mOTUs: Profiling Taxonomic Composition, Transcriptional Activity and Strain Populations of Microbial Communities
Hans-Joachim Ruscheweyh, Hans-Joachim Ruscheweyh, Alessio Milanese, Alessio Milanese, Lucas Paoli, Lucas Paoli, Anna Sintsova, Anna Sintsova, Daniel R. Mende, Daniel R. Mende, Georg Zeller, Georg Zeller, Shinichi Sunagawa, Shinichi Sunagawa
Abstract
The mOTU profiler, or mOTUs for short, is a software tool that enables the profiling of microbial communities in terms of their taxonomic composition, relative abundance of metabolically active members, and diversity of strain populations. To this end, it maintains a database of single-copy phylogenetic marker gene sequences, which are used as a reference to which short read metagenomic and metatranscriptomic reads are mapped for the identification and quantification of microbial taxa. Here, we describe the most common use cases of the mOTU profiler in two basic protocols. Additional supporting protocols provide information on its installation and in-depth guidance on adjusting its settings for increasing or decreasing the stringency with which taxa are detected and quantified, as well as for customizing the output file format. Guidelines for understanding the profiling results are provided, along with additional information on unique features, methodological details, and the development history of the tool. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
[Correction added on May 17, 2022, after first online publication: CSAL funding statement has been added.]
Basic Protocol 1 : Metagenomic and metatranscriptomic mOTU profiling
Basic Protocol 2 : Metagenomic SNV profiling
Support Protocol 1 : Installing mOTUs
Support Protocol 2 : Profiling pipeline–step by step
Support Protocol 3 : The mOTUs profiling routine using advanced parameters
Support Protocol 4 : Metagenomic SNV calling: advanced parameters
INTRODUCTION
Microorganisms and the complex communities they form have an impact on the health status of humans, animals, and plants. They also drive the global cycling of energy, nutrients, and biomass on Earth. To better understand the intricate interactions between microbial communities and their hosts, as well as their role in driving and maintaining the biogeochemical homeostasis on our planet, it is critical to study the composition of microbial communities across diverse environments.
To this end, we developed mOTUs: a software tool and database for profiling taxonomic composition, transcriptional activity, and strain populations of species in complex microbial community samples using shotgun sequencing data. These can either be the result of community DNA (metagenomic) or RNA/cDNA (metatranscriptomic) sequencing (Milanese et al., 2019). The mOTUs approach is based on a sequence database of single-copy phylogenetic marker genes (MGs) that are present in all organisms (Ciccarelli et al., 2006; Sorek et al., 2007). Clustering of these MGs by sequence similarity can be used to accurately delineate prokaryotic species (Mende, Sunagawa, Zeller, & Bork, 2013). Species-level clusters of MGs that were identified in both sequenced reference genomes and in metagenomes can thus be used as operational taxonomic units (mOTUs). These enable the identification and quantification of both “known” and “unknown” species, i.e., species for which no genome sequence is available yet, in metagenomic data sets (Sunagawa et al., 2013).
In most use cases, metagenomic data are used as an input to obtain a species-level taxonomic abundance profile as an output. In addition, mOTUs can be applied to metatranscriptomic data. Since most of the MGs represent ‘housekeeping’ genes, that is, genes that are constitutively expressed and show low expression variation across physiological/environmental conditions, the resulting profiles are highly correlated with taxonomic abundance profiles. Metatranscriptomic mOTU profiles can thus be used to estimate the relative abundances of the metabolically active fraction of microbial communities (Milanese et al., 2019). Finally, due to the phylogenetically conserved nature of the MGs, they can also be used as reference sequences for single nucleotide variation (SNV) analyses. This approach represents a computationally efficient alternative to generating SNV profiles based on whole genome sequences (Milanese et al., 2019). The obtained data allows researchers to study, for example, similarities between strain populations across samples.
Here, we describe two basic protocols for the main routines of mOTUs, namely metagenomic and metatranscriptomic community profiling (Basic Protocol 1) and metagenomic SNV profiling (Basic Protocol 2). We furthermore provide four support protocols. Support Protocol 1 describes how to install mOTUs. Support Protocols 2 and 3 explain how to customize the individual steps of the profiling pipeline, introducing a rich set of options to, e.g., change sensitivity or report results in different community-accepted formats. Support Protocol 4 discusses the impact of different parameter settings on the metagenomic SNV profiling function.
STRATEGIC PLANNING
Hardware/Software
The mOTU profiler is a command-line tool that requires a 64-bit Linux or MacOS system with at least 16 GB of RAM. Installation instructions are provided in Support Protocol 1. The runtime of the tool scales almost linearly with the number of input sequences. For example, ∼250,000 reads/min are processed by mOTUs with a 2.8-GHz processor. Parallelization on up to 16 compute cores improves runtime almost linearly; however, the use of additional cores tends to yield sublinear speed-ups. It is possible to use mOTUs on a personal desktop computer. To analyze large volumes of sequencing data, its use in a high-performance computing environment is recommended.
Input Files/Format
The mOTU profiler operates on metagenomic and metatranscriptomic sequencing data (tested on reads varying in length between 45 and 300 bp; also see Support Protocol 3) provided as quality-controlled (see below), FASTQ-formatted sequencing read files (compressed or uncompressed). The tool can be used to profile individual or multiple files from either paired-end and/or single-end libraries. Multiple input files can be freely combined and will result in one profile per execution. Profiles from multiple runs, e.g., from several samples, can be merged into a single table.
Paired-End Sequencing Reads
The most common input for mOTUs are paired-end (e.g., Illumina) sequencing read files. In each sequencing run, usually two compressed (gzip or bgzip) FASTQ-formatted, quality-controlled (see below) read files (referred to as *1.fq.gz and *2.fq.gz) are generated. Paired-end reads are derived from sequencing a DNA fragment (insert) from both ends. The reads in *1.fq.gz correspond to the insert sequence from one end in forward orientation, and the reads in *2.fq.gz correspond to the insert sequence from the other end and opposite strand in reverse orientation. In this case, the number of reads in both files is the same and the insert association is denoted in the header line of each read.
It is possible to process paired-end sequencing read files from multiple runs at once if the following rules are followed:
- The counting algorithm of mOTUs is based on inserts. Therefore, the names of reads in *1.fq.gz are required to match the names of reads in the *2.fq.gz file;
- The mOTU profiler can profile multiple paired-end runs from the same sample at a time if matching paired-end files are provided in matching order for the respective optional arguments of the mOTUs command;
- Files with interleaved reads (forward and reverse reads in the same file) are not supported. Interleaved read files should be separated, e.g., with BBMap (reformat.sh sample.in=interleaved.fq.gz out1=sample.1.fq.gz out2=sample.2.fq.gz), before using mOTUs;
- In the case of overlapping paired-end reads that were merged into a single read, the resulting files have to be provided as unpaired sequencing reads as an input to mOTUs (see below).
Unpaired Sequencing Reads
Users can also submit unpaired read files in which each read is counted as an independent event for taxonomic profiling. These reads are generated in single-end sequencing mode, were merged prior to processing, or are singletons that lost their paired read during quality control. If multiple read files are provided as input, they will be processed as one sample and result in a single output profile.
Other Formats
Library layouts other than the paired-end and single-end sequencing are currently not supported, nor is long-read sequencing. Sequencing read files in a different format than FASTQ should be translated, e.g.:
- BBMap can be used to translate FASTA-formatted read files to FASTQ-formatted read files: reformat.sh in=sample.1.fasta out=sample.1.fastq
- BedTools can be used to translate SAM/BAM-formatted read files to FASTQ-formatted read files: bamToFastq -i sample.1.bam -fq sample.1.fastq
Quality Control of Input Data
Quality control of input sequencing data is recommended for taxonomic profiling. This typically comprises multiple steps, such as the removal of contamination (e.g., PhiX), trimming of artificial sequences (adapter and barcode sequences), and removal of low-quality bases. The last step has the highest impact on the performance of mOTUs, as it influences the quality of the alignment when comparing reads against the database (e.g., by discarding longer reads with a low quality 3′-end). As a guideline, we recommend that all bases have a Phred quality score of at least 20 and that sequencing reads be at least 75 bases in length. If the length criteria cannot be met, please refer to Support Protocol 3 to adjust settings.
Basic Protocol 1: METAGENOMIC AND METATRANSCRIPTOMIC mOTU PROFILING
This protocol describes the basic profiling routine of mOTUs for metagenomic and metatranscriptomic short-read (e.g., Illumina) sequencing data with default settings. For metagenomic data, the output is a profile of microbial community members and their relative abundances. By default, the microbial community compositions are summarized into abundances of mOTUs, i.e., phylogenetic marker gene-based operational taxonomic units at species-level resolution (Mende et al., 2013; Sunagawa et al., 2013). For metatranscriptomic data, the output follows the same format, but reflects the relative taxonomic composition of transcriptionally active community members.
Necessary Resources
Hardware/Software
- The mOTU profiler is a command-line tool that requires a 64-bit Linux or MacOS system with at least 16 GB of RAM and its runtime scales almost linearly with the number of input sequences (see Strategic Planning). Installation instructions are provided in Support Protocol 1.
Input Files
- Paired-end or single-end short-read sequencing data (see Strategic Planning for more details) in FASTQ format. For demonstration purposes we have deposited sequencing files on Zenodo (https://zenodo.org/record/5012695).
Output Files
- The output of mOTUs is a two-column, tab-separated table. The first column contains taxonomic identifiers, by default at the rank of mOTUs (i.e., species level), and the values in the second column correspond to their relative abundances in the profiled sample. The profile can also be generated at other taxonomic ranks (see Support Protocol 3). Profiles from multiple samples can be merged into a single file with taxonomic identifiers in the first column and abundances of the profiled samples in the following columns.
1.Install mOTUs as described in Support Protocol 1.
2.Create a work folder and download the sequencing reads from Zenodo:
- mkdir basic_protocol_1
- cd basic_protocol_1
- wget https://zenodo.org/record/5012695/files/protocol_profile.zip
- unzip protocol_profile.zip
- cd protocol_profile
3.Generate a mOTUs profile with paired-end sequencing read files from a single sample:
-
motus profile -f input/ERR479298s.1.fq.gz -r
-
input/ERR479298s.2.fq.gz -n ERR479298s -o ERR479298s-default.motus
4.The output of mOTUs is a two-column, tab-separated table. The first column lists the identifiers of all mOTUs and the second column shows the relative abundance of each mOTU in the profiled sample. For example, meta_mOTU_v3_12429 has a relative abundance of 16%:
- grep "meta_mOTU_v3_12429" ERR479298s-default.motus
- Prevotella species incertae sedis [meta_mOTU_v3_12429] 0.1597971398
5.Individual profiles can be merged into a single file to facilitate comparative downstream analyses using the motus merge command. Here, we merge the profile created in the previous step with a precomputed profile:
- motus merge -i ERR479298s-default.motus,bp1_precomputed/ERR479045- default.motus -o merged.motus
6.The format of the merged profile is identical to the single sample profile with additional columns for each sample added:
- grep "meta_mOTU_v3_12429" merged.motus
- Prevotella species incertae sedis [meta_mOTU_v3_12429] 0.1597971398 0.0000000000
7.The profiling and merging routines have finished successfully if and when the output file (e.g., ERR479298s-default.motus or merged.motus) is created and the tool reports that no error has occurred. A guideline on how to interpret the profiling results can be found in Guidelines for Understanding Results.
Basic Protocol 2: METAGENOMIC SNV PROFILING
The mOTU profiler can be used to generate metagenomic single nucleotide variation (SNV) profiles of the mOTUs marker genes and provides a computationally efficient alternative to generating SNV profiles from whole-genome sequences. These profiles are subsequently used to produce between-sample distances for individual mOTUs, providing access to strain population diversity patterns for as many as 33,750 species. To this end, mOTUs will first align metagenomic sequencing reads against its marker gene database and then process the alignments using the metaSNV package (Costea et al., 2017).
Necessary Resources
Hardware/Software
- The mOTU profiler is a command-line tool that requires a 64-bit Linux or MacOS system with at least 16 GB of RAM and its runtime scales almost linearly with the number of input sequences (see Strategic Planning). Installation instructions are provided in Support Protocol 1.
Input Files
- Metagenomic SNV profiling is a pipeline split into two parts:
- motus map_snv requires paired-end or single-end short read sequencing data (see Strategic Planning for more details) in FASTQ format
- The BAM alignment files produced by motus map_snv serve as input for the motus snv_call routine
- For demonstration purposes we have deposited sequencing and alignment files on Zenodo (https://zenodo.org/record/5012568)
Output Files
- The motus map_snv command will create one BAM file per execution
- The motus snv_call command will create an output folder with four files/directories:
- Two files containing the read coverage information for each mOTU, both horizontal, i.e., the percentage of bases covered by at least one read (.perc.tab file), and vertical, i.e., the average number of reads covering each base (.cov.tab file)
- Per mOTU filtered allele frequencies of identified SNVs across samples (filtered-* directory)
- Per mOTU genetic distances between samples (distances-* directory)
- For the files containing distance information, both regular Manhattan (*mann.dist files) and dominant allele (*allele.dist files) distances are provided. The dominant allele distance is a Manhattan distance that only takes positions with a frequency change above 50% (i.e., change of the dominant allele) into account.
The SNV profiling routine is split into two steps (motus map_snv and motus snv_call) that need to be executed sequentially. The first step aligns sequencing reads of a single sample against a reduced mOTUs centroid database. The second step uses alignments from multiple samples to calculate metagenomic SNV profiles and distances using metaSNV.
1.Install mOTUs as described in Support Protocol 1.
2.Create a work folder and download the sequencing reads from Zenodo:
- mkdir basic_protocol_2
- cd basic_protocol_2
- wget https://zenodo.org/record/5012568/files/protocol_snv.zip
- unzip protocol_snv.zip
- cd protocol_snv
3.Execute the motus map_snv command to align reads from a paired-end sequencing sample against the mOTUs centroid database:
- motus map_snv -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-default.bam
4.The motus snv_call command takes a folder containing BAM files as input. Execute the following command to run motus snv_call:
- motus snv_call -d bp2_precomputed/bams/ -o motus-snvcall-default
5.Execute the following command to inspect the marker gene-based distances between metagenomic SNV profiles, which can be found in the distances-m2-d5-b80-c5-p0.9 folder:
- cat motus-snvcall-default/distances-m2-d5-b80-c5-
- p0.9/meta_mOTU_v3_12235.filtered.mann.dist
- SAMEA2466942-default.bam SAMEA2467017-default.bam
| SAMEA2466942-default.bam | 0.0 | 0.6728930543446672 |
| SAMEA2467017-default.bam | 0.6728930543446672 | 0.0 |
6.The motus map_snv step has finished successfully when the output alignment file is created (e.g., ERR479298-default.bam). The motus snv_call step has finished successfully when all result files are present and the tool reports “Successfully finished” in the command line. A guideline on how to interpret the SNV results can be found in Guidelines for Understanding Results.
Support Protocol 1: INSTALLING THE mOTU PROFILER
The mOTU profiler is written in Python 3 and can be executed on a 64-bit Linux or MacOS system. There are external dependencies that need to be pre-installed. This can be done either by manual installation or, preferably, by using the conda package manager.
Necessary Resources
Hardware
- As described in Strategic Planning
Software
- BWA, to align sequencing reads against the mOTUs database. Tested with version BWA-0.7.17 (r1188); https://github.com/lh3/bwa.
- SAMtools, to read the alignment files produced by BWA. Tested with version 1.6 or higher; https://github.com/samtools/samtools.
- Python as the runtime for the mOTU profiler. Tested with version 3.6.5 or higher; https://www.python.org/downloads/.
- Optional : metaSNV, for SNV calling and processing. Tested with version 1.0.3; http://metasnv.embl.de/. The metaSNV package is only needed for the snv_call command (Basic Protocol 2 and Support Protocol 4).
- Optional : miniconda, for installation using conda; https://docs.conda.io/en/latest/miniconda.html
Files
- The latest mOTUs package (tar.gz file); https://github.com/motu-tool/mOTUs
Option A: Installation using the Conda Package Manager
1a. The installation using the conda package manager is generally preferable, as it encapsulates the entire installation process into a single command once conda is installed. Execute the following command to install conda:
- curl -O https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
- sh Miniconda3-latest-Linux-x86_64.sh
- conda config --add channels defaults
- conda config --add channels bioconda
- conda config --add channels conda-forge
2a. Install the latest version of mOTUs using conda:
- conda install -c bioconda motus=3.0.0
Option B: Manual installation
1a. Both mandatory dependencies (BWA and SAMtools) can be downloaded from their respective GitHub repositories. Up-to-date installation instructions are provided by the developers. The metaSNV source code can be downloaded from http://metasnv.embl.de/ along with installation instructions. Python (version 3.6.5 or higher) should be installed following the instructions on the official website (https://www.python.org/downloads/).
2a. The manual installation consists of three steps. In step 1, the release files are downloaded from the official GitHub repository. The installation script in step 2 will download the database. The test routine in step 3 checks whether dependencies are installed correctly and whether profiling a test dataset yields the expected results. Execute the following commands to download and install mOTUs:
-
wget https://github.com/motu-tool/mOTUs/archive/refs/tags/3.0.0.tar.gz
-
tar -xzvf 3.0.0.tar.gz
-
cd mOTUs-3.0.0
-
python setup.py
-
python test.py
-
echo export PATH=
pwd:$PATH >> ∼/.bashrc -
source ∼/.bashrc
Support Protocol 2: PROFILING PIPELINE: STEP BY STEP
The motus profile routine is an analysis pipeline that consists of three steps, namely map_tax, calc_mgc, and calc_motu, which are internally executed in that order. Each step produces the output files that are needed for the next step, and therefore it can be useful to run the commands individually, e.g., to test different parameter settings, without the need to run the entire pipeline. This is especially useful, as the first step (map_tax) accounts for most of the CPU time needed to calculate profiles.
Necessary Resources
Hardware/Software
- The mOTU profiler is a command-line tool that requires a 64-bit Linux or MacOS system with at least 16 GB of RAM and its runtime scales almost linearly with the number of input sequences (see Strategic Planning). Installation instructions are provided in Support Protocol 1.
Input Files
- Paired-end or single-end short-read sequencing data (see Strategic Planning for more details) in FASTQ format. For demonstration purposes we have deposited sequencing files on Zenodo (https://zenodo.org/record/5012695).
Output Files
- The output of mOTUs is a two-column, tab-separated table. The first column contains taxonomic identifiers, by default at the rank of mOTUs (i.e., species level), and the values in the second column correspond to their relative abundances in the profiled sample. The profile can also be generated at other taxonomic ranks (see Support Protocol 3). Profiles from multiple samples can be merged into a single file with taxonomic identifiers in the first column and abundances of the profiled samples in the following columns.
- The intermediate output files produced by mOTUs contain:
- alignments of reads against the mOTUs database in BAM format
- marker gene abundances in a two-column, tab-separated table
1.Install mOTUs as described in Support Protocol 1.
2.Create a work folder and download the sequencing reads from Zenodo:
- mkdir support_protocol_2
- cd support_protocol_2
- wget https://zenodo.org/record/5012695/files/protocol_profile.zip
- unzip protocol_profile.zip
- cd protocol_profile
3.Execute the following command to align paired-end sequencing reads against the mOTUs database and store alignments in a BAM file:
- motus map_tax -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-default.bam
4.Parse the alignments in the BAM file and produce an intermediate file that assigns each insert to its marker gene cluster:
- motus calc_mgc -i ERR479298s-default.bam -n ERR479298s -o ERR479298s-default.mgc
5.The output of the calc_mgc step is a two-column, tab-separated table with the first column representing the name of the marker gene cluster and the second column containing the insert count (see Background Information and Fig. 1):
- head -n 10 ERR479298s-default.mgc
-
map_tax 3.0.0 | gene database: nr3.0.0 | 84 | calc_mgc 3.0.0 -y insert.scaled_counts -l 75
| ERR479298s | |
| COG0495.mOTU.v25.0000273 | 476.9980996151712 |
| COG0552.mOTU.v2.0001907 | 52.97275139363133 |
| COG0533.mOTU.v2.0001890 | 312.27285022688824 |
| COG0525.mOTU.v25.0000325 | 363.7120130887338 |
| COG0215.ext_mOTU_v3_003085 | 48.85735226138667 |
| COG0172.mOTU.v2.0002099 | 101.54979310332041 |
| COG0525.mOTU.v2.0001728 | 3.4031882043690063 |
| COG0525.mOTU.v25.0000424 | 2.255959601538731 |
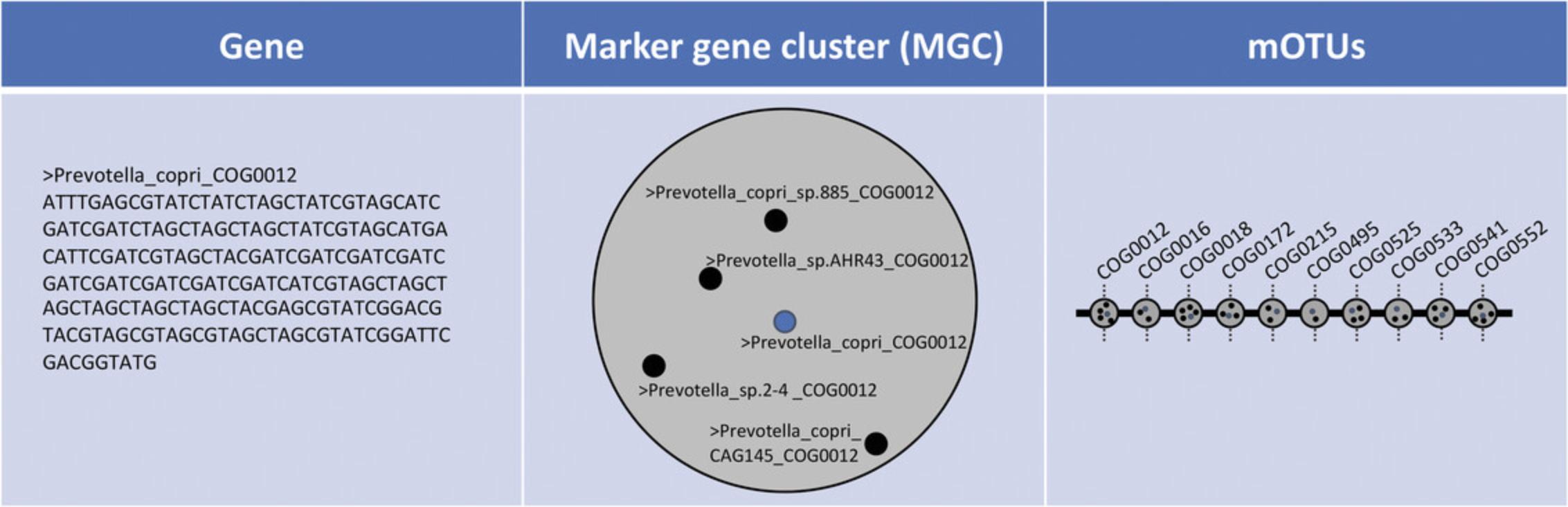
6.To calculate the abundance of each mOTU, execute the motus calc_motu command, which generates the mOTUs output file:
- motus calc_motu -i ERR479298s-default.mgc -n ERR479298s -o ERR479298s-default.motus
7.The output of mOTUs is a two-column, tab-separated table. The first column lists the identifiers of all mOTUs and the second column shows the relative abundance of each mOTU in the profiled sample. For example, meta_mOTU_v3_12429 has a relative abundance of 16%:
- grep "meta_mOTU_v3_12429" ERR479298s-default.motus
- Prevotella species incertae sedis [meta_mOTU_v3_12429] 0.1597971398
8.The routines finished successfully if and when the output file (e.g., ERR479298s-default.motus) is created and the tool reports that no error has occurred. A guideline on how to interpret the profiling results can be found in Guidelines for Understanding Results.
Support Protocol 3: THE mOTUs PROFILING ROUTINE USING ADVANCED PARAMETERS
Basic Protocol 1 and Support Protocol 2 illustrate the core functionality of the profiling routine using default settings for the most common metagenomic short read sequencing approaches. These settings were chosen to provide a balanced tradeoff between sensitivity and specificity. However, mOTUs offers flexibility in changing these settings according to user preference. This section describes all available options and their effects on the resulting profiles. It also introduces how profiles can be exported in BIOM format, and how results can be aggregated at different taxonomic ranks. The last section of this protocol shows how user-generated profiles can be merged with over 11,000 precomputed profiles from publicly available metagenomes and metatranscriptomes.
Necessary Resources
Hardware/Software
- The mOTU profiler is a command-line tool that requires a 64-bit Linux or MacOS system with at least 16 GB of RAM and its runtime scales almost linearly with the number of input sequences (see Strategic Planning). Installation instructions are provided in Support Protocol 1.
Input Files
- Paired-end or single-end short-read sequencing data (see “Strategic Planning” for more details) in FASTQ format. For demonstration purposes we have deposited sequencing files on Zenodo (https://zenodo.org/record/5012695).
Output Files
- The output of mOTUs is a two-column, tab-separated table. The first column contains taxonomic identifiers, by default at the rank of mOTUs (i.e., species level), and the values in the second column correspond to their relative abundances in the profiled sample. The profile can also be generated at other taxonomic ranks (see Support Protocol 3). Profiles from multiple samples can be merged into a single file with taxonomic identifiers in the first column and abundances of the profiled samples in the following columns.
- Alternative output formats supported by mOTUs are Bioboxes (http://bioboxes.org/) and BIOM (https://biom-format.org/).
- The intermediate output files produced by mOTUs contain:
- alignments of reads against the mOTUs database in BAM format
- marker gene abundances in a two-column, tab-separated table
The optional arguments of the motus profile command modify its runtime, the way sequencing reads are filtered and counted, and the format of the reported output.
NOTE : This protocol describes how to set optional arguments to change the default behavior of mOTUs. Each analysis step in this protocol is therefore self-contained and does not rely on output data of any other analysis step.
1.Install mOTUs as described in Support Protocol 1.
2.Create a work folder and download the sequencing reads from Zenodo:
- mkdir support_protocol_3
- cd support_protocol_3
- wget https://zenodo.org/record/5012695/files/protocol_profile.zip
- unzip protocol_profile.zip
- cd protocol_profile
3.Runtime : Most of the runtime of the motus profile command is spent computing alignments of input sequences against the marker gene database using the BWA aligner. Using multiple cores for the alignment step will lead to a near-linear decrease in runtime if the number of cores is lower than 16.Further increasing the number of cores will still improve runtime but at a lower rate. The number of cores used by mOTUs for the alignment step can be adjusted with the -t option. Here we run mOTUs with 1, 8, or 16 cores, which leads to a decrease in runtime (1600 s, 300 s, 191 s):
- time motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-t_1.motus -t 1
- time motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-t_8.motus -t 8
- time motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-t_16.motus -t 16
4.Length filtering : The length and similarity of a sequencing read aligned to a marker gene reference sequence determine whether the read is considered for the subsequent counting step. The default minimum length cutoff is set to 75 bases. If a substantial fraction of the sequencing reads is shorter than 75 bases, the -l option can be used to reduce the minimum alignment length to increase the fraction of mapped reads. Here we set the -l argument to a minimum alignment length of 50:
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-l_50.motus -l 50
5.Presence/absence : The mOTU profiler provides an option (-g) to adjust the precision and recall of species identification. Each mOTU consists of a minimum of 6 and up to 10 marker gene clusters (MGCs, see Fig. 1). To avoid the spurious detection of taxa, the number of detected MGCs can be used to adjust the confidence level. The -g option in motus profile (and in motus calc_motu) controls the minimum number of detected MGCs (to which reads are mapping) required to call a mOTU as being present in a sample. The default value is set to 3, representing half the minimum number of MGCs per mOTU. Execute the following commands to profile with different values for the -g option:
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-g_3.motus -g 3
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-g_1.motus -g 1
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-g_6.motus -g 6
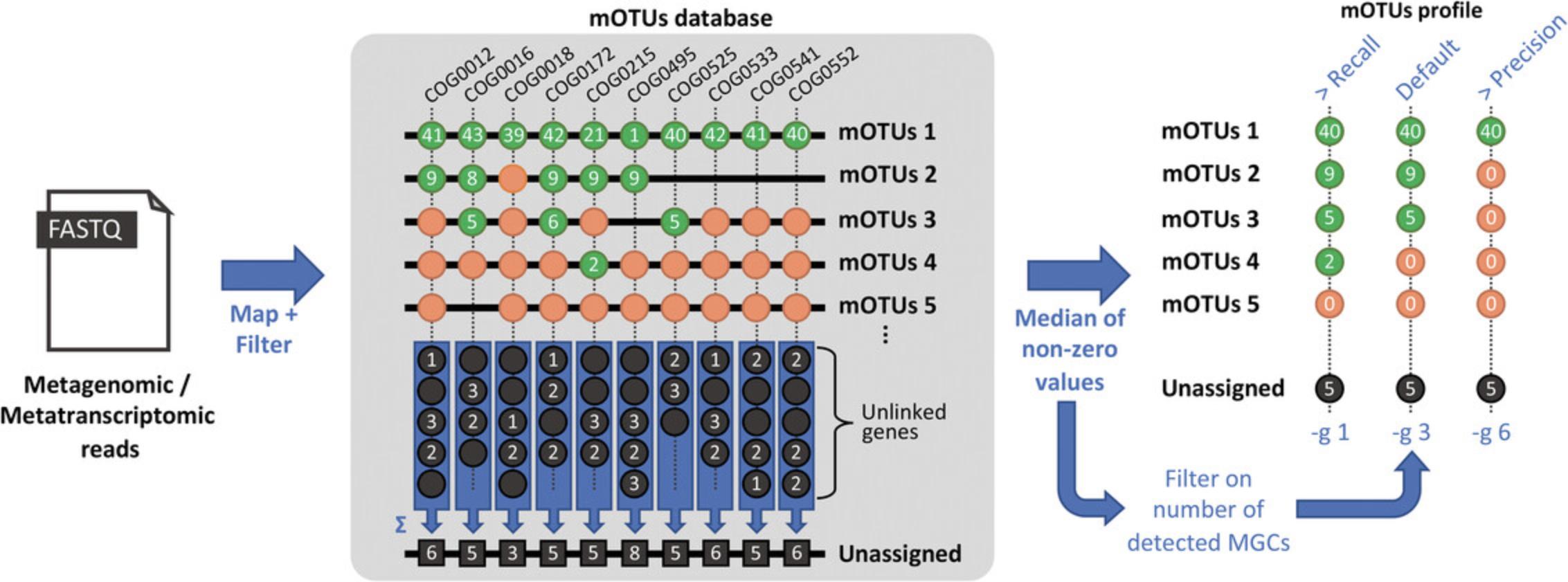
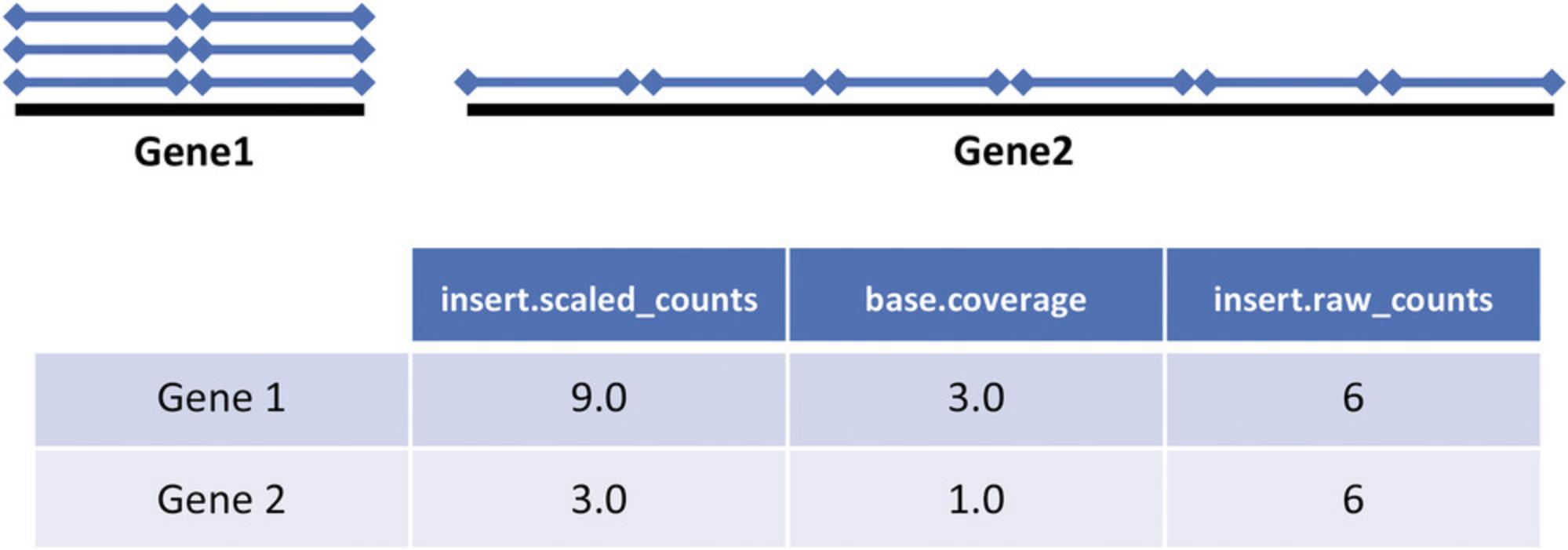
6.Quantification : There are three modes to quantify the abundance of mOTUs, namely base.coverage, insert.raw_counts, and insert.scaled_counts (Fig. 3). These alternatives can be selected using the -y option in the motus profile (and motus calc_mgc) command. The abundance of each mOTU is calculated as the median across all its MGCs with non-zero values. Execute the following commands to profile with different values for the -y option.
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-y_insert.raw_counts.motus -y insert.raw_counts
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-y_base.coverage.motus -y base.coverage
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-y_insert.scaled_counts.motus -y insert.scaled_counts
7.Output formats : The command motus profile creates by default a tab-separated file in which the columns correspond to the taxonomic annotation of each mOTU and its abundance. The mOTU profiler also supports the BIOM (https://biom-format.org/) output format, which can be invoked using the flag -B. The option -C can be used to generate an output file compliant with the CAMI/Bioboxes (http://bioboxes.org/) format. Execute the following commands to generate profiles with either BIOM or Bioboxes format:
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-B.biom -B
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-C_precision.cami -C precision
8.Taxonomy : The mOTU profiler uses the standard taxonomic hierarchy with seven ranks (kingdom, phylum, class, order, family, genus, and mOTU), where the typical species level is substituted by the equivalent mOTU level, which is reported by default. The flags -p and -q will add the NCBI taxonomy ID to the profile or report the full taxonomy, respectively. The profiles can be further aggregated at a different taxonomic level using the -k option. Execute the following command to add the associated NCBI taxonomy ID (here: 1262806) as a new column to the profile:
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-p.motus -p
- grep "meta_mOTU_v3_12886" ERR479298s-p.motus
- Clostridium sp. CAG:433 [meta_mOTU_v3_12886] 1262806 0.0184576392
Execute the following command to report the full taxonomy path:
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-q.motus -q
- grep "meta_mOTU_v3_12886" ERR479298s-q.motus
- meta_mOTU_v3_12886 k__Bacteria|p__Firmicutes|c__Clostridia|o__Clostridiales|f__Clostridiaceae|g__Clostridium|s__Clostridium sp. CAG:433 0.0184576392
Execute the following command to aggregate the taxonomic path at a different taxonomic level (here: phylum):
- motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-k_phylum.motus -k phylum
- grep "Actinobacteria" ERR479298s-k_phylum.motus
- Actinobacteria 0.0079714597
- ...
9.Counts/relative abundance : motus profile reports, by default, relative abundances of each mOTU. The relative abundances are directly calculated from the counts (bases or inserts; see Quantification). The output format can be modified using the -c flag. Execute the following commands to either report relative abundances (default) or to report counts (-c) instead:
-
motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-default.motus
-
grep "meta_mOTU_v3_12886" ERR479298s-default.motus
-
Clostridium sp. CAG:433 [meta_mOTU_v3_12886] 0.0184576392
-
motus profile -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-c.motus -c
-
grep "meta_mOTU_v3_12886" ERR479298s-c.motus
-
Clostridium sp. CAG:433 [meta_mOTU_v3_12886] 40
10.Merge profiles : Users who generated profiles in the default mOTUs output format and using the -c flag (reporting counts instead of relative abundances, see above) can add their profiles to precomputed profiles to contextualize and compare the taxonomic profiles of their samples with previously published ones. For the release of version 3.0.0 of mOTUs, we provided precomputed profiles from more than 11,000 public metagenomes and metatranscriptomes that are directly accessible via the motus merge command. Execute the following commands to merge the precomputed profile with either all mouse gut profiles, all mouse and dog gut profiles, or all public profiles, respectively.
- motus merge -i sp3_precomputed/ERR479298s-c.motus -a mouse -o merged.mouse.motus
- motus merge -i sp3_precomputed/ERR479298s-c.motus -a mouse,dog -o merged.mouse.dog.motus
- motus merge -i sp3_precomputed/ERR479298s-c.motus -a all -o merged.all.motus
Support Protocol 4: METAGENOMIC SNV CALLING: ADVANCED PARAMETERS
Basic Protocol 2 explains the core functionality of the metagenomic SNV profiling mode using default settings. The preconfigured settings filter both metagenomic samples and genomic positions to offer a good tradeoff between sensitivity and specificity. However, some use cases may require custom settings instead. This section provides an in-depth explanation of all options and describes their effects to the resulting profiles.
Necessary Resources
Hardware/Software
- The mOTU profiler is a command-line tool that requires a 64-bit Linux or MacOS system with at least 16 GB of RAM and its runtime scales almost linearly with the number of input sequences (see Strategic Planning). Installation instructions are provided in Support Protocol 1.
Input Files
- Metagenomic SNV profiling is a pipeline split into two parts:
- motus map_snv requires paired-end or single-end short read sequencing data (see Strategic Planning for more details) in FASTQ format
- The BAM alignment files produced by motus map_snv serve as input for the motus snv_call routine
- For demonstration purposes we have deposited sequencing and alignment files on Zenodo (https://zenodo.org/record/5012568)
Output Files
- The motus map_snv command will create one BAM file per execution
- The motus snv_call command will create an output folder with four files/directories:
- Two files containing the read coverage information for each mOTU, both horizontal, i.e., the percentage of bases covered by at least one read (.perc.tab file), and vertical, i.e., the average number of reads covering each base (.cov.tab file)
- Per mOTU filtered allele frequencies of identified SNVs across samples (filtered-* directory)
- Per mOTU genetic distances between samples (distances-* directory).
- For the files containing distance information, both regular Manhattan (*mann.dist files) and dominant allele (*allele.dist files) distances are provided. The dominant allele distance is a Manhattan distance that only takes positions with a frequency change above 50% (i.e., change of the dominant allele) into account.
The SNV profiling routine is composed of two steps: motus map_snv first aligns sequencing reads of a single sample against the reduced mOTUs centroid database, and motus snv_call subsequently processes alignments from multiple samples to calculate metagenomic SNV profiles and genetic distances. Details are described in Basic Protocol 2.
NOTE : This protocol describes how to set optional arguments to change the default behavior of mOTUs. Each analysis step in this protocol is therefore self-contained and does not rely on output data of any other analysis step.
1.Install mOTUs as described in Support Protocol 1.
2.Create a work folder and download the sequencing reads from Zenodo:
- mkdir support_protocol_4
- cd support_protocol_4
- wget https://zenodo.org/record/5012568/files/protocol_snv.zip
- unzip protocol_snv.zip
- cd protocol_snv
3.Runtime : Most of the runtime of the SNV profiling routine is spent computing alignments of input sequences against the marker gene database using the BWA aligner. Using multiple cores for the alignment step will lead to a near-linear decrease in runtime (up to 16 cores, further increases will still improve runtime, but at a lower rate). Execute the following command to use 16 cores (-t 16) for alignment:
- motus map_snv -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-default.bam -t 16
4.Length filtering : The length and similarity of a sequencing read aligned to a marker gene reference sequence determine whether the read is considered for the subsequent counting step. The default minimum length cutoff is set to 75 bases. If a substantial fraction of the sequencing reads is shorter than 75 bases, the -l option can be used to reduce the minimum alignment length to increase the fraction of mapped reads. Execute the following command to set the -l option to a minimum alignment length of 50:
- motus map_snv -f input/ERR479298s.1.fq.gz -r input/ERR479298s.2.fq.gz -o ERR479298s-l_50.bam -l 50
5.Sample support : The first step of the motus snv_call routine will calculate the vertical (average number of reads covering each base) and horizontal (percentage of bases covered by at least one read) coverage of each mOTU for each sample and then select only those mOTUs that fulfil the coverage-filtering criteria. By default these thresholds are 5× vertical coverage and 80% horizontal coverage, and can be controlled by the -fd and -fb options, respectively. In addition, a mOTU will be selected only if at least a given number of samples meet the coverage criteria (2 samples by default). This number can be controlled with the -fm option. Execute the following commands to alter filtering criteria:
- motus snv_call -d bp2_precomputed/bams/ -o motus-snvcall-fd_10 -fd 10
- motus snv_call -d bp2_precomputed/bams/ -o motus-snvcall-fb_90 -fb 90
- motus snv_call -d bp2_precomputed/bams/ -o motus-snvcall-fm_3 -fm 3
6.Base coverage : The previous filtering step selected mOTUs and filtered samples based on coverage values across whole mOTUs. This second filtering step will retain genomic positions for SNV calling within an individual mOTU. To that end, only positions in a mOTU that have enough coverage in at least a given proportion of filtered samples are retained. The base coverage threshold is controlled with the -fc option, and is set to a minimum of 5× coverage. The proportion of samples is controlled with the -fp option and is set to a minimum of 90% of the samples by default. Execute the following commands to alter filtering criteria:
- motus snv_call -d bp2_precomputed/bams/ -o motus-snvcall-fc_10 -fc 10
- motus snv_call -d bp2_precomputed/bams/ -o motus-snvcall-fp_1.0 -fp 1.0
7.Keep intermediate files : The arguments -K and -v allow users to control the reporting of the SNV profiling routine. First, -K will provide the user with all the intermediary files created by metaSNV. Second, -v controls the verbosity of the motus snv_call command according to four standardized levels of specificity. Execute the following command to keep intermediate files and enable verbose logging:
- motus snv_call -d bp2_precomputed/bams/ -o motus-snvcall-k-v_4 -K -v 4
GUIDELINES FOR UNDERSTANDING RESULTS
Understanding the mOTUs profiling output
Taxonomic profiling aims to accurately identify and estimate the relative abundance of organisms in a microbial community sample. Two common approaches to achieve this goal using metagenomic sequencing reads are to match them either to (i) k-mer spectra of reference genome sequences, or (ii) universal (or clade-specific) marker genes (Sun et al., 2020). For the latter approach, only metagenomic read mappings to marker genes are required for taxonomic profiling. The mOTU profiler uses 10 such marker genes, which reduces the reference sequence space for species level profiling to ∼15,000 bases. The fraction of reads from a given sequencing run mapping to MGs is typically <5% (see https://github.com/motu-tool/mOTUs/wiki/FAQ for more details). This reduction in sequence space represents a sensitivity tradeoff (higher if whole genome sequences are used); however, marker-gene-based tools are computationally more efficient and generally achieve a higher level of precision (Meyer et al., 2021). Furthermore, universal MGs from assembled metagenomes that cannot be merged into existing mOTUs or do not meet the criteria for forming a new mOTU are included in the database. This inclusion allows for estimating the cumulative relative abundance of organisms that are present in the sample, but not represented by any mOTU in the database, which further reduces quantification biases, and thus increases the accuracy of taxonomic profiles (Milanese et al., 2019).
Understanding the metagenomic SNV profiling output
The mOTU profiler enables population-level analysis of microbial strain diversity between samples. This can be achieved using the SNV profiling routine, which aligns metagenomic reads to centroid sequences of marker gene clusters (MGCs) of the mOTUs database. In prior work, this approach was demonstrated to represent a computationally efficient alternative to using whole genome sequences for computing strain population distances (Milanese et al., 2019).
The output of the SNV profiling routine provides three layers of information. Firstly, there are two tables for different types of coverage (vertical and horizontal) that can be explored to assess the abundance distribution of mOTUs across the profiled samples and to guide the subsequent selection of parameters for SNV calling according to Support Protocol 4. Second, for each mOTU meeting the selection criteria, the filter-* folder will contain a file with frequencies of identified SNVs across samples. Each row in this file represents a variable position and the corresponding SNV frequency across samples. The first column of this file provides colon-separated strings of information, including the marker gene identifier, the position of the SNV on that gene, and the synonymy of the SNV (i.e., synonymous vs. nonsynonymous substitution), as well as the reference and variant codons. Note that for positions with more than two detected alleles, the non-reference variants are represented on separate lines. The remaining columns are named after the input files and contain the frequency of each non-reference variant across selected samples. Finally, the SNV frequencies are used to compute between-sample distances. For each selected mOTU, two distance files are generated (distances-* folder) using different approaches. Specifically, the first file contains Manhattan distances between samples based on the frequency profiles of all variable positions, while in the second file only dominant alleles are considered. A change in the dominant allele corresponds to a frequency change above 50% (such that the dominant strain changes). Both distances are normalized by MG lengths to compute the average genetic variation per nucleotide. Higher coverage of a mOTU in a given sample leads to more reliable results, since more extensive sampling of the population reduces noise in the determination of SNV frequencies.
More details regarding the interpretation of results, including intermediate ones, can be found online (https://github.com/motu-tool/mOTUs/wiki), and examples of downstream analyses are showcased in Milanese et al. (2019).
COMMENTARY
Background Information
The development of mOTUs is rooted in the identification and phylogenomic application of a set of 40 single-copy, phylogenetic marker genes (MGs) that are found in all organisms and only rarely horizontally transferred (Ciccarelli et al., 2006; Sorek et al., 2007). These properties make the MGs ideally suitable for the de novo reconstruction of phylogenetic trees (Ciccarelli et al., 2006), for the placement of new organisms into precomputed reference trees (Stark, Berger, Stamatakis, & von Mering, 2010), and for the calibration of sequence identity cutoffs of MGs between microbial strains to globally group prokaryotes into species-level clusters (Mende et al., 2013).
Historically, it was the increased application of metagenomic sequencing for the study of natural microbial communities since the early 2010s that inspired the development of mOTUs. The idea was to design a tool that would make use of MGs to estimate the relative taxonomic abundance (i.e., profile) of microbial community members (Sunagawa et al., 2013). A subset (n = 10) of these MGs can be readily identified not only in reference genomes, but also in genomic fragments (contigs) that were directly assembled from metagenomes. As the genomic linkage of these fragments is not always apparent from the assembly, it needs to be inferred. Through abundance correlation, different MGs from the same species can be “linked,” and, once incorporated into the mOTUs database, used to represent and quantify “unknown” species in microbial communities. MGs that were identified but not linked can be used to estimate the abundance of organisms that are present in a sample but not represented by reference genomes or linked MGs. This feature enables mOTUs to reduce bias in the estimation of relative taxonomic abundances. Other tools, which solely depend on the availability of reference genomes, often tend to overestimate the relative abundance of the taxa represented in their databases. Furthermore, the 10 or 40 MGs can be used to reconstruct the phylogenetic relationships between the profiled species, which may not be possible using clade-specific genes or more difficult with k-mer based methods or solely based on average nucleotide identities.
The mOTU profiler can also be used for metatranscriptomic analyses, in particular for normalizing whole-community gene copy number estimates to obtain abundances on a per cell basis, for integrating metagenomics and metatranscriptomics data sets (Salazar et al., 2019), and for studying gene expression changes relative to the basal expression level of MGs representing housekeeping genes.
In summary, mOTUs is a versatile software tool for microbiome profiling, which is characterized by highly competitive performance (Meyer et al., 2021) and a small computational footprint.
Critical Parameters
Support Protocol 3 and Support Protocol 4 describe the critical parameters in-depth and explain the impact on the resulting profile file. Another factor that will change the result is the underlying marker gene database, which is being updated on a regular basis (1.0.0 in 2013, 2.0.0 and 2.5.1 in 2019, and 2.6.1 and 3.0.0 in 2021). Profiles from different database versions are not directly comparable and can therefore not be merged. Note that the motus merge command (see Basic Protocol 1) will only finish successfully if all profiles were generated with compatible versions of mOTUs.
Troubleshooting
Most issues that have been reported revolve around understanding the output of mOTUs, which are covered in Guidelines for Understanding Results. As a first step to troubleshooting, we recommend browsing the more frequently updated Wiki and Issues pages of the GitHub repository at https://github.com/motu-tool/mOTUs.
Acknowledgments
The authors acknowledge funding from the Swiss National Foundation through project grants 205321_184955, the NCCR Microbiomes (51NF40_180575), and the Strategic Focal Area “Personalized Health and Related Technologies (PHRT)” of the ETH Domain (#2018-521) to S.S. G.Z. was supported by the European Molecular Biology Laboratory and the German Federal Ministry of Education and Research (BMBF, the de.NBI network, grant no. 031A537B, and grant no. 031L0181A). The authors thank Chris Field, Quentin Clayssen, and other members of the Sunagawa Lab for their input on draft versions of this manuscript. The authors thank Renato Alves for the help with the bioconda integration.
Open access funding provided by Eidgenossische Technische Hochschule Zurich.
Supporting Information
Input and output file examples used in the Basic Protocol 1, Support Protocol 2, and Support Protocol 3 can be found at Zenodo (https://zenodo.org/record/5012695). Input and output file examples used in the Basic Protocol 2 and Support Protocol 4 can be found at Zenodo (https://zenodo.org/record/5012568).
Author Contributions
Hans-Joachim Ruscheweyh : data curation, formal analysis, investigation, methodology, resources, software, validation, writing original draft, writing review and editing; Alessio Milanese : data curation, formal analysis, investigation, methodology, resources, software, validation, visualization, writing original draft, writing review and editing; Lucas Paoli : data curation, formal analysis, investigation, methodology, resources, software, validation, writing original draft, writing review editing; Anna Sintsova : validation, writing original draft, writing review and editing; Daniel R. Mende : data curation, formal analysis, methodology, validation, writing original draft, writing review and editing; Georg Zeller : conceptualization, funding acquisition, investigation, supervision, writing original draft, writing review and editing; Shinichi Sunagawa : conceptualization, funding acquisition, investigation, project administration, supervision, validation, writing original draft, writing review and editing.
Conflicts of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing not applicable: no new data generated.
Literature Cited
- Ciccarelli, F. D., Doerks, T., von Mering, C., Creevey, C. J., Snel, B., & Bork, P. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science , 311(5765), 1283–1287. doi: 10.1126/science.1123061.
- Meyer, F., Lesker, T.-R., Koslicki, D., Fritz, A., Gurevich, A., Darling, A. E., … McHardy, A. C. (2021). Tutorial: Assessing metagenomics software with the CAMI benchmarking toolkit. Nature Protocols , 16(4), 1785–1801. doi: 10.1038/s41596-020-00480-3.
- Salazar, G., Paoli, L., Alberti, A., Huerta-Cepas, J., Ruscheweyh, H.-J., Cuenca, M., Field, C. M.,… Sunagawa, S. (2019). Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell , 179(5), 1068−1083. doi: 10.1016/j.cell.2019.10.014.
- Sorek, R., Zhu, Y., Creevey, C. J., Francino, M. P., Bork, P., & Rubin, E. M. (2007). Genome-wide experimental determination of barriers to horizontal gene transfer. Science , 318(5855), 1449–1452. doi: 10.1126/science.1147112.
- Stark, M., Berger, S. A., Stamatakis, A., & von Mering, C. (2010). MLTreeMap–accurate Maximum Likelihood placement of environmental DNA sequences into taxonomic and functional reference phylogenies. BMC Genomics , 11(1), 1–11. doi: 10.1186/1471-2164-11-461.
- Sun, Z., Huang, S., Zhang, M., Zhu, Q.-Y., Haiminen, N., Carrieri, A.-P., … Liu, Y.-Y. (2020). Challenges in benchmarking metagenomic profilers. bioRxiv , 2020.11.14.382994. doi: 10.1101/2020.11.14.382994
Key References
- Costea, P. I., Munch, R., Coelho, L. P., Paoli, L., Sunagawa, S., & Bork, P. (2017). metaSNV: A tool for metagenomic strain level analysis. PloS One , 12(7), e0182392. doi: 10.1371/journal.pone.0182392.
This article describes the development of metaSNV, which is a dependency for the SNV profiling routine of mOTUs.
- Mende, D. R., Sunagawa, S., Zeller, G., & Bork, P. (2013). Accurate and universal delineation of prokaryotic species. Nature Methods , 10(9), 881–884. doi: 10.1038/nmeth.2575.
This article describes the determination of marker gene-specific sequence similarity cutoffs for the delineation of prokaryotic species.
- Milanese, A., Mende, D. R., Paoli, L., Salazar, G., Ruscheweyh, H.-J., Cuenca, M., … Sunagawa, S. (2019). Microbial abundance, activity and population genomic profiling with mOTUs2. Nature Communications , 10(1), 1014. doi: 10.1038/s41467-019-08844-4.
This article describes an extension of the marker gene database and addition of metatranscriptomic and SNV profiling functionality to the mOTUs tool.
- Sunagawa, S., Mende, D. R., Zeller, G., Izquierdo-Carrasco, F., Berger, S. A., Kultima, J. R., … Bork, P. (2013). Metagenomic species profiling using universal phylogenetic marker genes. Nature Methods , 10(12), 1196–1199. doi: 10.1038/nmeth.2693.
This article describes the original development and implementation of the mOTUs tool.
Internet Resources
GitHub repository of the mOTU profiler.
Website site of the mOTU profiler.
Citing Literature
Number of times cited according to CrossRef: 5
- Sizhe Qiu, Hong Zeng, Zhijie Yang, Wei‐Lian Hung, Bei Wang, Aidong Yang, Dynamic metagenome‐scale metabolic modeling of a yogurt bacterial community, Biotechnology and Bioengineering, 10.1002/bit.28492, 120 , 8, (2186-2198), (2023).
- Hans-Joachim Ruscheweyh, Alessio Milanese, Lucas Paoli, Nicolai Karcher, Quentin Clayssen, Marisa Isabell Keller, Jakob Wirbel, Peer Bork, Daniel R. Mende, Georg Zeller, Shinichi Sunagawa, Cultivation-independent genomes greatly expand taxonomic-profiling capabilities of mOTUs across various environments, Microbiome, 10.1186/s40168-022-01410-z, 10 , 1, (2022).
- Rachel Shadoff, Mary Anne Panoyan, Nicole Novroski, Microbial Forensics: A Present to Future Perspective on Genomic Targets, Bioinformatic Challenges, and Applications, Forensic Genomics, 10.1089/forensic.2022.0007, 2 , 2, (42-64), (2022).
- Shen Jin, Daniela Wetzel, Melanie Schirmer, Deciphering mechanisms and implications of bacterial translocation in human health and disease, Current Opinion in Microbiology, 10.1016/j.mib.2022.102147, 67 , (102147), (2022).
- Birge D Özel Duygan, Jan R van der Meer, Recent advances in microbial community analysis from machine learning of multiparametric flow cytometry data, Current Opinion in Biotechnology, 10.1016/j.copbio.2022.102688, 75 , (102688), (2022).

