complete set of BridGE output files from the Parkinson’s disease IPDGC cohort (dbGaP study accession: phs000918.v1.p1 ). The Excel files include information about significant pathway-level interactions (FDR <0.25) discovered under the corresponding disease model (RR, DD, RD and combined) and pairwise SNP interaction lists and associated statistics (DD disease model only), one for BPM and one for WPM. The PDF file provides a network visualization.

