ADE 2022 Day 2: PCR
Tom Little
Abstract
DAY 2: ADE field practical instructions for PCR
Steps
Retrieve your four tubes from the PCR machine.
Add 24ul of PCR mix to the second tube in your strip. The PCR mix includes buffer, enzyme and primers to amplify the ~650 base fragment of cytochrome oxidase I (COX1) used as a "universal" DNA barcode for animals. Your tubes will now look like this, with the clear liquid (LYS) in tube 1, and a clear liquid (PCR) in tube 2, followed by two empty tubes. Make sure your sequencing identifier is still clearly visible
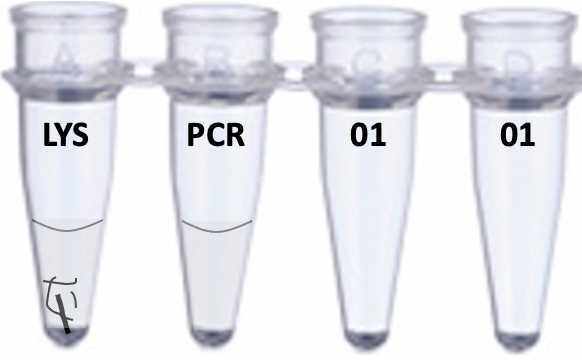
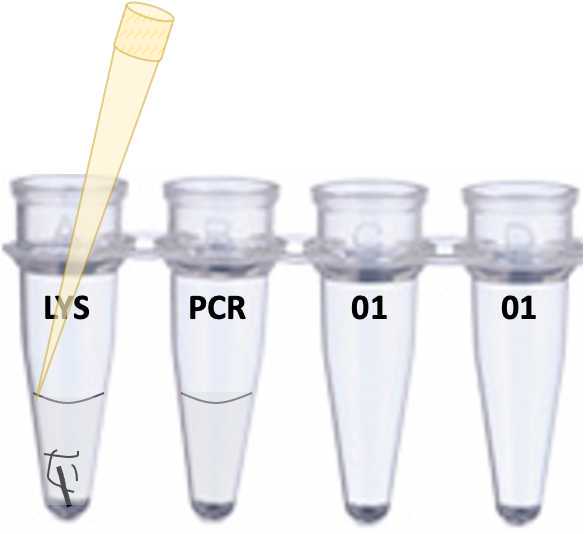
Put a new yellow tip on a p10 pipette and transfer 1ul of the clear LYS liquid in tube 1 to the PCR mixture in tube 2. Avoid picking up any lumps of tissue that might still be present, as these will stop the PCR from working. The best way to do this is to set your pipette to 1ul, depress the plunger, then slide the tip of your yellow tip down the side of the tube until it enters the liquid at the meniscus. At this point, gently raise the plunger to pick up 1ul and move it to the PCR mixture in tube 2. Make sure you are holding the tubes up to eye level, so you can see what is happening. For example, it is really helpful if you see the 1ul in the tip, and can see that you have actually transferred it to the PCR mix. Seeing is believing. Discard the tip when you are done.
Get a new yellow tip on a P10 or P20, set it to 10ul and gently mix the PCR solution by slowly plunging up and down.
Put the lids on your strip tube and check the labelling is clear. Take your strip tube to the PCR machine and put it in the block.
Once the PCR machine is full, the demonstrators will set the PCR amplification cycles running. The PCR machine cycles through 94°C (to denature DNA into single strands), 44°C (to allow the primers to anneal) and 72°C (to allow the DNA polymerase to copy the DNA strand). The PCR proceeds for 30 cycles, and will take about 2.5 hrs, but you do not have to wait for the PCR to finish.
This is a good opportunity to review how PCR works by watching https://www.youtube.com/watch?v=2KoLnIwoZKU
The "primers" mentioned in the video are a key part of what we are doing. We have added specific primers: two different ~20 nucleotide long bits of DNA that are an exact match to sequences on the COX1 gene, 650bp apart. This is why our PCR will amplify only the particular 650bp of the COX1 gene that works as a barcode to distinguish species. We had the primers synthesised by a company that specialises in this. We sent them the sequence of the primers, and they synthesised the primers in an appropriate concentration for PCR
Clean up your work area, replacing the tools in their bag, and discarding any used tips. You may now have time to specimens using the dichotomous keys, but the next session is entirely dedicated to this.
Long after you have left, the demonstrators, and Tom and Andrew will retrieve the tubes from the machine. We will prepare the DNA for sequencing on the Oxford Nanopore Minion. Time permitting, this will be live streamed to you, details in the next protocol (DNA sequencing)

