BUSCO: Assessing Genomic Data Quality and Beyond
Mosè Manni, Mosè Manni, Matthew R. Berkeley, Matthew R. Berkeley, Mathieu Seppey, Mathieu Seppey, Evgeny M. Zdobnov, Evgeny M. Zdobnov
Abstract
Evaluation of the quality of genomic “data products” such as genome assemblies or gene sets is of critical importance in order to recognize possible issues and correct them during the generation of new data. It is equally essential to guide subsequent or comparative analyses with existing data, as the correct interpretation of the results necessarily requires knowledge about the quality level and reliability of the inputs. Using datasets of near universal single-copy orthologs derived from OrthoDB, BUSCO can estimate the completeness and redundancy of genomic data by providing biologically meaningful metrics based on expected gene content. These can complement technical metrics such as contiguity measures (e.g., number of contigs/scaffolds, and N50 values). Here, we describe the use of the BUSCO tool suite to assess different data types that can range from genome assemblies of single isolates and assembled transcriptomes and annotated gene sets to metagenome-assembled genomes where the taxonomic origin of the species is unknown. BUSCO is the only tool capable of assessing all these types of sequences from both eukaryotic and prokaryotic species. The protocols detail the various BUSCO running modes and the novel workflows introduced in versions 4 and 5, including the batch analysis on multiple inputs, the auto-lineage workflow to run assessments without specifying a dataset, and a workflow for the evaluation of (large) eukaryotic genomes. The protocols further cover the BUSCO setup, guidelines to interpret the results, and BUSCO “plugin” workflows for performing common operations in genomics using BUSCO results, such as building phylogenomic trees and visualizing syntenies. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
[Correction added on May 16, 2022, after first online publication: CSAL funding statement has been added.]
Basic Protocol 1 : Assessing an input sequence with a BUSCO dataset specified manually
Basic Protocol 2 : Assessing an input sequence with a dataset automatically selected by BUSCO
Basic Protocol 3 : Assessing multiple inputs
Alternate Protocol : Decreasing analysis runtime when assessing a large number of small genomes with BUSCO auto-lineage workflow and Snakemake
Support Protocol 1 : BUSCO setup
Support Protocol 2 : Visualizing BUSCO results
Support Protocol 3 : Building phylogenomic trees
INTRODUCTION
With the advances of high-throughput sequencing technologies and their decreasing costs, the production of genomic data is increasing at an exponential rate. Large-scale genome projects such as the Earth BioGenome Project (Lewin et al., 2018), the Darwin Tree of Life Project (Threlfall & Blaxter, 2021), the 5,000 Arthropod Genomes Project (i5K Consortium, 2013), the 10,000 Plant Genomes Project (10KP; Cheng et al., 2018), the GEM (Genomes from Earth's Microbiomes) catalog (Nayfach et al., 2021), and the European Reference Genome Atlas (ERGA) aim at sequencing hundreds of thousands of new genomes in the coming years from all kingdoms of life. Additionally, many other individual studies are generating genomic resources at an accelerated pace, including hundreds of thousands of microbial metagenome-assembled genomes (MAGs). Despite the advances in computational methods, the generation of genomic resources can still be challenging (Richards, 2018; Sohn & Nam, 2018). Contamination and technical issues during sample preparation or assembly procedures can occur and complicate the generation of high-quality genomic resources—for example, in the case of highly complex and heterogeneous metagenomes, species with large genome sizes, or highly heterozygous samples. Intuitive metrics, along with easy-to-use tools, are needed to perform quality assessments on such a deluge of genomic resources. Quality assessments of draft data are required while generating these resources to timely identify possible issues to be solved before finalizing the data, for example, to identify redundancies in a draft genome assembly due to technical issues that can occur during de novo assembly procedures. Knowledge of the quality state of genomic resources is also required before attempting to perform subsequent or comparative analyses, to ensure that the quality of the input data is sufficient for the analysis of interest. This ensures an unbiased interpretation of the results and allows possible caveats and limitations to be considered if sub-optimal resources are included in the analyses. Quality control is therefore of paramount importance. For genome assemblies, metrics such as contigs/scaffold counts and contig/scaffold N50 values (meaning that half of the total assembly span is made up of contigs/scaffolds of length N50 or longer) only offer a summary technical view of genome assembly contiguity. Therefore, there is a necessity for tools that can accurately and quickly assess the readiness of genetic resources being generated for downstream analyses. In an effort to provide researchers with the ability to evaluate their genomic resources, the Benchmarking Universal Single-Copy Orthologue (BUSCO) tool provides evolutionarily sound measures of completeness and redundancy in terms of expected gene content (Manni, Berkeley, Seppey, Simão, & Zdobnov, 2021; Simão, Waterhouse, Ioannidis, Kriventseva, & Zdobnov, 2015; Waterhouse et al., 2018). This is achieved by considering the presence of a predefined and expected set of single-copy marker genes as a proxy for genome-wide completeness. BUSCO implements such quantifications not only for assembled genomes but also for transcriptomes and annotated protein-coding gene sets, and they can be applied to both prokaryotic and eukaryotic data. BUSCO identifies matches to sets of genes that are present as single-copy orthologs in a given taxonomic group, i.e., genes that are evolving under “single-copy control”, and thus expected to be present as single-copy genes in the majority of any newly sequenced species from the same taxonomic group. This expectation is defined empirically by identifying near-universal single-copy orthologs in major species clades with numerous sequenced and annotated genomes using the OrthoDB (Zdobnov et al., 2021) catalog of orthologs (http://www.orthodb.org). Exceptions are expected to occur due to gene duplications or losses that can take place along the evolution of a single species within a specific clade. When generating BUSCO datasets, the thresholds used to filter for genes that are almost always present as single-copy genes across a given clade are set to account for these exceptions and to further consider the incompleteness of most of the currently available assembled genomes and predicted gene sets, such as genes that are missing exclusively due to technical reasons. The latest versions of the BUSCO datasets (*_odb10; Manni et al., 2021) include 67 eukaryotic, 83 bacterial, 16 archaeal, and 27 viral datasets (see Supplementary Table 1 in Supporting Information). The datasets are bound to the software version so that the odb10 version datasets are intended to be used with BUSCO v4/v5 (Manni et al., 2021). The protocols in this manuscript describe the step-by-step commands to use BUSCO on various data types from different taxonomic origins and describe the use of the novel workflows introduced in BUSCO v4/v5.
Based on the input sequence type (genome assembly, annotated gene set, or transcriptome assembly) and the taxonomic domain of origin (Bacteria, Archaea, or Eukaryota), different workflows built upon several third-party tools, each performing one step of the global analysis, are executed. In this article, we have detailed the protocols for running all the currently implemented BUSCO workflows. Basic Protocol 1 describes the steps for the assessment of a single input file (either a genome assembly, annotated gene set, or transcriptome assembly) with a known taxonomic origin. Basic Protocol 2 describes the analysis of an input sequence without specifying a dataset for the assessment, which enables the evaluation of sequences with unknown taxonomic origin. Basic Protocol 3 describes the extension of Basic Protocols 1 and 2 to analyze multiple inputs, and can also be applied to metagenomic bins or MAGs from both prokaryotic and eukaryotic species. The Alternate Protocol describes alternative steps to increase the speed of the analysis described in Basic Protocol 3 by using the Snakemake workflow management system (Mölder et al., 2021). Support Protocol 1 describes the various ways to set up a BUSCO installation. Support Protocol 2 is intended for the visualization of BUSCO results, e.g., for plotting the position of the identified single-copy markers on genome assemblies, or visualizing syntenies between multiple assemblies. Support Protocol 3 describes the use of single-copy genes identified with BUSCO as markers for building phylogenomic trees. The input files and scripts required to follow these protocols are available at https://gitlab.com/ezlab/busco_protocol.
Basic Protocol 1: ASSESSING AN INPUT SEQUENCE WITH A BUSCO DATASET MANUALLY SPECIFIED
Basic Protocol 1 describes the scenario in which BUSCO is used to assess a single input, either a genome, gene set, or transcriptome, with a dataset manually specified by the user. This protocol assumes that the taxonomic origin of the input sequence is known. Before running BUSCO assessments, you need to first set up the BUSCO software and its dependencies (see Support Protocol 1). Users are also encouraged to visit the website and read the user guide for further details and up-to-date information (http://busco.ezlab.org).
Necessary Resources
Hardware
- A Unix-based workstation. BUSCO has been tested on several Linux platforms, and it is recommended to use a Linux machine. BUSCO and its dependencies may work on Macintosh machines and operating systems, but this has not been thoroughly tested.
- Optional, but recommended : The machine needs to have access to the Internet for downloading the BUSCO datasets and files.
Software
- The BUSCO package and its dependencies (see Support Protocol 1 for various ways of setting up BUSCO). For plotting scores: R, the ggplot2 library, and the script generate_plot.py from the BUSCO repository.
Files
- An assembled genome or transcriptome, or an annotated protein-coding gene set in standard FASTA format.
Preparing the input files and choosing a dataset
1.Download the content of the testing repository (https://gitlab.com/ezlab/busco_protocol), where you can find the inputs used for the examples. You can clone the repository using Git. If not available on your system, you can easily install Git by following the instructions at https://git-scm.com/. Run:
- $ git clone https://gitlab.com/ezlab/busco_protocol.git
In general, a command to run a BUSCO assessment with a dataset manually specified looks like the following:
- $ busco -i <SEQUENCE_FILE> -l
-o <OUTPUT_NAME> -m
with four main mandatory arguments:
- -i (or --in): a path to your FASTA file, which is either a nucleotide fasta file or a protein fasta file, depending on the BUSCO mode of assessment. From v5.1.0 the input argument can also be a directory containing fasta files to run the analysis on multiple inputs (see Basic Protocol 3).
- -l (or --lineage): the manually picked dataset for the assessment. It can be a dataset name, i.e., bacteria_odb10, or a relative (e.g., ./bacteria_odb10) or full (e.g., /home/user/bacteria_odb10) path. It is recommended to use the dataset name alone, as in this way BUSCO will automatically download and check the version of the dataset. In the case of a path, the dataset found in the given path will be used. Note that this argument is ignored when running with the auto-lineage workflow (see Basic Protocol 2).
- -m (or --mode): which analysis mode to run, either genome (or geno), proteins (or prot), or transcriptome (or tran).
- -o (or --out): the name of the output directory that identifies the analysis run and that contains all results, logs, and intermediate data.
The BUSCO command can take several additional arguments (also see Critical Parameters and Advanced Parameters in Commentary. The arguments can also be specified in a config file, which can be passed to BUSCO using the --config argument. If all parameters are set in the config.ini file, the command for launching BUSCO will look like:
- $ busco --config /path/to/config/config.ini
The arguments passed through the command line will overwrite the arguments in the config file. Remember to uncomment the lines in the config file to enable them (i.e., remove the semicolon in front of the line). For additional instructions on how to use the config file, see Advanced Parameters.
In the following examples, we are going to analyze the genome assembly and annotated gene set of the yeast Torulaspora globosa (assembly accession: GCF_014133895.1). We first need to select an appropriate dataset according to the taxonomy of this species. The lineage datasets used for BUSCO assessments are not packaged with the software. BUSCO will automatically download the required dataset when running the assessment. On the first BUSCO run, a busco_downloads/ folder will be created, containing the necessary lineage datasets within the subdirectory lineages/ (see Critical Parameters for more details on BUSCO datasets).
2.Manually select a dataset for the analysis. The most specific BUSCO dataset available for the yeast Torulaspora globosa is saccharomycetes_odb10. You can explore all available odb10 datasets by running:
- $ busco --list-datasets
Here we are using the most specific dataset to perform the assessment with the highest possible resolution. You could also assess this genome with all the other datasets matching the lineage of the organism. In this case, according to NCBI taxonomy (Schoch et al., 2020), the full taxonomic lineage of Torulaspora globosa is “cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Torulaspora”. On the basis of the currently available BUSCO odb10 datasets, besides the saccharomycetes_odb10 dataset, you could use the ascomycota_odb10, fungi_odb10, or eukaryota_odb10 datasets for the assessment. As a rule of thumb, it is always better to use the most specific dataset because it allows the highest-resolution analysis (i.e., more clade-specific markers are covered and scored). For example, saccharomycetes_odb10 is made of 2137 markers, while the ascomycota_odb10, fungi_odb10, and eukaryota_odb10 datasets are made of 1706, 758, and 255 markers, respectively. However, in some cases, it is preferable or necessary to use a lower-resolution dataset, e.g., for obtaining a set of markers shared among multiple species for building a phylogenomic tree (see Support Protocol 3). In such case, a less specific dataset might need to be selected, as the marker genes for building the alignments must be shared across all the species included in the tree. For example, for building a phylogenomic tree of fungal species that includes five budding yeasts (for which the most specific BUSCO dataset is saccharomycetes_odb10) and five boletales species (for which the most specific BUSCO dataset is boletales_odb10), the more general fungi_odb10 dataset could be used.
Running BUSCO on a single genome assembly with known taxonomy
In this example, we are going to assess the genome assembly of T. globosa using the saccharomycetes_odb10 dataset. In the downloaded repository, you can find this gene set at BUSCO_protocol/protocol1/Tglobosa_GCF_014133895.1_genome.fna.
3.Enter the busco_protocol testing folder:
- $ cd /{path_to_busco_protocol_folder}/
where {path_to_busco_protocol_folder} is the path to the busco_protocol/ testing folder you downloaded.
4.To launch an assessment of a genome assembly (which can be in the form of contigs, scaffolds, or chromosomes) the genome mode needs to be specified. Run the following command:
- $ busco -i ./protocol1/Tglobosa_GCF_014133895.1_genome.fna -l saccharomycetes_odb10 -m geno -o busco_out_Tglob_genome -c 12
You can specify a relative or full path to the input file, e.g., if the file is located in /usr/user_name/busco_protocol/protocol1/Tglobosa_GCF_014133895.1_genome.fna, and you are in the busco_protocol/ folder, you can specify the relative path with -i ./protocol1/Tglobosa_GCF_014133895.1_genome.fna, where ./ stands for the current working directory. Otherwise, you can specify the full path with -i /usr/user_name/busco_protocol/protocol1/Tglobosa_GCF_014133895.1_genome.fna. Additional parameters can be specified to change the default values of various settings. Here, for example, we specify the number of CPUs (central processing units) to be used with the --cpu (or -c) argument. By default, the --download_path argument, with which you can specify the location on your machine where you wish to store the downloaded datasets and additional files required to run the software, and the --out_path argument with which you can specify the location on your machine where you wish to store the output folder, are set to the current working directory. Feel free to change these paths to your preferred location on your machine. For directions on using additional parameters, see the Critical Parameters and Advanced Parameters sections in Commentary.
- $ tar -xfv arthropoda_odb10.2020-09-10.tar.gz
Running this assessment on 12 CPUs with otherwise default options should take approximately 1 min. After BUSCO has completed the run, the summary scores are printed to the standard output and reported in the short_summary*.txt file that you can find in the main output folder, in this example busco_out_Tglob_genome/. The summary file is named after the dataset used for the assessment and the output name. In this case, it will be named short_summary.specific.saccharomycetes_odb10.busco_out_Tglob_genome.txt, and will be similar to the following:
-
BUSCO version is: 5.2.2
-
The lineage dataset is: saccharomycetes_odb10 (Creation date: 2020-08-05, number of genomes: 76, number of BUSCOs: 2137)
-
Summarized benchmarking in BUSCO notation for file /data/manni/busco_protocol/protocol1/Tglobosa_GCF_014133895.1_genome.fna
-
BUSCO was run in mode: genome
-
Gene predictor used: metaeuk
- ***** Results: *****
- C:99.6%[S:99.5%,D:0.1%],F:0.1%,M:0.3%,n:2137
- 2129 Complete BUSCOs (C)
- 2126 Complete and single-copy BUSCOs (S)
- 3 Complete and duplicated BUSCOs (D)
- 3 Fragmented BUSCOs (F)
- 5 Missing BUSCOs (M)
- 2137 Total BUSCO groups searched
-
Dependencies and versions:
- hmmsearch: 3.1
- metaeuk: 4.a0f584d
This text file contains the classification of the identified BUSCO markers into categories of Complete (C), Complete and single-copy (S), Complete and duplicated (D), Fragmented (F), and Missing (M) BUSCOs as percentages and counts, and additional information such as the dataset used and the versions of the dependencies. In our example, BUSCO evaluates this genome assembly as of high quality, i.e., containing almost all the expected single-copy genes with a low duplication score. In the output folder, you can also find the logs/ subfolder containing the logs of BUSCO and its dependencies, and the run_<odb_dataset_name>/ folder (in this case run_saccharomycetes_odb10) containing all the other outputs and intermediate files. See Guidelines for Understanding Results for details on all BUSCO outputs and their interpretation.
When assessing a prokaryotic species (i.e., by using a prokaryotic dataset) or virus, BUSCO uses the gene predictor Prodigal (Hyatt et al., 2010) to predict genes. For eukaryotic species, as in this example, two alternative workflows, BUSCO_Metaeuk and BUSCO_Augustus, employing two different gene predictors, are available. From BUSCO v5 the default genome mode uses the BUSCO_Metaeuk workflow which employs the gene predictor MetaEuk (Levy Karin, Mirdita, & Söding, 2020) for predicting genes. In the command above, we used the BUSCO_Metaeuk workflow by default. To use the alternative BUSCO_Augustus workflow, which employs the gene predictor AUGUSTUS (Hoff & Stanke, 2019), you need to add the --augustus flag:
- $ busco -i ./protocol1/Tglobosa_GCF_014133895.1_genome.fna -l saccharomycetes_odb10 -m geno -o busco_out_Tglob_genome_Augustus -c 12 --augustus
Note that this takes much longer to run (∼11 min) compared to the default workflow (∼1 min) using 12 CPUs. Feel free to use more CPUs if available on your system. The BUSCO_Augustus workflow requires a working installation of tBLASTn and AUGUSTUS (see Support Protocol 1 for details on these dependencies), and BUSCO needs to know the location of the AUGUSTUS configuration directory. So check that the correct path is set for this variable by typing:
{AUGUSTUS_CONFIG_PATH}
If the variable is not set or you need to change it, you can declare it as follows:
- $ export AUGUSTUS_CONFIG_PATH="/path/to/AUGUSTUS/augustus-x.x.x/config/"
When using the BUSCO_Augustus workflow, one important optional argument to consider is the choice of AUGUSTUS pretrained species-specific gene prediction parameters. Each BUSCO lineage dataset has a predefined default selection, e.g., for the diptera_odb10 dataset the default species is “fly”, meaning AUGUSTUS gene prediction parameters pretrained on the fruit fly, Drosophila melanogaster. Also see Critical Parameters and Advanced Parameters in Commentary for further details on AUGUSTUS-specific parameters.
The output folder obtained from the BUSCO_Augustus workflow contains the same main output files as seen for the default BUSCO_Metaeuk workflow, and only differs for the intermediate output files generated by the different tools in the workflow. See Guidelines for Understanding Results for a detailed description of all the output files and folders generated with different workflows.
On medium to large eukaryotic genomes, and especially on very large genomes, e.g., greater than a few gigabase pair (Gbp), the BUSCO_Metaeuk workflow is much faster than the BUSCO_Augustus workflow. For example, running a genome assessment on the 10-Gbp genome assembly of the wheat Triticum dicoccoides with the poales_odb10 dataset (4896 markers) using 50 CPUs takes approximately 9 hr with the BUSCO_Metaeuk workflow, whereas it needs several days using the BUSCO_Augustus alternative. The two workflows yield comparable but not identical results, as they are based on different methodologies; for details see Manni et al. (2021). Depending on the input size and your specific needs, you may opt for the BUSCO_Augustus workflow, e.g., if you need to obtain AUGUSTUS retraining parameters (also see Suggestions for Further Analyses).
Running BUSCO on a single annotated protein-coding gene set with known taxonomy
The command for analyzing an annotated gene set is similar to that used for genome assemblies, except for the --mode argument, which is set to proteins (or prot). The input file is a FASTA file of the amino acid translations of the protein-coding genes. To properly evaluate the amount of BUSCO gene duplications (which can be due to technical artifacts or true duplications), isoforms must be filtered from the gene set before running the assessment. Otherwise, each BUSCO gene with isoforms will score as a duplicated BUSCO. There are different criteria to select isoforms; for our purposes we selected the longest isoform per gene (steps in Support Protocol 3 also describe how to select the longest isoform per gene using a GFF file and the corresponding genome).
5.For analyzing the gene set of the yeast T. globosa using the manually specified dataset, run:
- $ busco -i ./protocol1/Tglobosa_GCF_014133895.1_geneset.faa -l saccharomycetes_odb10 -m prot -o busco_out_Tglob_geneset -c 12
The result folder looks similar to the one obtained for assessing genome assemblies, without the intermediate files related to the gene prediction steps. The gene set analysis is faster than the analysis on the corresponding genome assembly.
Running BUSCO on a single transcriptome assembly with known taxonomy
Transcriptome assessments are launched with the same four mandatory options for assessing genome assemblies and gene sets. You just need to change the value of the --mode (or -m) argument to transcriptome (or tran) and provide a transcriptome assembly as input file. The transcriptome mode requires MetaEuk when assessing eukaryotes and tBLASTn when assessing prokaryotes (see Support Protocol 1 for more details on BUSCO dependencies). As for the gene set, to obtain true estimates of the numbers of duplicated BUSCOs for transcriptomes, these should be pre-processed to select just one representative transcript per gene. Here we analyze an assembled transcriptome from the female reproductive tract of the Asian tiger mosquito Aedes albopictus (NCBI BioProject: PRJNA223166; Boes et al., 2014) using the diptera_odb10 dataset. For this analysis, the transcriptome was not pre-processed to remove isoforms, and thus we may obtain a high number of duplicated BUSCOs which just reflects the presence of different isoforms.
6.To launch the transcriptome assessment, run:
- $ busco -i ./protocol1/Aalbopictus_transcriptome.fna -l diptera_odb10 -m tran -o busco_out_Aalbopictus_transcriptome -c 12
The main content of the result folder is similar to the one obtained for assessing genome assemblies, with some differences (see the BUSCO output subsection in the Guidelines for Understanding Results). If the transcriptome comes from a specific organ/tissue or time point, it may contain only a fraction of the full BUSCO sets. Our test transcriptome harbors approximately 38% of the 3285 diptera_odb10 markers, which is not surprising given the specialized function of the tissue under consideration. See Guidelines for Understanding Results for details on the output files and their interpretation.
Plotting BUSCO scores
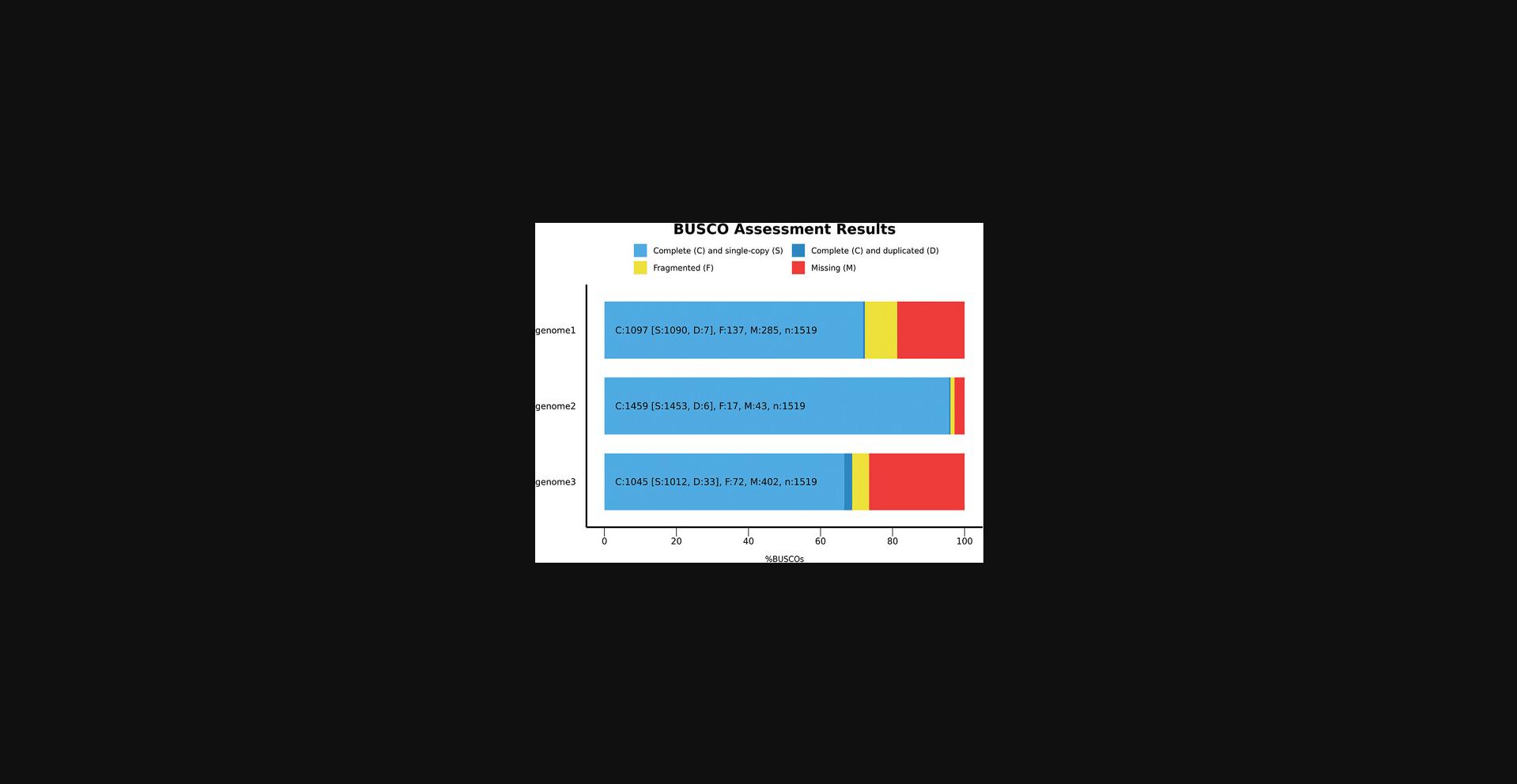
It is common to plot BUSCO scores as a bar chart from a single run, or from different runs side-by-side to visualize like-for-like comparisons, e.g., of different species/strains or assembly versions. To encourage the use of a standard and distinctive color scheme in publications, BUSCO includes a dedicated R (R Core Team, 2020) script to produce a figure and its source code that can be further edited, allowing one to customize the resulting bar chart (labels, fonts, axes, etc.). To run the script, Python is required. To produce the image, R and the ggplot2 library need to be available on the system. To plot the scores of one or multiple runs, it is sufficient to provide in a single folder the short_summary.txt files of each BUSCO run.
7.Collect the short_summary*.txt files in one folder, e.g.:
- $ mkdir BUSCO_summaries/
- $ cp OUT1/short_summary.*.lineage_odb10.OUT1.txt ./BUSCO_summaries/
- $ cp OUT2/short_summary.*.lineage_odb10.OUT2.txt ./BUSCO_summaries/
8.Then, simply run the generate_plot.py script available in the BUSCO repository (https://gitlab.com/ezlab/busco/scripts), providing the path to the directory containing the summary files:
- $ python3 generate_plot.py -wd BUSCO_summaries
A *.png image file and the corresponding R source code file, which can be further edited to make cosmetic adjustments to the plot, will be produced in the same folder containing the BUSCO summaries. By default, the run name is used as the label for each plotted result, and this is automatically extracted from the short summary file name: so, for short_summary.generic.lineage_odb10.OUT1.txt, the label would be “OUT1”. You can modify this in the file name as long as you keep the naming convention, e.g.: short_summary.generic.lineage_odb10.[edit_name_here].txt, or you can simply edit the R source code file to change any plotting parameters and produce a personalized bar chart by running the code manually in your R environment. However, we suggest keeping the same color scheme. By adding the --no_r flag to the command, the script will simply produce the R script required to reproduce the plot, which can then run on any system with R and ggplot2 installed. Figure 1 shows an example of the resulting default plot.
Basic Protocol 2: ASSESSING AN INPUT SEQUENCE WITH A DATASET AUTOMATICALLY SELECTED BY BUSCO
This protocol describes the case in which you do not specify a BUSCO dataset for the assessment. Instead, BUSCO attempts to automatically select the most appropriate dataset on the basis of a phylogenetic placement onto precomputed trees of marker genes extracted from the input file. This workflow is activated by using the --auto-lineage flag, and can be applied to all data types (genomes, gene sets and transcriptomes). With this option, no assumptions are made on the taxonomic origin of the input sequences, and it can be useful for inputs with unknown taxonomic origin, as in the case of metagenome-assembled genomes (MAGs), or on large sets of inputs where specifying each dataset manually can be tedious (see Basic Protocol 3 for running this workflow on multiple inputs). If the user knows the domain of origin of the input, the --auto-lineage-prok or --auto-lineage-euk flags can be specified for prokaryotic and eukaryotic species, respectively. BUSCO will run faster when one of these two options is selected instead of the full --auto-lineage. In general, if the taxonomy of the species is known, it is preferable to specify the dataset manually, as the time and resources needed increase substantially with the phylogenetic placement. Also, the placement procedure may resort to a broader, less-specific dataset for the assessment if not enough markers are placed on the tree to select the most specific one. In this case, the selection may roll back to one of the general domain datasets (i.e., archaea_odb10, bacteria_odb10, or eukaryota_odb10). See the Guidelines for Understanding Results for more details on the automatic selection of the datasets.
Necessary Resources
Hardware
- As described in Basic Protocol 1.Note that, usually, the RAM required for using the --auto-lineage option will not exceed 13 GB when assessing prokaryotic species; thus, the assessment can also run on a laptop. If the placement involves eukaryotic species, you might need more memory to run the assessment.
Software
- As described in Basic Protocol 1. Note that for running the auto-lineage workflow you need a working installation of SEPP and pplacer.
Files
- As described in Basic Protocol 1. The auto-lineage workflow can be applied to genome assemblies, gene sets, and transcriptomes. To perform the phylogenetic placement procedure, additional files are required, e.g., the precomputed super-alignments and trees. These are automatically downloaded by BUSCO during the analysis if a connection to the Internet is available. If not, you will have to download and put these files manually in the busco_downloads/ folder as described in the annotation to step 4 of Basic Protocol 1.
Running BUSCO on one input without specifying a lineage dataset
In this case, you do not need to specify a lineage dataset. BUSCO will first run the three root datasets (bacteria_odb10, archaea_odb10, and eukaryota_odb10) to figure out the domain of the input file, and then attempt to phylogenetically place the markers extracted in this first run on a precomputed superalignment and phylogenetic tree.
1.In this example, we are going to assess the same genome assembly of T. globosa assessed in Basic Protocol 1, but this time without specifying the saccharomycetes_odb10 dataset. Enter the BUSCO_protocol testing folder downloaded in Basic Protocol 1:
- $ cd {path_to_busco_protocol_folder}/
2.Run the command as:
- $ busco -i ./protocol1/Tglobosa_GCF_014133895.1_genome.fna -m geno -o busco_out_Tglob_genome_auto -c 12 --auto-lineage
Running this assessment on 12 CPUs with otherwise default options should take approximately 4 min, which is longer than the time needed to run the assessment described in Basic Protocol 1 when the dataset was manually specified (∼1 min), as additional steps are required here for the phylogenetic placement procedure. You can enter the output directory busco_out_Tglob_genome_auto/ to check the results and inspect which dataset was automatically selected for the assessment. If the run was successful, you should find the summary file short_summary.specific.saccharomycetes_odb10.busco_out_Tglob_genome_auto.txt, named by the same rule described in Basic Protocol 1. As you can see, BUSCO was able to select the correct and most specific dataset for the assessment (saccharomycetes_odb10), corresponding to the manual choice we made in Basic Protocol 1. Additionally, you will find the file short_summary.generic.eukaryota_odb10.busco_out_Tglob_genome_auto.txt, which corresponds to the short_summary file obtained by running the parent “root” domain dataset (in this case eukaryota_odb10) as the first step of the auto-lineage workflow. See Guidelines for Understanding Results for more information on the outputs produced by the auto-lineage workflow and how to interpret the results. In this example, you could specify the --auto-lineage-euk option instead of --auto-lineage, as we already know that our input file belongs to a eukaryotic species. You can apply the auto-lineage workflow to all the input types selecting the different modes covered in Basic Protocol 1.
Basic Protocol 3: ASSESSING MULTIPLE INPUTS
BUSCO v5 and higher can be run in “batch” mode on a collection of input files. These must be present in a single folder and must be of the same type, i.e., being either all genomes assemblies, transcriptomes or gene sets. As for analyzing a single input file, there are two ways to run BUSCO on multiple files: a) with a BUSCO dataset manually specified (described in Basic Protocol 1), and in this case the same dataset will be applied to all the input files, e.g., a set of insect genomes analyzed with the insecta_odb10 dataset; or b) in combination with the auto-lineage workflow (also see Basic Protocol 2). In this case each input file is analyzed independently with a specific dataset automatically selected by BUSCO, and thus can be applied on a set of taxonomically heterogeneous inputs, such as metagenomic data that include MAGs of both eukaryotic and prokaryotic origin.
Necessary Resources
Hardware
- As described in Basic Protocols 1 and 2.
Software
- As described in Basic Protocols 1 and 2.
Files
- As described in Basic Protocols 1 and 2.
Running BUSCO on multiple inputs using the same dataset specified manually
Collect all the input files you want to analyze in a folder, and simply specify this folder as input. Here we analyze a set of bacterial genomes with the mycoplasmatales_odb10 dataset. You can find the folder containing the input genomes in the subfolder ./protocol3/bact_genomes/.
1.Open the terminal and enter the following command:
- $ busco -i ./protocol3/bact_genomes -l mycoplasmatales_odb10 -m geno -o busco_out_mycoplas_genomes -c 12
BUSCO will figure out automatically that it must run on multiple files. For each run, a standard BUSCO result folder named after the input file will be created in the main output folder. In the main output folder, an additional text file summarizing the score of all the runs is provided. This batch mode can be applied to the other two data types by changing the attribute of the -m option.
Running BUSCO on multiple inputs without specifying a lineage dataset
As with the previous command, collect all the input files you want to analyze in a folder. This protocol is suitable for estimating the quality of metagenomic bins or “metagenome-assembled genomes” (MAGs) of unknown taxonomic origin, e.g., obtained from binner programs such as MetaBat (Kang et al., 2019), MaxBin2 (Wu & Singer, 2021), and MEGAN (Bağcı, Patz, & Huson, 2021; Huson et al., 2018). In such cases, the input file is a folder containing one FASTA file per bin. Here we analyze a mix of prokaryotic and eukaryotic genomes with known taxonomy, so we can compare the dataset selected by BUSCO with the ground truth represented by the taxonomic lineage of the inputs. As each input sequence can be either coming from a prokaryotic or eukaryotic species, we use the more general --auto-lineage flag to cover both cases.
2.Uncompress the fasta files:
- $ cd ./protocol3/genomes_mix/ && gunzip *.gz && cd ../../
3.Run the auto-lineage workflow by entering:
- $ busco -i ./protocol3/genomes_mix -m geno -o busco_out_mix -c 12 --auto-lineage
As with the previous example, the result folders for each run are written to the specified output directory (here busco_out_mix) and named after the corresponding input file name. An additional text file summarizing the scores of all the runs is also written in the main output directory. Table 1 shows an example of the summary file listing the BUSCO results for all the inputs analyzed. The first column reports the input file name; the second reports the most specific dataset that was selected by BUSCO on the basis of the placement procedure; the third to eighth columns report the standard BUSCO scores as percentages; and the ninth column reports the total number of markers for the corresponding dataset. In our example, the inputs are a mix of bacterial genomes, one green algal and two fungal genomes. The datasets selected by BUSCO are in agreement with the taxonomic lineages of the inputs. The table additionally reports the score obtained by running the three main domain datasets used to establish the most likely domain of the input file during the first step of the workflow. These scores might be useful for spotting heavy cross-domain contamination issues. However, note that these “by-products” scores need to be carefully interpreted (see Guidelines for Understanding Results for how to interpret these scores). This workflow can be run on the other data types (gene sets and transcriptomes) by changing the attribute of the -m argument (see Basic Protocol 1). This analysis should take approximately 30 min to complete when using 12 CPUs. On small to medium-sized genomes, increasing the number of CPUs will not speed up the analysis substantially, as the current implementation of parallelization in BUSCO is not optimized on small genomes. To speed up the analysis when analyzing a large number of prokaryotic or small eukaryotic genomes, you can take advantage of a workflow management system as described in Alternate Protocol.
| Input_file | Dataset | C | S | D | F | M | n | Scores_archaea_odb10 | Scores_bacteria_odb10 | Scores_eukaryota_odb10 |
|---|---|---|---|---|---|---|---|---|---|---|
| GCF_002215565.1_ASM221556v1_genomic.fna | sulfolobales_odb10 | 95.5 | 95.1 | 0.4 | 0.6 | 3.9 | 1244 | C:95.4%[S:95.4%,D:0.0%],F:2.1%,M:2.5%,n:194 | C:14.5%[S:14.5%,D:0.0%],F:10.5%,M:75.0%,n:124 | C:7.9%[S:7.5%,D:0.4%],F:5.1%,M:87.0%,n:255 |
| GCA_900452565.1_34347_D01_genomic.fna | legionellales_odb10 | 95.6 | 95.6 | 0.0 | 2.6 | 1.8 | 772 | C:17.0%[S:16.5%,D:0.5%],F:4.1%,M:78.9%,n:194 | C:91.1%[S:91.1%,D:0.0%],F:6.5%,M:2.4%,n:124 | C:4.7%[S:4.3%,D:0.4%],F:2.0%,M:93.3%,n:255 |
| GCA_003366055.1_ASM336605v1_genomic.fna | bacteria_odb10 | 63.7 | 63.7 | 0.0 | 8.9 | 27.4 | 124 | C:7.2%[S:7.2%,D:0.0%],F:1.5%,M:91.3%,n:194 | C:63.7%[S:63.7%,D:0.0%],F:8.9%,M:27.4%,n:124 | C:0.8%[S:0.8%,D:0.0%],F:1.2%,M:98.0%,n:255 |
| GCA_003367175.1_ASM336717v1_genomic.fna | burkholderiales_odb10 | 99.5 | 98.5 | 1.0 | 0.4 | 0.1 | 688 | C:18.0%[S:17.0%,D:1.0%],F:5.7%,M:76.3%,n:194 | C:100.0%[S:100.0%,D:0.0%],F:0.0%,M:0.0%,n:124 | C:5.5%[S:4.7%,D:0.8%],F:3.5%,M:91.0%,n:255 |
| GCA_003353085.1_ASM335308v1_genomic.fna | alteromonadales_odb10 | 99.5 | 99.1 | 0.4 | 0.2 | 0.3 | 820 | C:21.1%[S:21.1%,D:0.0%],F:4.6%,M:74.3%,n:194 | C:99.2%[S:96.0%,D:3.2%],F:0.8%,M:0.0%,n:124 | C:4.7%[S:3.9%,D:0.8%],F:2.0%,M:93.3%,n:255 |
| GCF_000182965.3_ASM18296v3_genomic.fna | saccharomycetes_odb10 | 98.6 | 98.0 | 0.6 | 0.8 | 0.6 | 2137 | C:58.8%[S:55.2%,D:3.6%],F:6.2%,M:35.0%,n:194 | C:30.6%[S:29.0%,D:1.6%],F:16.9%,M:52.5%,n:124 | C:94.9%[S:93.3%,D:1.6%],F:2.4%,M:2.7%,n:255 |
| GCF_002968355.1_ASM296835v1_genomic.fna | entomoplasmatales_odb10 | 95.5 | 94.6 | 0.9 | 1.5 | 3.0 | 332 | C:3.6%[S:3.6%,D:0.0%],F:2.6%,M:93.8%,n:194 | C:81.5%[S:81.5%,D:0.0%],F:1.6%,M:16.9%,n:124 | C:1.2%[S:1.2%,D:0.0%],F:0.4%,M:98.4%,n:255 |
| GCF_000226975.2_ASM22697v3_genomic.fna | natrialbales_odb10 | 98.7 | 98.3 | 0.4 | 0.3 | 1.0 | 1368 | C:99.5%[S:99.5%,D:0.0%],F:0.5%,M:0.0%,n:194 | C:21.0%[S:19.4%,D:1.6%],F:9.7%,M:69.3%,n:124 | C:7.5%[S:7.1%,D:0.4%],F:3.1%,M:89.4%,n:255 |
| GCA_001560045.1_GG12_C01_07_genomic.fna | sulfolobales_odb10 | 94.5 | 94.2 | 0.3 | 0.7 | 4.8 | 1244 | C:93.3%[S:93.3%,D:0.0%],F:3.1%,M:3.6%,n:194 | C:13.7%[S:13.7%,D:0.0%],F:11.3%,M:75.0%,n:124 | C:7.9%[S:7.5%,D:0.4%],F:5.1%,M:87.0%,n:255 |
| GCA_900036045.1_Methanoculleus_sp_MAB1_genomic.fna | methanomicrobiales_odb10 | 87.3 | 87.2 | 0.1 | 5.8 | 6.9 | 882 | C:88.6%[S:88.1%,D:0.5%],F:7.2%,M:4.2%,n:194 | C:17.7%[S:16.9%,D:0.8%],F:12.9%,M:69.4%,n:124 | C:5.1%[S:5.1%,D:0.0%],F:3.9%,M:91.0%,n:255 |
| GCA_003018975.1_ASM301897v1_genomic.fna | synechococcales_odb10 | 88.1 | 86.5 | 1.6 | 4.1 | 7.8 | 788 | C:18.0%[S:17.5%,D:0.5%],F:5.7%,M:76.3%,n:194 | C:83.8%[S:80.6%,D:3.2%],F:12.1%,M:4.1%,n:124 | C:6.3%[S:5.9%,D:0.4%],F:3.9%,M:89.8%,n:255 |
| GCF_002220235.1_ASM222023v1_genomic.fna | chlorophyta_odb10 | 97.1 | 96.7 | 0.4 | 0.7 | 2.2 | 1519 | C:54.1%[S:50.5%,D:3.6%],F:6.2%,M:39.7%,n:194 | C:68.6%[S:61.3%,D:7.3%],F:14.5%,M:16.9%,n:124 | C:76.9%[S:76.5%,D:0.4%],F:7.1%,M:16.0%,n:255 |
Alternate Protocol: DECREASING ANALYSIS RUNTIME WHEN ASSESSING A LARGE NUMBER OF SMALL GENOMES WITH BUSCO AUTO-LINEAGE WORKFLOW AND SNAKEMAKE
This protocol describes the use of a workflow management system, specifically Snakemake, to increase the speed of a BUSCO auto-lineage analysis on multiple genomes, and it is an alternative to Basic Protocol 3.Currently, BUSCO parallelization is not optimized for small genomes; therefore, using a workflow management system such as Snakemake with multiple CPUs can considerably reduce the runtime for analyses involving a large number of inputs, e.g., when assessing metagenomic bins or MAGs. Note that this Alternate Protocol is intended for analyzing prokaryotic genomes, small eukaryotic genomes, or a combination of both. It is not intended to be used on medium/large genomes, as for these the parallelization is already optimized in the standard BUSCO run.
Necessary Resources
Hardware
- As described in Basic Protocol 3.
Software
- As described in Basic Protocol 3. Conda and Snakemake (≥ v5.30), available at https://snakemake.readthedocs.io/en/stable/getting_started/installation.html. A clone of the BUSCO “plugins” GitLab repository is available at https://gitlab.com/ezlab/plugins_buscov5.git.
Files
- As described in Basic Protocol 1.
Running multiple BUSCO assessments with the auto-lineage workflow and Snakemake
To prevent collisions of simultaneous BUSCO runs attempting to write to the same location when using this alternate protocol, BUSCO will be run with the --offline flag, which does not allow BUSCO to connect to the Internet. Therefore, before running this workflow, you need to make sure to have all the required BUSCO datasets and files downloaded and updated on your machine.
In this example, we assess a mix of prokaryotic genomes, so we can just download the prokaryotic datasets.
1.To download the datasets in bulk, enter the location on your machine where you want to store the them:
- $ cd {/path_where_to_store_BUSCO_downloads/}
2.And run:
- $ busco --download prokaryota
If you plan to analyze a mix of prokaryotic and eukaryotic genomes, you will need all the datasets. These can be downloaded with busco --download all, which will take approximately 25 min to complete. The size of all extracted datasets is ∼127 GB (∼11 GB for the prokaryotic datasets only).
3.In the part of your system where you want to run BUSCO, clone the BUSCO “plugins” repository. Here we clone the repository into the directory of the repository you downloaded in Basic Protocol 1:
- $ cd /{path_to_busco_protocol_folder}/
- $ git clone https://gitlab.com/ezlab/plugins_buscov5.git
4.Then enter the BUSCO_batch_analysis_with_snakemake/ folder:
- $ cd plugins_buscov5/BUSCO_batch_analysis_with_snakemake
5.You first need to change some parameters in the YAML configuration file that, from the current working directory, you can find in ./config/config.yaml. You need to change the paths to the inputs and the busco_downloads/ folder using Vim or your preferred text editor, e.g.:
- $ vim config/config.yaml # and manually edit the file
If you have exactly followed the previous commands, the relative paths will be as follows:
-
wdir_path: "."
-
data_path: "../../alternate_protocol1/genomes_mix" # specify the path to the folder
-
file_suff: ".fna" # specify the suffix of your input files
-
out_path: "busco_out_mix_alter_protocol" # specify the output folder
-
configuration
-
BUSCO_threads: 5
-
autolineage: "--auto-lineage-prok" # or "--auto-lineage" or "--auto-lineage-euk"
-
download_path: "{path/where/you/stored/BUSCO_downloads}" # specify the path of the busco_downloads folder where the datasets and files are stored
6.In this example, we are going to use a job submission management system to submit our Snakemake workflow to a cluster. In the following example, we use SLURM, but a similar command can be launched with other submission management systems such as PBS. From the current working directory (i.e., BUSCO_batch_analysis_with_snakemake/), you may run:
- $ snakemake --cluster "sbatch --ntasks-per-node=1 --cpus-per-task=5 --job-name=busco_runs" --jobs 6
Here the --jobs argument specifies the total number of jobs to launch simultaneously. If in the config.yaml file we specify 5 CPUs for each BUSCO analysis, the overall command will use 30 CPUs in total. We suggest restricting the number of CPUs to 5-8 for each BUSCO analysis, as it is the optimal setting for small genomes (e.g., <10 Mbp) in the majority of cases.
Alternatively, you can run the workflow without using a job submission management system, as in the next command (where we also specify the --use-conda flag), e.g., with:
- $ snakemake --cores 30 --use-conda
Here -–cores defines the maximum number of CPUs that can be used by the workflow. With 5 CPUs per BUSCO analysis, as defined in the config.yaml file, the workflow should run six BUSCO analyses at the time.
The outputs of the analysis will be written to the busco_out_mix_alter_protocol/ folder. For the 25 prokaryotic genomes of this example, the analysis submitted through SLURM on a cluster using 30 CPUs completes in approximately 4 min.
Support Protocol 1: BUSCO SETUP
This protocol describes the different ways to install BUSCO and its dependencies. BUSCO was implemented using Python 3 and tested on Linux operating systems. It is therefore recommended to use a Linux machine for running BUSCO. There are currently three options to obtain BUSCO: using the Docker container, through Bioconda, or by installing BUSCO and its dependencies manually.
Necessary Resources
Hardware
- As described in Basic Protocol 1.
Software
- Python 3.3 or higher, the BUSCO tool, and its dependencies (refer to step 4, below). Optionally, a working installation of Docker or Singularity if using the Docker container option. Optionally, a working installation of conda if using the Bioconda option.
Installing BUSCO from Bioconda
To install BUSCO through Bioconda you need a working installation of conda. See how to install conda at https://docs.conda.io/projects/conda/en/latest/user-guide/install/index.html. Ensure you have a conda version >=4.8.4.Enter conda -V to check the version. If necessary, update conda by entering:
- $ conda update -n base conda
1.Install the latest BUSCO version. It is recommended to install BUSCO in a separate conda environment instead of your conda base environment. Enter:
- $ conda create -n <your_env_name> -c conda-forge -c bioconda busco=x.x.x
Conda can be a bit slow sometimes. You can instead use the mamba package manager (https://github.com/mamba-org/mamba), which is an extremely fast and popular conda replacement. You can install mamba with conda install -n base -c conda-forge mamba. To install BUSCO using mamba, simply replace conda with mamba in the previous command: mamba create -n <your_env_name> -c conda-forge -c bioconda busco=x.x.x.
where your_env_name is the name you want to assign to your new environment, and x.x.x refers to the BUSCO version, which is 5.2.2 as of writing of this manuscript. For more updated versions, change the version number to the latest one. You can check the last BUSCO version available on Bioconda by searching for the “BUSCO” recipe on the Bioconda search bar.
Now, in order to start working in the new environment, type:
- $ conda activate <your_env_name>
Double check the BUSCO version with:
- $ busco -v
You can exit the environment by typing:
- $ conda deactivate
Using the BUSCO Docker container
For each new release, a BUSCO container that wraps everything required to run a BUSCO analysis is also created. Currently, the container is made available on Docker Hub. Before getting the BUSCO container, you need to have Docker installed on your system. See the Docker user guide for details on installing and using Docker (Merkel, 2014) (https://www.docker.com/). Using the Docker container has the advantage of streamlining the setup and installation, and it also allows one to easily track all software versions used in the analyses (the version number, e.g., v5.2.1_cv1, identifies all components at once). It also guarantees that only dependency versions compatible with the most up-to-date BUSCO code are used.
2.With Docker installed, just fetch the BUSCO container with:
- $ docker pull ezlabgva/busco:vx.x.x_cv1
where x.x.x refers to the BUSCO version and _cv1 to the container version for a given BUSCO version. There should be only one container per version unless an issue with the container was fixed without changing the BUSCO code.
3.To run the BUSCO container, you will need to control the user that is run within the container. You can pass your own user id with the -u argument, e.g., -u $(id -u). Note that if you do not specify a user, the container will create files under a different user you do not control. With the mount option -v you specify the location on your filesystem on which you can write and transfer files between your filesystem and the container filesystem. In our example, we will use the current working directory from where you are launching the assessment. You could use another folder, but be careful not to specify a host folder that does not exist, as Docker will create it using the root account and without asking for confirmation. It is safer to use the current directory as in our example. Run the container as follows:
(id -u) -v $(pwd):/busco_wd ezlabgva/busco:vx.x.x_cv1 busco -i genome.fna <OTHER_OPTIONS>
To use a custom config.ini file to set run parameters, you need to extract this file from the container as follows:
(pwd):/busco_wd ezlabgva/busco:v5.2.1_cv1 cat /busco/config/config.ini > config.ini
You can then edit this newly created config.ini file with your preferred text editor and pass it to the BUSCO command using the --config argument, e.g.:
(pwd):/busco_wd ezlabgva/busco:v5.2.1_cv1 busco -i genome.fna --config=/busco_wd/myconfig.ini
If you want to run the BUSCO container in an HPC system that does not support Docker but supports Singularity, which is another containerization program commonly used on HPC clusters, you can convert the Docker container into a Singularity Image Format (see https://quay.io/repository/singularity/docker2singularity). Always consult the documentation of your specific HPC environment first.
Manually installing BUSCO and its dependencies
4.First, manually install the required BUSCO dependencies.
A fully functional BUSCO setup will require:
1.Python 3.3+ 2.BioPython (https://biopython.org/) and pandas (https://pandas.pydata.org/) Python modules. 3.HMMER: To evaluate amino acid sequences using profile HMMs, all modes of BUSCO require HMMER, version 3.1b2 or higher, which can be obtained from http://hmmer.org/. 4. MetaEuk: Required to assess eukaryotic genome assemblies with the default BUSCO_Metaeuk workflow. MetaEuk can be obtained from https://github.com/soedinglab/metaeuk. 5.Prodigal: Required to assess prokaryotic genomes. This can be obtained from https://github.com/hyattpd/Prodigal. 6.SEPP (Mirarab, Nguyen, & Warnow, 2011) and pplacer (Matsen, Kodner, & Armbrust, 2010): Required to run BUSCO with the auto-lineage workflow, i.e., to perform the phylogenetic placement of input sequences to automatically select a BUSCO dataset for the assessment. SEPP can be retrieved from https://github.com/smirarab/sepp/ and pplacer from https://github.com/matsen/pplacer. 7.tBLASTn from NCBI BLAST+ (Camacho et al., 2009): Only needed if you want to perform genome assessments of eukaryotic species using the BUSCO_Augustus workflow or transcriptome assessments on prokaryotic sequences. It can be downloaded from https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+. There is an issue with tBLASTn versions 2.4-2.10.0 when using more than one CPU. The issue was fixed in version 2.10.1+, so make sure you have at least version 2.10.1+ installed. 8.AUGUSTUS (v3.3.3 or above): Required if you want to use the BUSCO_Augustus workflow (by specifying the --augustus flag in the command). BUSCO supports versions 3.3.3 or higher, and the software can be obtained from http://bioinf.uni-greifswald.de/augustus/. It includes multiple PERL scripts (https://www.perl.org/), and you should refer to the most up-to-date AUGUSTUS documentation for its PERL requirements. The executables required by BUSCO are: augustus, etraining, gff2gbSmallDNA.pl, new_species.pl, and optimize_augustus.pl. Additional environment variables have to be set as follows:
PATH PATH - $ export AUGUSTUS_CONFIG_PATH=/path/augustus-3.x.x/config/
- R and ggplot2 library: These are required for plotting BUSCO results using the commands described in Basic Protocol 1.
Make sure that each software package listed above works independently of BUSCO before attempting to run any BUSCO assessment. The minimal versions of the dependencies compatible with the current BUSCO code are shown in Table 2. You can obtain information about future version compatibility on the BUSCO website (https://busco.ezlab.org/).
| Genome assessment (eukaryotes) | Genome assessment (prokaryotes and viruses) | Auto-lineage workflow | Auto-lineage prok workflow | Auto-lineage euk workflow | |
|---|---|---|---|---|---|
| Biopython and pandas modules | x | x | x | x | x |
| HMMER v3.1b2+ | x | x | x | x | x |
| MetaEuk v4-*+ | x | x | x | ||
| Prodigal v2.6.3+ | x | x | x | ||
| SEPP v4.3.10+/ Pplacer v1.1.alpha13+ | x | x | x | ||
| NCBI BLAST+ v2.10.1+ |
x (only if using the --augustus flag) |
x (only if using the --augustus flag) |
x (only if using the --augustus flag) |
||
| Augustus v3.3.3+ |
x (only if using the --augustus flag) |
x (only if using the --augustus flag) |
x (only if using the --augustus flag) |
5.It is recommended to install BUSCO in a separate virtual environment. To do this, first create a Python virtual environment using Python ≥3.3 and the “venv” module (from version 3.3, Python should already include the venv module). Create a virtual environment with:
- $ python3 -m venv <ENV_NAME>
where <ENV_NAME> is the name you want to assign to the environment. You might name the environment after the BUSCO version you are installing, e.g., busco5.2.2.
6.Enter the folder that has been created:
- $ cd <ENV_NAME>
7.Activate the environment with:
- $ source ./bin/activate
8.Install the Biopython and Pandas modules:
- $ pip3 install biopython pandas
9.Retrieve the BUSCO source code from the GitLab repository. You can download and extract the BUSCO repository manually from the GitLab release page (https://gitlab.com/ezlab/busco/-/releases), or using Git, clone the repository with:
- $ git clone https://gitlab.com/ezlab/busco.git
10.Enter the busco/ folder:
- $ cd busco
11.Execute the setup.py script using python to install BUSCO:
- $ python3 setup.py install
12.Check that BUSCO has been installed and verify its version by entering the busco command with the help argument:
- $ busco -h
If you do not want to create a separate virtual environment, you can install BUSCO using the same procedure described above excluding the commands from steps 2 to 4. In this case, from the busco/ folder, you need to execute the setup.py script with:
- $ python3 install setup.py –user #(with only user privileges)
or
- $ sudo python3 setup.py install #(with root privileges)
Support Protocol 2: VISUALIZING BUSCO RESULTS
This protocol describes how to use some of the BUSCO results to create charts for displaying the location of markers on the input genome and visualizing syntenies between genomes.
Necessary Resources
Hardware
- Any machine supporting an installation of Python and R.
Software
- Python, conda (and optionally mamba), Snakemake, R, Rstudio, and the RIdeogram library.
Files
- One or multiple full_table.tsv files for plotting markers and visualizing syntenies.
Visualizing BUSCO markers on genomes
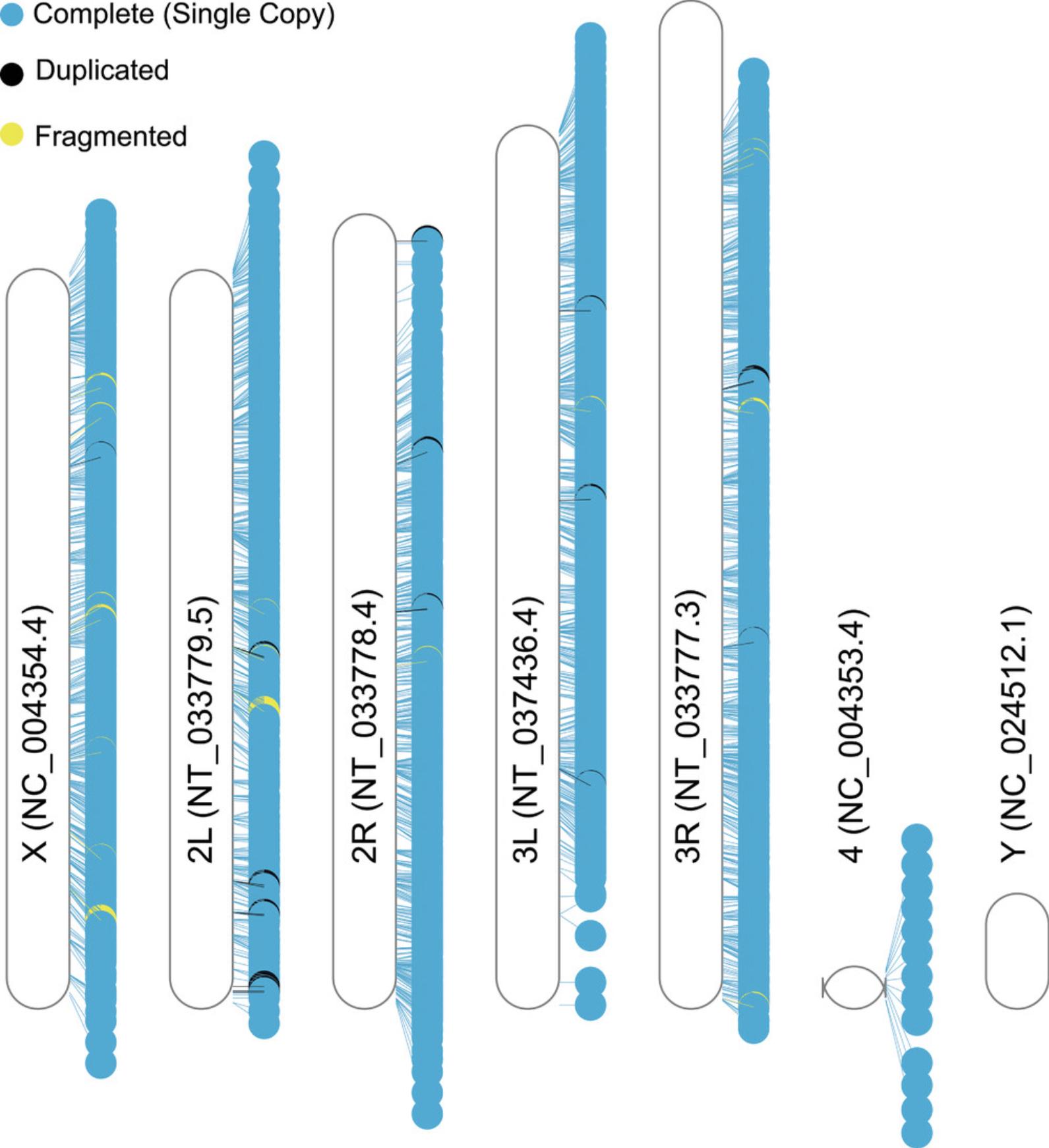
Sometimes it can be useful to visualize the location of BUSCO markers on the input genome, for example to show the distribution of markers, or highlight problematic regions, e.g., those with a high density of duplicated markers. This can be performed by extracting the BUSCOs’ coordinates from the full_table.tsv file and using a program for plotting genomic coordinates. There are several options available to plot these; here we use the R package RIdeogram (Hao et al., 2020).
1a. If not done already, download the busco_protocol:
- $ git clone https://gitlab.com/ezlab/busco_protocol
2a. Enter the support_protocol2/plot_markers subfolder:
- $ cd busco_protocol/support_protocol2/plot_markers
To generate the plot, you need to provide the path to the full_table.tsv file generated from a BUSCO run performed on a genome assembly. A file, {sample_name}_karyotype.txt, with karyotype information is also required. You can generate this file yourself, following this example (see also the RIdeogram documentation):
| Chr | Start | End | species | size | color |
| NC_004354.4 | 1 | 23542271 | Dmel | 12 | 25252 |
| NT_033779.5 | 1 | 23513712 | Dmel | 12 | 25252 |
| NT_033778.4 | 1 | 25286936 | Dmel | 12 | 25252 |
| NT_037436.4 | 1 | 28110227 | Dmel | 12 | 25252 |
| NT_033777.3 | 1 | 32079331 | Dmel | 12 | 25252 |
| NC_004353.4 | 1 | 1348131 | Dmel | 12 | 25252 |
| NC_024512.1 | 1 | 3667352 | Dmel | 12 | 25252 |
Alternatively, you can provide the path to the genome assembly used in the assessment, and the workflow will generate this file automatically from the assembly. Here we are using a full_table.tsv obtained by running the diptera_odb10 dataset on the D. melanogaster genome. You can find this file at support_protocol2/plot_markers/data/full_table/Dmel_full_table.tsv in the BUSCO protocol repository. Note that when using the workflow on your inputs, you need to name the full_table.tsv using the convention {sample_name}_full_table.tsv, and keep the same {sample_name}keyword across all the corresponding files related to the same sample. For example, if the full_table file is named Dmel_full_table.tsv, the karyotype file (if provided by the user) must be named Dmel_karyotype.txt, and the genome (if provided), Dmel.fna. You will need to edit the config.yaml file, which you can find in the plot_markers/ folder, to specify the paths to your own inputs. In this config file, you can also provide the path to a text file with the IDs of a subset of sequences you wish to plot, named as {sample_name}_selected_sequences.txt. If not provided, the script will attempt to plot all the sequences present in the input genome. Bear in mind that for a clean visualization, it is not possible to plot too many sequences, e.g., this approach will not work on a fragmented genome with hundreds or thousands of scaffolds. Alternatively, you can select some key sequences of interest to plot.
3a. To run the workflow on the test data, just enter:
- $ snakemake –cores 2 –use-conda
This workflow will: (a) extract the status, sequence IDs, and coordinates of each marker from the full_table.tsv; (b) extract the sequence lengths from the genome file to build a karyotype.txt file, if this file is not provided by the user.
4a. The plotting step is performed in the Rstudio environment where you can easily customize the appearance according to your needs and readily visualize the changes (you can also generate the plot from the command line using the Rscript command). Open Rstudio and set the working directory to the support_protocol2/plot_markers/ folder of the BUSCO plugins repository. From the Rstudio console run:
-
setwd("/path/to/support_protocol2/plot_markers/")
5a. Open the plot_markers/scripts/plot_markers2.R script within Rstudio. The paths to source the necessary files are already set for the test data. In this script, you may customize the R code to change colors and labels. Run the commands of this script within Rstudio to produce the plot. You can do this by selecting all the commands of the script and clicking on the “Run” button.
A *.png and a *.svg file (here Dmel.svg and Dmel.png) are written to the plot_markers/ folder. Figure 2 shows the resulting plot for this example. As you can see all D. melanogaster chromosomes are covered by complete and single-copy BUSCO markers, except for the Y chromosome.
Visualizing syntenies between genomes using BUSCO markers
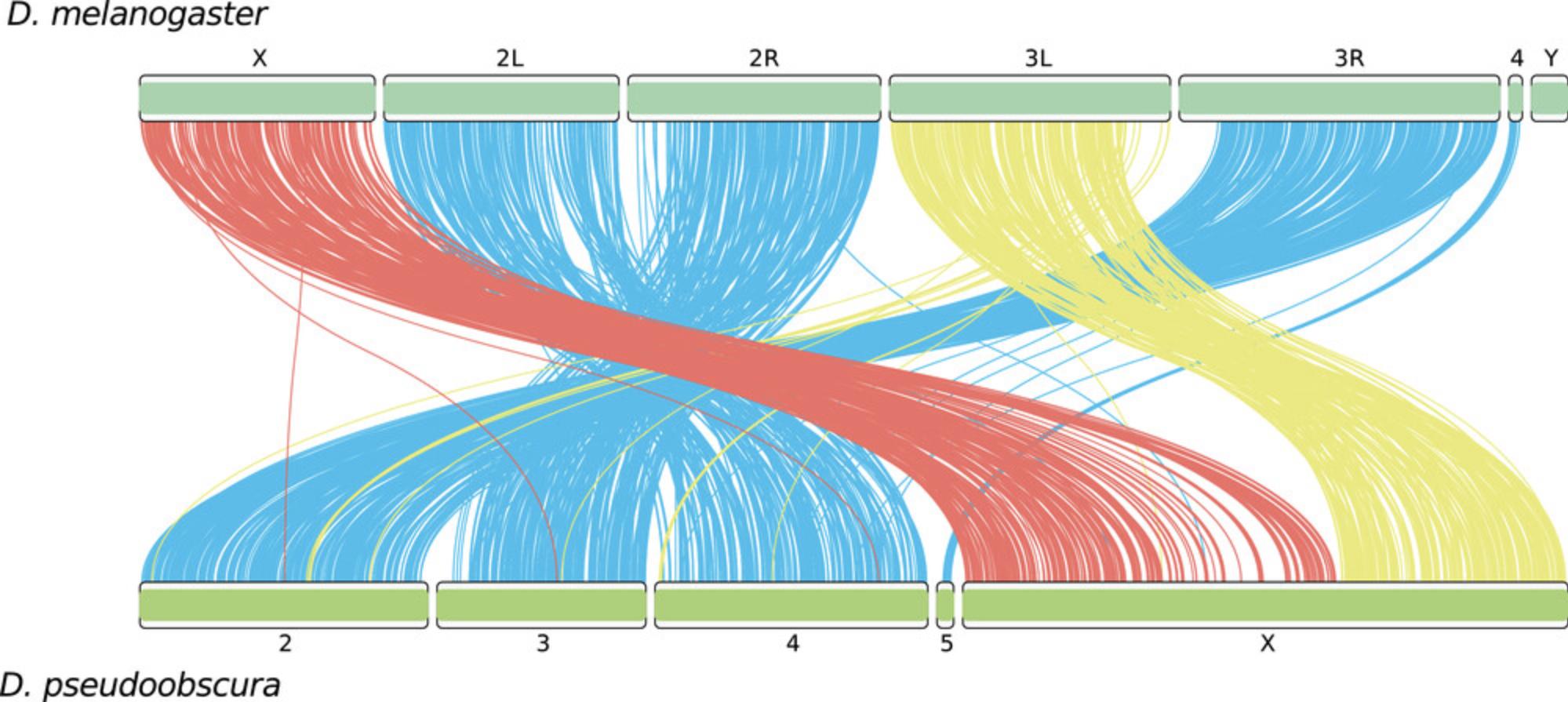
The genomic position of each single-copy ortholog identified by BUSCO on two or more genomes can be used to infer syntenies between genomes. For visualizing syntenies between two or more genomes, you will need the full_table.tsv files obtained by running BUSCO on the input genomes. Note that you need to use the same dataset for all the assessments. In this example, we infer syntenic relationships between Drosophila melanogaster and D. pseudoobscura chromosomes using the diptera_odb10 single-copy orthologs. There are various programs to plot syntenies; here we use the R package RIdeogram as in the previous example.
1b. If not done already, download the busco_protocol repository from the GitLab:
- $ git clone https://gitlab.com/ezlab/busco_protocol
2b. Enter the support_protocol2/plot_syntenies subfolder:
- $ cd busco_protocol/support_protocol2/plot_syntenies
You can find the two full_table.tsv files obtained by running BUSCO with the diptera_odb10 dataset on D. melanogaster and D. pseudoobscura assemblies in the plot_syntenies/data/full_table/ folder. To plot your own data, remove these files and add your full_table.tsv files.
3b. Then, run the script for creating the files required for generating the plot with:
- $ snakemake –cores 2 --use-conda
The script will pre-process the full_table.tsv files to create the file required for plotting the syntenies with RIdeogram. A karyotype.txt file can be provided to the command, in case you want to plot only a subset of sequences from the input files. You need to create this file with following format:
| Chr | Start | End | fill | species | size | color |
| 1 | 1 | 23542271 | 8AFFBB | Dmela | 12 | 7adb3d |
| 2 | 1 | 23513712 | 8AFFBB | Dmela | 12 | 7adb3d |
| 3 | 1 | 25286936 | 8AFFBB | Dmela | 12 | 7adb3d |
| 4 | 1 | 28110227 | 8AFFBB | Dmela | 12 | 7adb3d |
| 5 | 1 | 32079331 | 8AFFBB | Dmela | 12 | 7adb3d |
| 6 | 1 | 1348131 | 8AFFBB | Dmela | 12 | 7adb3d |
| 7 | 1 | 3667352 | 8AFFBB | Dmela | 12 | 7adb3d |
| 1 | 1 | 32422566 | 95FF7D | Dpseudo | 12 | 6c72f0 |
| 2 | 1 | 23510042 | 95FF7D | Dpseudo | 12 | 6c72f0 |
| 3 | 1 | 30706867 | 95FF7D | Dpseudo | 12 | 6c72f0 |
| 4 | 1 | 1881070 | 95FF7D | Dpseudo | 12 | 6c72f0 |
| 5 | 1 | 68158638 | 95FF7D | Dpseudo | 12 | 6c72f0 |
4b. Open Rstudio, and set the working directory to the support_protocol2/plot_syntenies/ folder inside the busco_protocol directory. From the Rstudio console, run:
-
setwd("/path/to/support_protocol2/plot_syntenies/")
5b. Open the plot_syntenies/scripts/plot_syntenies2.R script within Rstudio. The paths to source the necessary files are already set. Here you can customize the R code to change colors and labels. Now run the commands of this script within Rstudio to produce the plot. You can do this by selecting all the commands of the script and clicking on the “Run” button. A *.png and *.svg file should be written in the plot_syntenies/ folder. Figure 3 shows the chart of the syntenic relationships between the D. melanogaster and D. pseudoobscura chromosomes obtained with the test data.
Support Protocol 3: BUILDING PHYLOGENOMIC TREES
BUSCOs, being near-universal single-copy genes, can represent reliable markers to be used in phylogenomics studies. BUSCO assessments on multiple organisms can be used to quickly and easily identify single-copy markers to generate multiple sequence alignments (MSA) from which to infer the species phylogeny. BUSCO datasets can identify large sets of genes from genomic data of variable quality, avoiding the tedious and possibly biased manual selection of orthologs. In this protocol we illustrate a workflow to build a phylogenomic tree from annotated gene sets (with a GFF and genome assembly available for each species). In this case, we concatenate the MSAs into a superaligment and use a Maximum Likelihood (ML) approach to build the phylogenomic tree. The analyses presented here are by no means exhaustive, and you should choose the most suitable methods and parameters (e.g., using partitions, different models etc.) according to your specific goals and inputs.
Necessary Resources
Hardware
- As described in Basic Protocol 1.
Software
- As described in Basic Protocol 1 and additionally: conda (and optionally mamba), Snakemake, AGAT, MAFFT, TrimAl, IQ-TREE2, AMAS. Note that these tools will be automatically installed by running the Snakemake workflow (otherwise you can install these manually if you prefer).
Files
- A set of genome assemblies and their corresponding GFF files.
Building phylogenomic tree from a set of genome assemblies and GFF files
1.If not done already, download the busco_protocol repository from GitLab:
- $ git clone https://gitlab.com/ezlab/busco_protocol
2.Enter the support_protocol3/ folder:
- $ cd support_protocol3/
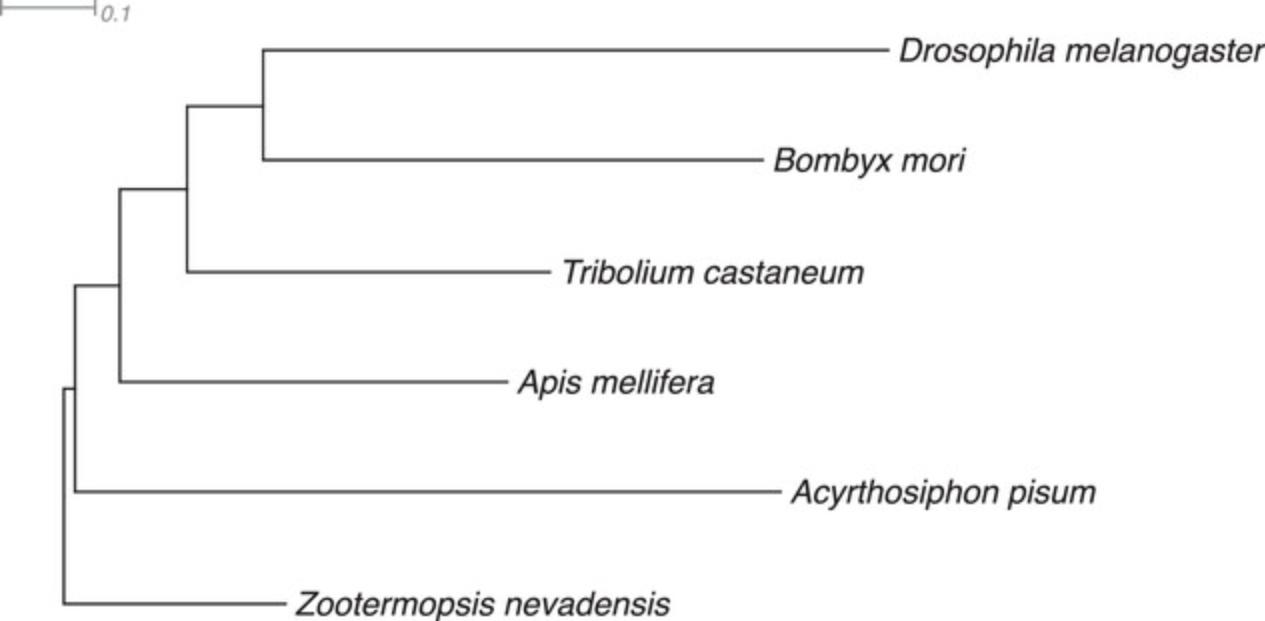
3.Collect the GFF and the genome assembly files you want to analyze in the GFFs/ and assemblies/ folders, respectively. In this example, we are going to build a small species tree using six insect species selected from various published sources: Drosophila melanogaster (GCF_000001215.4; Hoskins et al., 2015), Bombyx mori (GCF_014905235.1); Apis mellifera (GCF_003254395.2; Wallberg et al., 2019); Tribolium castaneum (GCF_000002335.3) (Herndon et al., 2020), Acyrthosiphon pisum (GCF_005508785.1; Li, Park, Smith, & Moran, 2019) and Zootermopsis nevadensis (GCF_000696155.1; Terrapon et al., 2014). Using the assembly accessions, you can download these with the NCBI “datasets” command-line tool (https://www.ncbi.nlm.nih.gov/datasets/), e.g., datasets download genome accession GCF_014905235.1 or manually from the NCBI portal or the ftp site using the wget command, e.g.:
-
$ cd genomes/
-
$ wget -O Drosophila_melanogaster.fna.gz
-
$ wget -O Bombyx_mori.fna.gz
-
$ wget -O Apis_mellifera.fna.gz
-
$ wget -O Tribolium_castaneum.fna.gz
-
$ wget -O Acyrthosiphon_pisum.fna.gz
-
$ wget -O Zootermopsis_nevadensis.fna.gz
-
$ gunzip *.gz && cd ../GFFs
-
$ wget -O Drosophila_melanogaster.gff.gz
-
$ wget -O Bombyx_mori.gff.gz
-
$ wget -O Apis_mellifera.gff.gz
-
$ wget -O Tribolium_castaneum.gff.gz
-
$ wget -O Acyrthosiphon_pisum.gff.gz
-
$ wget -O Zootermopsis_nevadensis.gff.gz
-
$ gunzip *.gz && cd ../
4.Edit the config/config.yaml file in which you need to specify the paths to the GFFs and genomes folders, and additional parameters such as which BUSCO dataset you want to use to identify the single-copy orthologs that encompass all species. If you exactly follow the steps in this protocol, all parameters in the config.yaml file are already set for this example. Analyses with higher-resolution datasets identify more markers for inferring the species phylogeny. In our example, we are using the insecta_odb10 dataset.
5.Run the Snakemake workflow, which comprises all the steps required for obtaining the species phylogeny. Briefly, the workflow consists of the following steps:
1.For each species, isoforms are first filtered to keep only the longest protein-coding sequence for each gene according to the corresponding GFF file. This is achieved using the AGAT toolkit (Dainat J.), which also produces a useful report that can be used to verify there are no issues with the GFF entries. 2.BUSCO is then run in protein mode on the resulting filtered gene sets using the dataset specified in the config.yaml file. 3.The single-copy genes identified from each run are extracted and selected based on user-defined parameters specified in the config.yaml file, e.g., to select only genes that are shared across 100% of the species and with no duplicates across all species. 4.For each orthologous group, proteins are aligned using MAFFT (Katoh & Standley, 2013) and trimmed with trimAl (Capella-Gutiérrez, Silla-Martínez, & Gabaldón, 2009). 5.Alignments are concatenated with AMAS (Borowiec, 2016). 6. The resulting superalignment is used to infer a maximum likelihood phylogeny using IQ-TREE 2 (Minh et al., 2020). To run the workflow, enter:
- $ snakemake –cores 16 --use-conda
Several intermediate files and the final phylogenetic.nwk tree file are written in the output folder. Assessments of the six insect species with the insecta_odb10 dataset identified 1154 complete and single-copy BUSCOs found in all species. Note that in the config.yaml file, you can also specify to use markers found as duplicated (the best scoring duplicate will be selected) and markers missing in a certain number of species, e.g., missing in 10% of the inputs. To visualize the Newick file, you can use any program for viewing phylogenetic trees, e.g., Dendroscope (Huson & Scornavacca, 2012). Figure 4 shows the resulting tree obtained in this example.
GUIDELINES FOR UNDERSTANDING RESULTS
BUSCO attempts to provide a quantitative assessment of completeness in terms of expected gene content, and can be applied to genome assemblies, assembled transcriptomes, and annotated protein-coding gene sets. The main results are simplified into categories of Complete and single-copy , Complete and duplicated , Fragmented , or Missing BUSCOs. BUSCO completeness results make sense only in the context of the biology of your organism and your data types. For example, you need to understand whether missing or duplicated genes are due to biological or technical reasons. For instance, a high level of duplication may be explained by a recent whole duplication event (biological) or redundancies due to technical issues, e.g., when the assembly procedure fails to correctly collapse (or separate) haplotypes. Transcriptomes and protein sets that are not filtered for isoforms may lead to a high proportion of duplicates. These and other aspects to consider when interpreting the BUSCO scores are described in this section, along with a detailed list of the outputs produced by each assessment workflow.
BUSCO outputs
Every BUSCO assessment produces an output folder named according to the value specified under the --out (or -o) argument. This folder contains all the results, consisting of several subfolders with lots of files including intermediate results. These are kept for your benefit, so that you can examine the results in more detail and, if needed, use the data for downstream analyses. The content of the result folder can change according to the workflow that has been run, but a successful assessment will always contain the following files and folders:
- short_summary*.txt file: a simple summary result file that reports the percentages and counts of “complete” (“single-copy” and “duplicated”), “fragmented”, and “missing” BUSCOs. This file is named after the dataset used for the assessment and the name given to the output folder, e.g., if the dataset used is diptera_odb10 and the output name out_fly the file will be named short_summary.specific.diptera_odb10.out_fly.txt. The keywords “specific” or “generic” are added to distinguish the most specific assessment from the generic assessment with the domain-level dataset (that is created only when using the auto-lineage workflow). The following is an example of the content of a short_summary*.txt file:
# BUSCO version is: 5.2.0 # The lineage dataset is: saccharomycetes_odb10 (Creation date: 2020-08-05, number of genomes: 76, number of BUSCOs: 2137) # Summarized benchmarking in BUSCO notation for file /data/manni/BUSCO_PROTOCOL/protocol1/Tglobosa_GCF_014133895.1_genome.fna # BUSCO was run in mode: genome # Gene predictor used: metaeuk
***** Results: ***** C:99.6%[S:99.5%,D:0.1%],F:0.1%,M:0.3%,n:2137 2129 Complete BUSCOs (C) 2126 Complete and single-copy BUSCOs (S) 3 Complete and duplicated BUSCOs (D) 3 Fragmented BUSCOs (F) 5 Missing BUSCOs (M) 2137 Total BUSCO groups searched
Dependencies and versions:
hmmsearch: 3.1 metaeuk: 4.a0f584d
The file also contains the version of the dependencies used to run the assessment.
- logs/ folder: containing the BUSCO log file and logs of the dependencies. Take some time to check the log files; these are there for your benefit in order to highlight potential problems that may have occurred during your BUSCO run. This is the first place to look when a run crashes. If you need help, submit a request on the GitLab issue board attaching the BUSCO log file or part of it.
- run_<dataset_odb10>/ folder: a folder containing all results and intermediate files relative to the assessment performed with the corresponding dataset <dataset_odb10>, e.g., for an assessment performed with the diptera_odb10 dataset, this folder will be named run_diptera_odb10.
- full_table.tsv file: the detailed list of all BUSCO genes and their predicted status in the genome, gene set, or transcriptome. It can be found in the run_<dataset_odb10>/ folder. This file is slightly different for the three modes. For “genome” mode, it displays the BUSCO orthologous group ID, followed by the status of the gene identified in the input sequence, along with its position (sequence name, start/end coordinates and strand), score, and length. The table additionally reports a keyword description and a link to the OrthoDB webpage of the corresponding orthologous group. For the protein mode, the identifier of the sequence is displayed and there are no start/end coordinates. With the transcriptome mode, it contains the identifier of the transcript, and the start and end coordinates refer to the start and end of the predicted match on the transcript. The full_table.tsv from a genome run looks like the following:
# BUSCO version is: 5.2.0 # The lineage dataset is: saccharomycetes_odb10 (Creation date: 2020-08-05, number of genomes: 76, number of BUSCOs: 2137)
| # Busco id | Status | Sequence | Gene Start | Gene End | Strand | Score | Length | OrthoDB url | Description |
| 0at4891 | Complete | NC_050729.1 | 306917 | 321619 | + | 4303.0 | 3383 | https://www.orthodb.org/v10?query=0at4891 | Midasin |
- busco_sequences/ folder: containing the amino acid sequences in FASTA format of all BUSCO genes identified by the assessment procedure, and organized in three subfolders (single_copy_busco_sequences/, fragmented_busco_sequences/ and multi_copy_busco_sequences/) according to the gene status.
- hmmer_output/ folder: contains a tabular format of each HMMER output, one for each candidate protein evaluated, named after the BUSCO profile HMM used, e.g., 25101at4891.out.1. Note that these are all initial candidates, including those that are not retained as positive matches in the final classification.
- missing_busco_list.txt file: listing the identifiers of missing BUSCOs.
If BUSCO is run on multiple inputs (i.e., a directory is provided to the input argument), the top-level output directory contains a number of subdirectories corresponding to each input file and named after the input file name. These directories contain the same output files as described above. An additional file is written in the top-level directory, listing all the resulting scores for each input file. A single busco.log file for the entire batch analysis is stored in the logs/ folder in the top-level directory. The result folder on a run assessing two input files look like the following:
output_batch_analysis/ ├── batch_summary.txt ├── GCF_000222975.1_ASM22297v1_genomic.fna ├── GCF_014133895.1_ASM1413389v1_genomic.fna └── logs/
The result folder also contains the outputs produced in each intermediate step depending on the workflow that has been run. Specifically, for genome assessments using the BUSCO_Metaeuk workflow or for the transcriptome assessments on eukaryotic species:
- metaeuk_output/ folder: results of the MetaEuk gene predictor.
In v5, BUSCO takes advantage of MetaEuk also to find BUSCO markers in eukaryotic transcriptomes. However, when assessing prokaryotic transcriptomes, BUSCO uses the previous transcriptome mode that relies on tBLASTn. The pipeline consists of a tBLASTn run taking BUSCO amino acid consensus sequences as queries and the input transcripts as a database to obtain a subset of sequences harboring potential matches to each BUSCO gene. In this case, a six-frame translation is blindly performed over the transcript length (which might include untranslated regions) and HMMER is run to assign a score to the candidate amino acid sequences. The output folder contains the hmmer_output/ directory, the short_summary.txt, the missing_buscos_list.tsv, and the full_table.tsv files similar to those of the eukaryotic transcriptome mode. Additionally, there will be the blast_output/ and translated_proteins/ folders containing the six-frame translations of every transcript having a match to a BUSCO marker during the tBLASTn analysis, including discarded candidates not present in the final classification. These sequences should not be considered genuine protein sequences without a thorough validation using a dedicated method.
For the genome assessment using the BUSCO_Augustus workflow the output contains the following specific folders:
- blast_db/ folder: containing the database created from the input file for the tBLASTn search.
- blast_output/ folder: containing the raw output of the two tBLASTn runs and the corresponding coordinates as defined by BUSCO to represent candidate regions.
- augustus_output/ folder: containing the results of the AUGUSTUS gene predictor. It includes the list of single-copy genes that were used to retrain AUGUSTUS, and, in the predicted_genes/ subfolders, one gene model for each candidate region evaluated named after the BUSCO block profile used, e.g., 241586at4751.out.1. In the extracted_proteins/ subfolder, there is one nucleotide and one amino acid sequence for each gene model, e.g., 241586at4751.fna.1 and 241586at4751.faa.1. The sequences in these subfolders represent all the initial candidates, including those that were not included in the final classification. The retraining parameters produced by BUSCO to be used by the second AUGUSTUS run are stored in the retraining_parameters/ subfolder. Other intermediate files that are produced during the analysis can be found in the remaining folders, e.g., gb/ and gff/ folders containing the files in GenBank and General Feature Format (GFF), respectively.
For the prokaryote/virus genome assessment:
- prodigal_output/ folder: containing the results of the Prodigal gene predictor.
Results obtained by using the auto-lineage workflow additionally include the files related to the assessments using the three generic “root” datasets and the outputs from the phylogenetic placement in the following subfolder:
- auto_lineage/ folder: containing the BUSCO results of the assessment using the three generic “root” datasets. They represent either incorrect or less specific choices of lineage. A symbolic link in the main output folder is made to the generic “root” dataset that was selected as most likely, in the following example, eukaryota_odb10/. This folder also contains the placement_files/ subfolder with the files of the phylogenetic placement of the extracted eukaryotic markers onto the precomputed eukaryota tree (not displayed in the tree-like structure below).
For example, the structure of the main output folder for an auto-lineage assessment on an eukaryotic genome looks like:
main_out/ ├── auto_lineage/ │ ├── run_archaea_odb10/ │ ├── run_bacteria_odb10/ │ └── run_eukaryota_odb10/ ├── logs/ │ ├── busco.log │ ├── hmmsearch_err.log │ ├── hmmsearch_out.log │ ├── metaeuk_err.log │ ├── metaeuk_out.log │ ├── prodigal_err.log │ ├── prodigal_out.log │ ├── sepp_err.log │ └── sepp_out.log ├── prodigal_output/ │ └── predicted_genes/ ├── run_eukaryota_odb10 -> path_to/main_out/auto_lineage/run_eukaryota_odb10 ├── run_saccharomycetes_odb10/ │ ├── busco_sequences/ │ ├── full_table.tsv │ ├── hmmer_output/ │ ├── metaeuk_output/ │ ├── missing_busco_list.tsv │ └── short_summary.txt ├── short_summary.generic.eukaryota_odb10.main_out.txt └── short_summary.specific.saccharomycetes_odb10.main_out.txt
In general, these intermediate outputs may be useful for other subsequent analyses or for in-depth investigation of the results.
How to report BUSCO results
You can report results in simple BUSCO notation, e.g.: C:89.0%[S:85.8%,D:3.2%],F:6.9%,M:4.1%,n:194, along with the name of the dataset. In addition, always report the BUSCO command and versions of the dependencies used for the run as supplementary information. If you use the BUSCO container, specifying the container version is sufficient to identify the versions of the dependencies and to safely reproduce a run. You can use the generate_plot.py script to produce a simple bar chart summarizing the results of one or multiple BUSCO runs (see Basic Protocol 1). Remember to report the versions of the genome assemblies, annotated gene sets, or transcriptomes assessed in the analyses.
Considerations on BUSCO score interpretation
To judge whether a score is satisfactory, the user will have to consider the type of sequence assessed and the biological context of the species of interest. A very good genome assembly should contain the majority of the BUSCO genes that were not lost during evolution. The number of gene losses for each specific species inside a clade cannot be precisely defined a priori , and it is possible that a small fraction of the expected single-copy genes is missing in certain species because of true gene loss, or present as multi-copy genes due to true gene duplications.
When comparing BUSCO scores of a genome assembly and its corresponding gene set, differences in the resulting scores may be due to several factors. When results from genome assembly assessments appear to be better than for their gene sets, it likely indicates that the approach taken by BUSCO to identify the subset of single-copy genes on the genome has produced better gene predictions than the one used in the annotation pipeline (at least for the subset of genes that make up the BUSCO lineage dataset). In this case, it may be possible to improve the annotation pipeline using different approaches or additional evidence. Conversely, if a gene set appears more complete than its corresponding genome, this may suggest that the custom annotation pipeline, which is usually tailored to that specific organism (e.g., by using multiple sources of evidence such as RNA-seq data), has resulted in generally better annotations than the general approach implemented in BUSCO. The user should assess both the assembly and the annotation results to judge whether the gene prediction strategy can be improved in light of the expected gene content in the genome. It is important that the user should never complete the annotated gene set with the BUSCO genes recovered by the genome mode prior to assessing it, as it would bias the evaluation of true annotation efficiency.
For the evaluation of transcriptome assembly, you need to consider the source material of the transcriptome. Not all BUSCO genes are necessarily expressed in the tissue/organ under consideration or at specific time points; therefore, transcriptomes are expected to show varying degrees of completeness depending on the sample. For example, with assessment of transcriptomes derived from specific tissues or life stages where the repertoire of transcripts is expected to be highly specialized, a low completeness score can be expected, and would in fact offer support that such targeted sampling was achieved.
Generally, a good genome assembly and its corresponding gene set should show low levels of duplications. Duplication of a few BUSCO genes in a genome is compatible with biological reality, as their evolution under single-copy control may be relaxed in some sublineages or specific species. However, a high duplication score could denote technical issues during the assembly procedure, e.g., indicating that the procedure has failed to correctly collapse (or separate) haplotypes, resulting in numerous pairs of highly similar duplicated genes. High duplication scores can also signal the presence of contaminant species, e.g., the presence of sequences from a species related to the target one. In such cases the single-copy markers from both species will score, and appear as duplicated markers. High duplication scores could also reflect biological reality, as for example with whole-genome duplication events. As mentioned above it is important to know the evolutionary context of the species under analysis to correctly interpret the results.
Users should be aware that the classification procedure that results in labeling the expected single-copy genes as “complete”, “fragmented”, and “missing”, is based by necessity on classification approximations that reflect the most likely state of the gene. For example, the label “missing” is applied to BUSCOs with no matches (probably truly absent from the dataset), but also to matches that do not meet the HMM score cutoffs. These could be in fact present as fragments that fail to align or that do not reach these cut-offs, therefore not having enough matching sequence to classify the partial match as “fragmented”. Classification of each BUSCO gene as complete is also based on a simplified classification based on the comparison of the length of the candidate sequence to length thresholds determined based on the distribution of gene lengths in the species used to build the datasets. It may be possible that a gene that is an outlier in terms of length or score will be classified as fragmented when it is in fact complete, or vice versa. Nonetheless, a high rate of fragmented BUSCO markers likely indicates issues in the sequencing or assembly procedures, or the inability of the annotation pipeline to fully capture the complexity of some gene models. Turning fragmented BUSCO gene matches into complete matches is a good indicator of a significant improvement of the quality of an assembly, especially when supported by changes in other metrics such as N50 values.
Other incongruences can arise when close homologs to BUSCO genes are present in the sequences under analysis. In such cases, the BUSCO classifier will give a better score to the true copies and therefore be able to discard the other sequences. However, if the actual BUSCO is missing, close homologs may sometimes reach a sufficient score to be considered as positive matches to a BUSCO gene that is in fact not present. An experienced user, manually investigating the outputs, may be able to spot such situations. Nevertheless, the overall quality of the input data will be clear from the overall score. Generally, inputs with low completeness scores and high duplication scores would warrant further investigation to determine if these are due to technical reasons that can be resolved by employing different assembly or annotation strategies.
Consideration on the automated selection of the dataset
The BUSCO auto-lineage workflow introduced in v4 first runs BUSCO on the three most generic datasets (archaea_odb10, bacteria_odb10, and eukaryota_odb10) to determine the most likely domain of origin of the input sequence. Once the most likely domain is selected, BUSCO automatically attempts to find the most appropriate BUSCO dataset to be used for the final assessment by performing a phylogenetic placement of the markers identified and extracted from the initial run. BUSCO evaluations are valid when an appropriate dataset is used, i.e., the dataset belongs to the lineage of the species to test. It is possible that the automated placement procedure selects suboptimal datasets for the assessment, e.g., when not enough markers are placed to confidently select a higher-resolution dataset. In that case, the BUSCO assessment is still valid but just at a lower resolution, e.g., when a bacterial species from the Actinobacteria clade is assessed with the most generic bacteria_odb10 dataset rather than the most specific actinobacteria_odb10 dataset. Though an infrequent scenario, it is also possible that BUSCO automatically selects, on the basis of the placement procedure, an incorrect dataset for the assessment. In this case the BUSCO scores might not be reliable. In our tests and benchmarks, we see that this event occurs rarely, and in the majority of such cases it is due to existing incongruences between the official taxonomy of the species and its actual phylogenetic signal.
When running the auto-lineage workflow on multiple files, the scores related to the assessments with the three general datasets (run to determine the domain of origin of the input file during the first step of the placement procedure) are also shown along with the final scores in the summary table listing the results of individual runs. These scores are there for your benefit, as they may help identify heavy contamination from other domains. However, you need to interpret these values with extra care. BUSCO matches in another domain do not necessarily mean that your genome/proteome contains sequences from that domain. These can be due to a certain fraction of overlapping markers or spurious matches among markers from different domains. However, high completeness scores in multiple domains might help you spot possible contamination issues.
COMMENTARY
Background Information
The BUSCO assessment process consists of running a computational pipeline to identify and then classify sets of genes expected to be found as single-copy in the majority of species within a certain taxonomic group. BUSCO matches can be identified from genome assemblies, annotated gene sets, or transcriptomes, and are classified based on custom sets of HMM (hidden Markov models) profiles using HMMER. For genome assessments, gene models are first built using the gene predictor MetaEuk with a collection of single-copy protein-coding genes as evidence (default BUSCO_Metaeuk workflow) or using ab initio gene prediction with AUGUSTUS on the potential matches identified by tBLASTn searches (BUSCO_Augustus workflow, available by specifying the --augustus flag). Matches that meet the BUSCO HMM score cutoffs are classified as “complete” if their lengths fall within BUSCO profile length expectations, and if found more than once they are classified as “duplicated”. Those that do not meet the length requirements are considered as partial matches and are classified as “fragmented”. BUSCOs without matches passing the thresholds are classified as “missing”. The assessments provide an intuitive quantification of the completeness of different data types in terms of expected gene content. BUSCO v4/v5 features novel workflows that can deal with large eukaryotic genomes and sequences of unknown origin, and can be applied to metagenome-assembled genomes.
Critical Parameters
BUSCO datasets
The collection of single-copy markers, built starting from the OrthoDB data, are organized in datasets corresponding to specific lineages. BUSCO versions 4 and 5 come with a total of 193 BUSCO odb10 datasets (Manni et al., 2021; also see Supplementary Table 1 in Supporting Information), representing major species groups. They are identified by the name of the taxon they represent and a version number, e.g., “eukaryota_odb10”. It is important to note that *_odb10 datasets are compatible with BUSCO versions 4 and 5 (old *_odb9 datasets are not compatible with these most recent versions). Each BUSCO marker has a unique identifier, e.g., 21160at4891, where the number preceding “at” represents a unique identifier within the level at which the orthologs were computed, which is indicated by the number (TaxID) following the “at” (in this example the Saccharomycetes level). The content of a standard BUSCO dataset is the following:
- info/ folder: containing the list of species used to create the set and additional information on the orthologous groups that make up the dataset.
- hmms/ folder: containing one profile HMM file for each BUSCO marker, and is required by HMMER.
- ancestral file: a FASTA file containing a consensus of the extant sequences for each BUSCO marker; it is required by tBLASTn.
- ancestral_variants file: a FASTA file containing a consensus and variants of the extant sequences. It is required by tBLASTn.
- refseq_db.faa file: a FASTA file containing a collection of representative sequences for each BUSCO marker. It is required by MetaEuk.
- prfl/ folder: containing one block profile file for each BUSCO marker; required by AUGUSTUS.
- dataset.cfg file: information about the dataset, including the number of BUSCO markers that make up the dataset and the number of species used to build the dataset. It also contains dataset-specific parameters to use with MetaEuk (such as the max intron length and max sequence length) and AUGUSTUS (specifically, the default species used by AUGUSTUS among those available with the tool, e.g., “fly” for the insecta_odb10 dataset).
- scores_cutoff file: minimal HMMER scores to reach for each gene to be considered as orthologous to BUSCO markers.
- lengths_cutoff file: minimal length values for BUSCO matches to be classified as complete.
- links_to_ODB10.txt file: description and link to the corresponding orthologous group web page on the OrthoDB database.
- missing_in_parasitic.txt file: an optional file containing the list of markers missing in a particular subclade within a parent lineage (e.g., for microsporidians in fungi_odb10 dataset).
BUSCO datasets are not distributed with the software. If your system is connected to the Internet, you do not need to retrieve the datasets; BUSCO will download the latest version of the required dataset(s) automatically when running the assessment if the name of the dataset is passed to the --lineage (or -l) argument, e.g., -l bacteria_odb10, or if the auto-lineage option is used. If a dataset has been downloaded previously, BUSCO will check that the local version corresponds to the latest version. By default, if a new version of a file is available, BUSCO will issue a warning to update the dataset, but the analysis will proceed with the local version of the dataset. The dataset will be updated directly if the flag --update-data is passed to the BUSCO command. BUSCO will replace the current file or folder with the latest version and archive the previous one. If a relative or full path to a dataset is passed to BUSCO, e.g., -l /my/own/path/bacteria_odb10 or -l ./path/bacteria_odb10, this automated management will be disabled. When running the auto-lineage workflow (no datasets specified), BUSCO will also automatically obtain the datasets, and additional files required to run the placement procedure.
If you are running BUSCO in an environment that does not have access to the Internet, you can pass the --offline argument to prevent BUSCO from attempting any download. You will have to download and unpack all files manually from the dataset repository (currently https://busco-data.ezlab.org/v5/data/) and place them in the BUSCO download folder. The location of this folder should then be passed using the command line flag, e.g.: --download_path /path/to/datasets. Optionally, you can download one or multiple datasets independently from a BUSCO assessment by passing a list of datasets to the --download argument, e.g.: busco --download bacteria_ob10 eukaryota_odb10. Four keywords are also available for bulk downloads: all, prokaryota, eukaryota, and virus. A valid busco_downloads/ folder looks as follows:
busco_downloads ├──file_versions.tsv ├──information │ ├──lineages_list.2019-11-27.txt │ └──virus_datasets.2020-11-26.txt ├──lineages │ ├──acidobacteria_odb10 │ ├──saccharomycetes_odb10 │ ├──viridiplantae_odb10 └──placement_files
├──list_of_reference_markers.bacteria_odb10.2019-12-16.txt ├──mapping_taxid-lineage.bacteria_odb10.2019-12-16.txt ├──mapping_taxids-busco_dataset_name.bacteria_odb10.2019-12-16.txt ├──supermatrix.aln.bacteria_odb10.2019-12-16.faa ├──tree.bacteria_odb10.2019-12-16.nwk ├──tree_metadata.bacteria_odb10.2019-12-16.txt
Choice of the dataset
If the taxonomic origin of the input sequence is known, the first decision that has to be made is which dataset should be used for the assessment among those available (see Basic Protocol 1). A good rule of thumb is to select the most specific lineage the species belongs to, as it will provide the best resolution possible for the evaluation. However, in specific cases the user may select a more generic dataset, representing a higher taxonomic level. Reducing the runtime of the analysis might be a reason for choosing a lower-resolution dataset, as the runtime is proportional to the number of genes included in the dataset, which tends to increase in lower taxonomic levels. Runtimes are less of an issue with the newly introduced BUSCO_Metaeuk workflow, as it can complete an assessment in a few hours even on large assemblies and large datasets. Long runtimes can still occur when using the BUSCO_Augustus workflow, which can take several days to complete on a combination of large assemblies and large datasets. In some cases, a less specific dataset that encompasses all the species under analysis must be selected, e.g., for extracting markers for building phylogenomic trees.
Troubleshooting
If you encounter problems while attempting to install or run BUSCO, please raise them on the “issues” page of the BUSCO GitLab repository at https://gitlab.com/ezlab/busco/-/issues. The following are some common issues that may arise when running BUSCO.
Installation issues
We encourage you to use the BUSCO Docker container or the BUSCO Bioconda package, as this will make installation and updates to future versions much simpler and easier to manage.
Issues with input files
Files that contain non-standard nucleotides or amino acids in the sequence lines, or non-ASCII characters in the header lines, may cause errors, and therefore these should be avoided wherever possible.
tBLASTn multithreading
When running BUSCO using multiple cores, a problem was noticed with tBLASTn versions 2.4-2.10.0. The problem was fixed in version 2.10.1+. Make sure you have at least this version or higher installed.
AUGUSTUS setup
You need to install AUGUSTUS only if you plan to assess eukaryotic genome assemblies using the BUSCO_Augustus workflow. As AUGUSTUS has dependencies of its own, e.g., Perl, you should consult the AUGUSTUS documentation for the correct installation procedures. If you are working on a system where AUGUSTUS has already been installed by an administrator and you do not have “write permission” to the AUGUSTUS “config” folder, you can simply copy the entire “config” folder to a location where you have “write permission” and then reset the “config” path variable to this location:
$ cp -r /path/to/AUGUSTUS/augustus-3.3.3/config /my/home/augustus/config; export AUGUSTUS_CONFIG_PATH="/my/home/augustus/config/".
Long runtimes
The analysis runtime will vary according to the exact system setup, computational resources, and input file(s). Assessments of genome assemblies require the initial steps of first identifying genomic regions that potentially harbor BUSCO matches, and predicting gene models. These are computationally intensive tasks, and therefore genome assembly assessments will take longer than gene set assessments. However, with the introduction of the BUSCO_Metaeuk workflow in BUSCO v5, the runtime is less of an issue even on very large genome assemblies (e.g., >10 Gbp). With the alternative BUSCO_Augustus workflow, users should take into account longer runtimes on medium and large-sized genomes.
High duplication scores in annotated gene sets or transcriptomes
These might be due to the presence of multiple isoforms per gene. For annotated gene sets, the transcript-to-gene relationships are defined in the General Feature Format (GFF) files, so you can select one isoform per gene before assessing a gene set. For example, you might keep the longest protein for each gene. For de novo transcriptomes, i.e., those without a reference genome, transcript-to-gene relationships are not defined, so you have two options: (a) run the assessments without pre-processing and acknowledge the fact that estimates of duplicated BUSCOs are likely to be inflated by the presence of multiple transcripts from the same gene, or (b) pre-process the transcriptome, for example by selecting just one representative transcript from the sets of highly similar transcripts clustered by identity/similarity.
Race condition when running multiple BUSCO analyses simultaneously
If you want to run multiple analyses simultaneously (e.g., as shown in Alternate Protocol), it is necessary to download all required datasets first and run the analysis with the --offline flag.
Dataset and BUSCO versions incompatibilities
The *_odb10 datasets are compatible with BUSCO versions 4 and 5. Older *_odb9 datasets are not compatible with these most recent versions.
Advanced Parameters
The full list of parameters can be printed by calling the help option: busco -h. Parameters can also be declared under the [busco_run] section of a config file (which is no longer mandatory in v5). The optional arguments for launching a BUSCO assessment give the user flexibility over many aspects of the run, all of which are described in full in the user guide. Some useful, general options to consider employing that do not affect the resulting estimates include:
- --force: this will overwrite the results from an analysis run with the same name.
- --tar: this will package and compress the results from steps that produce many output files.
- --restart: this restarts an incomplete run, preserving the output of each step that was successfully completed in the previous unfinished run.
BUSCO also offers multiple ways to fine-tune several aspects of the analysis that might influence the resulting scores. In particular, the gene predictors used in BUSCO, MetaEuk or AUGUSTUS, provide many options and these can be passed through BUSCO when running the genome mode using:
- --metaeuk_parameters and --metaeuk_rerun_parameters, or --augustus_parameters: these allow users to pass MetaEuk and AUGUSTUS-specific parameters for the gene prediction step.
However, it is important to note that the golden rule when using BUSCO would be not to change default parameters unless there is a biological or experimental reason to do so. In this way the scores remain comparable with the rest of the BUSCO user community. One biological justification for editing such parameters is for specifying a different codon usage. This can be changed with the MetaEuk parameter --translation-table or the AUGUSTUS parameter translation_table). If necessary, it may be relevant to explore the full set of parameters that can be passed to these gene predictors.
- MetaEuk sensitivity value ( -s ). The BUSCO_Metaeuk workflow for the assessment of eukaryotic genomes is already faster than the BUSCO_Augustus workflow. However, runtimes can be further decreased by using a smaller MetaEuk sensitivity value (-s), via the --metaeuk_parameters and --metaeuk_rerun_parameters BUSCO arguments described above. The default values are s=4.5 and s=6 for the first and second MetaEuk run, respectively, and were chosen as a trade-off between accuracy and runtimes.
In most settings, it is not advisable to change the sensitivity values, in order to keep BUSCO results comparable. In fact, changing the sensitivity values can have an impact on the estimates. Nevertheless, it can be convenient to use smaller sensitivity values when assessing very large genomes or for getting very quick BUSCO evaluations on draft/preliminary assemblies, e.g., when comparing multiple draft assemblies obtained by using different parameters or assembly pipelines.
- Augustus species parameters. AUGUSTUS comes with pre-trained gene prediction parameters for many species (see AUGUSTUS documentation for up-to-date details), so if parameter sets are available for the species to be assessed, then they should be selected. While BUSCO datasets are already configured with an AUGUSTUS species, the parameter set to be used can also be manually specified by adding the argument --augustus_species, e.g., --augustus_species camponotus_floridanus to the command. This only applies when using the BUSCO_Augustus workflow. For many other species, pretrained gene-prediction parameters are not yet available, so users should select the closest species for which such parameters are available, or run the assessment with the preselected default parameters. For the sake of reproducibility, it is important to specify which one was selected when reporting BUSCO results.
- Config file. BUSCO runs can also be configured using a user-editable configuration file (config.ini file). Up to BUSCO v4, the setup was controlled with this file. From v5, BUSCO will try to use the dependencies available in your environment, making the use of config.ini optional. This change was made to facilitate a better integration with Conda, where dependencies can be updated independently as new versions become available. You can still use this config file to override the path to the dependencies identified automatically by BUSCO and specify the paths to the dependencies you want to use (this simply tells BUSCO where they are installed on your system). You can find a template of the config.ini file in the config/ subfolder of the cloned BUSCO repository under the name config.ini.default. You can copy this file to config.ini before editing the file. To activate the config file, you can set the environment variable BUSCO_CONFIG_FILE with the path to the file, as follows:
$ export BUSCO_CONFIG_FILE="/path/to/myconfig.ini"
Alternatively, you can pass the path to your config file by using the --config argument in the BUSCO command:
$ busco –config /path/to/config.ini
All of the arguments that you can specify in a BUSCO command line can also be defined in the config file under the section [busco_run], usually -–param value on the command line corresponds to param=value in the config file, whereas --param corresponds to param=True. Note that the arguments specified through the command line will override those defined in the config.ini file.
Suggestions for Further Analysis
Although BUSCO's main function is to perform quality assessments of genomic data, the results can be used for performing other common genome analyses, such as identifying reliable markers for large-scale phylogenomics studies (covered in Support Protocol 3), visualizing syntenic blocks among genomes of different species/strains (covered in Support Protocol 2), and building training sets for gene predictors. We provide details on how to generate and use these training sets in the following paragraph.
Gene Predictor Training : To achieve the best results, gene-prediction tools such as AUGUSTUS, SNAP (Korf, 2004), and GENEID (Blanco, Parra, & Guigó, 2007) need to optimize their parameter configurations on each specific genome. BUSCO can provide high-quality gene model training data that can improve genome annotation procedures, especially in cases where there is a lack of supporting evidence such as native transcripts or orthologs from closely related species. For example, using BUSCO-trained parameters for gene prediction has resulted in improvements in the quality of the resulting gene model annotations over using available pre-trained parameters from other species (Waterhouse et al., 2018). Using the BUSCO_Augustus workflow, the pipeline automatically provides AUGUSTUS-ready parameters trained on BUSCO genes identified as complete and single copy. They are ideal for use during whole-genome annotation procedures that employ AUGUSTUS, especially when parameter sets for the species to be annotated or those of a close relative are not already available among those provided by AUGUSTUS. Moreover, the BUSCO_Augustus workflow can be run with the --long option to enable the optimization mode when retraining AUGUSTUS. This can further improve the resulting retraining parameters, but substantially increases the runtime. These AUGUSTUS retraining parameters are saved in the augustus_output directory in the output folder. To reuse the retraining parameters as a custom species with AUGUSTUS independently from BUSCO, the user needs to move the folder retraining_parameters/ to the $AUGUSTUS_CONFIG_PATH/species/ folder of their AUGUSTUS install and rename it to its original name, which can be deduced from its content. If the folder contains the file BUSCO_OUTPUT_NAME_xxx_parameters.cfg, the correct name to be used for naming the folder and identifying the species within AUGUSTUS is BUSCO_OUTPUT_NAME_xxx. Additionally, other gene predictors like SNAP can be trained by using as input the GenBank or GFF formatted gene models generated by BUSCO.
Acknowledgments
This work was supported by funding from the University of Geneva, Swiss National Science Foundation grant 310030_189062 and Swiss Institute of Bioinformatics SERI to E.Z.
Open access funding provided by Universite de Geneve.
Author Contributions
Mosè Manni : conceptualization, data curation, formal analysis, software, validation, visualization, writing original draft, writing review and editing; Matthew R. Berkeley : software, validation, writing review and editing; Mathieu Seppey : software, validation, writing review and editing; Evgeny M. Zdobnov : conceptualization, project administration, supervision, writing review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
BUSCO is licensed and freely distributed under the MIT Licence. The BUSCO source code is available through the GitLab project, https://gitlab.com/ezlab/busco, and it is also maintained on Bioconda and as a Docker container. BUSCO datasets are available at https://busco-data.ezlab.org/v5/data/lineages/. Versions and accessions of all the genome assemblies and gene sets analyzed in this protocol are reported in the text and are accessible through the NCBI database. All other data and code described in this manuscript are provided in a dedicated GitLab repository: https://gitlab.com/ezlab/busco_protocol.
Supporting Information
| Filename | Description |
|---|---|
| cpz1323-sup-0001-tableS1.pdf69.2 KB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Bağcı, C., Patz, S., & Huson, D. H. (2021). DIAMOND+MEGAN: fast and easy taxonomic and functional analysis of short and long microbiome sequences. Current Protocols , 1, e59. doi: 10.1002/cpz1.59.
- Blanco, E., Parra, G., & Guigó, R. (2007). Using geneid to identify genes. Current Protocols in Bioinformatics , 18, 4.3.1–4.3.28.
- Boes, K. E., Ribeiro, J. M. C., Wong, A., Harrington, L. C., Wolfner, M. F., & Sirot, L. K. (2014). Identification and characterization of seminal fluid proteins in the Asian tiger mosquito, Aedes albopictus. PLoS Neglected Tropical Diseases , 8, e2946. doi: 10.1371/journal.pntd.0002946.
- Borowiec, M. L. (2016). AMAS: A fast tool for alignment manipulation and computing of summary statistics. PeerJ , 4, e1660. doi: 10.7717/peerj.1660.
- Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., & Madden, T. L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics , 10, 421. doi: 10.1186/1471-2105-10-421.
- Capella-Gutiérrez, S., Silla-Martínez, J. M., & Gabaldón, T. (2009). trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics , 25, 1972–1973. doi: 10.1093/bioinformatics/btp348.
- Cheng, S., Melkonian, M., Smith, S. A., Brockington, S., Archibald, J. M., Delaux, P.-M., … Wong, G. K.-S. (2018). 10KP: A phylodiverse genome sequencing plan. GigaScience , 7, 1–9. doi: 10.1093/gigascience/giy013.
- Dainat, J. AGAT: Another Gff Analysis Toolkit to handle annotations in any GTF/GFF format. Available at https://www.doi.org/10.5281/zenodo.3552717.
- Gu, L., Reilly, P. F., Lewis, J. J., Reed, R. D., Andolfatto, P., & Walters, J. R. (2019). Dichotomy of dosage compensation along the Neo Z chromosome of the monarch butterfly. Current Biology , 29, 4071–4077.e3. doi: 10.1016/j.cub.2019.09.056.
- Hao, Z., Lv, D., Ge, Y., Shi, J., Weijers, D., Yu, G., & Chen, J. (2020). RIdeogram: Drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Computer Science , 6, e251. doi: 10.7717/peerj-cs.251.
- Herndon, N., Shelton, J., Gerischer, L., Ioannidis, P., Ninova, M., Dönitz, J., … Bucher, G. (2020). Enhanced genome assembly and a new official gene set for Tribolium castaneum. BMC Genomics , 21, 47. doi: 10.1186/s12864-019-6394-6.
- Hoff, K. J., & Stanke, M. (2019). Predicting genes in single genomes with AUGUSTUS. Current Protocols in Bioinformatics , 65, e57.
- Hoskins, R. A., Carlson, J. W., Wan, K. H., Park, S., Mendez, I., Galle, S. E., … Celniker, S. E. (2015). The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Research , 25, 445–458. doi: 10.1101/gr.185579.114.
- Huson, D. H., Albrecht, B., Bağcı, C., Bessarab, I., Górska, A., Jolic, D., & Williams, R. B. H. (2018). MEGAN-LR: New algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs. Biology Direct , 13, 6. doi: 10.1186/s13062-018-0208-7.
- Huson, D. H., & Scornavacca, C. (2012). Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Systematic Biology , 61, 1061–1067. doi: 10.1093/sysbio/sys062.
- Hyatt, D., Chen, G.-L., LoCascio, P. F., Land, M. L., Larimer, F. W., & Hauser, L. J. (2010). Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics , 11, 119. doi: 10.1186/1471-2105-11-119.
- i5K Consortium (2013). The i5K initiative: Advancing arthropod genomics for knowledge, human health, agriculture, and the environment. Journal of Heredity , 104, 595–600. doi: 10.1093/jhered/est050.
- Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., & Wang, Z. (2019). MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ , 7, e7359. doi: 10.7717/peerj.7359.
- Katoh, K., & Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Molecular Biology and Evolution , 30, 772–780. doi: 10.1093/molbev/mst010.
- Korf, I. (2004). Gene finding in novel genomes. BMC Bioinformatics , 5, 59. doi: 10.1186/1471-2105-5-59.
- Levy Karin, E., Mirdita, M., & Söding, J. (2020). MetaEuk—sensitive, high-throughput gene discovery, and annotation for large-scale eukaryotic metagenomics. Microbiome , 8, 48. doi: 10.1186/s40168-020-00808-x.
- Lewin, H. A., Robinson, G. E., Kress, W. J., Baker, W. J., Coddington, J., Crandall, K. A., … Zhang, G. (2018). Earth BioGenome Project: Sequencing life for the future of life. Proceedings of the National Academy of Sciences , 115, 4325–4333. doi: 10.1073/pnas.1720115115.
- Li, Y., Park, H., Smith, T. E., & Moran, N. A. (2019). Gene family evolution in the pea aphid based on chromosome-level genome assembly. Molecular Biology and Evolution , 36, 2143–2156. doi: 10.1093/molbev/msz138.
- Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., & Zdobnov, E. M. (2021). BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Molecular Biology and Evolution , 38, 4647–4654. doi: 10.1093/molbev/msab199.
- Matsen, F. A., Kodner, R. B., & Armbrust, E. V. (2010). pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics , 11, 538. doi: 10.1186/1471-2105-11-538.
- Merkel, D. (2014). Docker: Lightweight Linux containers for consistent development and deployment. Linux Journal , 2, 2.
- Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., & Lanfear, R. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution , 37, 1530–1534. doi: 10.1093/molbev/msaa015.
- Mirarab, S., Nguyen, N., & Warnow, T. (2011). SEPP: SATé-Enabled Phylogenetic Placement. In Biocomputing 2012 (pp. 247–258). Singapore: WORLD SCIENTIFIC.
- Mölder, F., Jablonski, K. P., Letcher, B., Hall, M. B., Tomkins-Tinch, C. H., Sochat, V., … Köster, J. (2021). Sustainable data analysis with Snakemake. F1000Research , 10, 33. doi: 10.12688/f1000research.29032.2.
- Nayfach, S., Roux, S., Seshadri, R., Udwary, D., Varghese, N., Schulz, F., … Eloe-Fadrosh, E. A. (2021). A genomic catalog of earth's microbiomes. Nature Biotechnology , 39, 499–509. doi: 10.1038/s41587-020-0718-6.
- R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at https://www.R-project.org/.
- Richards, S. (2018). Full disclosure: Genome assembly is still hard. PloS Biology , 16, e2005894. doi: 10.1371/journal.pbio.2005894.
- Schoch, C. L., Ciufo, S., Domrachev, M., Hotton, C. L., Kannan, S., Khovanskaya, R., … Karsch-Mizrachi, I. (2020). NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database , 2020, baaa062. doi: 10.1093/database/baaa062.
- Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., & Zdobnov, E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics , 31, 3210–3212. doi: 10.1093/bioinformatics/btv351.
- Sohn, J., & Nam, J.-W. (2018). The present and future of de novo whole-genome assembly. Briefings in Bioinformatics , 19, 23–40.
- Terrapon, N., Li, C., Robertson, H. M., Ji, L., Meng, X., Booth, W., … Liebig, J. (2014). Molecular traces of alternative social organization in a termite genome. Nature Communications , 5, 3636. doi: 10.1038/ncomms4636.
- Threlfall, J., & Blaxter, M. (2021). Launching the tree of life gateway. Available at https://wellcomeopenresearch.org/articles/6-125.
- Wallberg, A., Bunikis, I., Pettersson, O. V., Mosbech, M.-B., Childers, A. K., Evans, J. D., … Webster, M. T. (2019). A hybrid de novo genome assembly of the honeybee, Apis mellifera , with chromosome-length scaffolds. BMC Genomics , 20, 1–19. doi: 10.1186/s12864-019-5642-0.
- Waterhouse, R. M., Seppey, M., Simão, F. A., Manni, M., Ioannidis, P., Klioutchnikov, G., … Zdobnov, E. M. (2018). BUSCO applications from quality assessments to gene prediction and phylogenomics. Molecular Biology and Evolution , 35, 543–548. doi: 10.1093/molbev/msx319.
- Wu, Y.-W., & Singer, S. W. (2021). Recovering individual genomes from metagenomes using MaxBin 2.0. Current Protocols , 1, e128. doi: 10.1002/cpz1.128.
- Zdobnov, E. M., Kuznetsov, D., Tegenfeldt, F., Manni, M., Berkeley, M., & Kriventseva, E. V. (2021). OrthoDB in 2020: Evolutionary and functional annotations of orthologs. Nucleic Acids Research , 49, D389–D393. doi: 10.1093/nar/gkaa1009.
Citing Literature
Number of times cited according to CrossRef: 120
- Audrey Bourret, Christelle Leung, Gregory N. Puncher, Nicolas Le Corre, David Deslauriers, Katherine Skanes, Hugo Bourdages, Manon Cassista‐Da Ros, Wojciech Walkusz, Nicholas W. Jeffery, Ryan R. E. Stanley, Geneviève J. Parent, Diving into broad‐scale and high‐resolution population genomics to decipher drivers of structure and climatic vulnerability in a marine invertebrate, Molecular Ecology, 10.1111/mec.17448, 33 , 15, (2024).
- Janay A. Fox, David A. G. A. Hunt, Andrew P. Hendry, Lauren J. Chapman, Rowan D. H. Barrett, Counter‐gradient variation in gene expression between fish populations facilitates colonization of low‐dissolved oxygen environments, Molecular Ecology, 10.1111/mec.17419, 33 , 13, (2024).
- John T. Dobbs, Mee‐Sook Kim, Ned B. Klopfenstein, Jane E. Stewart, Comparative Genomics of Conifer‐Associated Fusarium spp., Forest Pathology, 10.1111/efp.12888, 54 , 5, (2024).
- Tetsushi Komoto, Kazuho Ikeo, Shunsuke Yaguchi, Takashi Yamamoto, Naoaki Sakamoto, Akinori Awazu, Assembly of continuous high‐resolution draft genome sequence of using long‐read sequencing, Development, Growth & DifferentiationDevelopment, Growth & DifferentiationDevelopment, Growth & Differentiation, 10.1111/dgd.12924, 66 , 4, (297-304), (2024).
- Daniel J. Nebauer, Leanne A. Pearson, Brett A. Neilan, Critical steps in an environmental metaproteomics workflow, Environmental Microbiology, 10.1111/1462-2920.16637, 26 , 5, (2024).
- Graham J. Etherington, Pei‐Shang Wu, Snezhana Oliferenko, Frank Uhlmann, Conrad A. Nieduszynski, Telomere‐to‐telomere Schizosaccharomyces japonicus genome assembly reveals hitherto unknown genome features, Yeast, 10.1002/yea.3912, 41 , 3, (73-86), (2024).
- Peter Sarkies, The curious case of the disappearing piRNAs, WIREs RNA, 10.1002/wrna.1849, 15 , 2, (2024).
- Deepu Mathew, Ravisankar Valsalan, The draft genome of Piper longum L., Crop Science, 10.1002/csc2.21313, 64 , 5, (2854-2862), (2024).
- Bradley Bushman, Matthew Robbins, Yinjie Qiu, Eric Watkins, Andrew Hollman, Nicole Mihelich, Dominic Petrella, Florence Breullin‐Sessoms, Ming‐Yi Chou, Paul Koch, Association of hard fescue (Festuca brevipila) stress tolerances with genome mapped markers, Crop Science, 10.1002/csc2.21155, 64 , 2, (1002-1014), (2024).
- Jill K. Olofsson, Torbjörn Tyler, Luke T. Dunning, Mats Hjertson, Åke Rühling, Anders J. Hansen, Morphological and genetic evidence suggest gene flow among native and naturalized mint species, American Journal of Botany, 10.1002/ajb2.16280, 111 , 2, (2024).
- Yanlong Wang, Jinpeng Zhen, Xiaoyu Che, Kang Zhang, Guowei Zhang, Huijuan Yang, Jing Wen, Jinxin Wang, Jiming Wang, Bo He, Ailong Yu, Yanhui Li, Zhigang Wang, Transcriptomic and metabolomic analysis of autumn leaf color change in Fraxinus angustifolia , PeerJ, 10.7717/peerj.15319, 11 , (e15319), (2023).
- Marta Ibañez-Lligoña, Sergi Colomer-Castell, Alejandra González-Sánchez, Josep Gregori, Carolina Campos, Damir Garcia-Cehic, Cristina Andrés, Maria Piñana, Tomàs Pumarola, Francisco Rodríguez-Frias, Andrés Antón, Josep Quer, Bioinformatic Tools for NGS-Based Metagenomics to Improve the Clinical Diagnosis of Emerging, Re-Emerging and New Viruses, Viruses, 10.3390/v15020587, 15 , 2, (587), (2023).
- Manfred Ritz, Nadim Ahmad, Thomas Brueck, Norbert Mehlmer, Comparative Genome-Wide Analysis of Two Caryopteris x Clandonensis Cultivars: Insights on the Biosynthesis of Volatile Terpenoids, Plants, 10.3390/plants12030632, 12 , 3, (632), (2023).
- Claudia Leoni, Caterina Manzari, Matteo Chiara, Pasqua Veronico, Giovanni Luigi Bruno, Graziano Pesole, Luigi R. Ceci, Mariateresa Volpicella, Chitinolytic Enzymes of the Hyperparasite Fungus Aphanocladium album: Genome-Wide Survey and Characterization of A Selected Enzyme, Microorganisms, 10.3390/microorganisms11051357, 11 , 5, (1357), (2023).
- Ning Xu, Huan Lu, Xueqian Yi, Simin Peng, Xiaohui Huang, Yu Zhang, Changzheng He, Potential of Alpha-(α)-Solanine as a Natural Inhibitor of Fungus Causing Leaf Spot Disease in Strawberry, Life, 10.3390/life13020450, 13 , 2, (450), (2023).
- Nadim Ahmad, Manfred Ritz, Anjuli Calchera, Jürgen Otte, Imke Schmitt, Thomas Brueck, Norbert Mehlmer, Biosynthetic Potential of Hypogymnia Holobionts: Insights into Secondary Metabolite Pathways, Journal of Fungi, 10.3390/jof9050546, 9 , 5, (546), (2023).
- Tao Zhang, Jianjv Feng, Wenni He, Xiaoting Rong, Hui Lv, Jun Li, Xinxin Li, Hao Wang, Lu Wang, Lixin Zhang, Liyan Yu, Genomic and Transcriptomic Approaches Provide a Predictive Framework for Sesquiterpenes Biosynthesis in Desarmillaria tabescens CPCC 401429, Journal of Fungi, 10.3390/jof9040481, 9 , 4, (481), (2023).
- Dmitry I. Karetnikov, Gennady V. Vasiliev, Stepan V. Toshchakov, Nikolay A. Shmakov, Mikhail A. Genaev, Mikhail A. Nesterov, Salmaz M. Ibragimova, Daniil A. Rybakov, Tatjana A. Gavrilenko, Elena A. Salina, Maxim V. Patrushev, Alex V. Kochetov, Dmitry A. Afonnikov, Analysis of Genome Structure and Its Variations in Potato Cultivars Grown in Russia, International Journal of Molecular Sciences, 10.3390/ijms24065713, 24 , 6, (5713), (2023).
- Aliya Yakupova, Andrey Tomarovsky, Azamat Totikov, Violetta Beklemisheva, Maria Logacheva, Polina L. Perelman, Aleksey Komissarov, Pavel Dobrynin, Ksenia Krasheninnikova, Gaik Tamazian, Natalia A. Serdyukova, Mike Rayko, Tatiana Bulyonkova, Nikolay Cherkasov, Vladimir Pylev, Vladimir Peterfeld, Aleksey Penin, Elena Balanovska, Alla Lapidus, Stephen J. OBrien, Alexander Graphodatsky, Klaus-Peter Koepfli, Sergei Kliver, Chromosome-Length Assembly of the Baikal Seal (Pusa sibirica) Genome Reveals a Historically Large Population Prior to Isolation in Lake Baikal, Genes, 10.3390/genes14030619, 14 , 3, (619), (2023).
- Shujing Wei, Zhao Song, Sisheng Luo, Yingxia Zhong, Yufei Zhou, Ruisen Lu, Transcriptome Analysis Reveals the Heat Stress Response Genes by Fire Stimulation in Michelia macclurei Dandy, Forests, 10.3390/f14030610, 14 , 3, (610), (2023).
- Shaojian Li, Zhenyu Wang, Meng Gao, Tong Li, Xiaowei Cui, Junhuai Zu, Suling Sang, Wanwan Fan, Haiyan Zhang, Intraspecific Comparative Analysis Reveals Genomic Variation of Didymella arachidicola and Pathogenicity Factors Potentially Related to Lesion Phenotype, Biology, 10.3390/biology12030476, 12 , 3, (476), (2023).
- Xiaobin Ou, Xiao Wang, Bing Zhao, Yi Zhao, Haiqing Liu, Yuankai Chang, Zhiwei Wang, Wenqi Yang, Xuebin Zhang, Ke Yu, Metabolome and transcriptome signatures shed light on the anti-obesity effect of Polygonatum sibiricum, Frontiers in Plant Science, 10.3389/fpls.2023.1181861, 14 , (2023).
- Lidiia Samarina, Songbo Wang, Lyudmila Malyukova, Alexandr Bobrovskikh, Alexey Doroshkov, Natalia Koninskaya, Ruset Shkhalakhova, Alexandra Matskiv, Jaroslava Fedorina, Anastasia Fizikova, Karina Manakhova, Svetlana Loshkaryova, Tsiala Tutberidze, Alexey Ryndin, Elena Khlestkina, Long-term cold, freezing and drought: overlapping and specific regulatory mechanisms and signal transduction in tea plant (Camellia sinensis (L.) Kuntze), Frontiers in Plant Science, 10.3389/fpls.2023.1145793, 14 , (2023).
- Maya Moshe, Chhedi Lal Gupta, Noa Sela, Dror Minz, Ehud Banin, Omer Frenkel, Eddie Cytryn, Comparative genomics of Bacillus cereus sensu lato spp. biocontrol strains in correlation to in-vitro phenotypes and plant pathogen antagonistic capacity, Frontiers in Microbiology, 10.3389/fmicb.2023.996287, 14 , (2023).
- Zhiyu Zhang, Mengfei Cui, Panting Chen, Juxing Li, Zhitao Mao, Yufeng Mao, Zhenjing Li, Qingbin Guo, Changlu Wang, Xiaoping Liao, Huanhuan Liu, Insight into the phylogeny and metabolic divergence of Monascus species (M. pilosus, M. ruber, and M. purpureus) at the genome level, Frontiers in Microbiology, 10.3389/fmicb.2023.1199144, 14 , (2023).
- Purva Gohil, Bhavya Nanavati, Kajal Patel, Vishal Suthar, Madhvi Joshi, Deepak B. Patil, Chaitanya G. Joshi, Assessing the efficacy of probiotics in augmenting bovine reproductive health: an integrated in vitro, in silico, and in vivo study, Frontiers in Microbiology, 10.3389/fmicb.2023.1137611, 14 , (2023).
- Sioly Becerra, Riccardo Baroncelli, Thaís R. Boufleur, Serenella A. Sukno, Michael R. Thon, Chromosome-level analysis of the Colletotrichum graminicola genome reveals the unique characteristics of core and minichromosomes, Frontiers in Microbiology, 10.3389/fmicb.2023.1129319, 14 , (2023).
- Jingjing Li, Haiyan Wang, Jianqing Zhu, Qi Yang, Yang Luan, Leming Shi, José Arturo Molina-Mora, Yuanting Zheng, De novo assembly of a chromosome-level reference genome of the ornamental butterfly Sericinus montelus based on nanopore sequencing and Hi-C analysis, Frontiers in Genetics, 10.3389/fgene.2023.1107353, 14 , (2023).
- Ekaterina Ponomareva, Ahmed Badiss, Tahera Sultana, Qing Yu, Hai D.T. Nguyen, Genome Characterization and Development of Real-Time PCR Assays for Ditylenchus dipsaci and D. weischeri , Journal of Nematology, 10.2478/jofnem-2022-0058, 54 , 1, (2023).
- Nina Vittorelli, Ricardo C. Rodríguez de la Vega, Alodie Snirc, Emilie Levert, Valérie Gautier, Christophe Lalanne, Elsa De Filippo, Pierre Gladieux, Sonia Guillou, Yu Zhang, Sravanthi Tejomurthula, Igor V. Grigoriev, Robert Debuchy, Philippe Silar, Tatiana Giraud, Fanny E. Hartmann, Stepwise recombination suppression around the mating-type locus in an ascomycete fungus with self-fertile spores, PLOS Genetics, 10.1371/journal.pgen.1010347, 19 , 2, (e1010347), (2023).
- Ryoma Sato, Yuri Kondo, Sakae Agarie, The first released available genome of the common ice plant (Mesembryanthemum crystallinum L.) extended the research region on salt tolerance, C3-CAM photosynthetic conversion, and halophism, F1000Research, 10.12688/f1000research.129958.1, 12 , (448), (2023).
- Nitin K. Singh, Jason M. Wood, Jose Patane, Livia Maria Silva Moura, Jonathan Lombardino, João Carlos Setubal, Kasthuri Venkateswaran, Characterization of metagenome-assembled genomes from the International Space Station, Microbiome, 10.1186/s40168-023-01545-7, 11 , 1, (2023).
- Shivam Aggarwal, Sathish Dorairaj, Nidhi Adlakha, Stoichiometric balance ratio of cellobiose and gentiobiose induces cellulase production in Talaromyces cellulolyticus, Biotechnology for Biofuels and Bioproducts, 10.1186/s13068-023-02296-1, 16 , 1, (2023).
- Bingyue Zhu, Pengyu Jin, Yiming Zhang, Yunxiao Shen, Wei Wang, Shuqiang Li, Genomic and transcriptomic analyses support a silk gland origin of spider venom glands, BMC Biology, 10.1186/s12915-023-01581-7, 21 , 1, (2023).
- Anne Caroline Mascarenhas dos Santos, Alexander Thomas Julian, Pingdong Liang, Oscar Juárez, Jean-François Pombert, Telomere-to-Telomere genome assemblies of human-infecting Encephalitozoon species, BMC Genomics, 10.1186/s12864-023-09331-3, 24 , 1, (2023).
- James E. Titus-McQuillan, Adalena V. Nanni, Lauren M. McIntyre, Rebekah L. Rogers, Estimating transcriptome complexities across eukaryotes, BMC Genomics, 10.1186/s12864-023-09326-0, 24 , 1, (2023).
- Shanmei Wang, Lei Wang, Jing Jin, Gang Li, Huanzhang Shao, Yang Song, Yuanzheng Sun, Yan Zhang, Jianjian Cheng, Lifeng Li, Genomic Epidemiology and Characterization of Carbapenem-Resistant Klebsiella pneumoniae in ICU Inpatients in Henan Province, China: a Multicenter Cross-Sectional Study, Microbiology Spectrum, 10.1128/spectrum.04197-22, (2023).
- Pablo De la Vega, Gaurav G. Shimpi, Bastian Bentlage, Genome Sequence of the Endosymbiont Endozoicomonas sp. Strain GU-1 ( Gammaproteobacteria ), Isolated from the Staghorn Coral Acropora pulchra (Cnidaria: Scleractinia) , Microbiology Resource Announcements, 10.1128/mra.01355-22, 12 , 3, (2023).
- Hussain Alattas, Julie Ardley, Rebecca Swift, Pragya Kant, Sarah Jackson, Ben Biddulph, Amanuel Bekuma, Jaco Zandberg, Sam Abraham, Ravi Tiwari, Wayne Reeve, Complete Genome Sequence of the Ice-Nucleation-Active Pseudomonas syringae pv. pisi Isolate MUP32, Isolated from Frost-Damaged Pea (Pisum sativum subsp. arvense cv. Dundale) in New South Wales , Microbiology Resource Announcements, 10.1128/mra.01276-22, 12 , 3, (2023).
- Hussain Alattas, Julie Ardley, Rebecca Swift, Sarah Jackson, Ben Biddulph, Amanuel Bekuma, Jaco Zandberg, Sam Abraham, Ravi Tiwari, Wayne Reeve, Complete Genome Sequence of the Ice-Nucleation-Active Pseudomonas syringae Strain MUP20, Isolated from Frost-Damaged Wheat (Triticum aestivum cv. Scepter) in Western Australia, Microbiology Resource Announcements, 10.1128/mra.01275-22, 12 , 3, (2023).
- Min-Jeong Kang, Eun-Kyung Bae, Eung-Jun Park, Kang-Hyeon Ka, Mi-Ra Son, Ki-Tae Kim, Jei-Wan Lee, Draft Genome Sequence for the Symbiotic Pine Mushroom Tricholoma matsutake, Microbiology Resource Announcements, 10.1128/mra.01271-22, (2023).
- Tilottama Mazumdar, Alexander Bartholomäus, Dino P. McMahon, Draft Genome of the Entomopathogenic Fungus Metarhizium robertsii DSM 1490, Microbiology Resource Announcements, 10.1128/mra.01267-22, 12 , 5, (2023).
- Hussain Alattas, Julie Ardley, Mark O’Dea, Rebecca Swift, Sarah Jackson, Ben Biddulph, Amanuel Bekuma, Jaco Zandberg, Sam Abraham, Ravi Tiwari, Wayne Reeve, Complete Genome Sequence of the Ice-Nucleation-Active Pseudomonas syringae Strain MUP17, Isolated from the Frost-Damaged Barley Cultivar Hordeum vulgare cv. La Trobe, Microbiology Resource Announcements, 10.1128/mra.01215-22, 12 , 3, (2023).
- Enoch B. Joel, Jessica L. Lenka, Bitrus Yakubu, Richard J. Kutshik, Aminu Tukur, Ishaya Y. Longdet, Metagenome-Assembled Genome Sequence of Stenotrophomonas maltophilia Strain UJ_SKK_5.5, Obtained from the Gut Microbiome of a Macrotermes bellicosus Termite Collected from Hot, Arid Nigeria, Microbiology Resource Announcements, 10.1128/mra.01060-22, 12 , 1, (2023).
- Stephen A. Rehner, Romina Gazis, Vinson P. Doyle, Willie A. S. Vieira, Philip M. Campos, Jonathan Shao, Genome Resources for the Colletotrichum gloeosporioides Species Complex: 13 Tree Endophytes from the Neotropics and Paleotropics, Microbiology Resource Announcements, 10.1128/mra.01040-22, 12 , 4, (2023).
- Jessica L. Lenka, Bitrus Yakubu, Richard J. Kutshik, Enoch B. Joel, Aminu Tukur, Ishaya Y. Longdet, Metagenome-Assembled Genome Sequence of a Strain of Burkholderia cepacia Isolated from the Gut of Macrotermes bellicosus in Nigeria, Microbiology Resource Announcements, 10.1128/mra.00777-22, 12 , 1, (2023).
- N. Kim, P. Vargas, K. Fortuna, J. Wagemans, H. Rediers, Draft Genome Sequences of 27 Rhizogenic Agrobacterium Biovar 1 Strains, the Causative Agent of Hairy Root Disease , Microbiology Resource Announcements, 10.1128/mra.00124-23, 12 , 5, (2023).
- Benedict H. Claxton Stevens, Bashir L. Rumah, Christopher E. Stead, Ying Zhang, A Complete Genome of the Alphaproteobacterial Methanotroph Methylocystis parvus OBBP, Microbiology Resource Announcements, 10.1128/mra.00076-23, 12 , 4, (2023).
- Wongsakorn Kwandee, Sirirat Boondireke, Wuttichai Mhuantong, Darika Kongrit, Draft Genome Sequence of Rhizobium sp. Strain AG207R, a Potential Bacteriocin Producer Isolated from Ginger Roots and Exhibiting Broad-Spectrum Antimicrobial Activity, Including against Multidrug-Resistant Enterococcus faecalis , Microbiology Resource Announcements, 10.1128/mra.00055-23, (2023).
- Anja Palandačić, Martin Kapun, Carola Greve, Tilman Schell, Sandra Kirchner, Luise Kruckenhauser, Nikolaus Szucsich, Nina Bogutskaya, From historical expedition diaries to whole genome sequencing: A case study of the likely extinct Red Sea torpedo ray, Zoologica Scripta, 10.1111/zsc.12632, 53 , 1, (32-51), (2023).
- Yanling Sun, Yang Liu, Jinfeng Shi, Lun Wang, Chengzhi Liang, Jun Yang, Jinfeng Chen, Mingsheng Chen, Biased mutations and gene losses underlying diploidization of the tetraploid broomcorn millet genome, The Plant Journal, 10.1111/tpj.16085, 113 , 4, (787-801), (2023).
- Leidys Murillo‐Ramos, Victoria Twort, Niklas Wahlberg, Pasi Sihvonen, A phylogenomic perspective on the relationships of subfamilies in the family Geometridae (Lepidoptera), Systematic Entomology, 10.1111/syen.12594, 48 , 4, (618-632), (2023).
- Yi Yuan, Xuan Li, Xu Yao, Xuehao Fu, Jie Cheng, Hongyan Shan, Xiaofeng Yin, Hongzhi Kong, Mechanisms underlying the formation of complex color patterns on Nigella orientalis (Ranunculaceae) petals, New Phytologist, 10.1111/nph.18681, 237 , 6, (2450-2466), (2023).
- Leilton W. Luna, Lisa M. Williams, Kenneth Duren, Reina Tyl, David P. L. Toews, Julian D. Avery, Whole genome assessment of a declining game bird reveals cryptic genetic structure and insights for population management, Molecular Ecology, 10.1111/mec.17129, 32 , 20, (5498-5513), (2023).
- Hisatake Ishida, Uwe John, Shauna A. Murray, Debashish Bhattacharya, Cheong Xin Chan, Developing model systems for dinoflagellates in the post‐genomic era, Journal of Phycology, 10.1111/jpy.13386, 59 , 5, (799-808), (2023).
- Hajar Ben Moussa, Jacques Pédron, Nicole Hugouvieux‐Cotte‐Pattat, Marie‐Anne Barny, Two species with a peculiar evolution within the genus Pectobacterium suggest adaptation to a new environmental niche, Environmental Microbiology, 10.1111/1462-2920.16479, 25 , 11, (2465-2480), (2023).
- Chien-Jui Huang, Ting-Li Wu, Po-Xing Zheng, Jheng-Yang Ou, Chao-Ling Ting, Yao-Cheng Lin, Complete Genome Sequence Resource of Pectobacterium colocasium Strain F1-1 that Causes Soft Rot Disease of Taro , Plant Disease, 10.1094/PDIS-09-22-2198-A, (2023).
- Yuhan Zhou, Jinsong Xiong, Ziqiang Shu, Chao Dong, Tingting Gu, Pengchuan Sun, Shuang He, Mian Jiang, Zhiqiang Xia, Jiayu Xue, Wasi Ullah Khan, Fei Chen, Zong-Ming Cheng, The telomere-to-telomere genome of Fragaria vesca reveals the genomic evolution of Fragaria and the origin of cultivated octoploid strawberry , Horticulture Research, 10.1093/hr/uhad027, 10 , 4, (2023).
- Fangping Li, Shiqiang Xu, Zitong Xiao, Jingming Wang, Yu Mei, Haifei Hu, Jingyu Li, Jieying Liu, Zhuangwei Hou, Junliang Zhao, Shaohai Yang, Jihua Wang, Gap-free genome assembly and comparative analysis reveal the evolution and anthocyanin accumulation mechanism of Rhodomyrtus tomentosa , Horticulture Research, 10.1093/hr/uhad005, 10 , 3, (2023).
- Kaixuan Zhao, Arong Luo, Qingsong Zhou, Wei Wei, Wenping Liu, Chaodong Zhu, Zeqing Niu, Zeyang Zhou, Dunyuan Huang, A Chromosome-Level Genome Assembly and Evolution Analysis of Andrena camellia (Hymenoptera: Andrenidae) , Genome Biology and Evolution, 10.1093/gbe/evad080, 15 , 5, (2023).
- Andrew N Black, Kristin J Bondo, Andrew Mularo, Alvaro Hernandez, Yachi Yu, Carleigh M Stein, Andy Gregory, Kent A Fricke, Jeff Prendergast, Dan Sullins, David Haukos, Michael Whitson, Blake Grisham, Zach Lowe, J Andrew DeWoody, A Highly Contiguous and Annotated Genome Assembly of the Lesser Prairie-Chicken ( Tympanuchus pallidicinctus ) , Genome Biology and Evolution, 10.1093/gbe/evad043, 15 , 4, (2023).
- Yuuki Kobayashi, Tomoko F Shibata, Hideki Hirakawa, Tomoaki Nishiyama, Akiyoshi Yamada, Mitsuyasu Hasebe, Shuji Shigenobu, Masayoshi Kawaguchi, The genome of Lyophyllum shimeji provides insight into the initial evolution of ectomycorrhizal fungal genomes , DNA Research, 10.1093/dnares/dsac053, 30 , 1, (2023).
- Samantha Bryce, Maureen Stolzer, Daniel Crosby, Ruijin Yang, Dannie Durand, Tina H. Lee, Human atlastin-3 is a constitutive ER membrane fusion catalyst, Journal of Cell Biology, 10.1083/jcb.202211021, 222 , 7, (2023).
- Massimiliano Molari, Christiane Hassenrueck, Rafael Laso-Pérez, Gunter Wegener, Pierre Offre, Stefano Scilipoti, Antje Boetius, A hydrogenotrophic Sulfurimonas is globally abundant in deep-sea oxygen-saturated hydrothermal plumes, Nature Microbiology, 10.1038/s41564-023-01342-w, 8 , 4, (651-665), (2023).
- Jaspreet S. Saini, Mosè Manni, Christel Hassler, Rachel N. Cable, Melissa B. Duhaime, Evgeny M. Zdobnov, Genomic insights into the coupling of a Chlorella-like microeukaryote and sulfur bacteria in the chemocline of permanently stratified Lake Cadagno, The ISME Journal, 10.1038/s41396-023-01396-y, 17 , 6, (903-915), (2023).
- Bo Yuan, Michelle F. Grau, Ramiro Mendonça Murata, Tamas Torok, Kasthuri Venkateswaran, Jason E. Stajich, Clay C. C. Wang, Identification of the Neoaspergillic Acid Biosynthesis Gene Cluster by Establishing an In Vitro CRISPR-Ribonucleoprotein Genetic System in Aspergillus melleus , ACS Omega, 10.1021/acsomega.2c08104, 8 , 19, (16713-16721), (2023).
- Maria Alvarez-Arevalo, Eva Baggesgaard Sterndorff, David Faurdal, Tue Sparholt Jørgensen, Anna-Sophie Mourched, Oliwia Vuksanovic, Subhasish Saha, Tilmann Weber, Extraction and Oxford Nanopore sequencing of genomic DNA from filamentous Actinobacteria, STAR Protocols, 10.1016/j.xpro.2022.101955, 4 , 1, (101955), (2023).
- Peng Wang, Fei Wang, A proposed metric set for evaluation of genome assembly quality, Trends in Genetics, 10.1016/j.tig.2022.10.005, 39 , 3, (175-186), (2023).
- Amanda Pacholak, Joanna Żur-Pińska, Artur Piński, Quynh Anh Nguyen, Marta Ligaj, Magdalena Luczak, Long D. Nghiem, Ewa Kaczorek, Potential negative effect of long-term exposure to nitrofurans on bacteria isolated from wastewater, Science of The Total Environment, 10.1016/j.scitotenv.2023.162199, 872 , (162199), (2023).
- Anna M. Roseboom, Quinten R. Ducarmon, Bastian V.H. Hornung, Céline Harmanus, Monique J.T. Crobach, Ed J. Kuijper, Rolf H.A.M. Vossen, Susan L. Kloet, Wiep Klaas Smits, Carriage of three plasmids in a single human clinical isolate of Clostridioides difficile, Plasmid, 10.1016/j.plasmid.2022.102669, 125 , (102669), (2023).
- Nickolas Menezes da Silva, Guilherme Fonseca Reis, Flávia de Fátima Costa, Maria Eduarda Grisolia, Marlon Roger Geraldo, Bruno Paulo Rodrigues Lustosa, Bruna Jacomel Favoreto de Souza Lima, Vinicius Almir Weiss, Emanuel Maltempi de Souza, Ruoyu Li, Yinggai Song, Mariana Machado Fidelis Nascimento, Diogo Robl, Renata Rodrigues Gomes, G. Sybren de Hoog, Vania Aparecida Vicente, Genome sequencing of Cladophialophora exuberans, a novel candidate for bioremediation of hydrocarbon and heavy metal polluted habitats, Fungal Biology, 10.1016/j.funbio.2023.03.003, 127 , 5, (1032-1042), (2023).
- J.A. Yoon, S.Y. Kwun, E.H. Park, M.D. Kim, Complete genome sequence data of Leuconostoc mesenteroides KNU-2 and Weissella hellenica MBEL1842 isolated from kimchi, Data in Brief, 10.1016/j.dib.2023.108919, 47 , (108919), (2023).
- Yavor Hadzhiev, Lucy Wheatley, Ledean Cooper, Federico Ansaloni, Celina Whalley, Zhelin Chen, Sara Finaurini, Stefano Gustincich, Remo Sanges, Shawn Burgess, Andrew Beggs, Ferenc Müller, The miR-430 locus with extreme promoter density forms a transcription body during the minor wave of zygotic genome activation, Developmental Cell, 10.1016/j.devcel.2022.12.007, 58 , 2, (155-170.e8), (2023).
- Nik Shazana Nik Mohd Sanusi, Rozana Rosli, Kuang-Lim Chan, Mohd Amin Ab Halim, Ngoot-Chin Ting, Rajinder Singh, Eng-Ti Leslie Low, Integrated consensus genetic map and genomic scaffold re-ordering of oil palm (Elaeis guineensis) genome, Computational Biology and Chemistry, 10.1016/j.compbiolchem.2022.107801, 102 , (107801), (2023).
- Xinchun Li, Yitian Bai, Zhen Dong, Chengxun Xu, Shikai Liu, Hong Yu, Lingfeng Kong, Qi Li, Chromosome-level genome assembly of the European flat oyster (Ostrea edulis) provides insights into its evolution and adaptation, Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 10.1016/j.cbd.2022.101045, 45 , (101045), (2023).
- Miryung Kim, Yong-Seok Kim, Chang-Jun Cha, Chryseobacterium paludis sp. nov. and Chryseobacterium foetidum sp. nov. Isolated from the Aquatic Environment, South Korea, Journal of Microbiology, 10.1007/s12275-022-00008-2, 61 , 1, (37-47), (2023).
- Frank M. You, Ismael Moumen, Nadeem Khan, Sylvie Cloutier, Reference Genome Sequence of Flax, The Flax Genome, 10.1007/978-3-031-16061-5_1, (1-17), (2023).
- Graham J. Etherington, Elisa Gomez Gil, Wilfried Haerty, Snezhana Oliferenko, Conrad A. Nieduszynski, Schizosaccharomyces versatilis represents a distinct evolutionary lineage of fission yeast, Yeast, 10.1002/yea.3919, 41 , 3, (95-107), (2023).
- Luiz Augusto Inojosa Ferreira, Rubem Silvério Oliveira, Jamil Constantin, Caio Brunharo, Evolution of ACCase‐inhibitor resistance in is conferred by a Trp2027Cys mutation in the herbicide target site, Pest Management Science, 10.1002/ps.7723, 79 , 12, (5220-5229), (2023).
- Lovely Mae F. Lawas, Mohamed O. Kamileen, C. Robin Buell, Sarah E. O'Connor, Courtney P. Leisner, Transcriptome‐based identification and functional characterization of iridoid synthase involved in monotropein biosynthesis in blueberry, Plant Direct, 10.1002/pld3.512, 7 , 7, (2023).
- Lauren Coombe, René L. Warren, Johnathan Wong, Vladimir Nikolic, Inanc Birol, ntLink: A Toolkit for De Novo Genome Assembly Scaffolding and Mapping Using Long Reads, Current Protocols, 10.1002/cpz1.733, 3 , 4, (2023).
- Yamkela Mgwatyu, Stephanie Cornelissen, Peter van Heusden, Allison Stander, Mary Ranketse, Uljana Hesse, Establishing MinION Sequencing and Genome Assembly Procedures for the Analysis of the Rooibos (Aspalathus linearis) Genome, Plants, 10.3390/plants11162156, 11 , 16, (2156), (2022).
- Nootjarin Jungkhun, Antonio Roberto Gomes de Farias, Jutatape Watcharachaiyakup, Nuttima Kositcharoenkul, Jong Hyun Ham, Sujin Patarapuwadol, Phylogenetic Characterization and Genome Sequence Analysis of Burkholderia glumae Strains Isolated in Thailand as the Causal Agent of Rice Bacterial Panicle Blight, Pathogens, 10.3390/pathogens11060676, 11 , 6, (676), (2022).
- Erica L. Fonseca, Sergio M. Morgado, Raquel V. Caldart, Ana Carolina Vicente, Global Genomic Epidemiology of Escherichia coli (ExPEC) ST38 Lineage Revealed a Virulome Associated with Human Infections, Microorganisms, 10.3390/microorganisms10122482, 10 , 12, (2482), (2022).
- Valeria Shapovalova, Elvira Shaidullina, Ilya Azizov, Eugene Sheck, Alexey Martinovich, Marina Dyachkova, Alina Matsvay, Yulia Savochkina, Kamil Khafizov, Roman Kozlov, German Shipulin, Mikhail Edelstein, Molecular Epidemiology of mcr-1-Positive Escherichia coli and Klebsiella pneumoniae Isolates: Results from Russian Sentinel Surveillance (2013–2018), Microorganisms, 10.3390/microorganisms10102034, 10 , 10, (2034), (2022).
- Lu Gan, Yuelan Yin, Qichen Niu, Xuebing Yan, Shuxia Yin, New Insights into the Mechanism of Trichoderma virens-Induced Developmental Effects on Agrostis stolonifera Disease Resistance against Dollar Spot Infection, Journal of Fungi, 10.3390/jof8111186, 8 , 11, (1186), (2022).
- Aleksandra E. Korenskaia, Olga D. Shishkina, Alexandra I. Klimenko, Olga V. Andreenkova, Margarita A. Bobrovskikh, Natalja V. Shatskaya, Gennady V. Vasiliev, Nataly E. Gruntenko, New Wolbachia pipientis Genotype Increasing Heat Stress Resistance of Drosophila melanogaster Host Is Characterized by a Large Chromosomal Inversion, International Journal of Molecular Sciences, 10.3390/ijms232416212, 23 , 24, (16212), (2022).
- Elisa Carotti, Federica Carducci, Samuele Greco, Marco Gerdol, Daniele Di Marino, Nunzio Perta, Anna La Teana, Adriana Canapa, Marco Barucca, Maria Assunta Biscotti, Transcriptional Contribution of Transposable Elements in Relation to Salinity Conditions in Teleosts and Silencing Mechanisms Involved, International Journal of Molecular Sciences, 10.3390/ijms23095215, 23 , 9, (5215), (2022).
- Min Hui, Qian Xin, Jiao Cheng, Zhongli Sha, Characterization and Comparison of Eye Development and Phototransduction Genes in Deep- and Shallow-Water Shrimp Alvinocaris longirostris and Palaemon carinicauda, Diversity, 10.3390/d14080653, 14 , 8, (653), (2022).
- Elisa Carotti, Federica Carducci, Adriana Canapa, Marco Barucca, Maria Assunta Biscotti, Transposable Element Tissue-Specific Response to Temperature Stress in the Stenothermal Fish Puntius tetrazona, Animals, 10.3390/ani13010001, 13 , 1, (1), (2022).
- Komwit Surachat, Patcharaporn Narkthewan, Chayanin Thotsagotphairee, Monwadee Wonglapsuwan, Walaiporn Thongpradub, The First Genome Survey and De Novo Assembly of the Short Mackerel (Rastrelliger brachysoma) and Indian Mackerel (Rastrelliger kanagurta), Animals, 10.3390/ani12141769, 12 , 14, (1769), (2022).
- Lina Reslan, George F. Araj, Marc Finianos, Rima El Asmar, Jaroslav Hrabak, Ghassan Dbaibo, Ibrahim Bitar, Molecular Characterization of Candida auris Isolates at a Major Tertiary Care Center in Lebanon, Frontiers in Microbiology, 10.3389/fmicb.2021.770635, 12 , (2022).
- Shishir K. Gupta, Özge Osmanoglu, Rashmi Minocha, Sourish Reddy Bandi, Elena Bencurova, Mugdha Srivastava, Thomas Dandekar, Genome-wide scan for potential CD4+ T-cell vaccine candidates in Candida auris by exploiting reverse vaccinology and evolutionary information, Frontiers in Medicine, 10.3389/fmed.2022.1008527, 9 , (2022).
- Qingfei Li, Na Wang, Chao Sui, Huadong Mao, Lu Zhang, Jinghua Chen, PacBio single molecule real-time sequencing of a full-length transcriptome of the greenfin horse-faced filefish Thamnaconus modestus, Frontiers in Marine Science, 10.3389/fmars.2022.1028231, 9 , (2022).
- Ismail Gumustop, Fatih Ortakci, Comparative Genomics of Lentilactobacillus parabuchneri isolated from dairy, KEM complex, Makgeolli, and Saliva Microbiomes, BMC Genomics, 10.1186/s12864-022-09053-y, 23 , 1, (2022).
- Xin Ma, Kexin Li, Fang Li, Jing Su, Weiwei Meng, Yanming Sun, Hui Sun, Jiazheng Sun, Yonghe Yuan, Yujia Lin, Songnian Hu, Xuefang Xu, Zilong He, Tracing Foodborne Botulism Events Caused by Clostridium botulinum in Xinjiang Province, China, Using a Core Genome Sequence Typing Scheme, Microbiology Spectrum, 10.1128/spectrum.01164-22, 10 , 6, (2022).
- Richard J. Kutshik, Bitrus Yakubu, Enoch B. Joel, Jessica L. Lenka, Aminu Tukur, Ishaya Y. Longdet, Assembly of Olivibacter sp. Strain UJ_SKK_5.1 Genome Sequence, Isolated from Metagenomes of Macrotermes bellicosus Guts Collected from Hot, Arid Nigeria , Microbiology Resource Announcements, 10.1128/mra.00895-22, 11 , 11, (2022).
- Ka Lip Chew, Rosemini Achik, Nurul Hudaa Osman, Sophie Octavia, Jeanette W. P. Teo, Genome Sequence of a Clinical Blood Isolate of Kodamaea ohmeri, Microbiology Resource Announcements, 10.1128/mra.00843-22, 11 , 12, (2022).
- Fardeen A. Siddiqui, Shoshana Trudel, Abigail E. Goen, Syeda A. Fatima, Kyle S. MacLea, Draft Genome Sequence of Xanthobacter aminoxidans ATCC BAA-299 T , Microbiology Resource Announcements, 10.1128/mra.00548-22, 11 , 8, (2022).
- Michelle J. Bauer, Anna Maria Peri, Lukas Lüftinger, Stephan Beisken, Haakon Bergh, Brian M. Forde, Cameron Buckley, Thom Cuddihy, Patrice Tan, David L. Paterson, David M. Whiley, Patrick N. A. Harris, Optimized Method for Bacterial Nucleic Acid Extraction from Positive Blood Culture Broth for Whole-Genome Sequencing, Resistance Phenotype Prediction, and Downstream Molecular Applications, Journal of Clinical Microbiology, 10.1128/jcm.01012-22, 60 , 11, (2022).
- See more

