A Low-Cost Tebuconazole-Based Screening Test for Azole-Resistant Aspergillus fumigatus
Amelie P. Brackin, Amelie P. Brackin, Jennifer M. G. Shelton, Jennifer M. G. Shelton, Alireza Abdolrasouli, Alireza Abdolrasouli, Matthew C. Fisher, Matthew C. Fisher, Thomas R. Sewell, Thomas R. Sewell
Abstract
The global emergence of azole resistance in Aspergillus fumigatus is resulting in health and food security concerns. Rapid diagnostics and environmental surveillance methods are key to understanding the distribution and prevalence of azole resistance. However, such methods are often associated with high costs and are not always applicable to laboratories based in the least-developed countries. Here, we present and validate a low-cost screening protocol that can be used to differentiate between azole-susceptible “wild-type” and azole-resistant A. fumigatus isolates. © 2020 The Authors.
Basic Protocol 1 : Preparation of Tebucheck multi-well plates
Basic Protocol 2 : Inoculation of Tebucheck multi-well plates
INTRODUCTION
Aspergillus fumigatus is an opportunistic fungal pathogen with an ecological niche of soil and composting vegetation, where it plays a vital role in ecosystem processes including decomposition and nutrient recycling (Rhodes, 2006; Tekaia & Latgé, 2005; Van De Veerdonk, Gresnigt, Romani, Netea, & Latgé, 2017). The fungus has a vast geographical range, facilitated by the rapid dispersion of airborne conidia that are able to tolerate a broad range of biotic stresses (Pringle et al., 2005; Sewell, Zhu, et al., 2019).
Humans inhale hundreds of conidia daily, which are frequently encountered in respiratory samples (Latgé, 1999; Mortensen et al., 2011). Typically, conidia are eliminated by the host's innate immune response, preventing attachment in the lung and subsequent infection (Balloy & Chignard, 2009). However, in immunocompromised individuals, conidia can become pathogenic resulting in a multitude of disorders including severe invasive aspergillosis (Brown et al., 2012). Current estimates indicate that more than 3 million people have invasive or chronic A. fumigatus infections (Bongomin, Gago, Oladele, & Denning, 2017). With the increase of immunocompromised individuals among the global population and rise in immunosuppressive therapies to treat human illnesses, successful management of globally emerging Aspergillus -related infections is largely dependent on early diagnosis and the initiation of effective antifungal treatment plans (Bernal-Martínez, Alastruey-Izquierdo, & Cuenca-Estrella, 2016; Denning et al., 2017; Walsh et al., 2008).
Triazoles are the most widely used class of antifungals owing to their efficacy and broad-spectrum activity (Price, Parker, Warrilow, Kelly, & Kelly, 2015). Their fundamental mode of action prevents fungal cell growth by inhibiting the lanosterol 14-α-demethylase enzyme in the ergosterol pathway, which is encoded by the cyp 51A gene (Bossche, Koymans, & Moereels, 1995). However, continual exposure to these compounds exerts selective pressures that can facilitate the evolution of azole-resistant A. fumigatus (AR Af). The recent emergence of two AR Af -associated cyp 51A alleles, TR34/L98H and TR46/Y121F/T289A, has raised concern due to their environmental association and increasing prevalence in azole-naive patients (Abdolrasouli et al., 2018; Chowdhary, Kathuria, Xu, & Meis, 2013; Mellado et al., 2007; Sewell, Zhu, et al., 2019; Snelders, Camps, et al., 2012; Stensvold, Jorgensen, & Arendrup, 2012; Van Der Linden et al., 2013; Zhang et al., 2017).
Several similar azole compounds belonging to the same structural class as the clinical triazoles, with near identical structures, are used extensively in agriculture for crop protection. Substrates with residual azoles, which effectively support the growth and reproduction of A. fumigatus , have the potential to become hotspots for azole resistance (Dunne, Hagen, Pomeroy, Meis, & Rogers, 2017; Prigitano et al., 2014; Schoustra et al., 2019). Environmental surveillance and resistance monitoring are needed to fully appreciate the prevalence and distribution of AR Af isolates in the environment (Fisher, Hawkins, Sanglard, & Gurr, 2018). Rapid diagnostics and surveillance methods will inevitably aid outbreak recognition, response, and prevention. However, current methods are targeted specifically for clinically derived samples, are associated with high costs, and are not widely available to laboratories based in low-income developing countries (Buil et al., 2017; Kahlmeter et al., 2006; White, Posso, & Barnes, 2017).
Here, we present a screening protocol to help identify AR Af isolated from the environment at a cost of ∼£0.70 per isolate. Our method is based on the agricultural azole tebuconazole, owing to it its use in the environment, its low cost, and its ability to differentiate between azole-susceptible “wild-type” A. fumigatus and AR Af (Alvarez-Moreno et al., 2017; Chowdhary, Kathuria, et al., 2012; Sewell, Zhang, et al., 2019; Snelders, Camps, et al., 2012; Verweij, Snelders, Kema, Mellado, & Melchers, 2009).
A. fumigatus is a Hazard Group 2 pathogen; therefore, suitable procedures and regulations for the handling of pathogenic microorganisms should always be followed. All open plate work should be carried out in a class 2 biological safety cabinet (BSC2) while wearing appropriate personal protective equipment. Extra care should be taken when handling due to the highly sporulating nature of the fungus.
Basic Protocol 1: PREPARATION OF TEBUCHECK MULTI-WELL PLATES
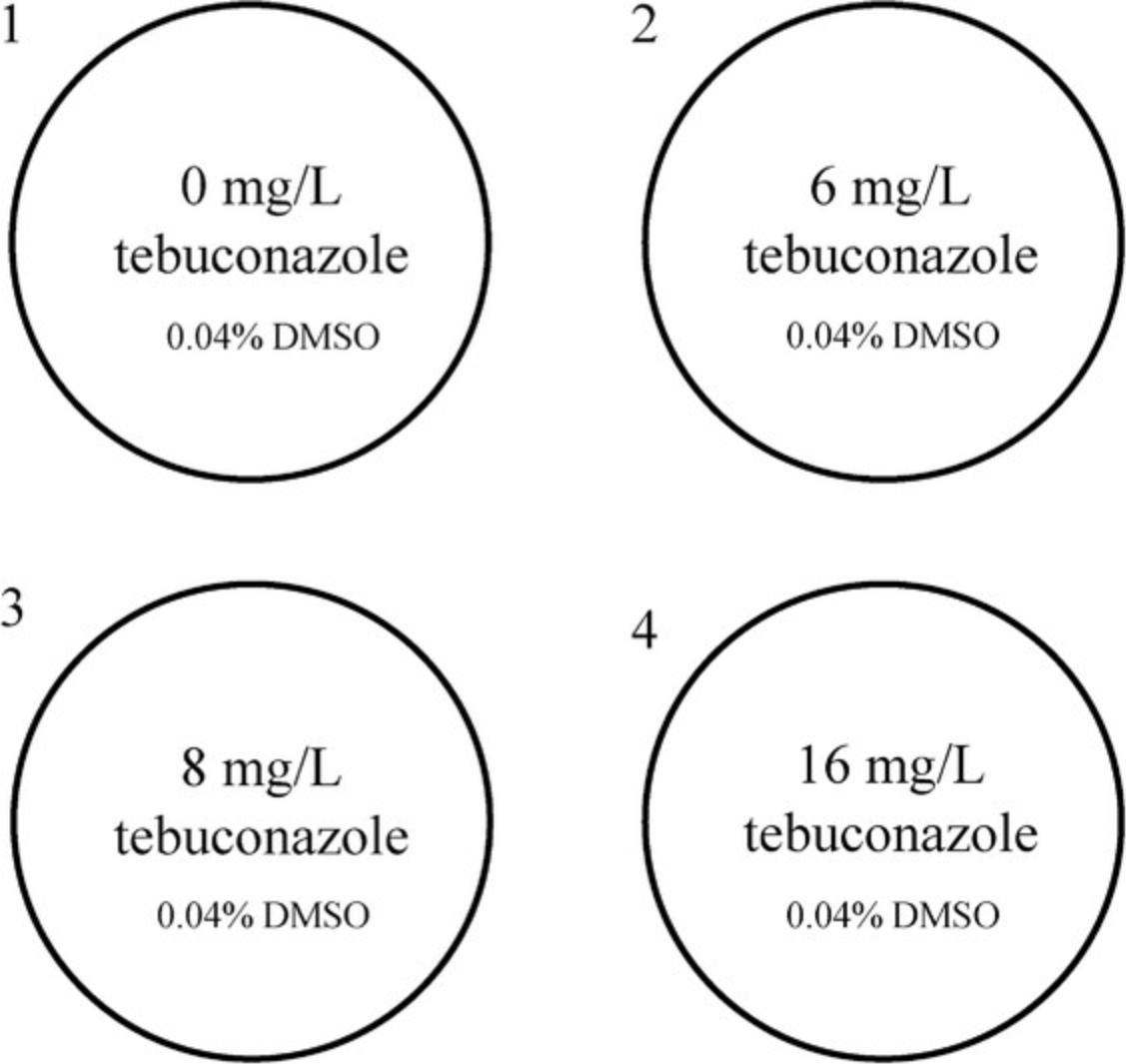
The first step in generating Tebucheck assay plates is to make the tebuconazole-incorporated agar at differing concentrations. Other culture media such as potato dextrose agar or malt extract agar can alternatively be used, although discrepancies in growth characteristics may be observed. Tebuconazole, 1-(4-chlorophenyl)-4,4-dimethyl-3-(1H-1,2,4-triazol-1-ylmethyl)pentan-3-ol (CAS no. 107534-96-3), is an agricultural triazole fungicide that inhibits sterol biosynthesis (demethylation inhibitor). The primary target of tebuconazole is lanosterol 14-α-demethylase (Kelly, Lamb, Corran, Baldwin, & Kelly, 1995), which is a key regulatory enzyme in the ergosterol biosynthetic pathway in fungi. Tebuconazole is classed as moderately toxic to mammals and can be dangerous in high doses (National Center for Biotechnology Information, 2020); consequently, all steps handling tebuconazole should be performed with care and while wearing single-use gloves. Table 1 provides the user with the required volumes/concentrations of tebuconazole and dimethyl sulfoxide (DMSO) for convenience. Tebuconazole stock solutions can be prepared in advance and stored in small-volume vials at −20°C for up to 1 month. To standardize the concentration of DMSO across the four wells of the assay, a supplementary volume must be added to give a final concentration of 0.032% (Table 1). Alternative methods for generating the tebuconazole-incorporated agar are acceptable; however, this may result in differential growth characteristics. The Tebucheck assay was developed using a 4-well multi-dish plate for standardization, reproducibility, and convenience. Other culture plates can be used, although discrepancies in growth characteristics may be observed. The stated volumes produce ∼32 4-well Tebucheck plates. Each well contains 1 ml Sabouraud dextrose agar (SDA) supplemented with three different concentrations of tebuconazole (Fig. 1). The first well is a drug-free control (0 mg/L tebuconazole), and wells 2, 3, and 4 contain SDA supplemented with 6, 8, or 16 mg/L tebuconazole, respectively. Tebucheck plates can be stored at 4°C for up to a month, although inspection of plates prior to use is recommended. The plates should be discarded if there is any obvious damage or contamination.
| Tube | Stocka tebuconazole (μl) | DMSO (µl) | Final tebuconazole concentration (mg/L) |
|---|---|---|---|
| 1 | 0 | 12.8 | 0 |
| 2 | 4.8 | 8 | 6 |
| 3 | 6.4 | 6.4 | 8 |
| 4 | 12.8 | 0 | 16 |
- a
Stock concentration = 50 mg/ml.
Materials
-
SDA (see recipe or purchase from a commercial vendor; e.g., Oxoid, cat. no. CM0041)
-
Tebuconazole, molecular biology grade, >98% purity (e.g., abcam, ab143703)
-
DMSO, molecular biology grade (e.g., Sigma-Aldrich, cat. no. D4540)
-
500-ml glass bottle (e.g., Duran, cat. no. 21 801 36 5)
-
Magnetic stir plate and stir bar
-
Benchtop autoclave
-
50°C water bath
-
50-ml conical tubes (e.g., Sarstedt, cat. no. 62.547.254)
-
Tube rack suitable for 50-ml tubes
-
50-ml serological pipette (e.g., Corning cat. no. CLS4490)
-
20-µl pipette and tips
-
4-well multi-dish plate (e.g., Thermo Fisher Scientific, cat. no. 176740)
-
1000-µl pipette and tips
Agar preparation
1.Add 19.5 g SDA to a 500-ml glass bottle.
2.Add 300 ml distilled water.
3.Mix gently on a magnetic stirrer for 5 min using a suitable-sized magnetic stir bar.
4.Autoclave in an automated benchtop autoclave to sterilize.
5.Allow molten medium to cool to ∼60°C inside a BSC2.
6.Move a suitable water bath inside the BSC2, and set to 50°C.
7.Label four 50-ml conical tubes 1 through 4, which correspond with the 4-well microplates used later.
8.Place tubes into the 50°C water bath using a 50-ml tube rack.
9.Using a serological pipette, transfer 40 ml molten SDA into the four conical tubes.
10.Allow SDA to equilibrate in the tubes for 5 min.
Incorporating tebuconazole
11.While the agar is equilibrating to temperature, prepare a 50 mg/ml stock solution of tebuconazole by dissolving 100 mg tebuconazole in 2 ml molecular biology–grade DMSO.
12.Using a 20-µl pipette, remove 12.8 µl SDA from each tube.
13.Using a 20-µl pipette, transfer the correct volume of tebuconazole stock for each corresponding tube (Table 1).
14.Gently mix each tube.
15.Using a 20-µl pipette, transfer the correct volume of DMSO into each corresponding tube (Table 1).
16.Gently mix each tube.
17.Leave tubes to infuse before immediately moving to Basic Protocol 2.
18.Using a 1000-µl pipette, transfer 1000 µl tebuconazole-incorporated agar from each tube into the corresponding well of the 4-well microplate (Fig. 1) starting with the control (0 µg/ml tebuconazole).
19.Once complete, allow plates to dry for 10 min in a BSC2.
Basic Protocol 2: INOCULATION OF TEBUCHECK MULTI-WELL PLATES
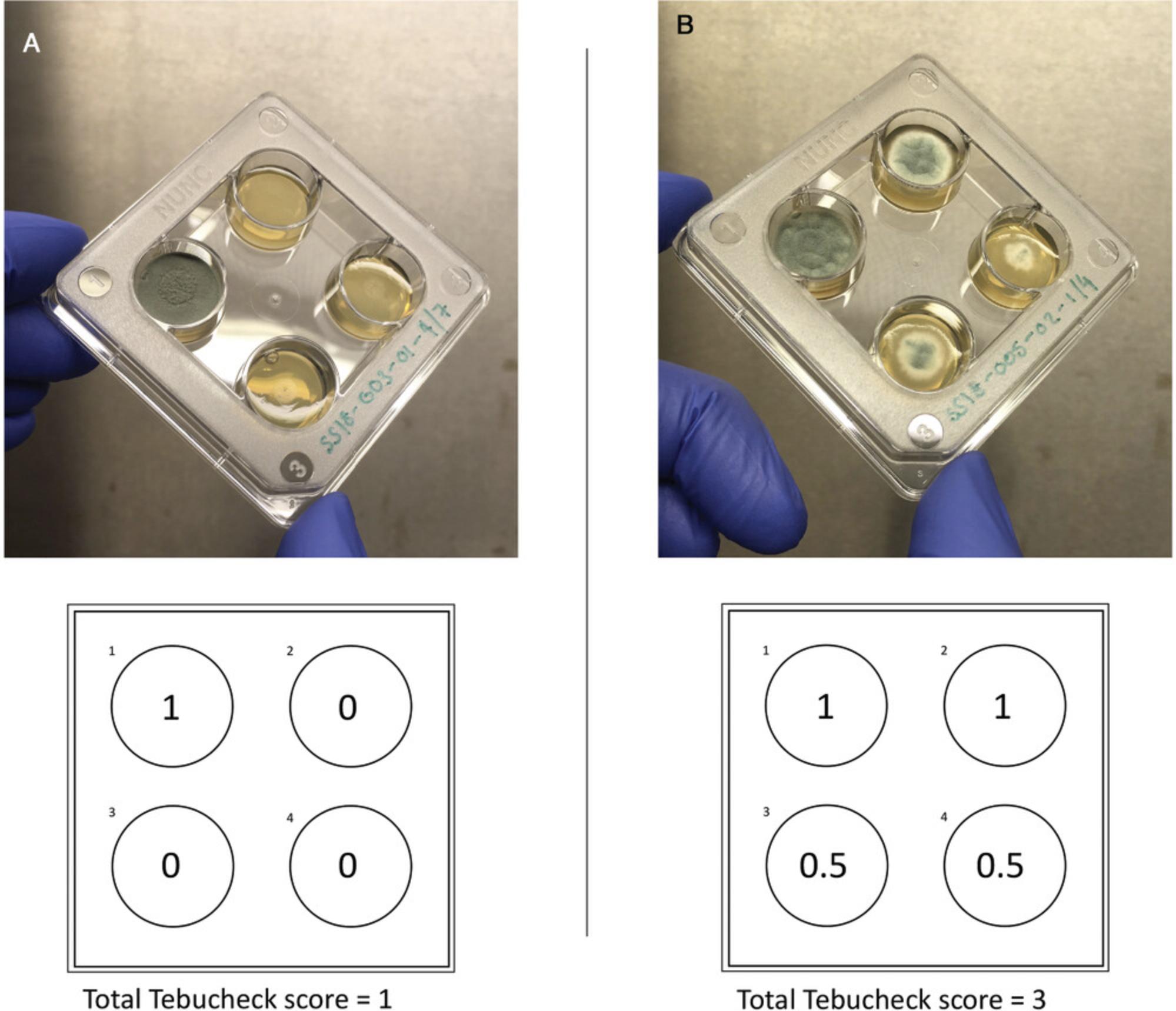
Inoculation of Tebucheck plates includes two incubation steps: (1) the initial culturing of A. fumigatus isolates to be tested and (2) incubation of the inoculated Tebucheck plates. The user must therefore allow 72 to 120 hr to complete this protocol. This time frame includes culture and preparation of pure single colonies plus the Tebucheck incubation period. Known wild-type A. fumigatus isolates can be purchased from CBS-KNAW Collections (http://www.wi.knaw.nl/Collections/) and used as a negative control. For positive controls, A. fumigatus isolates harboring the resistance alleles TR34/L98H and TR46/Y121F/T289A can be obtained by contacting the Fisher laboratory (https://www.fisherlab.co.uk). After incubation of Tebucheck plates, the presence or absence of growth on the surface of the four wells can be classified using the following scale: 0, no growth (0% to <10% growth coverage); 0.5, partial/visible growth (10% to <50% growth coverage); and 1, growth (50% to 100% growth coverage).
The end point of susceptibility should be recorded where visual growth was 0% to 10% coverage. For example, an isolate that grows vigorously in well 1, grows partially in well 2, and does not grow in wells 3 and 4 would receive a final Tebucheck score of 1.5.A population of Tebucheck A. fumigatus isolates can be analyzed together using a custom R script downloadable at https://github.com/abrackin/tebucheck.
Materials
-
SDA (see recipe or purchase from a commercial vendor; e.g., Oxoid, cat. no. CM0041)
-
A. fumigatus negative and positive controls
-
0.05% (v/v) Tween 80, sterile
-
Tebucheck plates (see Basic Protocol 1)
-
500-ml glass bottle (e.g., Duran, cat. no. 21 801 36 5)
-
Magnetic stir plate and stir bar
-
Benchtop autoclave
-
25-cm2 cell culture flask (e.g., Thermo Fisher Scientific, cat. no. 156340) or appropriate alternative
-
37°C cell culture incubator
-
Custom R script, available at https://github.com/abrackin/tebucheck
A. fumigatus culture
1.Add 26 g SDA to a 500-ml glass bottle.
2.Add 400 ml distilled water.
3.Mix gently on a magnetic stir plate for 5 min using a suitable-sized magnetic stir bar.
4.Autoclave in an automated benchtop autoclave to sterilize.
5.Allow molten medium to cool to ∼60°C inside a BSC2.
6.Carefully pour ∼15 ml molten SDA into a 25-cm2 cell culture flask or an appropriate alternative.
7.Place flasks horizontally (base down), and allow agar to solidify for 20 min.
8.Inoculate A. fumigatus sample (glycerol stock or agar punch) in the center of each cell culture flask.
9.Incubate at 37°C for 24 to 72 hr or when the colony has occupied ∼50% of the cell culture flask.
Tebucheck screening
10.Once fully grown, add 10 ml of 0.05% (v/v) Tween 80 to each cell culture flask.
11.Gently shake each flask to disrupt the spores.
12.Leave spore suspension for 5 min to allow any aerosolized spores to settle.
13.Inoculate each well of the Tebucheck plates with 5 μl spore suspension, taking care to introduce the suspension into the center of the well.
14.Incubate inoculated Tebucheck plate for 48 hr at 37°C.
15.Score Tebucheck end point by visually assessing presence or absence of growth on each well.
16.Analyze multiple isolates using the custom R script.
Sample data
Validation of Tebucheck was performed using a panel of 26 environmental A. fumigatus isolates (Table 2) selected from the Imperial College whole genome sequencing A. fumigatus culture collection (total collection >500 isolates). The panel represents the genetic population structure of the fungus, its vast geographical distribution, two of the most common environmentally associated azole antifungal-resistant genotypes (TR34/L98H [n = 8] and TR46/Y121F/T289A [n = 8]), and wild-type representatives (n = 10).
| Isolate no. | Isolate name | Mutation | Country | Date of isolation |
|---|---|---|---|---|
| 1 | AFU-379/E12/2 | Wild-type | India | 2012 |
| 2 | 47-154 | Wild-type | UK | 2012 |
| 3 | TF4-19 | Wild-type | Japan | 2013 |
| 4 | 47-181 | Wild-type | Hawaii | 1997 |
| 5 | 47-137 | Wild-type | South Africa | 2002 |
| 6 | 47-191 | Wild-type | Sri Lanka | 1991 |
| 7 | 47-151 | Wild-type | Australia | 1988 |
| 8 | 47-120 | Wild-type | Brazil | 1991 |
| 9 | 47-105 | Wild-type | Hungary | 1983 |
| 10 | AF293 (reference) | Wild-type | UK | 1993 |
| 11 | HYDE42 | TR34/L98H | UK | 2017 |
| 12 | HL102-1 | TR34/L98H | Taiwan | 2015 |
| 13 | D007 | TR34/L98H | Taiwan | 2014 |
| 14 | CDC2014730721 | TR34/L98H | USA | 2014 |
| 15 | BUU10 | TR34/L98H | Thailand | 2016a |
| 16 | BUU06 | TR34/L98H | Thailand | 2016a |
| 17 | NAN121 | TR34/L98H | Colombia | 2017a |
| 18 | ComPos-1 | TR34/L98H | UK | 2017 |
| 19 | NAN077 | TR46/Y121F/T289A | Colombia | 2017a |
| 20 | 302Wg | TR46/Y121F/T289A | Wales | 2015 |
| 21 | NAN087 | TR46/Y121F/T289A | Colombia | 2017a |
| 22 | DUB_48 | TR46/Y121F/T289A | Ireland | 2015 |
| 23 | DUB_54 | TR46/Y121F/T289A | Ireland | 2015 |
| 24 | DUB_50 | TR46/Y121F/T289A | Ireland | 2015 |
| 25 | E276 | TR46/Y121F/T289A | Germany | 2012 |
| 26 | E224 | TR46/Y121F/T289A | Germany | 2012 |
- a
Denotes publication date rather than isolation date.
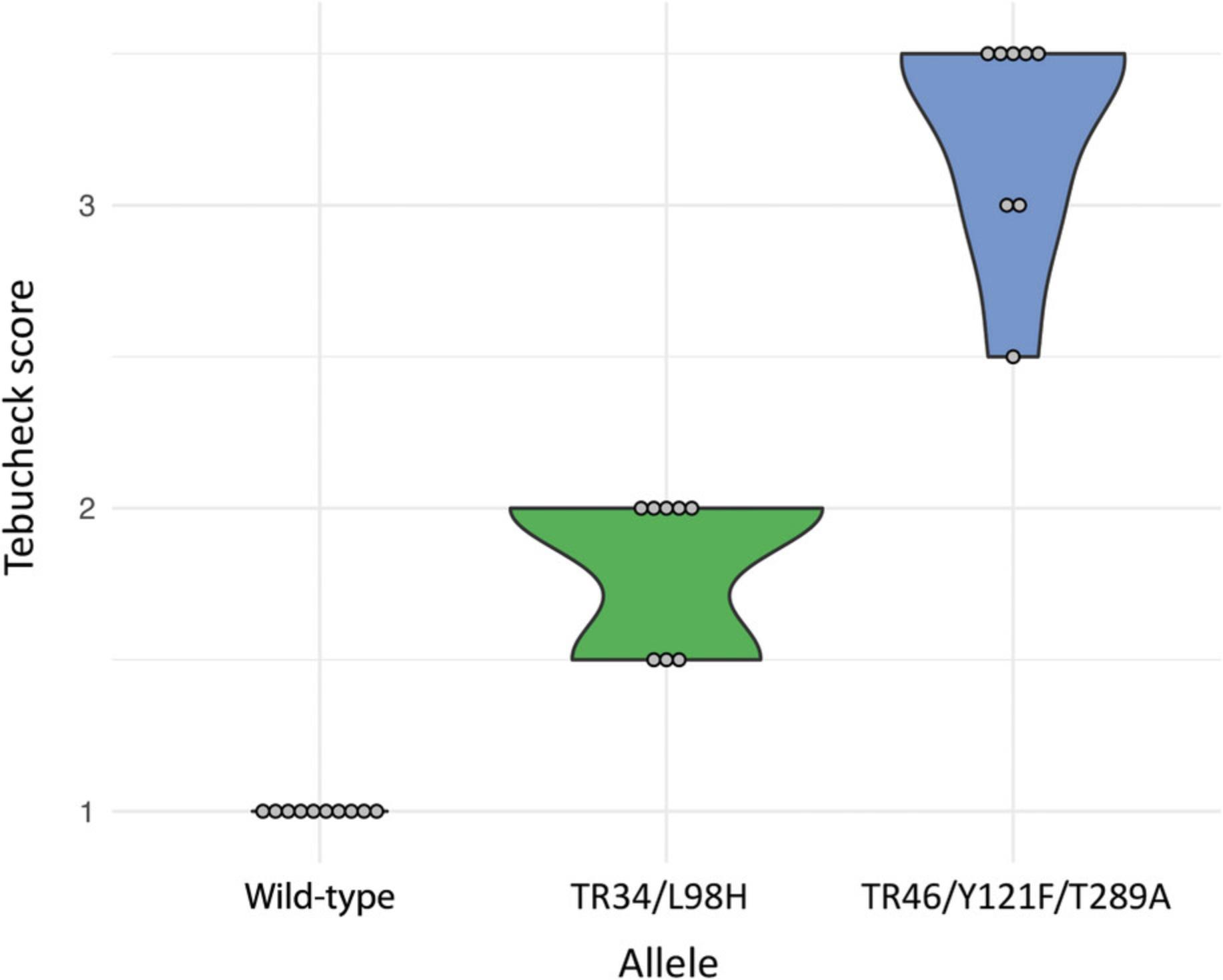
All wild-type isolates were susceptible to tebuconazole at 6 mg/L. Preliminary testing of the Tebucheck protocol used 4 mg/L, although this did not fully distinguish between wild-type and azole-resistant isolates. Isolates with known resistance alleles grew on the control well and exhibited various levels of growth in wells 2, 3, and 4 (Fig. 2). Of the isolates harboring the TR34/L98H resistance allele, 100% had growth in well 2, and 62.5% showed partial growth in well 3 (up to 8 mg/L tebuconazole susceptibility). All TR34/L98H isolates were scored 0 (0 to <10% growth coverage) in well 4 (16 mg/L), suggesting an end point of tebuconazole susceptibility at 8 mg/L. Of the isolates harboring the TR46/Y121F/T289A allele, 100% showed growth in well 2 (6 mg/L), 100% grew in well 3, and 62.5% grew in well 4 (16 mg/L), suggesting an end point of tebuconazole susceptibility ≥16 mg/L (Fig. 3).
REAGENTS AND SOLUTIONS
SDA
- 10 g/L mycological peptone
- 40 g/L glucose
- 15 g/L agar
- Adjust pH to 5.6 ± 0.2 when at 25°C
- Autoclave at 121°C for 15 min
- Store at room temperature for up to 3 months
COMMENTARY
Background Information
The emergence of environmental AR Af and its associated pathological risks to immunocompromised individuals is of global concern (Fisher et al., 2018). Currently, the majority of azole resistance monitoring studies are constrained to European countries, motivated primarily by the clinical emergence of AR Af in these regions (Snelders, van der Lee, et al., 2008; Van Der Linden et al., 2013; Verweij et al., 2009). However, there has been an increasing number of reports of AR Af in the least-developed countries (Meis, Chowdhary, Rhodes, Fisher, & Verweij, 2016) where, typically, financial assets, specialized equipment, trained staff, and testing capacity are limited. Precise estimates of prevalence are largely unknown in developing countries where knowledge is limited by a lack of surveillance and reporting. Without environmental screening it is difficult to determine whether variations in prevalence may be due to climatic conditions, fungal distributions, varying use of agrichemical products, or incomplete investigations. While advances in technology have resulted in a variety of diagnostic tools to save time and cost, they are not always widely available to some laboratories.
Molecular tools such as quantitative PCR, loop-mediated isothermal amplification (LAMP), and Sanger sequencing are effective diagnostic tools used in many research laboratories and are regularly used to characterize AR Af -associated resistance alleles (Sewell, Zhang, et al., 2019; White et al., 2017; Yu et al., 2019). Biological growth assays such as VIP-check™ (Buil et al., 2017) and EUCAST (Kahlmeter et al., 2006) are used to determine the organism's resistance phenotype. Many of these assays are targeted specifically at clinical AR Af isolates despite the environmental source of A. fumigatus. While these techniques are extremely effective and broadly applicable in many pathology and genomic research facilities, they can become costly when applied to large screening projects, particularly for those included in large-scale, longitudinal studies.
For many mycology laboratories, cost-effective culture-based diagnostics are paramount to research. Surveys conducted by the Asia Fungal Working Group comprising 241 mycology laboratories within seven Asian countries revealed that 89% had culture facilities, whereas only 17% performed DNA sequencing, and just 22% had molecular diagnostic facilities (Chindamporn et al., 2018). With the rise of antifungal resistance, routine susceptibility testing is essential, particularly in countries where the burden of fungal disease is high.
In order to address the issues outlined above, we present Tebucheck, a simple cost-effective method for the detection of AR Af using the agricultural antifungal tebuconazole. The method is intended for the use of determining the level of azole resistance in environmental samples and to aid in the forward planning of in-depth AR Af investigations. Affordable, easy-to-use identification tools such as Tebucheck can be used to identify AR Af hotspots and their implications on public health. It allows for screening and the subsequent selection of putative AR Af to be subjected to further downstream characterization and analysis, thus reducing the number (and cost) of high-precision testing.
Critical Parameters
Tebucheck has been designed to be a basic and easy-to-use assay, meaning it has very few parameters and a low chance of user error. However, a few critical conditions should be considered. Cell density is an important consideration when handling Tebucheck. If a very turbid spore suspension is used to inoculate the Tebucheck wells, uncharacteristic growth may occur. Users should aim to have a turbidity of ∼0.5 McFarland or a cell density reading at 600 nm between 0.08 and 0.13 using a standard spectrophotometer. However, if the guidelines set out in this protocol for the generation of A. fumigatus spore suspension are followed closely (Basic Protocol 2, steps 1 to 12), then there should not be an issue at this step, and cell density readings will not need to be verified. When creating the tebuconazole-incorporated medium, it is vitally important the user allows the tebuconazole stock solution to properly mix with the molten SDA. Gently mixing SDA and allowing it to equilibrate in the water bath for 5 min is enough to ensure a consistent concentration of tebuconazole is maintained across all wells of the Tebucheck assay plates. Growth rates between A. fumigatus isolates can vary and may affect the assessment and scoring after the mandatory 48-hr incubation period. If growth in the control well (0 µg/ml tebuconazole) is underdeveloped after 48 hr, then Tebucheck plates can be left to incubate to the point at which the control well exhibits ∼50% growth coverage (maximum 5-day incubation due to drug exhaustion).
Troubleshooting
Our findings suggest that Tebucheck could be a useful addition to the current range of AR Af diagnostics available. Here, we show that it can distinguish between resistant and susceptible isolates and putatively predicted, environmentally associated resistance genotypes. Despite this finding, other resistance mechanisms can also explain AR Af phenotypes, and there may be others yet undiscovered. It is important, therefore, that Tebucheck is predominantly used to distinguish between resistant and non-resistant environmentally acquired isolates and that any putative genotype calls be properly assessed with quantitative PCR, LAMP assay, or direct sequencing of the cyp 51A allele (Sewell, Zhang, et al., 2019; White et al., 2017; Yu et al., 2019).
Understanding Results
Tebucheck was validated using a panel of A. fumigatus isolates that were subjected to three concentrations of tebuconazole plus a control. Growth of all non-resistant isolates harboring the wild-type cyp 51A allele was restricted to the control well only, clearly defining azole-susceptible wild-type isolates from resistant isolates. All resistant isolates were able to grow on at least one concentration of tebuconazole-incorporated growth medium. By using the scoring system outlined in Basic Protocol 2, it is possible for users to analyze a population of A. fumigatus isolates and determine its level of resistance.
The four different concentrations of tebuconazole in the Tebucheck assay can also provide insight into the level of resistance the isolates exhibit, providing the user with a more detailed characterization of their AR Af samples. During validation, it became clear that isolates with different resistance genotypes had different growth characteristics on Tebucheck, with TR46/Y121F/T289A genotypes more likely to grow across all four wells of the assay and TR34/L98H restricted to two wells (Figs. 2 and 3). It is possible, therefore, to not only gauge the isolates level of resistance but also putatively suggest its cyp 51A genotype.
Time Considerations
Time considerations for this protocol depend on the number of A. fumigatus isolates the user wishes to process using Tebucheck. Considering a single set of Tebucheck plates (32 plates), Basic Protocol 1 should take ∼30 min, while Basic Protocol 2 can take between 3 and 5 days due to the incubation steps involved in prior culturing of A. fumigatus isolates and the mandatory 48-hr Tebucheck incubation period. Generally, the total time to perform the whole protocol, including incubation steps, is between 2 and 5 days.
Acknowledgments
The authors acknowledge joint Centre funding from the UK Medical Research Council and Department for International Development. T.R.S., A.P.B., J.M.G., and M.C.F. were supported by the Natural Environmental Research Council (NERC; NE/P001165/1), and all authors were supported by the Medical Research Council (MRC; MR/R015600/1). M.C.F. is a CIFAR Fellow in the Fungal Kingdom program.
Author Contributions
A.P.B., J.M.G.S., A.A., M.C.F., and T.R.S. conceived and designed the study. A.P.B., J.M.G.S., and T.R.S. tested the protocol and collected the data. A.P.B. and T.R.S. analyzed the data. A.P.B. and T.R.S. wrote the manuscript. A.P.B., J.M.G.S., A.A., M.C.F., and T.R.S. commented on the manuscript.
Literature Cited
- Abdolrasouli, A., Scourfield, A., Rhodes, J., Shah, A., Elborn, J. S., Fisher, M. C., … Armstrong-James, D. (2018). High prevalence of triazole resistance in clinical Aspergillus fumigatus isolates in a specialist cardiothoracic centre. International Journal of Antimicrobial Agents , 52, 637–642. doi: 10.1016/j.ijantimicag.2018.08.004.
- Alvarez-Moreno, C., Lavergne, R.-A., Hagen, F., Morio, F., Meis, J. F., & Le Pape, P. (2017). Azole-resistant Aspergillus fumigatus harboring TR 34 /L98H, TR 46 /Y121F/T289A and TR 53 mutations related to flower fields in Colombia. Scientific Reports , 7, 45631. doi: 10.1038/srep45631.
- Balloy, V., & Chignard, M. (2009). The innate immune response to Aspergillus fumigatus. Microbes and Infection , 11, 919–927. doi: 10.1016/j.micinf.2009.07.002.
- Bernal-Martínez, L., Alastruey-Izquierdo, A., & Cuenca-Estrella, M. (2016). Diagnostics and susceptibility testing in Aspergillus. Future Microbiology , 11, 315–328. doi: 10.2217/fmb.15.140.
- Bongomin, F., Gago, S., Oladele, R. O., & Denning, D. W. (2017). Global and multi-national prevalence of fungal diseases—estimate precision. Journal of Fungi , 3, 57. doi: 10.3390/jof3040057.
- Bossche, H. V., Koymans, L., & Moereels, H. (1995). P450 inhibitors of use in medical treatment: Focus on mechanisms of action. Pharmacology and Therapeutics , 67, 79–100. doi: 10.1016/0163-7258(95)00011-5.
- Brown, G. D., Denning, D. W., Gow, N. A. R., Levitz, S. M., Netea, M. G., & White, T. C. (2012). Hidden killers: Human fungal infections. Science Translational Medicine , 4, 165rv13. doi: 10.1126/scitranslmed.3004404.
- Buil, J. B., van der Lee, H. A. L., Rijs, A. J. M. M., Zoll, J., Hovestadt, J. A. M. F., Melchers, W. J. G., & Verweij, P. E. (2017). Single-center evaluation of an agar-based screening for azole resistance in Aspergillus fumigatus by using VIPcheck. Antimicrobial Agents and Chemotherapy , 61, e01250–17. doi: 10.1128/AAC.01250-17.
- Chindamporn, A., Chakrabarti, A., Li, R., Sun, P. L., Tan, B. H., Chua, M., … Chayakulkeeree, M. (2018). Survey of laboratory practices for diagnosis of fungal infection in seven Asian countries: An Asia Fungal Working Group (AFWG) Initiative. Medical Mycology , 56, 416–425. doi: 10.1093/mmy/myx066.
- Chowdhary, A., Kathuria, S., Xu, J., & Meis, J. F. (2013). Emergence of azole-resistant Aspergillus fumigatus strains sue to agricultural azole use creates an increasing threat to human health. PLoS Pathogens , 9, e1003633. doi: 10.1371/journal.ppat.1003633.
- Chowdhary, A., Kathuria, S., Xu, J., Sharma, C., Sundar, G., Singh, P. K., … Meis, J. F. (2012). Clonal expansion and emergence of environmental multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR34/L98H mutations in the cyp 51A gene in India. PLoS ONE , 7, e52871. doi: 10.1371/journal.pone.0052871.
- Denning, D. W., Perlin, D. S., Muldoon, E. G., Colombo, A. L., Chakrabarti, A., Richardson, M. D., & Sorrell, T. C. (2017). Delivering on antimicrobial resistance agenda not possible without improving fungal diagnostic capabilities. Emerging Infectious Diseases , 23, 177–183. doi: 10.3201/eid2302.152042.
- Dunne, K., Hagen, F., Pomeroy, N., Meis, J. F., & Rogers, T. R. (2017). Intercountry transfer of triazole-resistant Aspergillus fumigatus on plant bulbs. Clinical Infectious Diseases , 65, 147–149. doi: 10.1093/cid/cix257.
- Fisher, M. C., Hawkins, N. J., Sanglard, D., & Gurr, S. J. (2018). Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science , 360, 739–742. doi: 10.1126/science.aap7999.
- Kahlmeter, G., Brown, D. F. J., Goldstein, F. W., MacGowan, A. P., Mouton, J. W., Odenholt, I., & Stetsiouk, O. (2006). European Committee on Antimicrobial Susceptibility Testing (EUCAST) technical notes on antimicrobial susceptibility testing. Clinical Microbiology and Infection , 12, 501–503. doi: 10.1111/j.1469-0691.2006.01454.x.
- Kelly, S. L., Lamb, D. C., Corran, A. J., Baldwin, B. C., & Kelly, D. E. (1995). Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochemical and Biophysical Research Communications , 207, 910–915. doi: 10.1006/bbrc.1995.1272.
- Latgé, J.-P. (1999). Aspergillus fumigatus and aspergillosis. Clinical Microbiology Reviews , 12, 310–350. doi: 10.1128/CMR.12.2.310.
- Meis, J. F., Chowdhary, A., Rhodes, J. L., Fisher, M. C., & Verweij, P. E. (2016). Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philosophical Transactions of the Royal Society of London , 371, 20150460. doi: 10.1098/rstb.2015.0460.
- Mellado, E., Garcia-Effron, G., Alcázar-Fuoli, L., Melchers, W. J. G., Verweij, P. E., Cuenca-Estrella, M., & Rodríguez-Tudela, J. L. (2007). A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrobial Agents and Chemotherapy , 51, 1897–1904. doi: 10.1128/AAC.01092-06.
- Mortensen, K. L., Jensen, R. H., Johansen, H. K., Skov, M., Pressler, T., Howard, S. J., & Arendrup, M. C. (2011). Aspergillus species and other molds in respiratory samples from patients with cystic fibrosis: A laboratory-based study with focus on Aspergillus fumigatus azole resistance. Journal of Clinical Microbiology , 49, 2243–2251. doi: 10.1128/JCM.00213-11.
- National Center for Biotechnology Information. (2020). PubChem compound summary for CID 86102, tebuconazole. Retrieved June 16, 2020 from https://pubchem.ncbi.nlm.nih.gov/compound/Tebuconazole.
- Price, C. L., Parker, J. E., Warrilow, A. G. S., Kelly, D. E., & Kelly, S. L. (2015). Azole fungicides - Understanding resistance mechanisms in agricultural fungal pathogens. Pest Management Science , 71, 1054–1058. doi: 10.1002/ps.4029.
- Prigitano, A., Venier, V., Cogliati, M., De Lorenzis, G., Esposto, M. C., & Tortorano, A. M. (2014). Azole-resistant Aspergillus fumigatus in the environment of northern Italy, May 2011 to June 2012. Eurosurveillance , 19, 20747. doi: 10.2807/1560-7917.es2014.19.12.20747.
- Pringle, A., Baker, D. M., Platt, J. L., Wares, J. P., Latgé, J.-P., & Taylor, J. W. (2005). Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution , 59, 1886–1899. doi: 10.1111/j.0014-3820.2005.tb01059.x.
- Rhodes, J. C. (2006). Aspergillus fumigatus : Growth and virulence. Medical Mycology , 44, 77–81. doi: 10.1080/13693780600779419.
- Schoustra, S. E., Debets, A. J. M., Rijs, A. J. M. M., Zhang, J., Snelders, E., Leendertse, P., & Verweij, P. E. (2019). Environmental hotspots for azole resistance selection of Aspergillus fumigatus , the Netherlands. Emerging Infectious Diseases , 25, 1347–1353. doi: 10.3201/eid2507.181625.
- Sewell, T. R., Zhang, Y., Brackin, A. P., Shelton, J. M. G., Rhodes, J., & Fisher, M. C. (2019). Elevated prevalence of azole-resistant Aspergillus fumigatus in urban versus rural environments in the United Kingdom. Antimicrobial Agents and Chemotherapy , 63, e00548–19. doi: 10.1128/AAC.00548-19.
- Sewell, T. R., Zhu, J., Rhodes, J., Hagen, F., Meis, J. F., Fisher, M. C., & Jombart, T. (2019). Nonrandom distribution of azole resistance across the global population of Aspergillus fumigatus. mBio , 10, e00392–19. doi: 10.1128/mBio.00392-19.
- Snelders, E., Camps, S. M. T., Karawajczyk, A., Schaftenaar, G., Kema, G. H. J., van der Lee, H. A., & Verweij, P. E. (2012). Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS ONE , 7, e31801. doi: 10.1371/journal.pone.0031801.
- Snelders, E., van der Lee, H. A. L., Kuijpers, J., Rijs, A. J. M. M., Varga, J., Samson, R. A., & Verweij, P. E. (2008). Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Medicine , 5, 1629–1637. doi: 10.1371/journal.pmed.0050219.
- Stensvold, C. R., Jorgensen, L. N., & Arendrup, M. C. (2012). Azole-resistant invasive Aspergillosis : Relationship to agriculture. Current Fungal Infection Reports , 6, 178–191. doi: 10.1007/s12281-012-0097-7.
- Tekaia, F., & Latgé, J.-P. (2005). Aspergillus fumigatus : Saprophyte or pathogen? Current Opinion in Microbiology , 8, 385–392. doi: 10.1016/j.mib.2005.06.017.
- Van De Veerdonk, F. L., Gresnigt, M. S., Romani, L., Netea, M. G., & Latgé, J.-P. (2017). Aspergillus fumigatus morphology and dynamic host interactions. Nature Reviews Microbiology , 15, 661–674. doi: 10.1038/nrmicro.2017.90.
- Van Der Linden, J. W. M., Camps, S. M. T., Kampinga, G. A., Arends, J. P. A., Debets-Ossenkopp, Y. J., Haas, P. J. A., … Verweij, P. E. (2013). Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clinical Infectious Diseases , 57, 513–520. doi: 10.1093/cid/cit320.
- Verweij, P. E., Snelders, E., Kema, G. H. J., Mellado, E., & Melchers, W. J. G. (2009). Azole resistance in Aspergillus fumigatus : A side-effect of environmental fungicide use? The Lancet Infectious Diseases , 9, 789–795. doi: 10.1016/S1473-3099(09)70265-8.
- Walsh, T. J., Anaissie, E. J., Denning, D. W., Herbrecht, R., Kontoyiannis, D. P., & Marr, K. A., & Infectious Diseases Society of America (2008). Treatment of aspergillosis: Clinical practice guidelines of the Infectious Diseases Society of America. Clinical Infectious Diseases , 46, 327–360. doi: 10.1086/525258.
- White, P. L., Posso, R. B., & Barnes, R. A. (2017). Analytical and clinical evaluation of the PathoNostics AsperGenius assay for detection of invasive aspergillosis and resistance to azole antifungal drugs directly from plasma samples. Journal of Clinical Microbiology , 55, 2356–2366. doi: 10.1128/JCM.00411-17.
- Yu, L.-S., Rodriguez-Manzano, J., Malpartida-Cardenas, K., Sewell, T., Bader, O., & Armstrong-James, D. (2019). Rapid and sensitive detection of azole-resistant Aspergillus fumigatus by tandem repeat loop-mediated isothermal amplification. The Journal of Molecular Diagnostics , 21, 286–295. doi: 10.1016/j.jmoldx.2018.10.004.
- Zhang, J., van den Heuvel, J., Debets, A. J. M., Verweij, P. E., Melchers, W. J. G., Zwaan, B. J., & Schoustra, S. E. (2017). Evolution of cross-resistance to medical triazoles in Aspergillus fumigatus through selection pressure of environmental fungicides. Proceedings of the Royal Society B: Biological Sciences , 284, 20170635. doi: 10.1098/rspb.2017.0635.
Citing Literature
Number of times cited according to CrossRef: 4
- Benjamin C. Simmons, Johanna Rhodes, Thomas R. Rogers, Paul E. Verweij, Alireza Abdolrasouli, Silke Schelenz, Samuel J. Hemmings, Alida Fe Talento, Auveen Griffin, Mary Mansfield, David Sheehan, Thijs Bosch, Matthew C. Fisher, Genomic Epidemiology Identifies Azole Resistance Due to TR34/L98H in European Aspergillus fumigatus Causing COVID-19-Associated Pulmonary Aspergillosis, Journal of Fungi, 10.3390/jof9111104, 9 , 11, (1104), (2023).
- Luisa F. Gómez Londoño, Marin T. Brewer, Detection of azole-resistant Aspergillus fumigatus in the environment from air, plant debris, compost, and soil, PLOS ONE, 10.1371/journal.pone.0282499, 18 , 3, (e0282499), (2023).
- Jennifer M. G. Shelton, Johanna Rhodes, Christopher B. Uzzell, Samuel Hemmings, Amelie P. Brackin, Thomas R. Sewell, Asmaa Alghamdi, Paul S. Dyer, Mark Fraser, Andrew M. Borman, Elizabeth M. Johnson, Frédéric B. Piel, Andrew C. Singer, Matthew C. Fisher, Citizen science reveals landscape-scale exposures to multiazole-resistant Aspergillus fumigatus bioaerosols , Science Advances, 10.1126/sciadv.adh8839, 9 , 29, (2023).
- Yongjun Chen, Dongmei Yan, Jialin Xu, Hui Xiong, Shaorong Luan, Ciying Xiao, Qingchun Huang, The importance of selecting crystal form for triazole fungicide tebuconazole to enhance its botryticidal activity, Science of The Total Environment, 10.1016/j.scitotenv.2022.158778, 854 , (158778), (2023).

