Three-Dimensional Fruit Tissue Habitats for Culturing Caenorhabditis elegans
Malosree Maitra, Malosree Maitra, Aurélie Guisnet, Aurélie Guisnet, Sreeparna Pradhan, Sreeparna Pradhan, Michael Hendricks, Michael Hendricks
Abstract
Environmental factors influence many traits of biological interest, but reproducing an animal's natural habitat in a controlled laboratory environment is challenging. Environmental enrichment—adding complexity to the usually simplistic conditions under which laboratory animals are raised—offers a potential tool for better understanding biological traits while maintaining controlled laboratory conditions. For the model nematode Caenorhabditis elegans , the contrast between the natural environment and the laboratory conditions in which they are raised is enormous. Although several methods have been developed in an effort to complexify C. elegans laboratory conditions, there is still a need for an enriched controlled laboratory habitat in which C. elegans can be raised over several generations, the bacterial food availability is similar to that in traditional agar plates, and the animals are crawling as opposed to swimming or burrowing. To this end, we describe here a standardized protocol for creating controlled, reproducible, three-dimensional environments for multigenerational maintenance of C. elegans in the laboratory. These environments are derived from decellularized apple hypanthium tissue and have bacterial food uniformly distributed throughout. We also describe how traditional C. elegans methods of collecting synchronized eggs, cleaning contaminated stocks, and collecting animal populations are adapted to our scaffold environment. These methods can be adapted to host different bacteria or bacterial populations, and the resulting scaffolds can be used in a range of experimental designs for behavioral and phenotypical studies in C. elegans and other nematodes. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Decellularization and storage of apple tissue
Basic Protocol 2 : Preparation of plates from decellularized apple scaffolds
Basic Protocol 3 : Synchronization of eggs or animals and cleaning contaminated stocks from scaffold plates
Alternate Protocol : Collection of non-synchronized larvae and adults from scaffold plates
INTRODUCTION
The laboratory conditions in which Caenorhabditis elegans are classically grown differ greatly from the wild environments in which they thrive. Notably, wild C. elegans navigate three-dimensional habitats of plant stems or rotting fruits, feeding on bacteria and small eukaryotes while experiencing a range of changing biotic factors and predators (Félix & Braendle, 2010). In the lab, C. elegans are typically cultured on flat, homogeneous soft agar Petri dishes, constantly crawling in an abundant monoxenic bacterial food source under constant humidity and temperature conditions (Stiernagle, 2006). Although these simplified conditions empower the use of C. elegans as a model organism and allow for strongly standardized and controlled experimental designs, the importance of gene-environmental interactions suggests that these minimalistic conditions can affect gene expression patterns and behaviors in ways that limit our understanding of the phenomena being studied. As studying nematodes in the wild or perfectly reproducing their wild habitat in a controlled laboratory setting is challenging, we turn here to environmental enrichment, which involves adding features to laboratory habitats to provide sensory stimulation or promote activity and interaction in order to create a habitat that resembles the dimensionality and complexity of the wild habitat.
Previous approaches to culturing C. elegans in a three-dimensional or enriched environment such as liquid culture, gel culture, soil, molded agar, paper-based platforms, beads, or gauze have each presented challenges (Baird, 1999; Lee et al., 2011; Lee, Lee, Choi, Park, & Lee, 2015; Park et al., 2008; Pierce-Shimomura et al., 2008; Tahernia, Mohammadifar, & Choi, 2020). In particular, there is a need for an enriched culture method where animals can be maintained over several generations, environmental variables are fully controlled, and locomotion patterns are comparable to those observed in standard growing conditions (Petersen, Dirksen, & Schulenburg, 2015; Voelkl et al., 2020).
In this article, we describe how to create decellularized scaffolds from fruit tissue and use them to prepare three-dimensional enriched maintenance plates for C. elegans that are analogous in most ways to standard laboratory culture plates. The apple fruit hypanthium tissue was chosen for its average porosity, durable structural integrity, size and uniformity, availability, and previous success in mammalian cell growth (Modulevsky, Lefebvre, Haase, Al-Rekabi, & Pelling, 2014).
Due to the porosity and dimensionality of the decellularized scaffolds, standard C. elegans culture methods (as outlined in Stiernagle, 2006) do not allow bacteria to grow uniformly through the scaffold. Uniform growth of bacteria through the scaffold is essential to ensure that food is uniformly available to the population so that the effects of the enriched environment are not confounded by the effects of food availability. For this reason, we have devised our protocol to ensure uniform growth throughout the enriched scaffold habitat. With our protocols, C. elegans grown on scaffold plates can be maintained through multiple generations using traditional maintenance equipment (i.e., a compound microscope and platinum worm pick), and tailoring the enriched habitat to fit on traditional agar plates allows this method to be used without additional laboratory equipment. In addition, the food source and density can be adapted to the requirements of the experimental design, and the stiffness and porosity of the enriched habitat allow animals to crawl rather than swim or burrow. Overall, this enriched habitat allows for controlled and standardized culture of C. elegans in an enriched environment that is similar in most ways to traditional culture methods and plates.
In Basic Protocol 1, we describe how to decellularize apple tissue into extracellular matrix scaffolds and prepare the scaffolds for subsequent growth of bacterial food. In Basic Protocol 2, we describe how to use the prepared scaffolds to create enriched culture plates with monoxenic bacterial food throughout. Although our work uses the traditional Escherichia coli OP50 strain as a food source, we also outline the critical elements that must be fine-tuned for growth of other bacteria. In Basic Protocol 3, we describe how to collect synchronized eggs from animals cultured on scaffold plates and how to clean animal stocks from contaminated scaffold plates. Finally, in the Alternate Protocol, we describe how to collect non-synchronized animal populations from scaffold plates.
STRATEGIC PLANNING
To create scaffold and agar maintenance plates, both Basic Protocols 1 and 2 must be completed. These protocols are time-consuming and require several overnight steps that can impede experimental workflow. To alleviate workload when a large number of plates are needed, large batches of decellularized scaffold plates can be prepared in advance and stored as outlined in Basic Protocol 1.
For Basic Protocol 2, fresh apples, as opposed to stored apples, provide the best results. The apples should not be damaged or bruised, as these will create oxidized scaffolds with poor structural integrity. Apples should be kept at room temperature before use, as warming, cooling, or freezing them will also decrease the quality of the resulting scaffolds. Short but wide apples tend to have the most usable material.
Basic Protocol 2 calls for four NGM agar plates. These should be removed from storage and left at room temperature (preferably spread out, not stacked) for 2-3 days prior to use to allow excess moisture to evaporate. Plates should not be left outside of storage for more than 3 days, however, as low ambient air humidity leads to excess drying. It is also recommended to set out a handful of extra plates (four to six) because of sporadic contamination. Thus, to carry out Basic Protocol 2 as described, eight plates should be removed from storage 2 days before starting the protocol.
Basic Protocol 1: DECELLULARIZATION AND STORAGE OF APPLE TISSUE
This protocol describes methods for decellularizing, washing, and storing extracellular matrix scaffolds derived from apple tissue. Because the numerous washes in this protocol are time-consuming, it can be advantageous to make scaffolds in bulk and store them until use. At the end of this protocol, scaffolds should be rigid and translucent-white, not soft or oxidized (yellow-orange).
This protocol is described for a large but manageable batch of 20 apple slices, with each slice being used to prepare a single scaffold plate later in Basic Protocol 2.Details are provided for adjusting the protocol for more or fewer slices. The “wash, sterilize, and store slices” section can be carried out several times in one day by starting batches at 15-min intervals, so that transfers between washes are staggered and done continuously. To keep track of wash steps and incubation times, it is recommended to create a table of steps and fill it in as the steps are carried out. For additional details, see Critical Parameters.
CAUTION : Extreme care should be taken when manipulating large volumes of ethanol, as they are a significant fire hazard. Ethanol should be kept away from fire and heat sources. Flame-sterilized forceps should be totally cooled before any contact with ethanol.
CAUTION : Sodium dodecyl sulfate (SDS) should be kept away from heat and manipulated with appropriate personal protective equipment as it is flammable and causes irritation. SDS solution is also slippery. All working areas, shaking platforms, and flasks should be wiped clean of any spill to prevent falls.
Materials
-
Fresh MacIntosh apples (Malus domestica ‘MacIntosh’)
-
0.5% (w/v) sodium dodecyl sulfate (SDS, Fisher Bioreagents, cat. no. BP166500) in deionized water
-
Deionized water
-
100% ethanol and 70% (v/v) ethanol in deionized water
-
Mandolin slicer
-
4-cm-diameter round steel cookie cutter
-
4-L glass wide-mouth Erlenmeyer flasks, sterile
-
Aluminum foil
-
Shaking platform
-
Stainless steel offset broad flat-tip forceps (Thermo Fisher Scientific, cat. no. 16-100-116), sterile
-
Sterile sample bags (Thermo Fisher Scientific, cat. no. 01-812-201)
-
Vacuum sealer
Day 1: Cut and decellularize slices
NOTE : Complete these steps as quickly as possible, as prolonged exposure to air increases the risk of scaffold oxidation. Manipulate apples and slices with gloves, as contact with human skin also speeds up oxidation.
1.Make 3-mm-thick slices parallel to the core from two or three Macintosh apples using a mandolin slicer.
2.Cut out circles of hypanthium tissue from the slices using a round cookie cutter with a 4-cm diameter. Discard skin and slices containing ovary core.
3.Immediately soak slices in 2 L of 0.5% (w/v) SDS in a sterile 4-L glass wide-mouth Erlenmeyer flask. Cover the mouth of the flask with aluminum foil to prevent introduction of debris and leave overnight (12-24 hr) at room temperature on a shaking platform at 125 rpm.
Day 2: Wash, sterilize, and store slices
4.Add 2 L deionized water to a new sterile 4-L flask and transfer scaffolds to this flask. To do this, pour the liquid content of the flask into a waste container while using forceps (sterilized by flaming, but totally cooled) to catch the decellularized slices as they flow out of the flask and transfer them to the new flask.
5.Cover the mouth of the new flask with foil, then shake and twirl by hand for 30 s.
6.Repeat steps 4-5 for a total of three quick washes.
7.Repeat step 4, then cover the mouth with foil and shake for 30 min at room temperature and 125 rpm.
8.Repeat step 7.
9.Add 2 L of 70% (v/v) ethanol to a sterile 4-L flask and transfer scaffolds to the ethanol.
10.Cover the mouth with foil and shake for 1 hr at room temperature and 125 rpm.
11.Add 2 L of 100% ethanol to a sterile 4-L flask and transfer scaffolds into it.
12.Cover the mouth with foil and shake for 1 hr at room temperature and 125 rpm.
13.Proceed directly to Basic Protocol 2 or store if needed. To store, transfer to sterile sample bags, taking care not to transfer excessive amounts of ethanol. Vacuum-seal the bags and keep up to 3 weeks at 4°C.
Basic Protocol 2: PREPARATION OF PLATES FROM DECELLULARIZED APPLE SCAFFOLDS
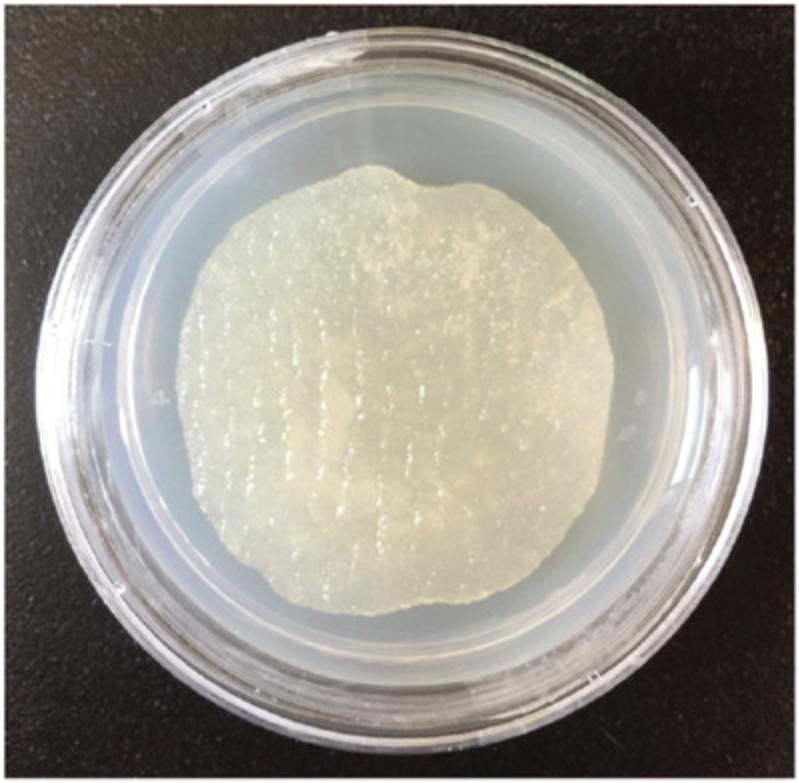
This protocol is designed to create three-dimensional, controlled monoxenic maintenance plates (Fig. 1) from decellularized scaffolds. Due to the porosity of the scaffolds, it is not possible to obtain uniform bacterial growth throughout the scaffolds using standard methods (as outlined in Stiernagle, 2006). This protocol was developed to ensure uniform bacterial growth through a series of washes and incubations.
For parallel culture of animals on standard agar plates, we also describe how to prepare standard agar plates with a comparable cross-sectional bacterial density to the scaffold plates through a modification of the standardized methods outlined in Stiernagle (2006). We describe how to produce two plates of each type at once, with additional details for adjusting the protocol to produce more plates.
NOTE : Scaffolds are more susceptible to contamination than traditional agar plates. Throughout this protocol and when using the resulting plates, always work under sterile conditions close to a flame, with sterile reagents, equipment, and working surfaces.
Materials
-
Decellularized scaffold slices (see Basic Protocol 1)
-
NGML (see recipe)
-
E. coli OP50 plated stock (e.g., Caenorhabditis Genetics Center; see Stiernagle, 2006)
-
6-cm NGM agar plates (see recipe)
-
Sterile water
-
250-ml glass wide-mouth Erlenmeyer flask
-
Aluminum foil
-
Shaking platform
-
Sterile scalpel (optional)
-
125-ml glass Erlenmeyer flasks
-
37°C shaking incubator
-
Sterile 6-cm Petri dishes (Thermo Fisher Scientific, cat. no. AS4051)
-
Kimwipes Delicate Task Wipers, 1-ply (Thermo Fisher Scientific, cat. no. 06-666-2)
-
Water-resistant tight-seal box
-
28°C incubator
-
Sterile stainless steel offset broad flat-tip forceps (Thermo Fisher Scientific, cat. no. 16-100-116)
-
Parafilm (Thermo Fisher Scientific, cat. no. 13-374-12)
Day 1: Prepare scaffolds and bacterial culture
1.Transfer two decellularized scaffold slices to 100 ml NGML in a sterile 250-ml glass wide-mouth Erlenmeyer flask under sterile conditions. Cover the mouth with foil and shake overnight (8-16 hr) at room temperature on a shaking platform at 145 rpm to dilute out the ethanol.
2.Inoculate three sterile 125-ml flasks containing 50 ml NGML with a single colony of E. coli OP50 from a plated stock. Cover flasks with foil and incubate on a shaking platform at 37°C overnight.
Day 2: Inoculate scaffolds and standard culture plates
3.Add 5 ml saturated OP50 culture to each of two sterile 6-cm Petri dishes.
4.Transfer one scaffold into each dish. Add more culture if necessary to completely submerge the scaffold.
5.Pipet 100 µl of the same culture onto each of two 6-cm NGM agar plates and shake briefly to spread. If there is condensation on the lid, wipe it off with delicate-task wipers.
6.Line the interior of a water-resistant tight-seal box with delicate-task wipers and saturate them with sterile water.
7.Place scaffold dishes and agar plates inside the box and close tightly. Make sure plates are flat and stable inside the box.
8.Incubate 5.5-6 hr at 28°C.
9.Take a scaffold with forceps (sterilized by flaming) and gently tap your wrist on the work surface to release 1-2 drops of excess culture. Deposit on a new NGM agar plate and gently lay the flat portion of the forceps over the scaffold to push out air from between the scaffold and the agar surface. Repeat with the second scaffold.
10.Seal scaffold plates and seeded agar plates with Parafilm and keep upside down in a closed box at room temperature for at least 24 hr before use. Store plates this way for up to 3 weeks.
Basic Protocol 3: SYNCHRONIZATION OF EGGS AND ANIMALS AND CLEANING CONTAMINATED STOCKS FROM SCAFFOLD PLATES
Several experimental designs require the use of synchronized eggs or animal populations. This protocol is adapted from Stiernagle (2006) for collection of synchronized eggs from scaffold plates. The inhabited scaffold is sectioned and washed to release the eggs, which are then collected in a pellet and can be used directly or added to a fresh plate to grow the synchronized population to the desired stage.
This protocol is not recommended for quantifying egg laying inside scaffolds. Eggs tend to adhere to the surface on which they are laid, and it is not possible to assess the proportion of eggs collected with this protocol due to the dimensionality of the scaffold.
It is not uncommon for C. elegans stocks on culture plates to develop a fungal or bacterial contamination, and scaffold plates are more prone to contamination than standard agar plates. To address this issue, we also describe how to clean contaminants from scaffold plate worm stocks using an adaptation of the bleach method from Stiernagle (2006). Contamination spreads easily within and between scaffold plates, so it is important to clean stocks or dispose of contaminated plates rapidly once contamination occurs. Using bleach to remove contaminants also dissolves hatched animals, but embryos are protected by the eggshell and will survive. The bleach is subsequently washed out to protect these animals as they hatch. It is thus essential to start the protocol with a plate that contains eggs or gravid adults. If all eggs have already hatched, it is preferable to transfer a few young contaminated animals to a fresh plate and proceed with the protocol when they have laid eggs, because excessive contamination can impede growth and egg laying. After cleaning contaminated stocks with bleach solution, a sterile pellet of eggs is collected in a small centrifuge tube and can be re-plated onto a fresh plate to resume animal culture.
Synchronization and decontamination use the same basic egg extraction method, and the slight differences between them are outlined below. The Alternate Protocol can be used to collect non-synchronized larval or adult worms from scaffold plates.
NOTE : If collected eggs or worms will be re-plated, carry out these protocols under sterile conditions close to a flame, with sterile reagents, equipment, and working surfaces.
Materials
-
Scaffold plates with C. elegans :
- late-L4 from any growing condition (for synchronizing)
- contaminated plate containing eggs (for cleaning)
-
Seeded scaffold plate (from Basic Protocol 2)
-
NGML (see recipe) or bleach solution (see recipe)
-
Sterile deionized, distilled water
-
Sterile platinum worm pick
-
Sterile scalpel
-
Sterile stainless steel offset broad flat-tip forceps (Thermo Fisher Scientific, cat. no. 16-100-116)
-
1.5-ml microcentrifuge tubes (Diamed, cat. no. BPSMCT150-N)
-
Compound microscope
1.For synchronizing only : Place ten late-L4 C. elegans larvae on a freshly seeded scaffold plate using a sterile platinum worm pick and keep overnight (8-12 hr) at 20°C.
2.Cut the scaffold in three parallel pieces of equal width using a sterile scalpel. Be careful not to cut or scratch the agar while cutting the scaffold as eggs and animals can get stuck in the interstices.
3.Use flat-tip forceps to lay the pieces on top of one another on one side of the dish using sterile forceps (Fig. 2).
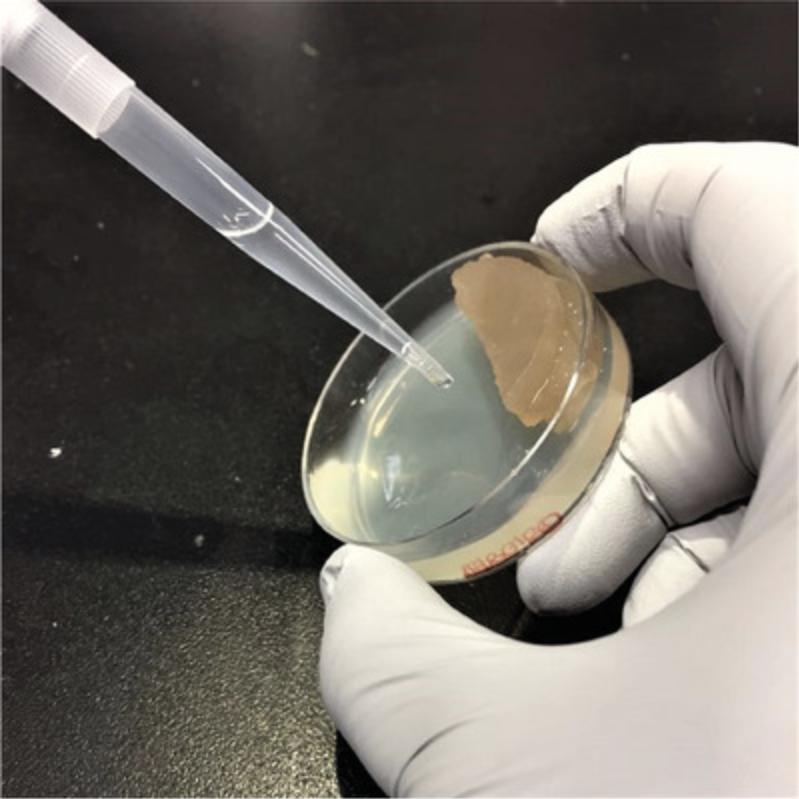
4.Holding the dish at a 30°-45° angle with the scaffold pieces at the top, wash 2 ml NGML over the scaffold pieces so that the eggs and animals are washed out and collected along the opposite edge (Fig. 2).
5.Collect as much liquid without eggs or animals as possible from the bottom of the dish by observing under a microscope, and wash this solution over the scaffold pieces again.
6.Repeat step 5 until no more eggs or animals are washed out of the scaffold pieces.
7.Transfer the collected liquid to a 1.5-ml microcentrifuge tube.
8.Centrifuge for 1 min at 2000 × g , room temperature, in a tabletop microcentrifuge.
9.Remove as much liquid (supernatant) as possible from the tube while being careful not to remove any eggs.
10.If eggs need to be washed completely of bacterial food before use, repeat steps 8-9 three to four times, filling the tube with sterile water each time.
Alternate Protocol: COLLECTION OF NON-SYNCHRONIZED LARVAE AND ADULTS FROM SCAFFOLD PLATES
Animals can be picked directly from the scaffold, but a large portion of the population remains inaccessible within the scaffold. If the user requires a large number of animals, but does not need eggs or a synchronized population, this protocol can be used to force animals out of the scaffold and is faster than Basic Protocol 3.By shaking scaffold pieces in liquid, the animals are forced to exit the scaffold and can be easily collected with a pipet.
Again, this protocol is not recommended to assess population size, as it is not possible to know whether all animals have exited the scaffold. It is also not recommended as an alternative for collecting eggs, as cutting and shaking the scaffold breaks off pieces that are as small as eggs and can be difficult to differentiate from eggs.
Additional Materials (also see Basic Protocol 3)
- Scaffold plate with growing C. elegans population
- Reusable 6-cm glass Petri dish (Thermo Fisher Scientific, cat. no. 08-747A)
- Glass Pasteur pipet
1.Clean a glass Petri dish with ethanol and pressurized air.
2.Fill dish ¾ with NGML (∼10 ml).
3.Use a scalpel to cut out 1- to 2-cm3 scaffold pieces from a plate of C. elegans and then use forceps to move them delicately to the Petri dish.
4.Release animals by shaking the scaffold pieces delicately in the liquid with the forceps.
5.Collect animals with a glass Pasteur pipet under a microscope.
6.If animals need to be washed completely of bacterial food before use, see Basic Protocol 3, steps 7-10.
REAGENTS AND SOLUTIONS
Bleach solution
Add 2 volumes household bleach to 1 volume of 5 M NaOH (Fisher Bioreagents, cat. no. BP359212) in a conical tube and vortex briefly to mix. Store up to 6 months at room temperature.
Cholesterol, 5 mg/ml
Add 75 mg cholesterol (BioShop, cat. no. CHL380) to 15 ml of 100% ethanol in a 25-ml conical screw-cap tube and vortex continuously until completely dissolved. Store up to 6 months at room temperature.
Nematode growth medium (NGM) agar plates
- 975 ml deionized, distilled water
- 3 g NaCl (Fisher Chemical, cat. no. S2713)
- 2.5 g peptone (Thermo Scientific, cat. no. LP0037B)
- 22 g agar (Sigma Life Science, cat. no. A1296)
- 1 ml 5 mg/ml cholesterol (see recipe)
- 25 ml 1 M potassium phosphate buffer (see recipe)
- 1 ml sterile 1 M CaCl2 (Fisher Bioreagents, cat. no. FLBP510500)
- 1 ml sterile 1 M MgSO4 (Fisher Chemical, cat. no. M65500)
Add the first five reagents in order to a 2-L flask with stir bar. Cover with foil, sterilize by autoclaving, and cool on a stir plate to 60°-70°C (warm to touch). Add the remaining three reagents in order while stirring. Pour 9 ml medium per 6-cm sterile Petri dish (Thermo Fisher Scientific, cat. no. AS4051) and let plates dry spread out (not stacked) for 36-48 hr at room temperature. Store up to 1 month at 4°C. Remove from storage 2 days before use to evaporate condensation (see Strategic Planning).
Nematode growth medium liquid (NGML)
- 487.5 ml deionized, distilled water
- 1.5 g NaCl (Fisher Chemical, cat. no. S2713)
- 1.25 g peptone (Thermo Scientific, cat. no. LP0037B)
- 500 µl 5 mg/ml cholesterol (see recipe)
- 12.5 ml 1 M potassium phosphate buffer (see recipe)
- 500 µl sterile 1 M CaCl2 (Fisher Bioreagents, cat. no. FLBP510500)
- 500 µl sterile 1 M MgSO4 (Fisher Chemical, cat. no. M65500)
Add the first four reagents in order to a 500-ml glass or Pyrex bottle with cap and stir bar. Screw cap on loosely, sterilize by autoclaving, and cool to 20°-25°C. Add the remaining three reagents in order. Stir 5 min, then screw cap on tightly. Store up to 1 month at room temperature.
Potassium phosphate buffer, 1 M
- For 12.5 ml :
- 10.85 ml sterile 1 M KH2PO4 (Fisher Chemical, cat. no. P2853)
- 1.65 ml sterile 1 M K2HPO4 (Fisher Chemical, cat. no. P288500)
- Prepare fresh before use
COMMENTARY
Background Information
Growing interest in the effects of gene-environment interactions demands reliable controlled techniques for environmental enrichment in model organisms such as C. elegans and other nematodes (Alfred & Baldwin, 2015; Gardner, Boitano, Mancino, & D'Amico, 1975). Current methods for culture of C. elegans in enriched conditions, such as liquid culture, gel culture, soil, molded agar, paper-based platforms, beads, or gauze, do not allow the level of control and flexibility required for robust and diverse experimental designs similar to traditional agar plates. Indeed, none of the current methods permit multigenerational culture of C. elegans , uniform distribution of bacterial food throughout the habitat, and an environment in which animals crawl, as opposed to swimming or burrowing. These are all characteristic features of traditional agar plate culture. In order to study the effects of environmental enrichment in C. elegans , an enriched habitat needs to be similar to traditional agar plates so that the effects of enrichment are not confounded by other factors such as food availability and animal locomotion. For instance, while the paper-like platforms presented by Tahernia et al. (2020) are three-dimensional and porous, they do not allow for multigenerational maintenance of stocks, and adult animals are too large to navigate freely as they do on traditional plates. Similarly, the Nematode Growth Tube 3D presented by Lee, Yoon, and Lee (2016) allows for great three-dimensional visualization of animals, but the soft agar living substrate forces animals to burrow instead of crawl, prevents the E. coli food source from growing uniformly (resulting in a patchy food environment), and does not allow for multigenerational stock maintenance.
Decellularized apple tissue offers several advantages as a substrate for C. elegans culture. First, it is porous enough for good liquid exchange and for C. elegans worms of all sizes to navigate it, yet strong enough not to sag, maintaining the structural integrity and dimensionality of the habitat. Second, the uniformity of the hypanthium tissue and the bulky shape of the fruit allows this protocol to be easily adapted for different scaffold shapes and sizes that are still similar in texture. Third, unlike many other fruits, apples are readily available year-round in our area and many others. Finally, successful growth of mammalian cells on decellularized apple tissue by Modulevsky et al. (2014) suggested that this scaffold could be a suitable medium for growing E. coli , the standard C. elegans laboratory food source.
Inspired by these factors, we developed this protocol to create three-dimensional enriched environments for C. elegans culture that is similar to standard culture. The Petri dish format allows animals to be easily maintained over generations using the same equipment as for traditional agar plates: a compound microscope and a platinum worm pick. Our protocol was specifically devised to ensure that scaffold plates have a similar cross-sectional bacterial density as traditional agar plates and that the bacterial food is grown uniformly throughout. Furthermore, the average porosity of the decellularized apple tissue allows animals to crawl freely in three dimensions, without swimming or burrowing. Decellularized fruit tissue scaffolds are also adaptable in size to different experimental designs and can be fine-tuned to host a diversity of bacterial cultures or nutrient solutions.
Despite these advantages, there are several considerations that users need to keep in mind. (1) Scaffolds seeded with a bacterial culture become contaminated more easily than their standard agar counterparts. (2) Due to the color and texture of scaffolds, animals cannot be observed or tracked if located more than 1 mm within the scaffold, which presents challenges for imaging. Depth of view can be improved by passing seeded scaffolds through a vacuum chamber, but this significantly increases the risk of contamination. (3) This protocol uses bacterial cultures grown in NGML. Typically, C. elegans is fed dense E. coli cultures made in lysogenic broth (LB), which is very favorable to bacterial growth (Fujikawa & Morozumi, 2005; Stiernagle, 2006). However, the orange coloration of LB makes it difficult to observe animals on seeded scaffolds. In addition, scaffolds need to be incubated for bacteria to grow throughout. Parallel incubation of standard agar plates seeded with LB bacterial culture resulted in agar plates with consistently higher cross-sectional bacterial densities than scaffolds. Hence, NGML, which is transparent and less favorable for bacterial growth, is used instead to diminish culture density and suppressing overgrowth on agar plates, yielding scaffold plates and agar plates with similar cross-sectional bacterial density. (4) Although animals can freely navigate the entire scaffold regardless of stage (L1-L4, adult) and are thus exposed to the same environmentally enriched conditions through their life, it is unknown whether theoretical differences in gas levels and transport through the scaffold, due to the porosity and humidity of the scaffold compared to traditional agar plates, are different enough to have an effect on cultured animals. (5) Although regular maintenance of C. elegans stocks is similar on scaffold plates and traditional plates, there is an additional time consideration to the preparation of the scaffold plates. Similarly, although it is faster, the standard drop-bleach method presented by Stiernagle (2006) is not recommended for cleaning scaffold plates, because the bleach drop is absorbed by the scaffold and can damage animals.
The suite of protocols presented here can also be adapted to several other applications that may be of interest to the user. For instance, as described by Guisnet, Maitra, Pradhan, and Hendricks (2021), they can be used to study nictation behavior in dauer animals. Nictation is a means of dispersal for several nematodes, but requires the living substrate to be textured and thus is not observable on flat standard agar plates unless they are extremely crowded or contaminated. Our scaffold habitat is the first method to allow observation of nictation in a controlled laboratory setting at the population level. We have observed nictation, group towering, and jumping behaviors at high frequency on our plates. In particular, this was the first observation of a jumping-type behavior in the C. elegans species, suggesting that the expression of some behaviors is limited or modified by the living environment.
These scaffolds can be used to study the effects of environmental enrichment on C. elegans behavior, phenotype, or gene expression. They can also be used for new experimental designs that are not possible with traditional agar plates, such as creating a habitat with a continuous concentration gradient of bacterial food or investigating different phenotypes of vertical navigation and chemotaxis. Moreover, they can be adapted to host different bacteria or bacterial populations. The heterogeneity and dimensionality of scaffolds can be beneficial to bacteria that thrive in more complex, texturized conditions. For example, some toxin-producing bacteria may have a more naturalistic reproductive fitness in our scaffold plates, allowing a more accurate assessment of the behavioral and physiological response of various nematodes to those bacteria in the wild.
Critical Parameters
Choice of apples
The choice of apple cultivar has a significant effect on the resulting scaffold plates. After trying a number of cultivars available in the Montreal area (MacIntosh, Lobo, Granny Smith, Cortland, Paula Red, Spartan, and Empire), MacIntosh apples were favored for their average porosity, slow oxidation, good toughness, infrequent contamination, yield per fruit, and year-round availability. If different cultivars are used, wash and incubation times may need to be adjusted to obtain similar results.
Wash volume
The specified ratios of slices to volume of liquid are essential to ensure that debris and solutions from previous washes are diluted out of the scaffolds. Using a higher ratio (more slices or less liquid) can lead to inconsistent results and oxidation.
Washing in batches
Basic Protocol 1 can be carried out several times in one day by starting batches at 15-min intervals, such that transfers between washes are staggered and can be done continuously. It is easy to lose track of the wash steps and incubation times, however, when several batches are run in parallel. Thus, a table of steps and times can be very useful (Table 1). It is best to fill out the table as the steps are carried out. It should not be filled in advance, as this will not account for transfer time or allow for complications (drying flasks, cleaning surfaces, etc.) that can delay timing. Transferring slices between washes takes only ∼1 min for 20 slices, but the time needed for cleaning and drying flasks (∼10 min) must be considered. If the flasks are reused for the different washes, flasks should be cleaned during wash wait times to minimize time between washes and to ensure that they are dry for the next wash. If complications arise or step timings between different batches coincide, prioritize batches in earlier steps and favor increasing wash times over shortening them. If a single shaking platform is shared for all batches, use the location of the flasks on the platform to keep track of batch identification, as markings or tape labels come off during cleaning.
| Batch A | Batch B | Batch C | ||
|---|---|---|---|---|
| Hand wash Ib | 9:45 a.m. √ | 10:00 a.m. √ | 10:15 a.m. √ | |
| Hand wash II | √ | √ | √ | |
| Hand wash III | √ | √ | √ | |
| Water I (30 min)c | Start | 10:00 a.m. | 10:15 a.m. | 10:30 a.m. |
| Finish | 10:30 a.m. | 10:45 a.m. | 11:00 a.m. | |
| Water II (30 min) | Start | 10:40 a.m. | 10:50 a.m. | 11:05 a.m. |
| Finish | 11:10 a.m. | 11:20 a.m. | 11:35 a.m. | |
| 70% ethanol (1 hr) | Start | 11:15 a.m. | 11:25 a.m. | 11:40 a.m. |
| Finish | 12:15 p.m. | 12:25 p.m. | 12:40 p.m. | |
| 100% ethanol (1 hr) | Start | 12:20 p.m. | 12:30 p.m. | 12:45 p.m. |
| Finish | 1:20 p.m. | 1:30 p.m. | 1:45 p.m. |
- a Batches A, B, and C are processed on the same day in parallel, staggered by 15 min.
- b Hand washes are quick and do not have wait times; their completion is marked only by a checkmark.
- c Although the first 30-min deionized water wash for batch A ends at the same time that this step starts for batch C, batch C is started before batch A is transferred to the next wash. Transfer time between washes varies (5-10 min) depending on whether wash flasks were cleaned beforehand.
Oxidation
The reaction that leads to scaffold oxidation (characterized by a yellow-orange color) fundamentally changes the oxidation state of the atoms that compose the scaffold matrix. Oxidized scaffold plates tear more easily, lose structural integrity, and sag. Oxidation cannot be reversed once it occurs. Cooled, heated, damaged, or bruised fruits oxidize very quickly and should not be used. Fruits that yield strong white slices at first but are oxidized the day after decellularization were likely storage fruits that were picked the previous season. Thus, storage fruits should also be avoided, if possible. If good apples were used, oxidation is most likely to happen before fruit slices are decellularized, and is usually noticeable within 1-2 min of when slices are first incubated to be decellularized. The best way to prevent oxidation is by getting slices shaking in SDS as quickly as possible. Oxidation is also likely to occur once scaffolds have been seeded if the plates are not properly sealed.
Bacterial density
Basic Protocol 2 was specifically fine-tuned to give a similar cross-sectional bacterial density between scaffold plates and standard agar plates so that the effects of the enriched habitat are not confounded with the effects of bacterial density when comparing behaviors and phenotypes of scaffold-grown animals and agar-grown animals. If a different slice thickness, apple cultivar, or bacterial culture is used, bacterial density may be affected and it is recommended to verify density and adjust temperature and incubation times, if necessary. To maintain scaffold integrity and sterility, other deviations from the protocol are not recommended.
Contamination
As mentioned above, scaffold plates are more prone to contamination than their standard agar counterparts. All manipulations following the last ethanol wash must be performed under sterile conditions using sterile materials: working surface, scalpel, forceps, flaks, solutions, plates, pick, and so on. It is also good practice to run several replicate batches on a given day rather than one large batch. For example, wash out ethanol in several small flasks rather than a single large flask, make smaller batches of NGML and use different bottles on the same experimental day, and use different bacterial cultures when preparing several scaffolds.
Stored scaffolds in which the ethanol has evaporated are very likely to render contaminated plates, although contamination is not grown and visible at that stage. It is important to ensure that sample bags are sealed tightly to prevent evaporation.
Troubleshooting
Fungal contamination is the most common contamination on scaffold plates and is likely to result from the use of non-sterile items such as forceps and picks, from leaving plates open too long during use, or from improperly sealed bags. Make sure that all material has been sterilized by autoclaving prior to use, and that manipulation tools are properly sterilized by flaming between uses. Keep scaffold plates open and unsealed only when in use. Bacterial contamination is not as frequent and is more likely to result from use of non-sterile solutions such as NGML or bacterial cultures. Make sure that sterile equipment and reagents are used and that reagents have been properly sterilized as described in Reagents and Solutions. In addition, prepare solutions and bacterial cultures in small batches to avoid opening and closing bottles frequently.
The most common problems with NGML solutions are contamination and salt precipitation. Contaminated NGML tends to be cloudy or have dense white filaments. Contamination is likely to occur when bottles are opened under non-sterile conditions. Make sure to use bottles close to a flame and prepare small volumes to avoid opening bottles often. White crystal deposits indicate salt precipitation, which happens if salts are added when the medium is still too warm. Make sure the medium is cooled to 20°-25°C after autoclaving before adding salts.
Contaminated bacterial cultures are sometimes very easy to detect as large floating masses or dense cloudiness throughout. Other more subtle contaminations include small (1-5 mm) floating particles or a darkened coloration, and can be detected by placing flasks against a light. Contaminated cultures are likely to result from contaminated materials or solutions. Make sure to work under sterile conditions near a flame, to sterilize all flasks by autoclaving before use, and to prepare and use solutions under sterile conditions.
Understanding Results
Decellularized and washed scaffolds should be white and rigid. They should not be soft or colored, and they should not bend or squash when held with forceps. These features should stay unchanged during the storage period.
Seeded scaffold plates should have a slight yellow tint due to the peptone in the NGML. Scaffolds should always keep their structural integrity and stay rigid. They should not tear or flatten easily under pressure from a worm pick, scalpel, or forceps. Because scaffolds are soaked and saturated in culture, it is difficult to prevent the culture from spreading beyond the scaffold to the bare agar around it. However, to encourage retention of animals inside the scaffold, the ring of culture around the scaffold should be no more than 1 mm wide. The spread of culture out of the scaffold is minimized by removing excess culture when the scaffold is plated (see Basic Protocol 2, step 9). Figure 1 provides an example of a seeded scaffold plate.
Time Considerations
It takes no more than 10 min to prepare the apple slices for overnight decellularization in SDS. Completing the washes and storage takes 4-5 hr for a single batch. For seeding, setting up overnight incubations of scaffolds and culture takes <10 min for a batch of two plates or ∼20 min for a batch of fifteen plates. Excluding the 5 hr incubation time, seeding scaffolds should take ∼1 hr. Collecting and washing eggs or animals from scaffolds should take no more than 30 min.
Acknowledgments
Strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by funding from the Natural Science and Engineering Research Council (NSERC, RGPIN/05117-2014), the Canada Foundation for Innovation (CFI, 32581), and the Canada Research Chairs Program (950-231541). A.G. is supported by funding from the Fonds de recherche du Québec—Nature et technologie (FRQNT, 300853).
Author Contributions
Aurélie Guisnet : Conceptualization, investigation, methodology, project administration, validation, visualization, writing –original draft, writing–review and editing; Malosree Maitra : Conceptualization, investigation, methodology, project administration, validation, writing–review and editing; Sreeparna Pradhan : Conceptualization, methodology, supervision, validation, writing–review and editing; Michael Hendricks : Conceptualization, funding acquisition, methodology, supervision, validation, writing–review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Literature Cited
- Alfred, J., & Baldwin, I. T. (2015). New opportunities at the wild frontier. eLife , 4, e06956. doi: 10.7554/eLife.06956.
- Baird, S. E. (1999). Natural and experimental associations of Caenorhabditis remanei with Trachelipus rathkii and other terrestrial isopods. Nematology , 1(5), 471–475. doi: 10.1163/156854199508478.
- Félix, M.-A., & Braendle, C. (2010). The natural history of Caenorhabditis elegans. Current Biology , 20(22), R965–R969. doi: 10.1016/j.cub.2010.09.050.
- Fujikawa, H., & Morozumi, S. (2005). Modeling surface growth of Escherichia coli on agar plates. Applied and Environmental Microbiology , 71(12), 7920–7926. doi: 10.1128/AEM.71.12.7920-7926.2005.
- Gardner, E. B., Boitano, J. J., Mancino, N. S., & D'Amico, D. P. (1975). Environmental enrichment and deprivation: Effects on learning, memory and exploration. Physiology & Behavior, 14(3), 321–327. doi: 10.1016/0031-9384(75)90040-2.
- Guisnet, A., Maitra, M., Pradhan, S., & Hendricks, M. (2021). A three-dimensional habitat for C. elegans environmental enrichment. PLoS One , 16(1), e0245139. doi: 10.1371/journal.pone.0245139.
- Lee, D., Lee, H., Choi, M.-K., Park, S., & Lee, J. (2015). Nictation assays for Caenorhabditis and other nematodes. Bio-protocol , 5, e1433. doi: 10.21769/BioProtoc.1433.
- Lee, H., Choi, M.-K., Lee, D., Kim, H.-S., Hwang, H., Kim, H., … Lee, J. (2011). Nictation, a dispersal behavior of the nematode Caenorhabditis elegans , is regulated by IL2 neurons. Nature Neuroscience , 15(1), 107–112. doi: 10.1038/nn.2975.
- Lee, T. Y., Yoon, K.-H., & Lee, J. I. (2016). NGT-3D: A simple nematode cultivation system to study Caenorhabditis elegans biology in 3D. Biology Open , 5(4), 529–534. doi: 10.1242/bio.015743.
- Modulevsky, D. J., Lefebvre, C., Haase, K., Al-Rekabi, Z., & Pelling, A. E. (2014). Apple derived cellulose scaffolds for 3D mammalian cell culture. PLoS One , 9(5), e97835. doi: 10.1371/journal.pone.0097835.
- Park, S., Hwang, H., Nam, S.-W., Martinez, F., Austin, R. H., & Ryu, W. S. (2008). Enhanced Caenorhabditis elegans locomotion in a structured microfluidic environment. PLoS One , 3(6), e2550. doi: 10.1371/journal.pone.0002550.
- Petersen, C., Dirksen, P., & Schulenburg, H. (2015). Why we need more ecology for genetic models such as C. elegans. Trends in Genetics , 31(3), 120–127. doi: 10.1016/j.tig.2014.12.001.
- Pierce-Shimomura, J. T., Chen, B. L., Mun, J. J., Ho, R., Sarkis, R., & McIntire, S. L. (2008). Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proceedings of the National Academy of Sciences of the United States of America , 105(52), 20982–20987. doi: 10.1073/pnas.0810359105.
- Stiernagle, T. (2006). Maintenance of C. elegans (February 11, 2006). WormBook, the C. elegans research community. doi: 10.1895/wormbook.1.101.1.
- Tahernia, M., Mohammadifar, M., & Choi, S. (2020). Paper-supported high-throughput 3D culturing, trapping, and monitoring of Caenorhabditis elegans. Micromachines , 11(1), 99. doi: 10.3390/mi11010099.
- Voelkl, B., Altman, N. S., Forsman, A., Forstmeier, W., Gurevitch, J., Jaric, I., … Würbel, H. (2020). Reproducibility of animal research in light of biological variation. Nature Reviews Neuroscience , 21(7), 384–393. doi: 10.1038/s41583-020-0313-3.
Citing Literature
Number of times cited according to CrossRef: 1
- Melis Toker-Bayraktar, Berkay Erenay, Burak Altun, Sedat Odabaş, Bora Garipcan, Plant-derived biomaterials and scaffolds, Cellulose, 10.1007/s10570-023-05078-y, 30 , 5, (2731-2751), (2023).

