ABI Sanger Sequencing of Avian Clock genes to elucidate markers for Migration Phenology
Louis-Stéphane Le Clercq, Desiré Lee Dalton, Antoinette Kotzé, Paul Grobler
Abstract
This protocol follows up on "PCR Amplification of Clock genes with EmeraldAmp® GT PCR Master Mix in Avian species" and is intended to provide the next steps used in the sanger sequencing of the produced amplicons. This protocol uses ABI BigDye reagents (but suitable alternatives exist). The same primers used to produce the PCR products, of the respective clock genes, are used individually in a forward and reverse sequencing reaction. Cycling conditions mimic those used for PCR. Sequencing reactions are purified and subsequently resolved on an ABI Genetic Analyzer. The sequence read data was used in a BLAST search and confirmed to be the genes and regions of interest for all tested species.
Before start
Thaw reagents On ice .* Wipe workspace with 10% volume Bleach, followed by 70% volume Ethanol, and ddH2O before (and after).
- UV the relevant laminar flow cabinets.
Attachments
Steps
Big Dye Master Mix setup
Prepare
*Sample information has been deposited to BioSample and associated to the BioProject (PRJNA737185) which used this protocol.
(An experiment template is included as an excel spreadsheet)
Prepare the following sequencing master mixes in duplicate, one for each primer (forward and reverse).
Master Mix:
| A | B | C | D |
|---|---|---|---|
| BigDye™ 3.1 Ready MM | X2.5 | X1 | 4 |
| Primer | 3.2 µM | 3.2 pM | 1 |
| Nuclease free water | - | - | 4 |
Components of sequencing reaction, indicating stock and final concentrations as well as the relative volume needed in microliters. (for a 20μL reaction you can double the volume of each component)

Add 9µL to 1µL to the individual wells of a 96-well PCR plate or thin-walled PCR tubes.
Cycle sequencing
Program and run the following cycle conditions on a thermal cycler, e.g.
Equipment
| Value | Label |
|---|---|
| SimpliAmp Thermal Cycler | NAME |
| PCR | TYPE |
| Applied Biosystems | BRAND |
| A24811 | SKU |
| Any standard PCR thermocycler will suffice | SPECIFICATIONS |
- Initial denaturation at
96°Cfor0h 1m 0s - 25 cycles of:
- Denaturation at
96°Cfor0h 0m 10s - Annealing at
50°Cfor0h 0m 5s - Extension at
60°Cfor0h 2m 0s
- Hold at
4°Cuntil next step.
Sequence reaction clean-up
Purify the sequencing products using
Vortex the bottle of BigDye XTerminator™ beads for 8 to 10 seconds before mixing with the SAM solution.
Transfer the indicated volume of bead mix (BigDye XTerminator™ bead solution and SAM solution) to each.
Vortex the 96-well plate/tubes at 1800rpm on a shaker, e.g.
Equipment
| Value | Label |
|---|---|
| IKA MS 3 Digital Vortex Mixer | NAME |
| Vortex mixer | TYPE |
| IKA | BRAND |
| 3319000 | SKU |
| Vortex mixing of plates | SPECIFICATIONS |
In a swinging bucket centrifuge, centrifuge the plate at 1000x g,21°C .
Capillary electrophoresis & Data capture
Load sequencing reactions to sequencing plate and set up a run on the genetic analyzer, e.g.

Equipment
| Value | Label |
|---|---|
| 3500 Genetic Analyzer | NAME |
| Sequence analyzer | TYPE |
| Applied Biosystems | BRAND |
| 4440470 | SKU |
| DNA sequence fragment analysis | SPECIFICATIONS |
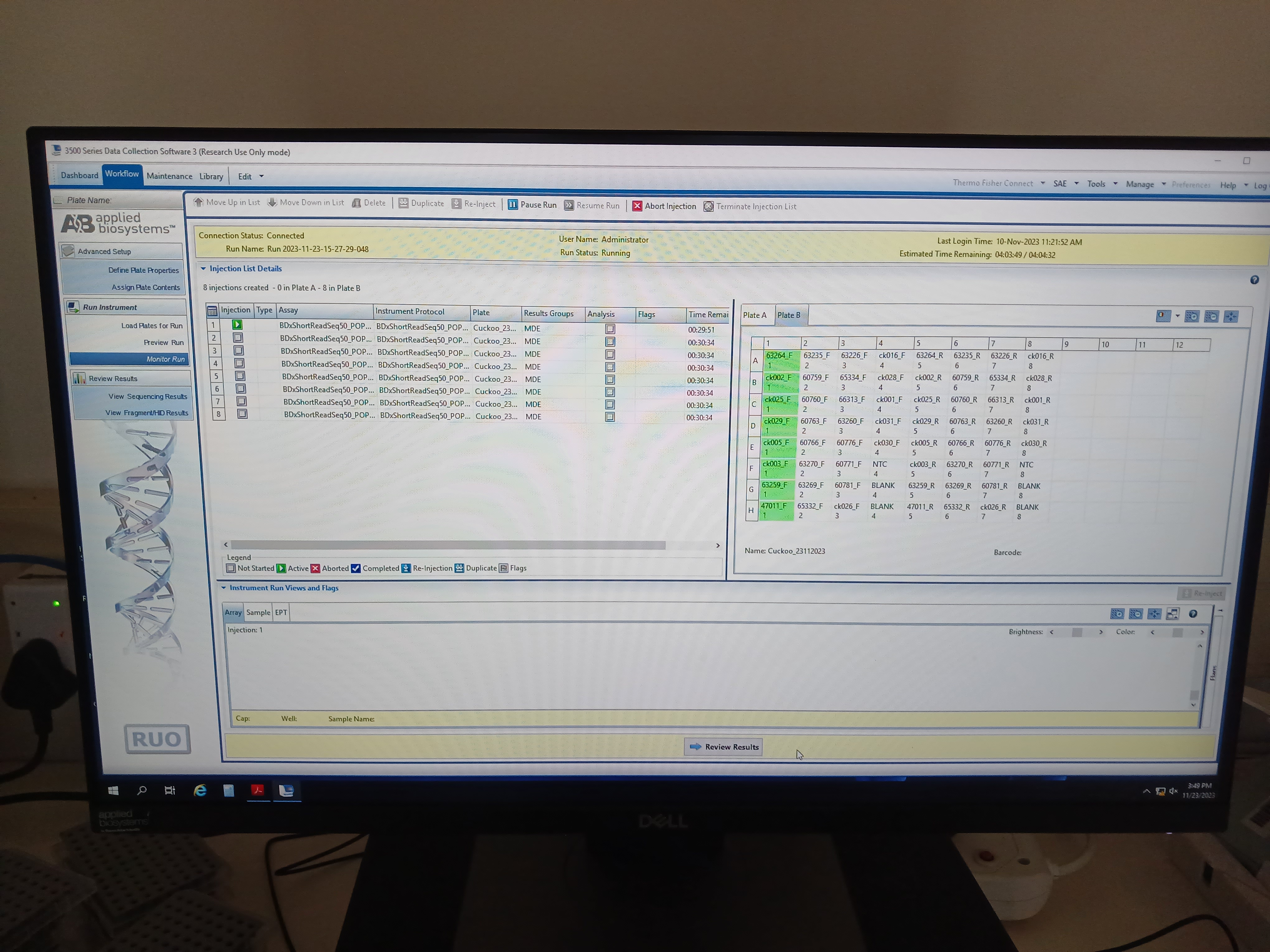
- Export sequence read trace files once done!
- Files can be opened with
Software
| Value | Label |
|---|---|
| BioEdit | NAME |
| Windows 10 | OS_NAME |
| 32-bit | OS_VERSION |
| Informer | REPOSITORY |
| Tom Hall | DEVELOPER |
| https://bioedit.software.informer.com/7.2/ | LINK |
| 7.2.6.1 | VERSION |
or
Software
| Value | Label |
|---|---|
| MEGA | NAME |
| https://www.megasoftware.net | LINK |
| X | VERSION |
or
Software
| Value | Label |
|---|---|
| Sequence Scanner | NAME |
| Windows 10 | OS_NAME |
| 32-bit | OS_VERSION |
| Informer | REPOSITORY |
| Life Technologies | DEVELOPER |
| https://sequence-scanner-software.software.informer.com/2.0/ | LINK |
| 2 | VERSION |



