Functional Assays Evaluating Immunosuppression Mediated by Myeloid-Derived Suppressor Cells
Kerem Ben-Meir, Kerem Ben-Meir, Nira Twaik, Nira Twaik, Yaron Meirow, Yaron Meirow, Michal Baniyash, Michal Baniyash, Or Reuven, Or Reuven, Ivan Mikula Jr., Ivan Mikula Jr., Hadas Ashkenazi-Preiser, Hadas Ashkenazi-Preiser, Leonor Daniel, Leonor Daniel, Guy Kariv, Guy Kariv, Mahdi Kurd, Mahdi Kurd
cell culture
chronic inflammation
flow cytometry
immunosuppression
inflammatory mouse model
MDSC
myeloid-derived suppressor cell
Abstract
Myeloid-derived suppressor cells (MDSCs) are heterogenous populations of immature myeloid cells that can be divided into two main subpopulations, polymorphonuclear (PMN) MDSCs and monocytic (M) MDSCs. These cells accumulate during chronic inflammation, characterizing an array of pathologies such as cancer, inflammatory bowel disease, and infectious and autoimmune diseases, and induce immunosuppression. The suppressive effects of MDSCs on the immune system are studied mainly when focusing on their features, functions, and impact on target cells such as T cells, natural killer cells, and B cells, among others. Herein, we describe methods for the analysis of MDSC immunosuppressive features and functions, measuring different mediators that contribute to their activities and how they impact on T cell function. The protocols described are a continuation to those in a companion Current Protocols article by Reuven et al. (2022), which uses a generated single-cell suspension and isolated cells to test their activity. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Evaluating MDSC suppressive features
Alternate Protocol 1 : Dichlorofluorescein diacetate–based reactive oxygen species detection
Support Protocol 1 : Detection of nitric oxide secretion
Support Protocol 2 : Measurement of arginase activity
Basic Protocol 2 : Evaluating MDSC suppressive function
Alternate Protocol 2 : In vitro effects of MDSCs on expression of T cell receptor complex during activation
Support Protocol 3 : Effect of MDSCs on interferon γ production
Basic Protocol 3 : Effect of MDSCs on T cell proliferation
Basic Protocol 4 : Effect of MDSCs on T cell cytotoxic activity
Alternate Protocol 3 : In vivo cytotoxicity assay
Basic Protocol 5 : Analysis of MDSC differentiation
INTRODUCTION
Myeloid-derived suppressor cells (MDSCs) are a heterogenous population of immature cells that can be divided to two subpopulations, polymorphonuclear (PMN)-MDSCs and monocytic (M)-MDSCs, based on their phenotype (described in a companion Current Protocols article by Reuven et al., 2022) and activity, as presented here. MDSC accumulation and associated immunosuppression are evident in diseases characterized by chronic inflammation. To test the function of the different MDSC subpopulations during chronic inflammation, we used a pathology-free chronic inflammation murine model system, which is enriched with elevated levels of MDSCs and displays a suppressed immune status. The model is referred to as the Bacille Calmette–Guérin (BCG) model and is described in detail in a Current Protocols article by Ben-Meir, Twaik, Meirow, & Baniyash (2022). To fully understand the contribution of MDSCs to the immunosuppressive environment, it is imperative to perform both in vivo and in vitro analyses.
In this article we present assays that characterize the immunosuppressive features of MDSCs by evaluating the secretion of various mediators, such as reactive oxygen species (ROS) and nitric oxide (NO), and by assessing arginase-1 activity. We also describe methods to assess the suppressive function of MDSCs by measuring their inhibitory effect on: (1) T cell proliferation, (2) T cell receptor (TCR) complex integrity, (3) cytotoxic activity, and (4) interferon (IFN)-γ production. Moreover, as MDSCs lose their suppressive function with differentiation, a method to investigate the differentiation state of the cells is presented as well.
This article describes a set of protocols testing the immunosuppressive features and functions of MDSCs, which are a continuation to the protocols described in detail in a companion Current Protocols article by Reuven et al. (2022), which focuses on the isolation and phenotypic characterization of MDSCs. The protocols presented herein were used successfully in numerous repetitive experiments and are in routine use by various laboratories including our own.
Basic Protocol 1 details methods to evaluate the suppressive features of MDSCs. The protocol describes detection methods of ROS production and secretion by flow cytometry and luminescence. Support Protocol 1 describes how to measure NO secretion in the medium of cultured MDSCs, and Support Protocol 2 describes measuring the activity of the enzyme arginase-1. Hence, the presented Basic Protocol 1 and Support Protocols allow the measurement of different mediators of immunosuppressive features of MDSCs and could contribute to pinpointing differences between MDSCs from different pathologies and tissue based on their suppressive features.
Basic Protocol 2 describes various methods used to evaluate the immunosuppressive function of MDSCs. In contrast to the methods describing measurements of intrinsic MDSC suppressive features, analysis of MDSC suppressive function is based on evaluating how they affect T cells, which are their primary targets. This protocol describes methods to isolate the target cells (T cells) and assess how they are affected by MDSCs. One indication is related to the changes MDSCs induce in the expression of TCR subunits. Measuring expression of various TCR subunits on T cells allows for distinguishing between activated and immunosuppressed cells (Alternate Protocol 2). An additional indication of the suppressive effect of MDSCs on T cells is the change they induce in the expression of IFN-γ by T cells, which is presented in this Support Protocol 3.
Basic Protocol 3 focuses on MDSC-induced proliferation inhibition of T cells. When T cells are activated via the TCR complex, MDSCs induce cell proliferation arrest that can be measured and quantified using flow cytometry. This protocol can be adapted for both isolated T cells and single-cell suspensions of splenocytes acquired using Basic Protocol 2 in this manuscript and in protocols presented in a companion Current Protocols article by Reuven et al. (2022).
Basic Protocol 4 and Alternate Protocol 3 present ex vivo and in vivo methods to test the ability of MDSCs to alter cytotoxic activity induced by T cells. The protocols use T cells derived from premelanosome protein (PMEL) transgenic mice and describe activation via the TCR complex with the gp100 peptide. The ex vivo assay relies on the expansion of naive isolated T cells from Basic Protocol 2, whereas the in vivo method uses adoptive transfer of both cytotoxic T cells (CTLs) and target and nontarget cells to the mouse.
Basic Protocol 5 focuses on the changes in MDSC phenotype in the presence of macrophage colony‒stimulating factor (M-CSF) and granulocyte-macrophage colony‒stimulating factor (GM-CSF), which are known to induce myeloid cell differentiation and proliferation. As MDSCs represent an undifferentiated and immature population of the myeloid lineage, they become less suppressive as they differentiate according to the change in their phenotype. The process of MDSC differentiation can be measured by tracking differentiation markers using flow cytometry. These markers are detailed in the protocol and allow for the evaluation of MDSC maturation toward macrophages and dendritic cells.
NOTE : All protocols using live animals must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of laboratory animals.
NOTE : All protocols involving humans and human samples must first be reviewed and approved by an Institutional Review Board (IRB) or must follow local guidelines for the use of human samples. All patients must provide informed consent.
NOTE : Perform procedures with cold solutions. Keep tissue and cells at 4°C until use unless stated otherwise. All solutions and equipment coming into contact with cells for culture must be sterile and endotoxin free, and steps should be performed using aseptic technique.
NOTE : All cell incubations use a cell culture incubator, maintaining a humidified 37°C and 5% CO2 environment.
Basic Protocol 1: EVALUATING MDSC SUPPRESSIVE FEATURES
Herein we describe a protocol for the measurement of ROS production from total MDSCs and isolated PMN-MDSCs. Production of ROS has been previously shown to induce DNA damage, leading to the contribution of cancerous transformation over time. Moreover, MDSC accumulation in the tumor microenvironment is well established, and they are known ROS producers. Thus, measuring ROS production by MDSCs represents an important method to assess their suppressive features.
This method uses luminol to detect initial and rapid intracellular and extracellular ROS accumulation kinetics over a period of 1 hr. For the detection of only extracellularly secreted ROS, replace luminol with isoluminol, which does not penetrate the cell membrane.
Materials
-
Isolated PMN-MDSCs or total MDSCs (see companion Current Protocols article by Reuven et al., 2022)
-
Hanks’ balanced salt solution (HBSS) with Ca2+ and Mg2+, without phenol red (e.g., Biolab, cat. no. 009015237500)
-
250 ng/ml lipopolysaccharide (LPS) from Escherichia coli O26:B6 (e.g., Sigma-Aldrich, cat. no. L3755)
-
ROS detection solution (see recipe)
-
Microplate reader capable of detecting luminescence between 370 and 700 nm (e.g., Tecan Spark)
-
96-well flat-bottom plate, white polystyrene (e.g., Millipore Sigma, cat. no. CLS3917-100EA)
Luminol-based reactive oxygen species production over time
1.Program ROS template in the microplate reader: Set temperature to 37°C. Define experimental wells, and read luminescence with a 1000-ms kinetics loop and 30-s interval over a 1-hr period.
2.Wash isolated PMN-MDSCs or MDSCs twice with HBSS, and bring concentration to 2–4 × 106 cells/ml in HBSS.
3.Load 100 μl cells (0.2–0.4 × 106 cells) in each sample well of a 96-well plate.
4.To each well, add 80 μl of 250 ng/ml LPS (100 ng/ml final), and 20 μl ROS detection solution.
5.Place plate in preheated microplate reader, and immediately read samples.
Analysis of results
6.Average replicates for each sample.
7.Plot average luminescence values for each sample as a function of time.
Alternate Protocol 1: DICHLOROFLUORESCEIN DIACETATE–BASED REACTIVE OXYGEN SPECIES DETECTION
Alternate Protocol 1 represents a second method for ROS detection using the substance 2′,7′-dichlorofluorescin diacetate (H2DCFDA), which penetrates and remains inside the cells, allowing detection of intracellular late-phase ROS formation by flow cytometry. H2DCFDA can also be used for ROS imaging using microscopy.
Materials
-
Splenocytes, total MDSCs, or isolated PMN-MDSCs (see companion Current Protocols article by Reuven et al., 2022)
-
RPMI 1640 full medium (see recipe)
-
80 ng/ml LPS from E. coli O26:B6 (e.g., Sigma-Aldrich, cat. no. L3755)
-
20 μM H2DCFDA (e.g., Sigma-Aldrich, cat. no. D6883)
-
96-well U-bottom cell culture plate (e.g., Grenier Bio-One, cat. no. 655180)
-
Cell culture incubator, 37°C, 5% CO2
-
70-μm mesh
-
Multicolor flow cytometer with a plate loader module equipped with 488-nm laser (e.g., Beckman Coulter CytoFLEX V-B-R series or equivalent)
-
Flow cytometry data analysis software (e.g., CytExpert, version 2.4 or similar)
1.Adjust cell concentration to 2–4 × 106 cells/ml in RPMI 1640 full medium.
2.Load 100 μl cells (0.2–0.4 × 106 cells) in each sample well of a 96-well plate.
3.Add 50 μl of 80 μg/ml LPS (20 μg/ml final) and 50 μl of 20 μM H2DCFDA (5 μM final) for a final volume of 200 μl/well.
4.Place plate in a cell culture incubator for 4 to 6 hr.
5.Resuspend and strain cells using a 70-μm mesh to ensure a single-cell suspension, and analyze immediately by flow cytometry.
6.Analyze results using flow cytometry data software. Create a fold-increase graph by extracting mean fluorescence intensity (MFI) and subtracting baseline control fluorescence intensity from each sample.
Support Protocol 1: DETECTION OF NITRIC OXIDE SECRETION
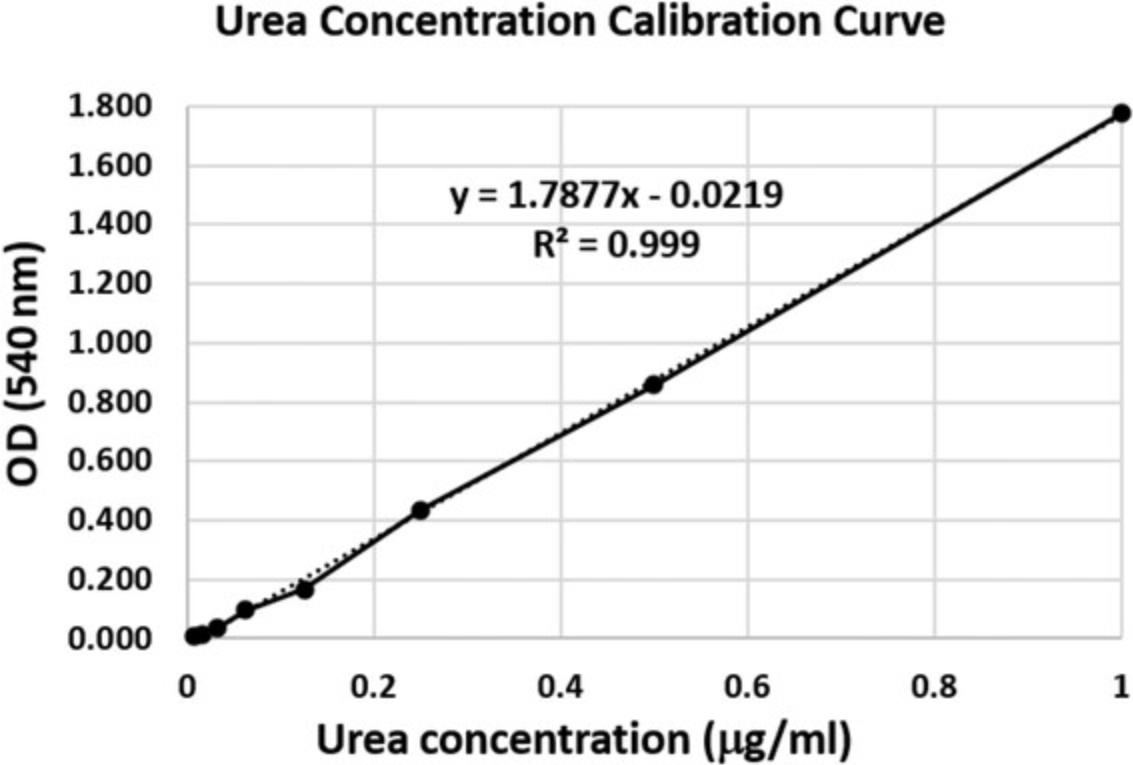
One of the suppressive mediators of MDSCs is NO, which is generated by the enzyme inducible NO synthase (iNOS). Elevated levels of iNOS detected within MDSCs are related primarily to M-MDSCs and are associated with increased production and secretion of NO. Elevated levels of NO can lead to alterations in T cells responses due to their direct effect on T cells or indirect effect of inhibiting antigen presentation to T cells by dendritic cells. In this assay, the activity of iNOS in MDSCs is measured using the Griess reagent, which creates a colorimetric reaction with nitrites and nitrates to produce magenta azo product. Creating a calibration curve using sodium nitrite allows for the calculation of nitrite concentration in the medium according to optical density (OD) values.
Materials
-
Isolated M-MDSCs or total MDSCs (see companion Current Protocols article by Reuven et al., 2022)
-
RPMI 1640 full medium (see recipe)
-
LPS from E. coli O26:B6 (e.g., Sigma-Aldrich, cat. no. L3755)
-
Griess reagent (see recipe)
-
100 mM sodium nitrite (e.g., Sigma-Aldrich, cat. no. S-2252)
-
Cell culture incubator, 37°C, 5% CO2
-
Centrifuge with plate adaptor
-
96-well flat-bottom transparent plate suitable for OD readings (e.g., Thermo Scientific, cat. no. 3355)
-
Microplate reader capable of reading absorbance (e.g., Tecan Spark)
Sample preparation
1.Plate isolated MDSCs in RPMI 1640 full medium with an activator such as 100 ng/ml LPS, and incubate for 48 to 72 hr.
2.After incubation of activated cells, centrifuge plate 8 min at 300 × g , 4°C.
3.Collect 100 μl supernatant from each sample well, and transfer to a new 96-well flat-bottom transparent plate suitable for OD readings.
4.Thaw Griess reagent aliquot at room temperature.
5.Add 100 μl Griess reagent (1:1 ratio) to each sample and calibration curve wells.
Preparation of calibration curve
6.Dilute 100 mM sodium nitrite stock with the same medium used for culturing the cells (e.g., RPMI 1640 full medium) at a ratio of 1:200 to final concentration of 500 μM.
7.Make ten serial dilutions until reaching a concentration of ∼1 μM.
8.Plate triplicates of each concentration of the calibration curve in the same plate containing samples.
9.Add 100 μl Griess reagent (1:1 ratio) to each sample and calibration curve wells.
Obtaining results
10.Read plate containing sample wells and calibration curve using a plate reader capable of detecting OD at wavelength of 540 nm.
11.Add trendline to the calibration curve samples, and use it to calculate the concentration of nitrites in the medium by converting OD values to concentration of nitrites.
Support Protocol 2: MEASUREMENT OF ARGINASE ACTIVITY
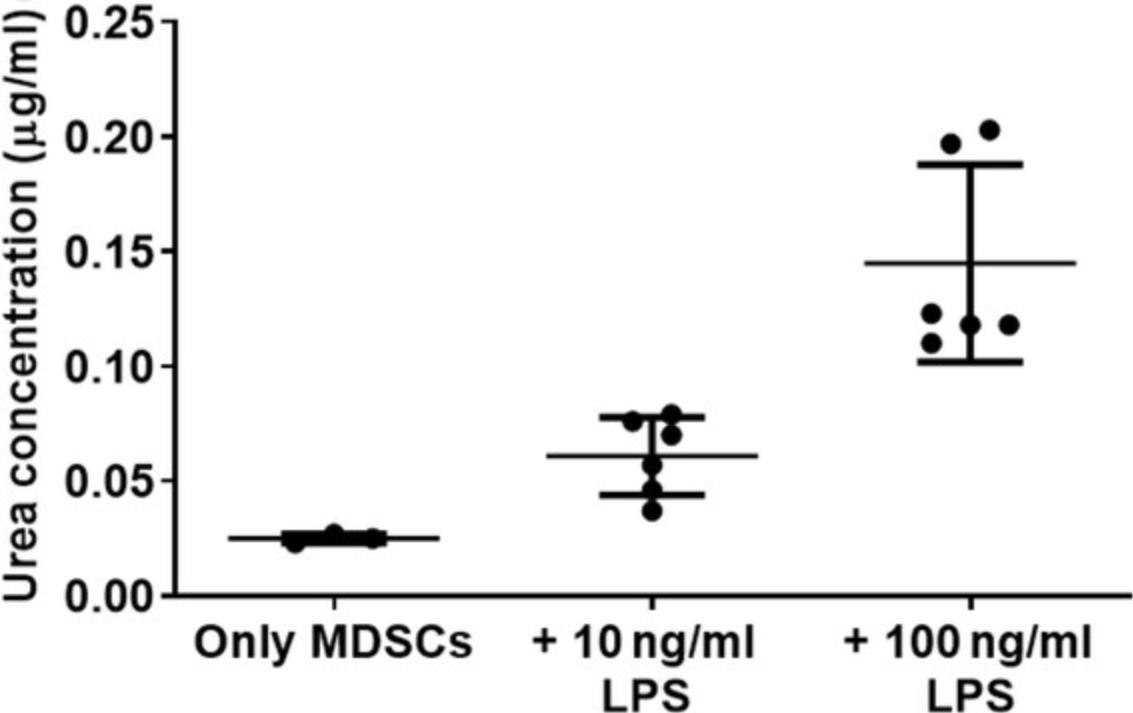
Increased levels and activity of the enzyme arginase-1 in MDSCs, primarily in the M-MDSC subpopulation, can lead to deprivation of arginine from the surrounding, thus altering T cells responses. In this assay, the activity of arginase-1 in MDSCs is measured by the ability of MDSC lysate to catalyze the conversion of L-arginine to urea and ornithine. Urea reacts with its substrate α-isonitrosopropiophenone (ISPF) to generate a colored product, in which the levels are proportional to the amount of urea produced. The obtained readout can be interpreted as the arginase-1 activity in the cell lysate.
Materials
-
Isolated M-MDSCs or total MDSCs (see companion Current Protocols article by Reuven et al., 2022)
-
Lysis buffer (see recipe)
-
25 mM Tris-HCl, pH 7.5 (e.g., Biolab, cat. no. 20092391)
-
10 mM manganese (II) chloride tetrahydrate (MnCl2•4H20; e.g., Sigma-Aldrich, cat. no. M8054-100G)
-
0.5 M L-arginine, pH 9.7 (e.g., Sigma-Aldrich, cat. no. A5131)
-
Acidic mixture (see recipe)
-
9% ISPF (see recipe)
-
1 mg/ml urea (e.g., Sigma-Aldrich, cat. no. U5128) in double distilled water
-
1.5-ml microcentrifuge tubes
-
Microcentrifuge
-
Variable temperature heating block
-
Vortex mixer
-
ELISA plates: 96-well flat-bottom transparent plate suitable for OD readings (e.g., Thermo Scientific, cat. no. 3355)
-
Microplate reader capable of reading absorbance (e.g., Tecan Spark)
-
Computer running Microsoft Excel or similar analysis software
Cell preparation and enzymatic reaction
1.Divide 1 × 105 isolated M-MDSCs or total MDSCs in 1.5-ml microcentrifuge tubes, and centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
2.Lyse cells in 50 μl lysis buffer, and incubate at room temperature for 30 min.
3.Add 50 μl of 25 mM Tris-HCl and 5 μl of 10 mM MnCl2 to the cell lysate.
4.Incubate in a heating block at 56°C for 10 min.
5.Add 100 μl of 0.5 M L-arginine and mix.
6.Incubate samples in a heating block at 37°C for 2 hr.
7.Prepare acidic mixture for stopping the reaction.
8.Add 900 µl acidic mixture to lysate.
9.Add 40 μl of 9% ISPF to mixture and vortex.
10.Incubate in a heating block at 95°C for 30 min.
Preparation of standard curve
11.Prepare serial dilutions of urea using double distilled water (1, 0.5, 0.25, 0.125, 0.062, and 0.03125 μg/ml).
12.Add 100 μl each urea concentration (prepared in step 11) to 450 μl acidic mixture and 20 μl of 9% ISPF.
13.For a blank sample, add 100 μl double distilled water to 450 μl acidic mixture and 20 μl of 9% ISPF.
14.Incubate in a heating block at 95°C for 30 min.
15.Load 100 μl sample (in triplicate) and 100 μl standard curve samples in ELISA plates.
16.Read plate at 540 nm using an ELISA plate reader.
Analysis of results
17.Average triplicate readings for each standard and sample.
18.Plot corrected absorbance values for each standard as a function of urea concentration, including blank readings.
19.Construct standard curve, and calculate trendline equation based on standard curve data using Excel or other similar analysis software.
20.Calculate amount of urea in the sample based on OD values using the trendline equation.
Basic Protocol 2: EVALUATING MDSC SUPPRESSIVE FUNCTION
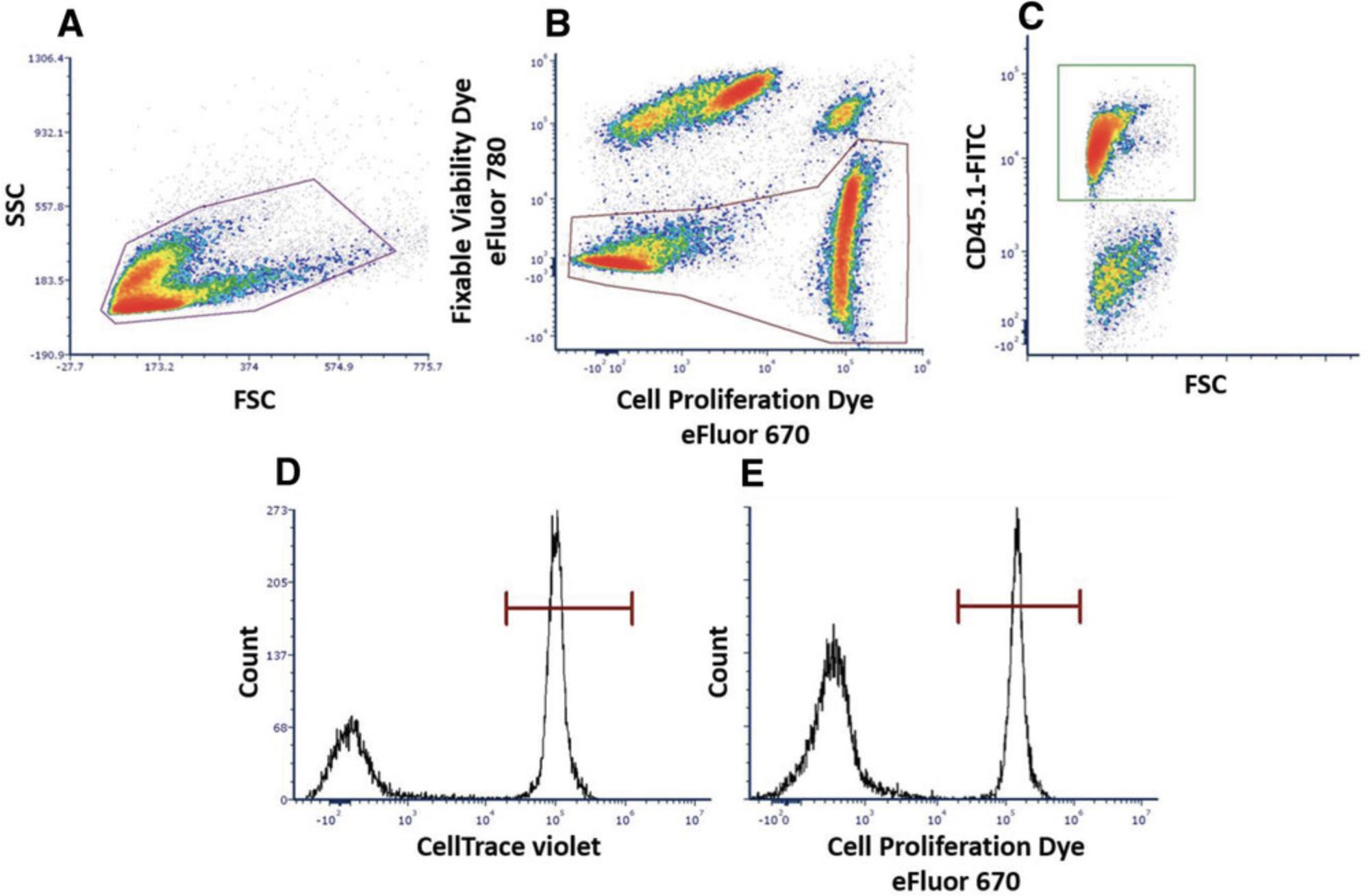
The cell surface TCR is composed of six different transmembrane subunits: the α and β subunits, which recognize peptide-major histocompatibility complexes (MHCs) present on the cell surface of antigen-presenting cells, and the CD3 ε, δ, and γ and CD247 ζ subunits, which serve as signaling molecules. Binding of the TCR to the peptide-MHC complex leads to activation of intracellular pathways within the T cell mediated by the signaling subunits that bear immunoreceptor tyrosine-based activation motifs (ITAMs). These motifs comprise consensus sequences containing the amino acid tyrosine, which is phosphorylated upon activation and ultimately leads to the activation of cells and internalization of the TCR. Of the TCR subunits, CD247 possesses a higher number of ITAMs and thus is considered vital for T cell activity. Under chronic inflammation, it was shown that expression of CD247 is downregulated by MDSCs, whereas levels of the rest of the TCR subunits, such as CD3ε, remain unchanged (Ezernitchi et al., 2006). Flow cytometry analyses measuring expression levels of CD3ε, TCR-β, and CD247 molecules can distinguish between a suppressive state of T cells due to MDSC activity and an activated state via the TCR. Moreover, IFN-γ production is associated with optimal CTL function and has been shown to be affected by MDSCs. Therefore, determining IFN-γ expression in T cells when surrounded by MDSCs could enable determination of MDSC suppressive activity that could then be translated for in vivo testing. In a similar manner, MDSC functions can be determined in vivo by testing T cell or natural killer (NK) cell cytotoxic capacity. As previously noted, the presence of MDSCs is closely associated with the severity of cancer, in particular, due to their effect on T and NK cell cytotoxic functions. Therefore, a fast and simple examination of the capacity of MDSCs to interfere with the activity of effector CD8 T cells and NK cells can clarify the impact of modifications to experimental models in vivo.
Select methods describe manipulation of the entire splenocyte population or of isolated T cells as described here (also see a companion Current Protocols article by Reuven et al., 2022). When harvesting other tissue, use appropriate tissue-specific methods, as additional use of enzymatic digestion and/or mechanical force might be required.
All steps in this protocol require sterile working conditions, except sample preparation for flow cytometry analysis.
Materials
-
Isolated splenocytes (see companion Current Protocols article by Reuven et al., 2022)
-
Separation buffer (see recipe)
-
EasySep Mouse T Cell Isolation Kit (e.g., StemCell Technologies, cat. no. 19851A) containing:
- Normal rat serum
- RapidSphere beads
- Isolation cocktail
-
Dulbecco's phosphate-buffered saline (DPBS), without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
-
Trypan blue
-
Fixable Viability Dye eFluor 780 (e.g., Thermo Fisher Scientific, cat. no. 65-0865-18)
-
4% (w/v) paraformaldehyde (e.g., Thermo Fisher Scientific, cat. no. AAJ61899AK) in DPBS
-
Permeabilization solution (see recipe)
-
Antibody mix containing:
- Purified anti-mouse CD16/32, Fc blocker, clone 93 (e.g., BioLegend, cat. no. 101302)
- APC anti-mouse CD90.2 (Thy-1.2), clone 30-H12 (e.g., BioLegend, cat. no 105311)
- PE anti-mouse CD3ε, clone 145-2C11 (e.g., BioLegend, cat. no. 100307)
- FITC anti-mouse CD247, clone H146-968 (e.g., Thermo Fisher Scientific, cat. no. MA5-17673)
-
FACS buffer (see recipe)
-
5-ml round-bottom tubes
-
Vortex mixer
-
EasyEights EasySep Magnet (e.g., StemCell Technologies, cat. no. 18103)
-
15-ml conical tubes
-
Centrifuge with plate adaptor
-
Hemocytometer
-
96-well U-bottom plate, tissue culture surface (e.g., Thermo Scientific, cat, no, 163320)
-
Multicolor flow cytometer with plate loader module (e.g., Beckman Coulter CytoFLEX V-B-R series or equivalent)
Isolation of mouse T cells
These steps are based on the commercial kit from StemCell Technologies, in which negative selection of T cells is performed from a single-cell suspension of control mouse splenocytes. The protocol is relatively quick and yields high purity, ranging from 90% to 98%. Isolated T cells could be used in several assays to evaluate MDSC suppressive activities. Isolation of T cells should be performed in a sterile environment for cell culture if required.
1.Wash isolated splenocytes twice with separation buffer.
2.Adjust concentration of splenocytes to 100 × 106 cells/ml using separation buffer.
3.Place samples in a 5-ml round-bottom tube, and add 50 µl rat serum per milliliter sample. Mix by pipetting gently.
4.Add 50 μl isolation cocktail per milliliter sample. Mix and incubate at room temperature for 10 min.
5.Vortex RapidSphere beads extensively for 30 s, and add 75 μl per milliliter sample.
6.Mix and incubate for 2.5 min at room temperature.
7.Bring volume to 2.5 ml using separation buffer and mix gently.
8.Place tube in magnet, and incubate for 5 min at room temperature.
9.Carefully aspirate suspension, avoiding touching the inside of the tube facing the magnet, and transfer suspension to a new 15-ml conical tube. Discard round-bottom tube containing the rest of the cells.
10.Add 5 to 7 ml separation buffer to the 15-ml tube, and centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
11.Resuspend cell pellet in DPBS, and count cells using a hemocytometer and trypan blue.
Staining for cell viability
12.Acquire splenocyte single-cell suspension (see companion Current Protocols article by Reuven et al., 2022), and keep in DPBS at 4°C.
13.Count single-cell suspension using a hemocytometer and trypan blue, and for each sample calculate volume needed to plate 0.1–1 × 106 live cells/well in a 96-well plate.
14.Centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
15.Add 100 µl Fixable Viability Dye 780 diluted in DPBS at a 1:1000 ratio. Gently mix by pipetting, and incubate at 4°C in the dark for 20 min.
16.Add 100 µl DPBS, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
17.Add 200 µl DPBS, and centrifuge again 5 min at 300 × g , 4°C, to ensure maximum removal of residual viability dye. Discard supernatant.
Staining of TCR subunits
18.Add 100 µl of 1% paraformaldehyde, and incubate at 4°C in the dark for 20 min.
19.Add 100 µl DPBS, and centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
20.Add 200 µl DPBS, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
21.Add 200 µl permeabilization solution, and incubate at room temperature in the dark for 10 min.
22.Centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
23.Add 50 µl antibody mix, and mix gently by pipetting. Incubate at 4°C in the dark for 30 min.
24.Add 150 µl permeabilization solution, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
25.Add 100 to 200 µl FACS buffer, depending on the number of live cells added in step 13.
26.Mix gently and read plate by flow cytometry.
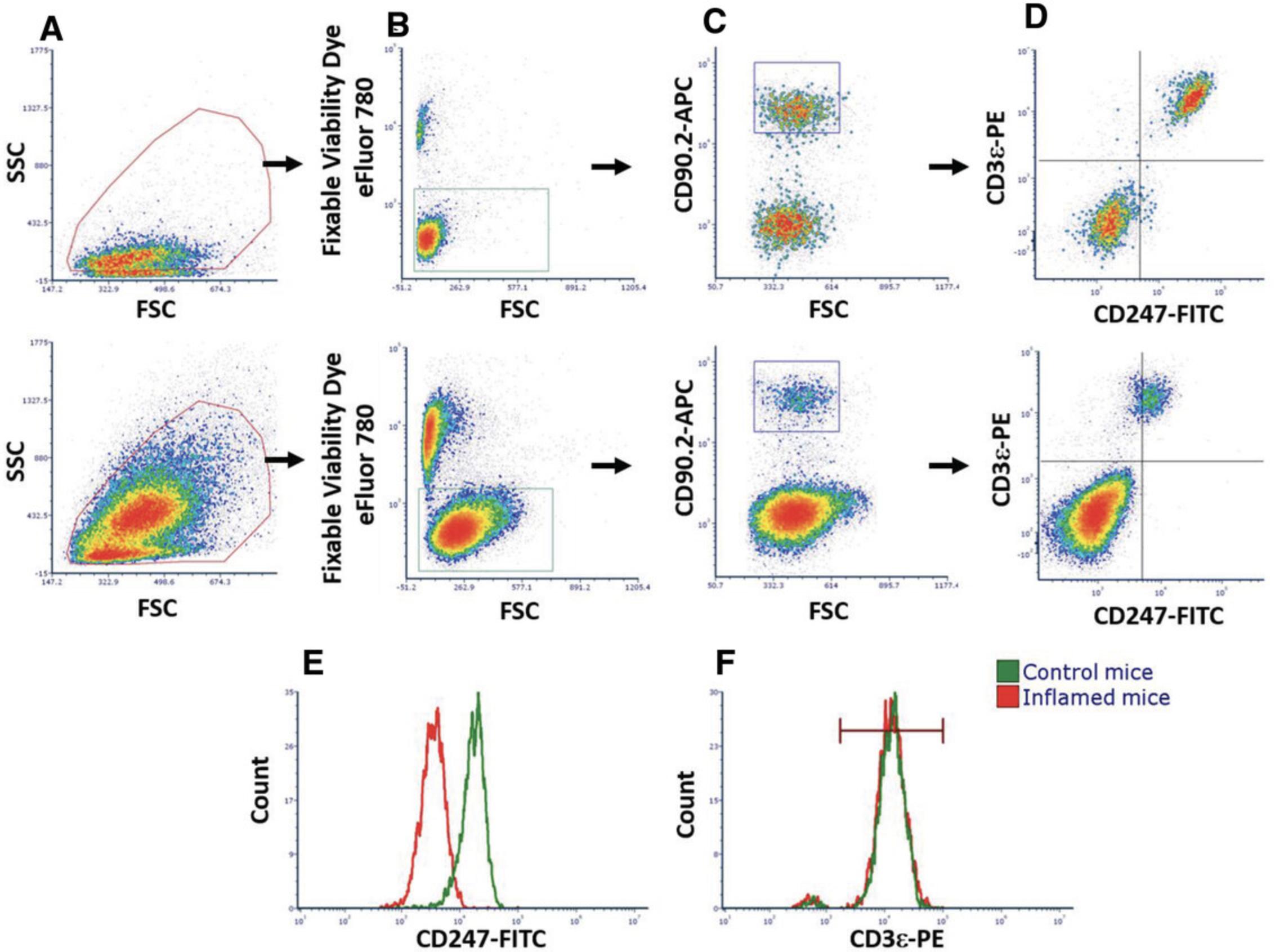
Alternate Protocol 2: IN VITRO EFFECTS OF MDSCs ON EXPRESSION OF T CELL RECEPTOR COMPLEX DURING ACTIVATION
Alternate Protocol 2 introduces a method to test the direct effects of MDSCs on expression of the TCR complex during activation of T cells through the TCR. Different MDSC sources can be used for this protocol to test the effects of MDSCs from various mouse genotypes.
Additional Materials (also see Basic Protocol 2)
- RPMI 1640 full medium (see recipe)
- Isolated MDSCs (see companion Current Protocols article by Reuven et al., 2022)
- Antibodies:
- Purified anti-mouse CD3ε antibody, clone 145-2C11 (e.g., BioLegend, cat. no. 100340)
- PE anti-mouse TCR-β chain antibody, clone H57-597 (e.g., BioLegend, cat. no. 109207)
- Cell culture incubator 37°C, 5% CO2
Sample preparation and plating
1.Acquire splenocyte single-cell suspension (see companion Current Protocols article by Reuven et al., 2022). Resuspend cells in RPMI 1640 full medium.
2.Count single-cell suspension using a hemocytometer and trypan blue, and for each sample adjust cell concentration to 6 × 106 cells/ml for splenocytes and 2 × 106 cells/ml for T cells using RPMI 1640 full medium.
3.Acquire isolated MDSCs (see companion Current Protocols article by Reuven et al., 2022). Resuspend in RPMI 1640 full medium, and keep at 4°C.
4.Count MDSCs using a hemocytometer and trypan blue, and adjust cell concentration to 6 × 106 cells/ml in RPMI 1640 full medium.
5.Prepare an activation cocktail of 0.4 μg/ml anti-CD3ε in RPMI 1640 full medium.
6.Gently mix splenocytes. Add 50 µl (0.3 × 106 cells) into all wells and mix.
7.Prepare sample wells by adding RPMI 1640 full medium as follows:
-
Add 150 µl RPMI 1640 full medium to negative control wells containing splenocytes only.
-
Add 100 μl RPMI 1640 full medium to wells designated to test the suppression by MDSC of nonactivated splenocytes.
-
Leave wells designated for T cell activation with MDSC coculture empty.
8.Add 100 μl activation cocktail (from step 5) to wells for splenocytes.
9.Gently mix MDSCs, and add 50 µl (0.3 × 106 cells) to wells designed for testing suppression.
10.Incubate plate in a cell culture incubator for 16 hr.
Obtaining results by flow cytometry
11.After 16 hr, remove plate from incubator.
12.Centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
13.Add 200 µl DPBS to each well, and repeat step 12 to ensure maximum removal of serum.
14.Add 100 µl Fixable Viability Dye 780 diluted in DPBS at a 1:1000 ratio. Gently mix by pipetting, and incubate at 4°C in the dark for 20 min.
15.Add 100 µl DPBS, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
16.Add 200 µl DPBS, and centrifuge again 5 min at 300 × g , 4°C, to ensure maximum removal of residual viability dye. Discard supernatant.
17.Add 100 µl of 1% paraformaldehyde, and incubate at 4°C in dark for 20 min.
18.Add 100 µl DPBS, and centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
19.Add 200 µl DPBS, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
20.Add 200 µl permeabilization solution, and incubate at room temperature in the dark for 10 min.
21.Centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
22.Add 50 µl antibody mix prepared in permeabilization solution. Mix gently and incubate at 4°C in dark for 30 min.
- 0.2 µg/ml anti-CD90.2 (Thy-1.2) APC
- 0.4 µg/ml anti-TCR-β PE
- 0.5 µg/ml anti-CD247 FITC
- 1 µg/ml unlabeled anti-CD16/32.
Since anti-CD3ε was used for the activation of T cells, it is recommended to instead use antibodies for TCR-β or other TCR subunits to detect the change in expression levels of a representative TCR subunit in addition to CD247.Although TCR-β is part of the TCR complex, it is not affected by MDSCs and mimics CD3ε kinetics in terms of expression.
23.Add 150 µl permeabilization solution, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
24.Add 100 to 200 µl FACS buffer, depending on the number of cells added per well.
25.Mix gently and read plate by flow cytometry.
![Details are in the caption following the image Gating strategy to visualize changes in CD247 and T cell receptor (TCR)-β expression in T cells from in vitro samples. These steps contain (A) a morphological gate represented by side scatter (SSC) and forward scatter (FSC), (B) gating of the T cell population using the T cell marker like CD90.2 (Thy-1), and (C) an option for gating a specific T cell population (e.g., CD8 T cells). (D) CD247 expression in nonactivated T cells (CD90.2; mean fluorescence intensity [MFI]; green histogram), activated T cells (MFI; black histogram), and activated T cells cocultured with myeloid-derived suppressor cells (MDSCs; MFI; red histogram). (E) TCR-β expression in nonactivated T cells (CD90.2; MFI; green histogram), activated T cells (MFI; black histogram), and activated T cells cocultured with MDSCs (MFI; red histogram). Testing the expression of CD247 and TCR-β demonstrates a suppressive effect of MDSCs on T cell activation. (D and E) CD247 and TCR-β are downregulated with the entire TCR complex owing to the activation state (MFI; black histograms) and when incubated with MDSCs. CD247 is the only TCR subunit to be downregulated, whereas TCR-β expression remains unchanged (MFI; red histograms) in nonactivated cells (MFI; green histogram). These results distinguish the states of activation and immunosuppression induced by MDSCs.](https://static.yanyin.tech/literature_test/cpz1557-fig-0005-m.jpg)
Support Protocol 3: EFFECT OF MDSCs ON INTERFERON γ PRODUCTION
This protocol presents conditions for testing IFN-γ by flow cytometry using a protein transport inhibitor, which inhibits protein transport from the endoplasmic reticulum to the Golgi complex and leads to the accumulation of secreted molecules within the cells. This protocol represents a continuation for the plate made using Alternate Protocol 2.
Additional Materials (also see Basic Protocol 2 and Alternate Protocol 2)
- Activated T cells (see Alternate Protocol 2)
- 1000× (5 mg/ml) brefeldin A (e.g., BioLegend cat. no. 420601)
- Antibodies:
- PE anti-mouse IFN-γ antibody, clone XMG1.2 (e.g., BioLegend, cat. no. 505807)
- Anti-CD8 PB
Activation and viability staining of T cells
1.Following activation of T cells as described in Alternate Protocol 2, add brefeldin A (1:1000 final dilution) to all wells designated for measuring IFN-γ production, and incubate for the last 4 to 6 hr of activation in the cell culture incubator.
2.Remove plate from incubator, and centrifuge 5 min at 300 × g , 4°C.
3.Add 200 µl DPBS to each well, and repeat step 2 to ensure maximum removal of serum.
4.Add 100 µl Fixable Viability Dye 780 diluted in DPBS at a 1:1000 ratio. Gently mix by pipetting, and incubate at 4°C in the dark for 20 min.
5.Add 100 µl DPBS, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
6.Add 200 µl DPBS, and centrifuge again 5 min at 300 × g , 4°C, to ensure maximum removal of residual viability dye. Discard supernatant.
Staining for the detection of IFN-γ production
7.Add 100 µl of 1% paraformaldehyde, and incubate at 4°C in the dark for 20 min.
8.Add 100 µl DPBS. Centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
9.Add 200 µl DPBS, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
10.Add 200 µl permeabilization solution. Mix gently and incubate at room temperature in the dark for 10 min.
11.Centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
12.Add 50 µl antibody mix prepared in permeabilization solution:
- 0.5 μg/ml anti-IFN-γ PE
- 1 μg/ml anti-CD8 PB
- 0.2 μg/ml anti-Thy-1 APC
- 1 μg/ml unlabeled anti-CD16/32.
13.Mix gently and incubate at 4°C in the dark for 30 min.
14.Add 150 µl permeabilization solution, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
15.Add 100 to 200 µl FACS buffer, depending on the number of cells cultured per well.
16.Mix gently and read plate by flow cytometry.
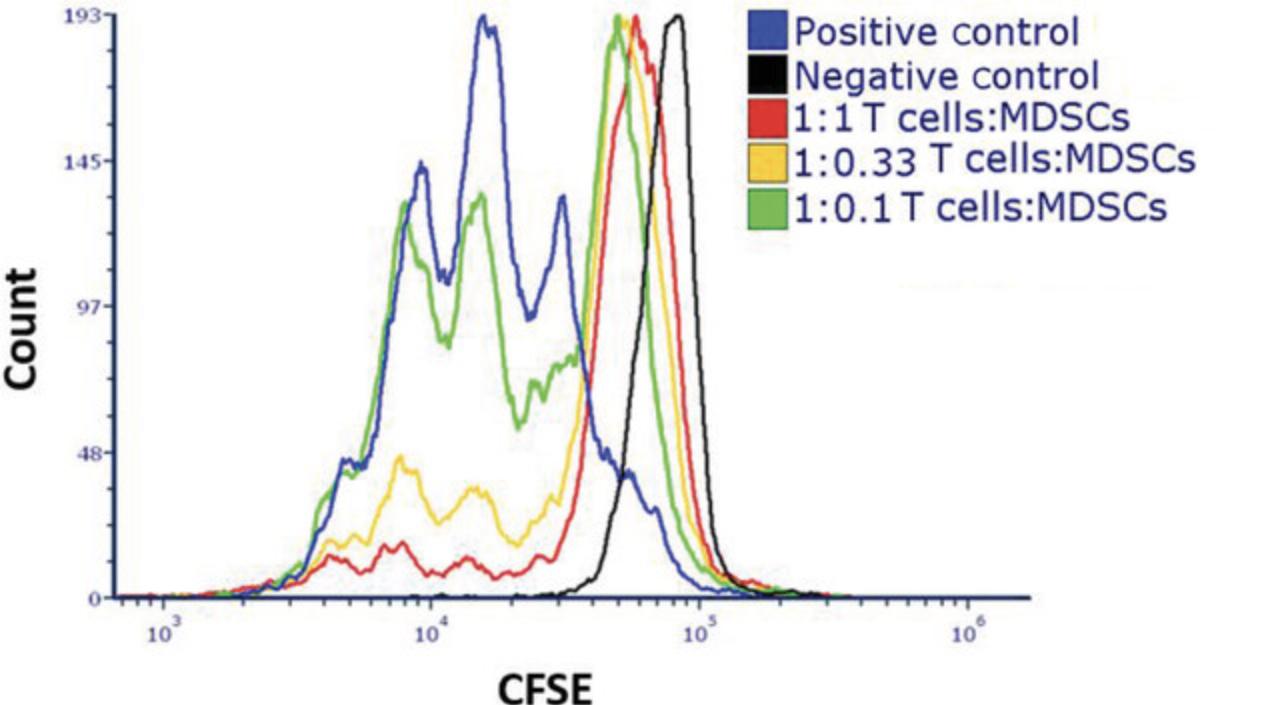
Basic Protocol 3: EFFECT OF MDSCs ON T CELL PROLIFERATION
One of the hallmarks of the suppressive functions of MDSCs is their ability to inhibit T cell responses to activation, which enables assessment by measuring T cell proliferation arrest. The in vitro activation methodology may differ depending on the experiment, but it must include two activating signals: one that works through the CD3ε subunit of the TCR complex and the second that works via the costimulatory receptor CD28.In our laboratory, two different methods for T cell activation have been calibrated: (1) splenocyte single-cell suspension activation using soluble anti-CD3ε antibody (with the costimulatory signal provided by antigen-presenting cells within the spleen) and (2) activation of isolated T cells using plate-bound anti-CD3ε and anti-CD28. The presented method describes a flow cytometry–based analysis to study the proliferation of T cells following their activation. The cells in this protocol are labeled with a fluorescent cell-permeable dye that covalently couples to intracellular molecules. Upon cell division, the dye is divided equally between the two daughter cells. Cells displaying lower fluorescence intensity reflect a successful proliferation process, and cells with higher intensities depict arrested nonproliferating cells.
Materials
-
50 mM borate buffer, pH 8.5, sterile
-
Antibodies:
- Purified anti-mouse CD3ε antibody (e.g., BioLegend, cat. no. 100340)
- Purified anti-mouse CD28 antibody (e.g., BioLegend, cat. no. 102116)
- APC anti-mouse CD90.2 (Thy-1.2), clone 30-H12 (e.g., BioLegend, cat. no 105311)
- Purified anti-mouse CD16/32, Fc blocker, clone 93 (e.g., BioLegend, cat. no. 101302)
-
RPMI 1640 full medium (see recipe)
-
Splenocyte single-cell suspension or isolated T cells (see Basic Protocol 2 and companion Current Protocols article by Reuven et al., 2022)
-
Trypan blue
-
DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
-
Carboxyfluorescein succinimidyl ester (CFSE) cell proliferation dye (e.g., Thermo Fisher Scientific, cat. no. C1157)
-
Heat-inactivated fetal calf serum (HI-FCS; e.g., Sigma-Aldrich, cat. no. F7524)
-
Isolated M-MDSCs or total MDSCs (see companion Current Protocols article by Reuven et al., 2022)
-
96-well U-shaped microplates, cell culture treated (e.g., Thermo Scientific, cat. no. 168136)
-
Sterile packaging (from microplates)
-
Hemocytometer
-
37°C water bath
-
Centrifuge with plate adaptor
-
Cell culture incubator, 37°C, 5% CO2
-
Flow cytometer and associated analysis software
NOTE : Cell proliferation dyes may have toxic effects on cells. Hence, we recommend CFSE staining, using a concentration that was optimized to the lowest nontoxic level.
NOTE : When calculating the number of wells in the 96-well plate, do not forget to include positive and negative controls (T cells only with or without activation, respectively).
Preparation of proliferation plate (day 0)
1.For each sample well, prepare 100 µl of 50 mM borate buffer containing 3 μg/ml anti-CD3ε and 3 μg/ml anti-CD28 antibodies.
2.Insert culture plate into sterile package and close tightly. Incubate at 4°C overnight.
3.Remove plate from 4°C (but keep in sterile package), and leave to equilibrate to room temperature for 1 hr.
4.Add 100 µl RPMI 1640 full medium to each sample well using a multichannel pipettor without touching the bottom of the well.
5.Aspirate 200 µl from the sample wells and discard liquid.
6.Add 200 µl RPMI 1640 full medium, and leave until T cells are ready for plating.
Labeling isolated T cells or total spleen cells (day 1)
7.Count single-cell suspension of splenocytes or isolated T cells using a hemocytometer and trypan blue, and adjust cell concentration to 20 × 106 cells/ml in DPBS (2× concentrated cells).
8.Prepare 2× CFSE in DPBS. For splenocytes prepare 2.5 μM CFSE, and for isolated T cells prepare 1.25 μM CFSE.
9.Mix 2× cells (from step 7) with 2× CFSE (from step 8) at a 1:1 ratio (i.e., 1 ml cells with 1 ml CFSE), and incubate in a 37°C water bath for 10 min. Gently shake tube every 2 to 3 min to mix the cells with the CFSE.
10.Add 1 vol. filtered FCS to the cells, and incubate at room temperature for 1 min.
11.Add RPMI 1640 full medium to fill the tube containing cells, and centrifuge cells 8 min at 300 × g , 4°C. Discard supernatant.
12.Resuspend cells in fresh RPMI 1640 full medium, and count using a hemocytometer and trypan blue.
Cell plating (day 1)
The coculture proliferation assay is different for isolated T cells and for total splenocytes with regard to the T cell activation procedure (steps 17a and 17b). Good experimental planning is imperative to reduce the risk of errors.
To ensure homogeneity of replicates within the experiment, it is recommended to prepare technical replicates in a microcentrifuge tube and then divide equal volumes into the sample wells. For example, for triplicates of 200 µl each, prepare one microcentrifuge tube with 700 μl cells and medium (100 µl excess), and divide 200 µl into each sample well).
13.Use a multichannel pipettor and aspirate the 200 µl medium from each well prepared in steps 1 through 6, without touching the bottom of the well.
14.Adjust concentration of cells prepared in step 7 to 2 × 106 cells/ml for isolated T cells and 6 × 106 cells/ml for splenocytes.
15.Plate 100 µl CFSE-labeled isolated T cells (0.2 × 106 cells/well) in each sample well.
16.Close plate tightly in a sterile package, and incubate at 4°C for 1 hr, allowing T cells to sediment before adding MDSCs.
17.Coculture cells with M-MDSCs:
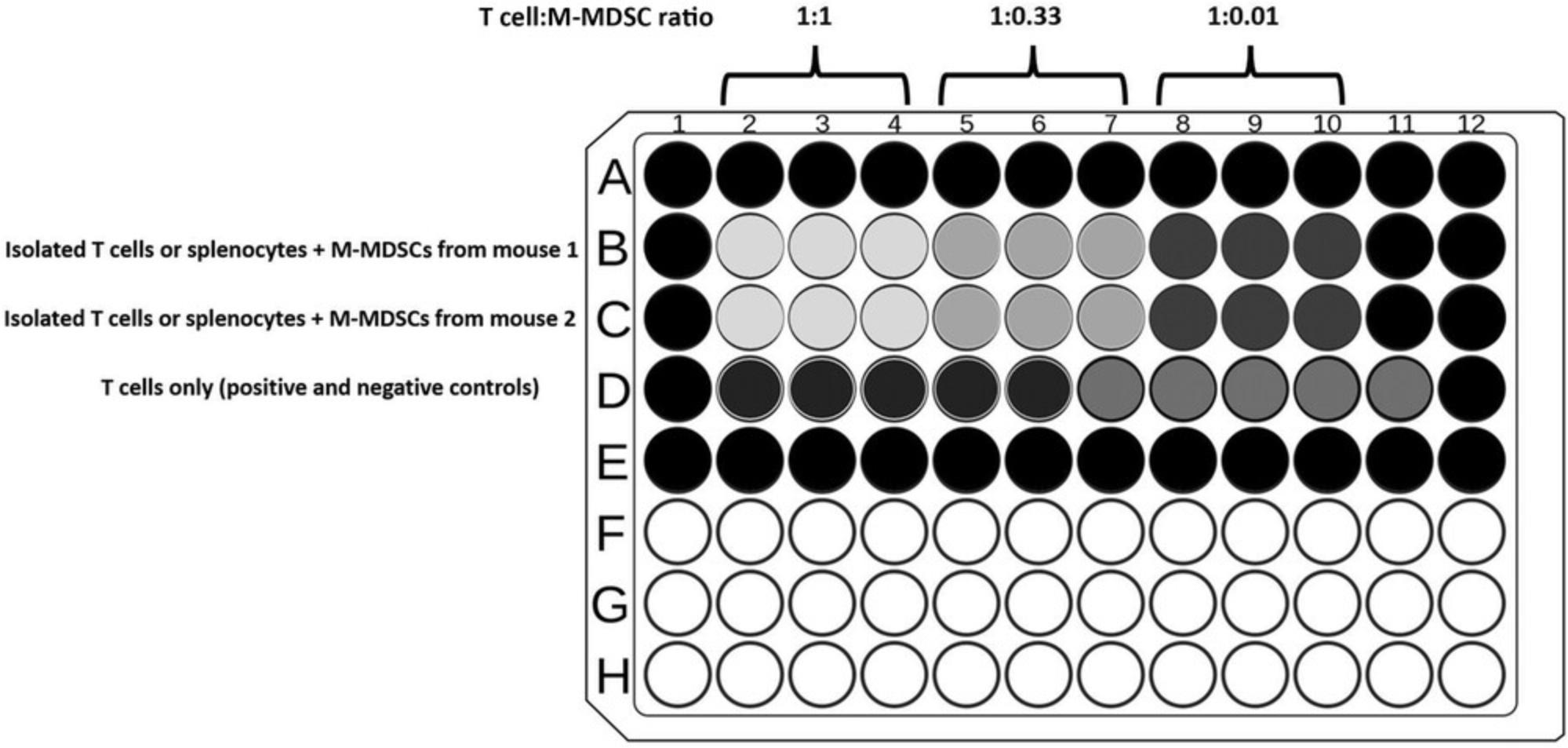
- T cells: After T cells have sedimented in the wells, add M-MDSCs by gently dispensing cells in order to reduce T cell contact with plate-bound antibodies. Prepare treatments in triplicate as described in Tables1and2.
Tables1and2represent examples for the preparation of experiments in which M-MDSCs isolated from two different mice are compared with regard to their suppressive function. The suppressive effect of M-MDSCs on T cells is determined and compared to activated T cells or activated splenocytes (positive control). The ratio between M-MDSCs and T cells or splenocytes, which are indicated in the example (presented in Tables1and2, respectively), are recommended for achieving experimental results that depict the immunosuppressive function of MDSCs and also allow for distinguishing among the features of MDSCs isolated from different mice (e.g., two different strains or models). This protocol could also be used for pan-MDSCs (isolated with anti-Gr-1), according to the cell isolation steps presented in a companion Current Protocols article by Reuven et al. (2022), and requires different ratios to determine suppression.
- Splenocytes: Coculture total splenocytes with M-MDSCs such that each sample well contains a final volume of 200 µl and final concentration of 1 μg/ml soluble monoclonal anti-CD3ε antibody.
Coculture of total splenocytes with M-MDSCs is simpler than isolated T cells, as it does not require the preparation of plate-bound antibodies for activation. The calculation of cell ratios assumes that the control mouse spleen contains ∼30% T cells. An example experimental design is presented in Table2for a proliferation assay using total splenocytes cultured with MDSCs.
| Sample | Tube replicateb | M-MDSC source 1 (4 × 106 cells/ml), μl | M-MDSCs source 2 (4 × 106 cells/ml), μl | RPMI 1640 full medium, μl |
|---|---|---|---|---|
| T cells + MDSC source 1c | B2-B4 | 375 | - | - |
| B5-B7 | 250 | - | 125 | |
| B8-B10 | 37.5 | - | 337.5 | |
| T cells + MDSC source 2c | C2-C4 | - | 375 | - |
| C5-C7 | - | 250 | 125 | |
| C8-C10 | - | 37.5 | 337.5 | |
| Positive controld | D2-6 | - | - | 375 |
| Negative controle | D7-11 | - | - | 375 |
-
CFSE, carboxyfluorescein succinimidyl ester; M-MDSC, monocytic myeloid-derived suppressor cell.
- a
Different treatments are prepared in microcentrifuge tubes and are divided into each well, as designated in the scheme, to reduce heterogeneity between replicates. The treatments are made in triplicate with excess for a total of 375 μl per treatment. Isolated T cells are already seeded in the plate (see Basic Protocol 3 step 3). A total of 100 μl content from each tube is transferred into each well. The final volume of each well is 200 μl.
- b
Refers to wells within the plate.
- c
Coculture of T cells with M-MDSCs is done at three different ratios: Wells B2-4 are at a 1:1 ratio of T cells:M-MDSCs. Wells B5-7 are a 1:0.33 ratio, and wells B8-10 are at a 1:0.1 ratio.
- d
Positive control wells are isolated T cells activated with plate-bound anti-CD3ε and anti-CD28 antibodies.
- e
Negative control wells are isolated T cells without activation.
| Sample | Tube replicateb | Splenocytes (6 × 106 cells/ml), μl | M-MDSC source 1 (8 × 106 cells/ml), μl | M-MDSC source 2 (8 × 106 cells/ml), μl | RPMI 1640 full medium with 12 mg/ml anti-CD3ɛ, μl | RPMI 1640 full medium, μl |
|---|---|---|---|---|---|---|
| Splenocytes + MDSC source 1c | B2-B4 | 125 | 562.5 | - | 62.5 | - |
| B5-B7 | 125 | 250 | - | 62.5 | 312.5 | |
| B8-B10 | 125 | 187.5 | - | 62.5 | 375 | |
| Splenocytes + MDSC source 2c | C2-C4 | 125 | - | 562.5 | 62.5 | - |
| C5-C7 | 125 | - | 250 | 62.5 | 312.5 | |
| C8-C10 | 125 | - | 187.5 | 62.5 | 375 | |
| Positive Controld | D2-6 | 125 | - | - | 62.5 | 562.5 |
| Negative Controle | D7-11 | 125 | - | - | - | 625 |
-
CFSE, carboxyfluorescein succinimidyl ester; M-MDSC, monocytic myeloid-derived suppressor cell.
- a
Different treatments are prepared in microcentrifuge tubes and are divided into each well, as designated in the scheme, to reduce heterogeneity between replicates. The treatments are made in triplicate with excess for a total of 750 μl per treatment. Isolated T cells are already seeded in the plate (see Basic Protocol 3 step 3). A total of 100 μl content from each tube is transferred into each well. The final volume of each well is 200 μl.
- b
Refers to wells within the plate.
- c
Coculture of total splenocytes with M-MDSCs is done at different ratios: Wells B2-4 are at a 1:1 ratio of splenocytes:M-MDSCs. Wells B5-7 are a 1:0.33 ratio, and wells B8-10 are at a 1:0.1 ratio.
- d
Positive control wells are total splenocytes activated with soluble anti-CD3ε antibody.
- e
Negative control wells are total splenocytes without activation.
18.Incubate plate for 72 hr in a cell culture incubator.
Analysis of T cell proliferation using flow cytometry (day 4)
19.After 72 hr of incubation, centrifuge plate with cells 8 min at 300 × g , 4°C. Discard supernatant.
20.Stain cells for flow cytometry as described in the companion Current Protocols article by Reuven et al. (2022), using the following suggested antibody mix:
- 1 μg/ml anti-CD90.2 (Thy-1.2) APC
- 1 μg/ml unlabeled anti-CD16/32.
As T cells internalize TCR upon activation, a different marker is required for distinguishing T cells. We recommend staining pan-T cells using anti-CD90.2 or CD90.1 (depending on which isoform is expressed on the cells) or anti-CD8α and anti-CD4.
When choosing the labeled antibody, do not forget that T cells are prestained with a florescent proliferation marker.
21.For accurate comparison of assay wells, set a gate in the flow cytometry software to distinguish T cells, and set reading parameters for ∼5000 gated T cells for each sample.
22.Set gain of 488-nm channel (for CFSE), and use nonactivated T cell samples, which are the samples with the highest florescence intensity, as the cells did not proliferate.
Basic Protocol 4: EFFECT OF MDSCs ON T CELL CYTOTOXIC ACTIVITY
Basic Protocol 4 describes the suppressive effect of MDSCs on the function of cytotoxic CD8 T cells isolated from PMEL mice and stimulated with the peptide gp100. CD8 T cells from PMEL mice recognize the specific peptide gp100, allowing for the selective activation of cells when introducing the peptide to the mice. Several conditions, such as peptide concentration and MDSC number, should be adjusted according to experimental specifications. Using this protocol, expansion of 2 × 106 naive PMEL CD8 T cells (at day 0) yields ∼50–60 × 106 cells (at day 7), with a maximum of 10% dead cells and purity of 98%. The initial duration of T cell activation (day 0 to 2) is an orientational time marker, which can be adjusted for convenience according to the time of isolation, with an average time of 24 to 36 hr.
To stimulate isolated T cells, plate-bound anti-CD3ε and anti-CD28 antibodies are used. When using total splenocytes, specific peptides (e.g., gp100) can be added directly into the cell culture for T cell activation. This protocol can be used to activate both CD8 and CD4 T cells depending on the mouse strain.
Materials
-
DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
-
Antibodies:
- Purified anti-mouse CD3ε, clone 145-2C11 (e.g., BioLegend, cat. no. 100340)
- Purified anti-mouse CD28, clone 37.51 (e.g., BioLegend, cat. no. 102116)
-
Isolated splenocytes and MDSCs (see companion Current Protocols article by Reuven et al., 2022)
-
EasySep Mouse CD8+ T Cell Isolation Kit (e.g., StemCell Technologies, cat. no. 19853)
-
Trypan blue
-
RPMI 1640 full medium (see recipe)
-
Recombinant murine IL-2 (e.g., PeproTech, cat. no. 212-12)
-
Cell Proliferation Dye eFluor 670 (ef670; e.g., Thermo Fisher Scientific, cat. no. 65-0840-85)
-
CellTrace Violet (CTv) Cell Proliferation Kit (e.g., Thermo Fisher Scientific, cat. no. C34557)
-
gp100 peptide
-
Fixable Viability Dye eFluor 780 (e.g., Thermo Fisher Scientific, cat. no. 65-0865-18)
-
FACS buffer (see recipe)
-
12-well plate, tissue culture treated (e.g., Thermo Scientific, cat. no. 150628)
-
Cell culture incubator, 37°C, 5% CO2
-
Hemocytometer
-
15-ml tube
-
Refrigerated centrifuge with swinging-bucket rotor
-
37°C water bath
-
Flow cytometer
-
96-well U-shaped microplates, cell culture treated (e.g., Thermo Scientific, cat. no. 168136)
NOTE : All steps performed in this protocol require sterile conditions, working in a laminar flow hood.
NOTE : This protocol describes the expansion of CD8 T cells in a 12-well plate. This can be changed to other plate sizes by adjusting the number of T cells and volume according to the surface of the wells used.
Day 0
1.Prepare antibody mix in DPBS containing 3 μg/ml anti-CD3ε and 3 μg/ml anti-CD28 antibodies.
2.Add 1 ml antibody mix to each well of the 12-well plate, and incubate for 2 to 4 hr in a cell culture incubator at 37°C or overnight at 4°C.
3.Obtain total splenocyte single-cell suspension as described in a companion Current Protocols article by Reuven et al. (2022), and resuspend cells in EasySep buffer. Isolate CD8 T cells according to the manufacturer's instructions. Keep cells at 4°C.
4.Count isolated CD8 T cells using a hemocytometer and trypan blue. Transfer 2 × 106 CD8 T cells to a new 15-ml tube.
5.Centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
6.Add 1 ml RPMI 1640 full medium, and keep cells at 4°C.
7.Remove antibody mix from wells in the assay plate by aspirating.
8.Wash each antibody-coated well two times by gently adding and then discarding 1 ml sterile DPBS.
9.Add 1 ml RPMI 1640 full medium and 1 ml isolated T cells (2 × 106 cells) to each well.
10.Incubate in a cell culture incubator for 2 days.
Day 2
11.Prewarm RPMI 1640 full medium in a 37°C water bath.
12.Carefully collect 2 ml medium containing activated CD8 T cells from the plate, and move to a new 15-ml tube.
13.Add 4 ml RPMI 1640 full medium to bring to 6 ml per tube.
14.Centrifuge 16 min at 50 × g , room temperature, to pellet live cells and to eliminate dead cells. Discard supernatant.
15.Resuspend cells in 2 ml RPMI 1640 full medium.
16.Prepare 300 U/ml (60 ng/ml; 3×) IL-2 in RPMI 1640 full medium and mix well.
17.To each well of a 12-well plate, add 1 ml RPMI 1640 full medium, 1 ml of 3× IL-2, and 1 ml RPMI 1640 full medium containing CD8 T cells.
18.Incubate in a cell culture incubator for 2 to 3 days.
Days 4 and 5
19.Collect 3 ml RPMI 1640 full medium containing cells from each individual well, and place into a separate 15-ml tube. Alternatively, pool two wells into a single tube to bring to 6 ml.
20.Adjust total volume to 6 ml using RPMI 1640 full medium, and centrifuge tubes 16 min at 50 × g , room temperature. Discard supernatant.
21.Resuspend cells in 1 ml RPMI 1640 full medium per well collected in the tube.
22.Count cells using a hemocytometer and trypan blue. Adjust cell concentration to 0.5–0.8 × 106 cells/ml.
23.Prepare 3× IL-2 as described in step 16.
24.To each well of a new 12-well plate, add 1 ml RPMI 1640 full medium, 1 ml of 3× IL-2, and 1 ml CD8 T cells (0.5–0.8 × 106 cells).
25.Incubate in a cell culture incubator for 2 to 3 days.
Day 7
Harvesting of CD8 T cells (CTLs)
26.Transfer 3 ml from each well containing cells into a separate 15-ml tube. Alternatively, pool two wells into a single tube to bring to 6 ml.
27.Adjust total volume to 6 ml using RPMI 1640 full medium, and centrifuge tubes 16 min at 50 × g , room temperature. Discard supernatant.
28.Resuspend cells in 1 to 2 ml RPMI 1640 full medium.
29.Count T cells using a hemocytometer and trypan blue. Adjust cells to desired concentration (e.g., 4 × 106 cells/ml).
Isolation of MDSCs and splenocytes
30.Obtain isolated MDSCs (see companion Current Protocols article by Reuven et al., 2022), and resuspend in RPMI 1640 full medium.
31.Count MDSCs using a hemocytometer and trypan blue. Adjust cells to desired concentration (e.g., 4 × 106 cells/ml).
32.Acquire splenocytes (see companion Current Protocols article by Reuven et al., 2022) from CD45.1 mice, and resuspend in DPBS. Keep cells at 4°C.
33.Count single-cell splenocyte suspension using a hemocytometer and trypan blue, and adjust cell concentration to 10 × 106 cells/ml in DPBS in two separate 15-ml tubes.
34.Stain one tube containing splenocyte suspension with 1 μM ef670, and stain the other tube with 1 μM CTv. Stain for 10 min in a 37°C water bath. Mix gently every few minutes.
35.Add 5× vol. RPMI 1640 full medium to each tube, and centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
36.Add 6 ml RPMI 1640 full medium, and centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
37.Count labeled cells using a hemocytometer and trypan blue.
38.Adjust concentration of stained splenocytes in tubes to 5 × 106 cells/ml in RPMI 1640 full medium.
Preparation of target and nontarget cells and cytotoxic assay
39.Add gp100 peptide to CTv-stained splenocytes to a 1 μg/ml final concentration (pulsed cells), which will serve as the target cells. Add the same volume of DPBS to ef670-stained splenocytes (nonpulsed cells), which will serve as a control (nontarget cells).
40.Incubate cells in a cell culture incubator for 1 hr, mixing every 10 min.
41.After incubation, centrifuge cells 5 min at 300 × g , 4°C. Discard supernatant.
42.Add 6 ml RPMI 1640 full medium and resuspend cells.
43.Count cells using a hemocytometer and trypan blue, and resuspend in RPMI 1640 full medium to the desired concentration.
44.Mix cells stained with CTv and ef670 at a 1:1 ratio, and confirm proper mixing by flow cytometry.
45.If needed, resuspend in RPMI 1640 full medium at desired concentration (e.g., 4 × 106 cells/ml).
46.Plate CTLs, MDSCs, and mixture of target + nontarget cells into a 96-well plate. See suggested plate plan in Figure 8.
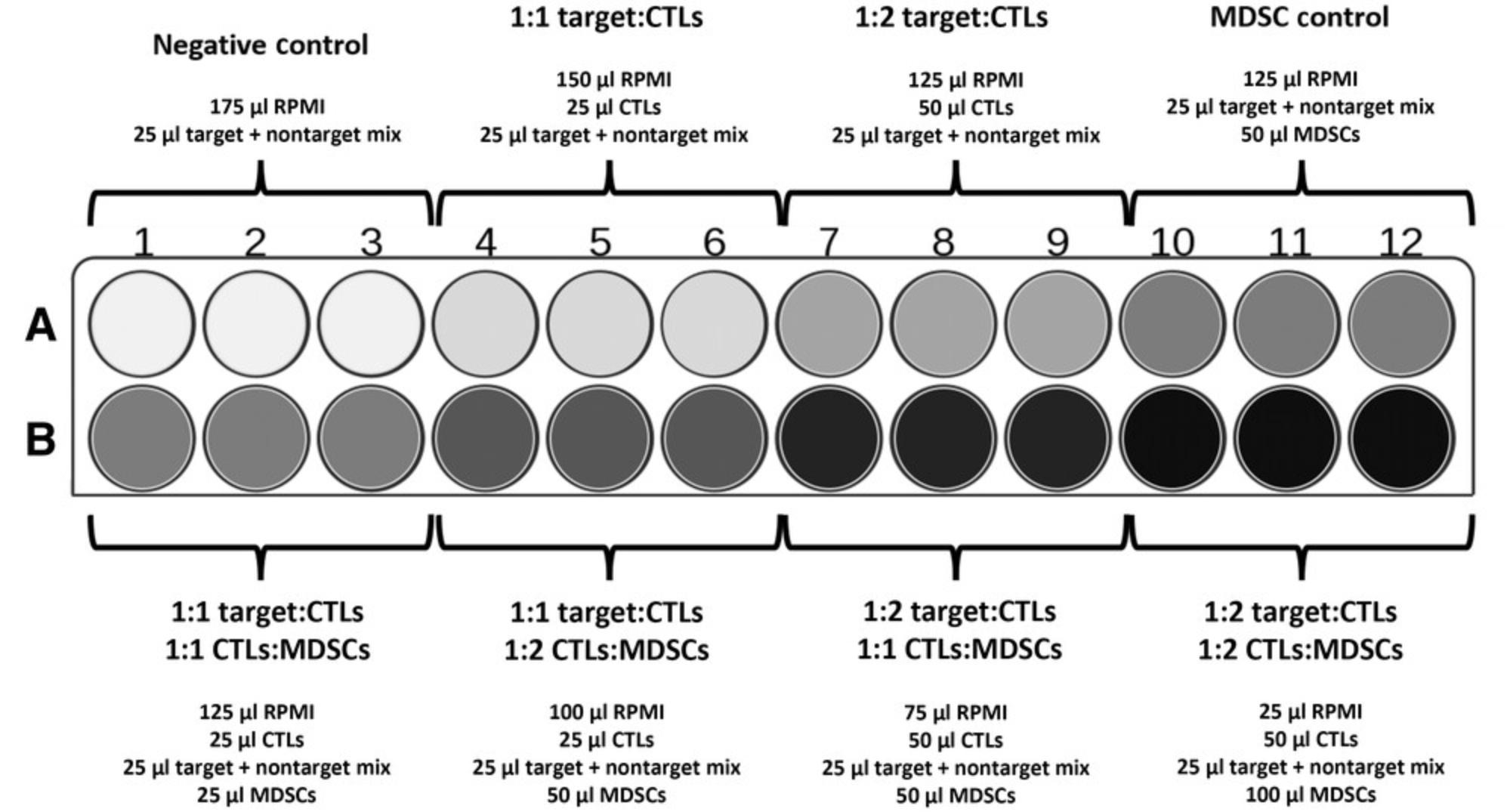
47.Incubate plate in a cell culture incubator at 37°C, 5% CO2 for 4 to 24 hr.
Days 7 and 8
48.Remove plate from incubator, and centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
49.Add 200 µl DPBS, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
50.Add 100 μl Fixable Viability Dye 780 diluted in DPBS at a 1:1000 ratio. Mix cells gently, and incubate at 4°C for 20 min.
51.Add 100 μl DPBS, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
52.Add 100 to 200 µl FACS buffer, depending on the number of cells, and mix gently.
53.Read plate by flow cytometry.
54.Calculate changes in killing capacity of CTLs under the effect of MDSCs using the following equation:
\begin{equation*}{\rm{Percent\ specific\ killing\ = \ }}\left( {{\rm{1;-\ }}\left[ {\frac{{ \def\eqcellsep{&}\begin{array}{@{}{1}{c}@{}} {{\it{(after\ seeding\ cells)}}}\\ {{\rm{target\ (pulsed)/nontarget\ (nonpulsed)}}} \end{array} }}{{ \def\eqcellsep{&}\begin{array}{@{}{1}{c}@{}} {{\rm{target\ (pulsed)/nontarget\ (nonpulsed)}}}\\ {{\it{(before\ seeding\ cells)}}} \end{array} }}} \right]} \right){\rm{\ \times \ 100}}{\rm{.}}\end{equation*}
Alternate Protocol 3: IN VIVO CYTOTOXICITY ASSAY
Alternate Protocol 3 describes an adaptation of the in vitro cytotoxicity assay for in vivo settings. The protocol involves obtaining CTLs and target and nontarget cells, as described in Basic Protocol 4 and the companion Current Protocols article by Reuven et al. (2022). These cells are adoptively transferred into recipient mice by intravenous (i.v.) injection, and the results are calculated according to the percentage of target cells versus nontarget cells within the spleen.
Additional Materials (also see Basic Protocol 4)
-
Control and chronically inflamed mice
-
Splenocyte single-cell suspension (see companion Current Protocols article by Reuven et al., 2022)
-
25-G needles
-
1-ml syringes
-
Mouse restrainer for tail vein injection
-
Additional reagents and equipment for mouse euthanasia (see Current Protocols article by Donovan & Brown, 2005)
CTL adoptive transfer
1.Obtain CTLs as described in Basic Protocol 4.
2.Resuspend cells in 5 ml RPMI 1640 full medium in a 15-ml tube.
3.Wash cells twice with 5 ml RPMI 1640 full medium by centrifuging 5 min at 300 × g , 4°C. Discard supernatant.
4.Resuspend cells in 5 ml RPMI 1640 full medium, and count using a hemocytometer and trypan blue.
5.Adjust cells to desired concentration for i.v. tail injection in DPBS (e.g., 6 × 106 cells/200 µl DPBS).
6.Perform i.v. injection of 200 µl cells in DPBS per each control and chronically inflamed mouse, and wait until MDSCs in the inflammatory environment affect T cells in vivo , which varies depending on the mouse model (e.g., 6 to 24 hr).
Nontarget and target cell injection
7.Obtain syngeneic mixture of target + nontarget cells as described in Basic Protocol 4.
8.Resuspend cells in 5 ml DPBS, and count using a hemocytometer and trypan blue.
9.Centrifuge 5 min at 300 × g , 4°C. Discard supernatant.
10.Resuspend cells to desired concentration for i.v. tail injection in DPBS (e.g., 6 × 106 cells/200 µl DPBS).
11.Perform i.v. injection in the tail by injecting 200 µl cells per mouse, and then wait the desired amount of time.
Sample preparations for flow cytometry
12.Euthanize mice and prepare a single-cell suspension of splenocytes according to the companion Current Protocols article by Reuven et al. (2022).
13.Add cells resuspended in DPBS to a 96-well plate (e.g., 1 × 106 cells/well).
14.Centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
15.Add 100 µl Fixable Viability Dye 780 diluted in DPBS at a 1:1000 ratio. Mix gently by pipetting, and incubate at 4°C in the dark for 20 min.
16.Add 100 µl DPBS, and centrifuge plate 5 min at 300 × g , 4°C. Discard supernatant.
17.Add 200 µl DPBS, and centrifuge again 5 min at 300 × g , 4°C. Discard supernatant.
18.Add 100 to 200 µl FACS buffer, depending on the number of cells per well.
19.Mix gently and read plate by flow cytometry.
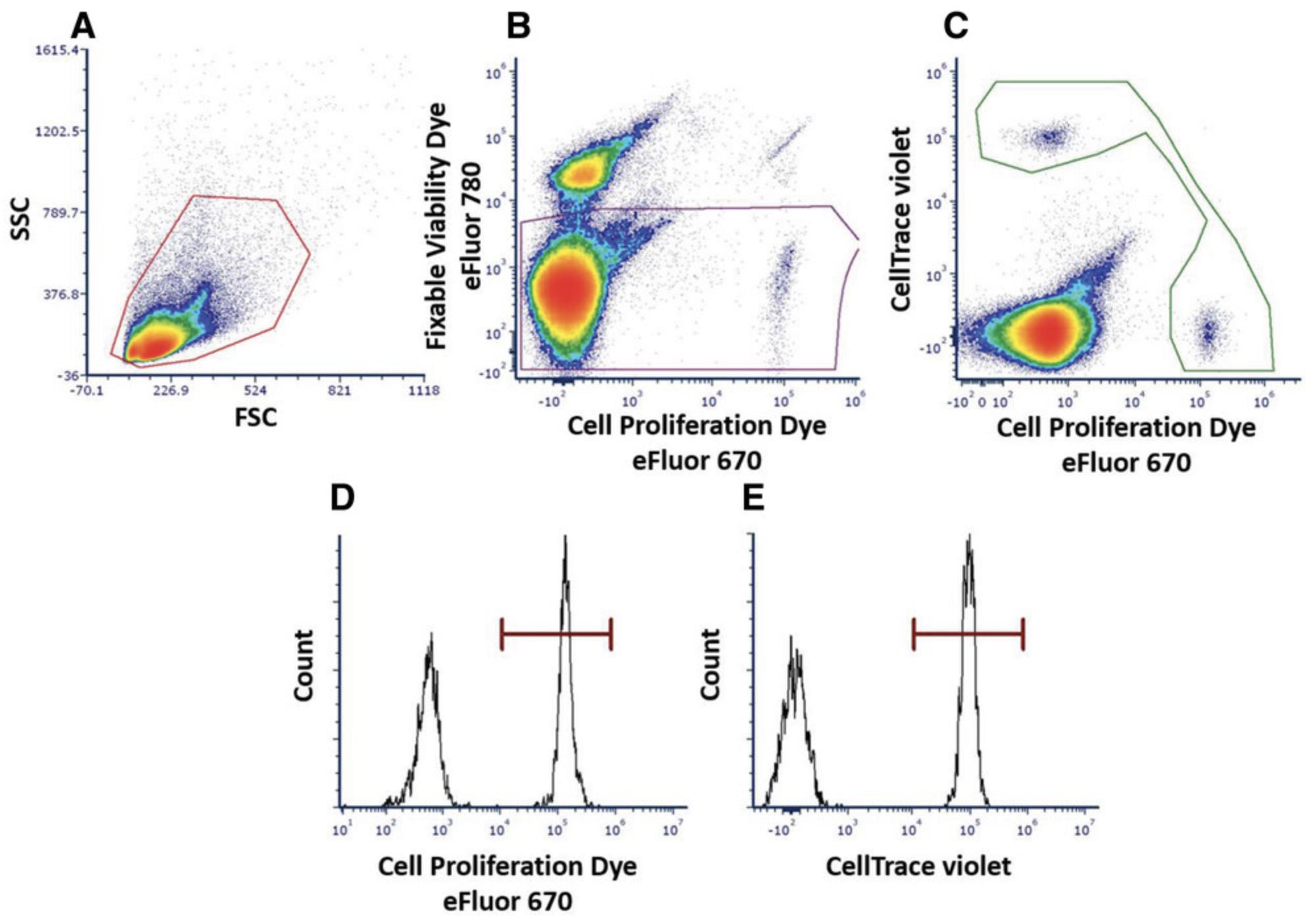
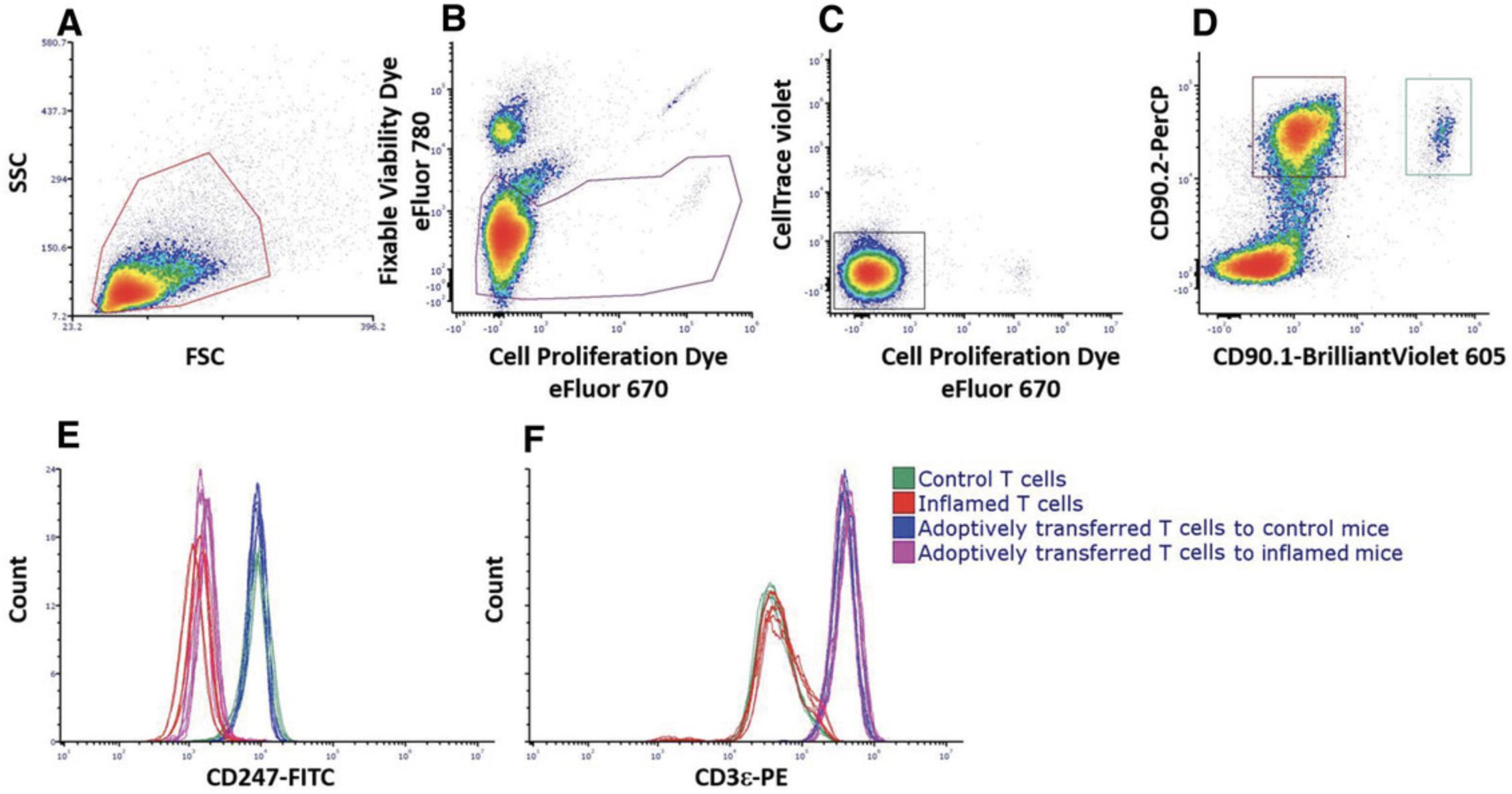
Basic Protocol 5: ANALYSIS OF MDSC DIFFERENTIATION
Under normal conditions, immature myeloid cells migrate to peripheral lymphoid organs and differentiate into mature cells such as neutrophils, macrophages, and dendritic cells. In contrast, under pathologic conditions, these cells undergo differentiation arrest, expand, and exhibit immunosuppressive functions. Investigating the mechanisms by which MDSCs stay in an undifferentiated state and expand, as well as finding new targets to modulate their differentiation and maturation, are key demands for understanding MDSC biology and how they could be manipulated. It has been suggested that promoting MDSC differentiation into mature myeloid cells can serve as a therapeutic strategy as they lose their suppressive features and functions. Herein, we propose an in vitro assay to study differentiation and the maturation state of the M-MDSC subpopulation using GM-CSF or M-CSF stimulation. GM-CSF was shown to drive bone marrow–derived monocytes into mature dendritic cells, whereas stimulation with M-CSF was shown to prime monocytes into macrophages.
Materials
-
Isolated M-MDSCs or total MDSCs (see companion Current Protocols article by Reuven et al., 2022)
-
RPMI 1640 full medium (see recipe)
-
Recombinant murine GM-CSF (e.g., PeproTech, cat. no. 315-03)
-
Recombinant murine M-CSF (e.g., PeproTech, cat. no. 315-02)
-
Optional: effector molecules of interest
-
DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
-
1× Accutase (e.g., Sigma-Aldrich, cat. no. A6964)
-
FACS buffer (see recipe)
-
Antibodies:
- Anti-MHCII FITC, clone M5/114.15.2 (e.g., TONBO, cat. no. 35-5321-u100)
- Anti-CD86 PE, clone P03 (e.g., BioLegend, cat. no. 105105)
- Anti-F4/80 APC, clone BM8 (e.g., BioLegend, cat. no. 123116)
- Anti-CD11c PE/Cy7, clone N418 (e.g., BioLegend, cat. no. 117318)
- Purified anti-mouse CD16/32 antibody, Fc blocker, clone 93 (e.g., BioLegend, cat. no. 101302)
-
96-well flat-bottom plate, tissue culture treated (e.g., Thermo Scientific, cat. no. 167008)
-
Cell culture incubator, 37°C, 5% CO2
-
Centrifuge with plate adaptor
-
96-well U-bottom plate
-
Multicolor flow cytometer with plate loader module (e.g., Beckman Coulter CytoFLEX V-B-R series or equivalent)
-
Flow cytometry data analysis software (e.g., CytExpert, version 2.4 or similar)
Plate preparation (day 0)
1.Obtain isolated M-MDSCs according to the companion Current Protocols article by Reuven et al. (2022).
2.Resuspend isolated M-MDSCs (1 × 106 cells/ml) in RPMI 1640 full medium.
3.Prepare 40 ng/ml GM-CSF or 40 ng/ml M-CSF in RPMI 1640 full medium.
4.Plate 100 μl/well (1 × 105 cells) cell suspension in a 96-well flat-bottom plate.
5.Add 100 μl/well GM-CSF or M-CSF (20 ng/ml final).
6.If investigating the effect of different molecules on M-MDSC differentiation and maturation, add molecules of interest to cell culture. Adjust volumes and concentration of cells and stock solutions to maintain GM-CSF and M-CSF at a 20 ng/ml final concentration. Incubate cells for 3 to 5 days at 37°C.
7.After 3 to 5 days harvest cells: Centrifuge plate 5 min at 300 × g , 4°C, and discard supernatant. Wash cells with DPBS and centrifuge again. Discard supernatant.
8.Add 100 μl of 1× Accutase to each well, and incubate for 30 to 60 min at 37°C until cells detach from the well.
9.Pipette cells and transfer cell suspension to a 96-well U-bottom plate.
10.Wash wells with 100 μl DPBS to detach remaining cells, and combine cells with the cells in the 96-well plate.
Flow cytometry staining (day 3 or 5)
11.Centrifuge plate with recovered cells 5 min at 300 × g , 4°C.
12.Prepare antibody mix solution in FACS buffer as follows:
- 1 µg/ml anti-MHCII FITC
- 0.5 µg/ml anti-CD86 PE
- 0.5 µg/ml anti-F4/80 APC
- 0.5 µg/ml anti-CD11c PE/Cy7
- 1 µg/ml unlabeled anti-CD16/32.
13.Add 50 µl antibody mix per well and resuspend cells.
14.Incubate cells at 4°C in the dark for 30 min.
15.Add 150 µl FACS buffer to each well to wash unbound antibodies.
16.Centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
17.Resuspend cells in 200 µl FACS buffer
18.Analyze cells by flow cytometry.
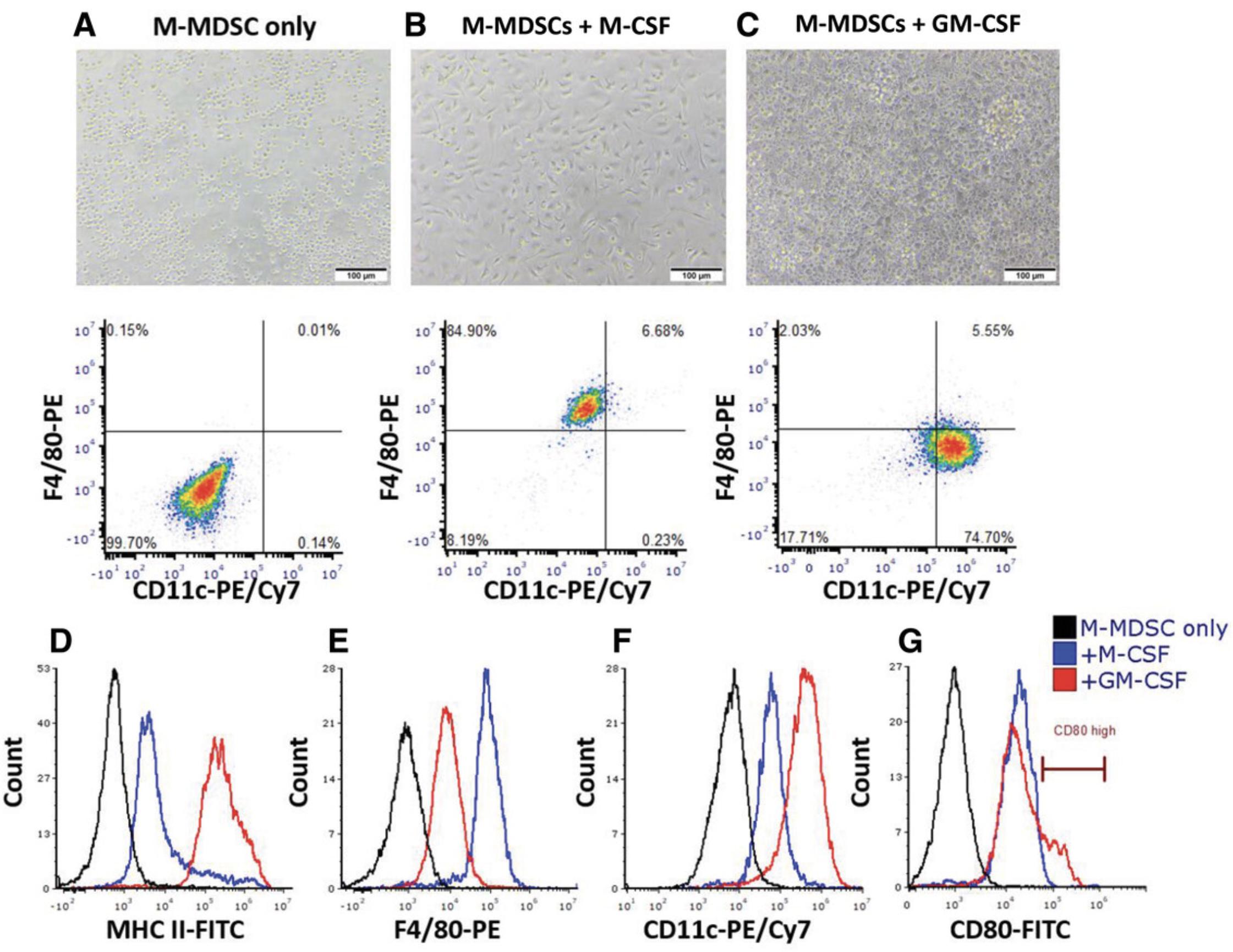
REAGENTS AND SOLUTIONS
Acidic mixture
- 1 ml of 96% H2SO4 (e.g., Millipore Sigma, cat. no. 101516)
- 3 ml of 85% H3PO4 ( e.g., Millipore Sigma, cat. no. 100573)
- 7 ml distilled deionized water
- Prepare fresh before use
FACS buffer
- 50 ml of 10× DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-5A)
- 5 ml of 0.1 M sodium azide (e.g., Sigma-Aldrich, cat. no. 199931)
- 7.5 ml HI-FCS (e.g., Sigma-Aldrich, cat. no. F7524) filtered through 0.45-µm mesh
- 437.5 ml double distilled water
- Store at 4°C for up to 1 month
Griess reagent
- 100 mg sulfanilamide (e.g., Sigma-Aldrich, cat. no. S-9251)
- 10 mg N -(1-naphthyl)ethyl-eneniamine (e.g., Millipore Sigma, cat. no. 1062370025)
- 9.7 ml ultrapure water
- 0.3 ml phosphoric acid (2.5% final; e.g., Sigma-Aldrich, cat. no. P-5811)
- Vortex until solution appears clear
- Store at −20°C for up to 3 months
ISPF, 9%
- 0.9 g ISPF (e.g., Sigma-Aldrich, cat. no. I3502)
- 10 ml of 100% ethanol
- Store at 4°C for up to 6 months
Lysis buffer
- 10 ml DPBS
- 10 μl Triton X-100 (0.1% [v/v] final; e.g., Sigma-Aldrich, cat. no. X100)
- 10 μl of Protease Inhibitor Cocktail Set III (1 ng/ml final; Millipore Sigma, cat. no. 539134)
- Prepare fresh before use
Permeabilization solution
- 45 ml FACS buffer (see recipe)
- 5 ml of 1% Saponin Quillaja sp. (0.1% [w/v] final; e.g., Sigma-Aldrich, cat. no. S4521) in DPBS
- Prepare fresh before use
ROS detection solution
- 10 μl of 50 mM luminol (e.g., Millipore Sigma, cat. no. 123072)
- 100 µl of 400 U/ml horseradish peroxidase (e.g., Millipore Sigma, cat. no. P8125)
- 890 μl HBSS with Ca2+ and Mg2+, without phenol red (e.g., Biolab, cat. no. 009015237500)
- Mix gently
- Prepare fresh before use, protecting from light and keeping on ice until use
RPMI 1641 full medium
- 500 ml RPMI 1640 medium (e.g., Biological Industries, cat. no. 01-100-1A)
- 40 ml HI-FCS (8% [v/v] final; e.g., Sigma-Aldrich, cat. no. F7524) filtered through 0.45-µm mesh
- 5 ml of 200 mM L-glutamine (2 mM final; e.g., Biological Industries, cat. no. 03-020-1B)
- 5 ml of 10,000 U/ml penicillin, 10 mg/ml streptomycin (e.g., Biological Industries, cat. no. 03-031-1B)
- 0.5 ml of 50 mM 2-mercaptoethanol (e.g., Thermo Fisher, cat. no. 31350010)
- 5 ml of 1 M HEPES (10 mM final; e.g., Biological Industries, cat. no. 03-025-1B)
- Store at 4°C for up to a month
Separation buffer
- 49 ml DPBS
- 1 ml HI-FCS (2% [v/v] final; e.g., Sigma-Aldrich, cat. no. F7524)
- 100 μl of 0.5 M EDTA, pH 8 (1 mM final; e.g., Biological Industries, cat. no. 01-862-1B)
- Strain through a 0.45-μm filter to a 50-ml tube
- Store at 4°C for up to 1 month
COMMENTARY
Background Information
MDSCs represent a population of heterogeneous, immature cells of myeloid lineage. Under pathologies characterized by chronic inflammation, MDSCs undergo differentiation arrest, expand in the bone marrow, and polarize toward immunosuppressive cells located in the periphery and sites of inflammation (Ben-Meir, Twaik, & Baniyash, 2018). MDSCs have been reported in various human cancer types and mouse models and display unique properties. These properties include increased total number during disease progression, enhanced suppressive features and functions, distinct homing to specific tissue, and diversity and plasticity (Ashkenazi-Preiser, Mikula, & Baniyash, 2021; Hou, Hou, Huang, Lei, & Chen, 2020). MDSC immunosuppressive features are characterized by elevated levels of secreted ROS and NO, production of proinflammatory cytokines, and enhanced activity of arginase-1, which results in the deprivation of arginine and cysteine from the environment. Their suppressive function is imposed on target immune cells including NK, B, dendritic, and T cells. This is exemplified by impaired cytotoxic activity of T and NK cells, reduced production of IFN-γ, arrest of T and B cell proliferation, and altered antigen-presenting function of dendritic cells. Because of these characteristics, MDSCs interfere with immune-based anticancer therapies, thus affecting treatment success rates (Sade-Feldman et al., 2016). Therefore, identifying novel markers for diagnosis, prognosis, and targeting of MDSCs is necessary to improving anticancer therapies.
In general, the protocols presented herein can be divided into methods studying the suppressive features and functions of MDSCs and methods to investigate the basic biology of MDSCs. The suppressive features of MDSCs rely on enzymatic reactions that produce mediators leading to altered immune cell responses of T and NK cells, DNA damage, and secreted cytokines and growth factors that affect immune responses and support tumor progression. The suppressive function of MDSCs is studied on target cells, mainly T and NK cells, focusing on how MDSCs alter T cells function such as proliferation, TCR CD247 and CD3/TCRαβ subunit expression, change in T and NK cell cytotoxic activity, and production of IFN-γ (Ugel et al., 2009). Moreover, the presented protocols allow for studying the biology of MDSCs in relation to their distribution in vivo in different tissue and their cell fate through differentiation arrest to maturation stages using flow cytometry and microscopy.
There are several known molecular pathways activated in MDSCs that contribute to their suppressive features and functions. The enzyme arginase-1, which is highly expressed in activated MDSCs, is known to catalyze the conversion of L-arginine to urea and ornithine. Since arginine is a precursor of polyamines, it has an important role in cellular proliferation. Many studies suggest that one of the mechanisms by which MDSC modify the TCR (CD247 downregulation) and inhibit T cell proliferation and effector function is by depleting L-arginine from the immediate environment (Gabrilovich, 2021). Inhibition of arginase-1 activity, as performed in several studies, has been shown to rescue T cell proliferation and suppress tumor growth (Vonwirth et al., 2021). Hence, measuring arginase activity in MDSCs appears to be important in studying the mechanisms of immunosuppression by these cells. L-arginine deprivation can also be induced by iNOS, of which levels are known to be upregulated in activated MDSCs. iNOS utilizes L-arginine and produces NO, thus leading to not only arginine deprivation in the environment but also generation of NO, which is known to impair the activity of antigen-presenting cells and thus serves as another mediator of MDSC immunosuppressive activity (Markowitz et al., 2017).
ROS secreted by MDSCs present within the inflammatory sites represent another form of MDSC immunosuppressive features, which can contribute to tumor development by causing DNA damage and inhibiting immune functions. ROS are known to lead to cell and tissue damage, further enhancing the inflammatory response that may result in recruitment of inflammatory immune cells. Persistent ROS secretion has been linked to various chronic inflammatory diseases such as cancer, chronic infection, rheumatic arthritis, and atherosclerosis (Ohl & Tenbrock, 2018).
Downregulation of CD247 in T and NK cells represents another major effect of MDSCs on the adaptive and innate arms of the immune system, respectively. Therefore, identification of CD247 expression in T and NK cells allows assessment of inflammatory and immunosuppressive states, which are correlated with the accumulation of MDSCs (Baniyash, 2004; Darcy et al., 2014; Ezernitchi et al., 2006). Hence, a mouse model for experimentation, especially in the field of chronic inflammation and cancer, may be assessed both for the phenotypic MDSC expression pattern and their effect on the CD247 expression in T and NK cells. Furthermore, expression of CD247 in T cells is tied to various T cell functions, which enables fast functional confirmation of isolated MDSCs, assessment of different molecules and cells affecting MDSCs–T cells interactions, and establishment of “appropriate” ex vivo and in vitro coculturing conditions mimicking in vivo conditions.
Critical Parameters and Troubleshooting
T cell proliferation in vitro as a response to activation is evident when cell culture parameters are ideal, which applies more specifically to isolated T cells. When activation parameters are not ideal, T cells may shift to an anergic state, resulting in inhibition of proliferation and other immune responses. For that reason, it is important to optimize cell culture conditions, such as amounts of activating antibodies or peptides and antigen-presenting cells, cells seeded per well, and well shape (i.e., flat or U shape). Another variable to consider when performing calibration of T cell proliferation assays is the FCS within the medium. In our laboratory, we order four to five different lots of FCS and use the one that supports the highest T cell proliferation rates.
Continual and proper mouse healthcare is important because mice harboring parasites or that carry inflammation signs for various reasons may give rise to exhausted T cells, which will enter an anergic state upon activation.
To ensure proximity of T cells and MDSCs in coculture, we have optimized the use U-bottom plates while maintaining a low number of cells in a low volume. Using a flat-bottom 96-well plate is also possible, but different conditions must be optimized before. Furthermore, to avoid allogeneic response of T cells toward MDSCs in coculture, isolated MDSCs need to originate from a syngeneic mouse strain, preferably of the same sex.
Time Considerations
Before performing an experiment, researchers should consider that different protocols require different time investments. Experiments involving isolation of cells from different organs or cell coculture take more time than simple experiments that use isolated cells directly without manipulation. When isolating MDSCs from BCG mice (described in detail in a Current Protocols article by Ben-Meir et al., 2022), the model first needs to be generated. Because endpoint is reached after 3 weeks, experiments using such BCG mice should be carefully planned. The time necessary to complete each protocol changes based on the number of mice used and cell numbers required. Preparation of CTLs takes 7 to 8 days before in vitro and in vivo cytotoxic activities are tested. Evaluating the suppressive features and functions of MDSCs requires the preparation of samples, which can take 2 to 8 hr, and an additional 16 to 72 hr until reaching the experimental endpoint, depending on the protocol used. For activation of isolated T cell and MDSC coculture experiments, culture plates with plate-bound antibodies should be prepared a day before the experiment. The time required to analyze the data depends on the type of results to be obtained.
Acknowledgments
The authors gratefully acknowledge the support of the Society of Research Associates of the Lautenberg Center and the Harold B. Abramson Chair in Immunology. The authors also thank the grant support from the Israel Science Foundation, the Israeli Ministry of Health, the Israel Cancer Research Fund, The Israel Ministry of Science and Technology, the Gross Foundation, the Bruce and Baila Waldholtz Funds, and the Joseph and Matilda Melnick Funds.
Author Contributions
Or Reuven : data curation, formal analysis, methodology, validation, writing–original draft, writing review and editing; Ivan Mikula : formal analysis, methodology, validation, writing–original draft; Hadas Ashkenazi-Preiser : formal analysis, methodology, validation, writing–original draft; Nira Twaik : formal analysis, methodology, validation, writing–original draft; Kerem Ben-Meir : formal analysis, methodology, validation, writing–original draft; Yaron Meirow : formal analysis, methodology, validation, writing–original draft; Leonor Daniel : validation, writing–review and editing; Guy Kariv : validation; Mahdi Kurd : validation; Michal Baniyash : conceptualization, funding acquisition, methodology, supervision, validation, writing–review and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data, tools, and materials that support the protocols of this article (or their source) are available from the corresponding author upon reasonable request.
Literature Cited
- Ashkenazi-Preiser, H., Mikula, I., & Baniyash, M. (2021). The diverse roles of myeloid derived suppressor cells in mucosal immunity. Cellular Immunology , 365, 104361. doi: 10.1016/j.cellimm.2021.104361
- Baniyash, M. (2004). TCR ζ-chain downregulation: Curtailing an excessive inflammatory immune response. Nature Reviews Immunology , 4, 675–687. doi: 10.1038/nri1434
- Ben-Meir, K., Twaik, N., & Baniyash, M. (2018). Plasticity and biological diversity of myeloid derived suppressor cells. Current Opinion in Immunology , 51, 154–161. doi: 10.1016/j.coi.2018.03.015
- Ben-Meir, K., Twait, N., Meirow, Y., & Baniyash, M. (2022). An in vivo mouse model for chronic inflammation–induced immune suppression: A “factory” for myeloid-derived suppressor cells (MDSCs). Current Protocols , 2, e558. doi: 10.1002/cpz1.558
- Darcy, C. J., Minigo, G., Piera, K. A., Davis, J. S., McNeil, Y. R., Chen, Y., … Woodberry, T. (2014). Neutrophils with myeloid derived suppressor function deplete arginine and constrain T cell function in septic shock patients. Critical Care , 18, 1–12. doi: 10.1186/cc14003
- Donovan, J., & Brown, P. (2005). Euthanasia. Current Protocols in Neuroscience , 33, A.4H.1–A.4H.4. doi: 10.1002/0471142301.nsa04hs33
- Ezernitchi, A. V., Vaknin, I., Cohen-Daniel, L., Levy, O., Manaster, E., Halabi, A., … Baniyash, M. (2006). TCR ζ down-regulation under chronic inflammation is mediated by myeloid suppressor cells differentially distributed between various lymphatic organs. The Journal of Immunology , 177, 4763–4772. doi: 10.4049/jimmunol.177.7.4763
- Gabrilovich, D. I. (2021). The dawn of myeloid-derived suppressor cells: Identification of arginase I as the mechanism of immune suppression. Cancer Research , 81, 3953–3955. doi: 10.1158/0008-5472.can-21-1237
- Hou, A., Hou, K., Huang, Q., Lei, Y., & Chen, W. (2020). Targeting myeloid-derived suppressor cell, a promising strategy to overcome resistance to immune checkpoint inhibitors. Frontiers in Immunology , 11, 783. doi: 10.3389/fimmu.2020.00783
- Markowitz, J., Wang, J., VanGundy, Z., You, J., Yildiz, V., Yu, L., … Carson, W. E. (2017). Nitric oxide mediated inhibition of antigen presentation from DCs to CD4+ T cells in cancer and measurement of STAT1 nitration. Scientific Reports , 7, 1–13. doi: 10.1038/s41598-017-14970-0
- Ohl, K., & Tenbrock, K. (2018). Reactive oxygen species as regulators of MDSC-mediated immune suppression. Frontiers in Immunology , 9, 2499. doi: 10.3389/fimmu.2018.02499
- Reuven, O., Mikula, I. Jr., Ashkenazi-Preiser, H., Twaik, N., Ben-Meir, K., Meirow, Y., … Baniyash, M. (2022). Phenotypic characterization and isolation of myeloid-derived suppressor cells. Current Protocols , 2, e561. doi: 10.1002/cpz1.561
- Sade-Feldman, M., Kanterman, J., Klieger, Y., Ish-Shalom, E., Olga, M., Saragovi, A., … Baniyash, M. (2016). Clinical significance of circulating CD33+ CD11bHLA-DR myeloid cells in patients with stage IV melanoma treated with ipilimumab. Clinical Cancer Research , 22, 5661–5672. doi: 10.1158/1078-0432.ccr-15-3104
- Ugel, S., Delpozzo, F., Desantis, G., Papalini, F., Simonato, F., Sonda, N., … Bronte, V. (2009). Therapeutic targeting of myeloid-derived suppressor cells. Current Opinion in Pharmacology , 9, 470–481. doi: 10.1016/j.coph.2009.06.014
- Vonwirth, V., Bülbül, Y., Werner, A., Echchannaoui, H., Windschmitt, J., Habermeier, A., … Munder, M. (2021). Inhibition of arginase 1 liberates potent T cell immunostimulatory activity of human neutrophil granulocytes. Frontiers in Immunology , 11, 3823. doi: 10.3389/fimmu.2020.617699
Citing Literature
Number of times cited according to CrossRef: 2
- Or Reuven, Ivan Mikula, Hadas Ashkenazi‐Preiser, Nira Twaik, Kerem Ben‐Meir, Yaron Meirow, Leonor Daniel, Guy Kariv, Mahdi Kurd, Michal Baniyash, Phenotypic Characterization and Isolation of Myeloid‐Derived Suppressor Cells, Current Protocols, 10.1002/cpz1.561, 2 , 10, (2022).
- Kerem Ben‐Meir, Nira Twaik, Yaron Meirow, Michal Baniyash, An In Vivo Mouse Model for Chronic Inflammation–Induced Immune Suppression: A “Factory” for Myeloid‐Derived Suppressor Cells (MDSCs), Current Protocols, 10.1002/cpz1.558, 2 , 10, (2022).

