The Dawn of Plant Molecular Biology: How Three Key Methodologies Paved the Way
Marc Somssich, Marc Somssich
Arabidopsis thaliana
CaMV 35S promoter
plant biotechnology
plant molecular biology
plant transformation
Abstract
The adoption of Arabidopsis thaliana in the 1980s as a universal plant model finally enabled researchers to adopt and take full advantage of the molecular biology tools and methods developed in the bacterial and animal fields since the early 1970s. It further brought the plant sciences up to speed with other research fields, which had been employing widely accepted model organisms for decades. In parallel with this major development, the concurrent establishment of the plant transformation methodology and the description of the cauliflower mosaic virus (CaMV) 35S promoter enabled scientists to create robust transgenic plant lines for the first time, thereby providing a valuable tool for studying gene function. The ability to create transgenic plants launched the plant biotechnology sector, with Monsanto and Plant Genetic Systems developing the first herbicide- and pest-tolerant plants, initiating a revolution in the agricultural industry. Here I review the major developments over a less than 10-year span and demonstrate how they complemented each other to trigger a revolution in plant molecular biology and launch an era of unprecedented progress for the whole plant field. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
[Correction added on May 16, 2022, after first online publication: CAUL funding statement has been added.]
INTRODUCTION
The beginnings of molecular biology
The term “ molecular biology ” was first circulated by William Thomas Astbury in his 1950 Harvey Lecture “Adventures in Molecular Biology” (Astbury, 1961; Astbury et al., 1952). Inspired by his recent protein x-ray diffraction studies, Astbury intended the term to cover such topics as the folding and unfolding of protein chains or the structure of nucleic acids (Astbury, 1961). To Astbury, molecular biology was primarily concerned with the descriptive study of molecular form and three-dimensional structure. However, throughout the 1950s and 60s, the field's focus shifted toward manipulating form and function, thanks to the identification of enzymes that could catalyze specific reactions on a molecule. This development eventually culminated in the invention of the gene cloning methodology (see next section), which started the molecular biology revolution of the 1970s and 80s (Fig. 1). One of the main triggers for this development was the identification of restriction enzymes, the functions of which were first described by Werner Arber and Daisy Dussoix in 1962 (Arber and Roulland-Dussoix, 1962; Roulland-Dussoix and Arber, 1962). In these back-to-back papers, Arber and Dussoix demonstrated that Escherichia coli could enzymatically cleave the DNA of a λ phage, thereby “ restricting ” its ability to infect the bacterium (Arber and Roulland-Dussoix, 1962; Roulland-Dussoix and Arber, 1962). Meselson and Yuan (1968), Smith and Wilcox (1970), and Mertz and Davis (1972) subsequently isolated and functionally described the first type I (Eco K) and II (Hind III) restriction enzymes and the first restriction enzyme that creates nucleotide overhangs, so-called “ sticky ends ” (Eco RI). The team of Daniel Nathans then went on to demonstrate that these restriction enzymes could be used to cleave the genome of the simian virus 40 into specific fragments, thereby allowing genome mapping (Adler and Nathans, 1973; Danna, Sack, and Nathans, 1973; Sack and Nathans, 1973). Concurrently, the teams of Peter Berg, Stanley Cohen, and Herbert Boyer demonstrated that restriction enzymes could be used to splice a gene into a bacterial plasmid and that this plasmid could then be reintroduced into a bacterial host, where it would be replicated and expressed, just like the regular bacterial genome (Cohen, Chang, Boyer, and Helling, 1973; Jackson, Symons, and Berg, 1972).
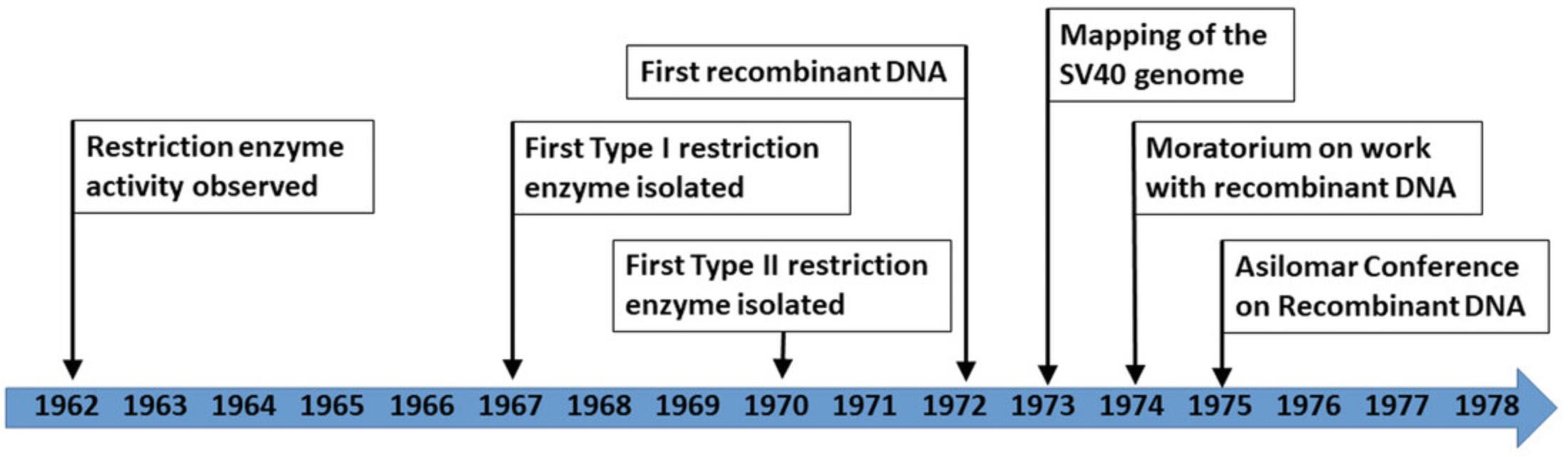
Aftermath of the development of molecular cloning
The invention of molecular cloning was followed by the discovery that DNA from different organisms could also be spliced, ligated, and transferred into a bacterial host. In one of the first such events, Peter Berg's team recombined DNA from E. coli and the λ phage into the circular genome of the simian virus 40 (Jackson et al., 1972). Annie Chang and Stanley Cohen transferred a plasmid containing Staphylococcus genes into E. coli the following year. (Chang and Cohen, 1974). For the first time, recombinant DNA had been created by scientists in a lab, and molecular cloning had been developed. The importance of these findings was recognized with Nobel prizes for Peter Berg in Chemistry (1980) and for Arber, Nathans, and Smith in Physiology and Medicine (1978) (Arber, 1979; Berg, 1981; Nathans, 1979; Smith, 1979). At the same time, it had not escaped the attention of scientists that by transferring DNA from one species into another, they had truly come to the shores of a completely new world and that they had not yet had the time to assess the risks of such work (Berg, Baltimore, and Boyer, 1974a, 1974b, 1974c). Therefore, the field agreed on a self-imposed moratorium on work with recombinant DNA to properly assess the potential biohazards and create adequate containment measures, guidelines, and regulations under which recombinant DNA work could be safely resumed (Berg, 2008). To discuss these matters, Paul Berg organized the Asilomar Conference on Recombinant DNA in 1975, where nearly 150 scientists (biologists, chemists, physicists), lawyers, and policy advisors came together (Berg, 2008; Berg, Baltimore, Brenner, Roblin, and Singer, 1975). The attendees agreed to move forward with strict application of the precautionary principle and stringent guidelines on how the work had to be contained, thereby minimizing any risk derived from this kind of research. And so, work on recombinant DNA was resumed in 1975, initiating the molecular biology revolution (Berg et al., 1975).
In the following years, scientists working on well-established model organisms, such as E. coli , yeast, or Drosophila, were racing ahead with new molecular biology methods and tools, which were now being developed at a rapid pace (Fig. 2). For instance, agarose gel electrophoresis was created in 1972 (3 years after SDS gel electrophoresis) (Johansson, 1972; Weber and Osborn, 1969), and Southern, northern, and western blotting techniques followed between 1975 and 1979 (Alwine, Kemp, and Stark, 1977; Southern, 1975; Towbin, Staehelin, and Gordon, 1979). From the late 60s to the late 70s, the prerequisites for the polymerase chain reaction (PCR) were developed, such as in vitro synthesis of short stretches of DNA, replication of short DNA strands via polymerases, or isolation and use of a heat-resistant DNA polymerase from the extreme thermophile Thermus aquaticus (Agarwal et al., 1970; Chien, Edgar, and Trela, 1976; Hutchison et al., 1978; Khorana, 1979; Kleppe, Ohtsuka, Kleppe, Molineux, and Khorana, 1971; Wells, Jacob, Narang, and Khorana, 1967). These innovations were eventually combined into an automated PCR machine in the 1980s (which resulted in another Nobel Prize in Chemistry, for Kary Mullis and Michael Smith, in 1994) (Mullis, 1994; Mullis et al., 1986; Saiki et al., 1988; Smith, 1994). DNA sequencing techniques were described in the mid-1970s, followed by direct genomic sequencing, the foundation for modern high-throughput sequencing techniques, in the 1980s (Nobel Prize in Chemistry, 1980, for Frederick Sanger and Walter Gilbert) (Church and Gilbert, 1984; Gilbert, 1981; Jay, Bambara, Padmanabhan, and Wu, 1974; Sanger, 1981; Sanger, Nicklen, and Coulson, 1977). Thanks to the development of immunofluorescence microscopy in the mid-1970s, it became possible to observe proteins in living cells for the first time (Fig. 2) (Lazarides and Weber, 1974; Somssich, 2021).
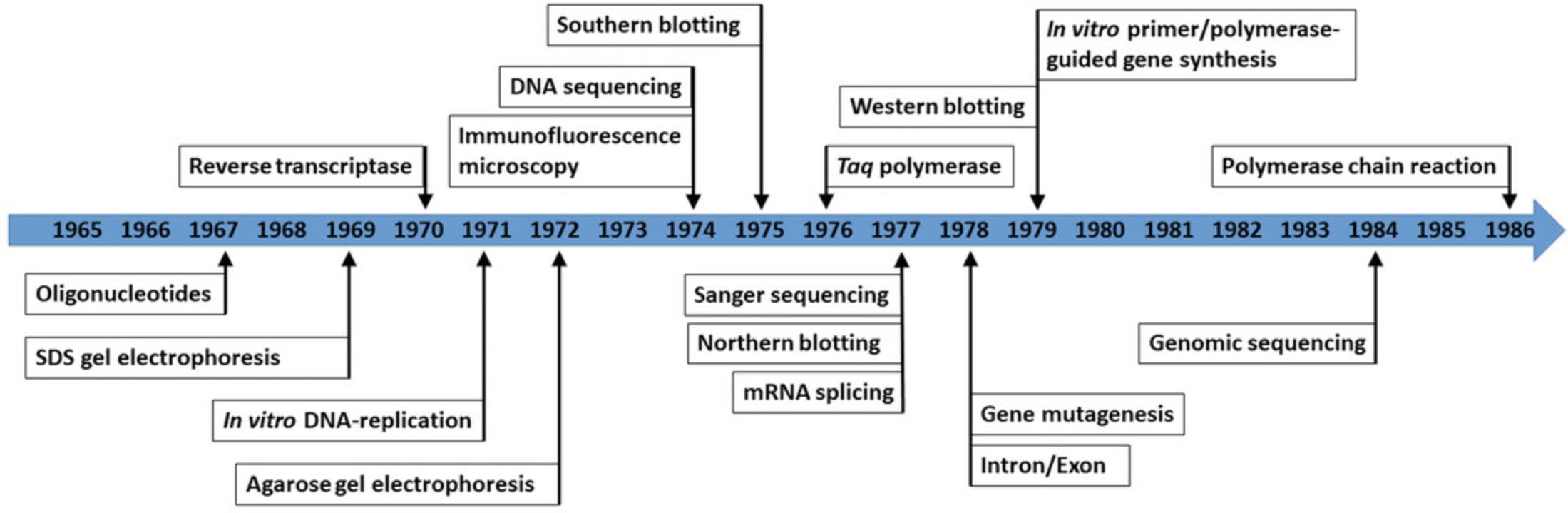
The origins of plant molecular biology
In contrast, the plant science community moved at a much slower pace, partly because different groups were working on various genetically complex crop plants. Working directly on a crop plant eliminated the translation step from a lab model to a field crop, and most funding agencies were unwilling to fund work on a model plant (Ausubel, 2018; Rédei, 1992). However, this also meant that the molecular biology methods developed in the animal and bacterial genetics fields now had to be adopted individually for each species of interest. It was becoming increasingly apparent that this approach was no longer appropriate. In addition, molecular biology work typically requires a genetically simple organism with a small genome and relatively low genetic redundancy, something common crop plants, such as rice, maize, or barley, are not. A standard plant system for molecular biology should furthermore be small in stature to allow, for example, large-scale mutagenesis screens. It should also have a short life cycle to progress through generations quickly. In addition, and quite importantly for plants, protoplast culture and plant regeneration from protoplasts (i.e., plant cells that have had their cell wall removed) should be possible. Thus, at a 1980 Cold Spring Harbor Laboratory (CSHL) plant biology meeting, Frederick Ausubel and a handful of colleagues implored their peers to adopt a universal model plant to allow implementation of the various new molecular biology techniques being developed in other communities and thereby advance the plant science field as a whole (Ausubel, 2018). His proposal was initially met with pushback from his peers, but Ausubel found an unexpected supporter in James Watson, one of the meeting's organizers. Eventually, and after intense discussion, “ most of the other attendees acknowledged the potential advantages of a model species for the plant community ” (Ausubel, 2018). In the following year, Ausubel and John Bedbrook organized the first CSHL “Frontiers in Plant Science” summer course (at the time, called “Molecular Biology of Plants”), focusing on the adoption of modern molecular biology techniques to a model plant, for which there were several candidates. Ausubel and Bedbrook's model of choice was Petunia hybrida (Ausubel, 2018). P. hybrida , like Nicotiana tabacum , was already regularly used in lieu of a single universal model. The main advantage of these species was that they were amendable to protoplasting, cell culture, and regeneration. P. hybrida is also relatively small and has a diploid genome (while N. tabacum has a complex allotetraploid genome) (Edwards et al., 2017; Gerats and Vandenbussche, 2005). However, one course participant, Leslie Leutwiler, from Elliot Meyerowitz's lab, argued for A. thaliana (Ausubel, 2018). Leutwiler presented her unpublished work on the genome size of A. thaliana , which she had determined to contain just around 100 megabases (Leutwiler, Hough-Evans, and Meyerowitz, 1984). To Ausubel and the other course participants, “ it was clear that the combination of a small genome and the advantages of Arabidopsis as a genetic model most likely made it unbeatable as a model laboratory plant for molecular genetic analysis ” (Ausubel, 2018). The adoption of A. thaliana as a universal plant model in the early 1980s thus allowed plant researchers to focus their resources and attention on adopting new techniques and methods to a single and genetically simple plant, speeding up the technology transfer from other research fields to the plant sciences.
This article chronicles the developments that led to the adoption of A. thaliana as a plant model, the development of plant transformation, and the description of the cauliflower mosaic virus 35S promoter. The latter two breakthroughs finally enabled plant researchers to create transgenics. Next to the scientific value of using transgenic organisms to study gene function, this also initiated the rise of the plant biotechnology sector. These major events, which all occurred concurrently in the early 1980s, fundamentally changed the field of plant biology and still impact our work today.
TOWARD PLANT MOLECULAR BIOLOGY AND BIOTECHNOLOGY
Arabidopsis thaliana (L.) Heynh. as model plant
While it took until the early 1980s for the plant science community to adopt A. thaliana as the model plant, work on A. thaliana goes back to the early 20th century (Somssich, 2018a). Botanist Friedrich Laibach had determined, for his PhD thesis in 1907, the number of chromosomes in different plant species (Laibach, 1965). Having found that A. thaliana had only five chromosomes—the smallest number among all the plants he had analyzed—he subsequently worked on establishing this little plant as a model for genetic and developmental studies (Laibach, 1965). His work resulted in a publication in which he called on the plant science community to adopt this plant as their model (Laibach, 1943). Few scientists, however, shared Laibach's vision and enthusiasm at the time because of the aforementioned focus on crop plants. And so, the A. thaliana research community remained small for the next few decades (the first A. thaliana symposium in 1965 was attended by just 25 participants) (Somerville and Koornneef, 2002). One scientist who did share Laibach's vision was György P. Rédei, who started work on A. thaliana after receiving seeds from Laibach in 1955 (Rédei, 1992). Rédei fled his native Hungary during the revolution of 1956 and relocated to the USA, where he remained the only scientist working on A. thaliana for the next two decades. During those years, he did pioneering work in the field, including establishing the A. thaliana Columbia and Landsberg erecta ecotypes (Rédei, 1962, 1992). In addition to Rédei in the USA, a small group of plant scientists working on A. thaliana was established in the Netherlands after Rédei passed on some Landsberg erecta seeds to Willem Feenstra from Groningen University in 1959. As a result, work in the USA was initially done primarily on the Columbia ecotype, while Landsberg erecta was dominant in Europe (Feenstra, 1964). Then, with the development of molecular cloning in the early 1970s, Rédei felt that it was time to reiterate Laibach's call to adopt A. thaliana as a universal plant model, specifically for genetic work. He published an article in 1975 summarizing all the advantages of A. thaliana as an ideal plant system, focusing on its simple genetics and small genome, mutability, availability of metabolic mutants, and ease of creating crosses (Rédei, 1975).
Contrary to Laibach's 1943 article, Rédei did have some success with his call. One scientist who read it was Maarten Koornneef in the Netherlands. Rédei's review, together with an article describing how phytohormones were essential for virtually every aspect of plant biology, inspired Koornneef to use A. thaliana as a genetic tool to study the role of these plant hormones (Koornneef, 2021). Starting with a gibberellin mutant isolated in his supervisor's lab (Jaap van der Veen), Koornneef quickly started to use chemical- and radiation-induced mutagenesis to screen for additional interesting mutants himself (Koornneef, 1978; Koornneef, Barbaro, and van der Veen, 1977). Koornneef was not content only with phenotypic analyses of his mutants. As reported by Rédei in his 1975 review, trisomics were available for A. thaliana and could be used to create linkage groups for different genetic markers to map them to the five chromosomes (Koornneef, 1983; Koornneef and van der Veen, 1983). Using the 76 mutants he had available as markers, Koornneef reported, in 1983, the first linkage map of all five chromosomes of A. thaliana (Koornneef et al., 1983).
Rédei's article not only reignited interest in A. thaliana in the Netherlands, it also finally popularized the plant in the USA. In 1978, while on holiday, Chris and Shauna Somerville read Rédei's article, as well as Mary-Dell Chilton's 1977 breakthrough report that Agrobacterium tumefaciens , a gram-negative soil bacterium and the causal agent of crown gall disease, could insert a piece of DNA into the plant genome (Pennisi, 2000). The two had just finished graduate school, with Shauna specializing in plant biology and Chris, in molecular biology, focusing on E. coli. After reading these two papers, the Somervilles decided to combine their expertise to spearhead a new field of plant molecular biology (Pennisi, 2000). Chris Somerville focused his efforts on photorespiration. In the absence of mutant plants, photosynthesis and photorespiration were primarily studied using biochemical and physiological techniques. However, the genetics of A. thaliana proved to be as simple as Rédei had promised in his review. So, by 1980, Chris Somerville and his mentor, William Ogren, had published two high-impact papers on A. thaliana photosynthesis mutants, identifying new components and pathways involved in photorespiration (Somerville and Ogren, 1979, 1980). Robert Pruitt and Leslie Leutwiler, students in Elliot Meyerowitz's Drosophila genetics lab, were impressed by this new genetics model and convinced Meyerowitz to let them analyze the genome of this plant. This resulted in two papers, which provided the first overview of the genomic content and structure of A. thaliana , and led to Leutwiler's attendance at the CSHL summer course, where her presentation convinced Frederick Ausubel and the other participants that A. thaliana was “unbeatable” as a model plant (Ausubel, 2018; Leutwiler et al., 1984; Pruitt and Meyerowitz, 1986). Coincidentally, Ausubel had also heard Chris Somerville present his work on the A. thaliana photosynthesis mutants at a Gordon Conference on Molecular Biology, and these two encounters made him “ decide on the spot that I would start switching my laboratory (…) to Arabidopsis ” (Ausubel, 2018). The same year, Caren Chang from the Meyerowitz lab had cloned the first gene from A. thaliana , an alcohol dehydrogenase (Chang and Meyerowitz, 1986). Based on these developments, Elliot Meyerowitz published an article in 1987 that, contrary to earlier papers, not merely called for the adoption of A. thaliana as a model plant but instead formally announced its arrival and establishment as a robust model system for plant molecular genetics, development, physiology, and biochemistry (Meyerowitz, 1987).
The value of A. thaliana as a universal plant model quickly became evident when other labs isolated mutants for their respective genes and pathways of interest. Chris Somerville neatly summarized this in his conference report for the 1989 Arabidopsis Meeting when he wrote that “ this year's poster is next year's breakthrough ,” as posters were often presentations of newly isolated genes and mutants (Somerville, 1989). This 1989 meeting was attended by approximately 400 scientists, a steep increase from the ∼25 attendees at the first (1965) and second (1975) International Arabidopsis Symposium, demonstrating how rapidly work on A. thaliana was taken up once it was accepted as a model in the early 1980s (Somerville, 1989; Somerville and Koornneef, 2002). Another massive accomplishment that demonstrated, once again, that A. thaliana was indeed the right choice for a universal plant model was the completion of the A. thaliana genome sequence in the year 2000 (Arabidopsis Genome Initiative, 2000). It was the highest quality genome sequence available, even containing difficult-to-sequence regions, such as the centromeres, which were avoided in the genome sequences of other organisms, such as humans (Arabidopsis Genome Initiative, 2000; Pennisi, 2000). The publication of the genome also brought the plant sciences into the genomics era, as large-scale “omics” techniques generally rely on the availability of a high-quality genome sequence. And while this was already a huge achievement, in the following years, the 1001 Genome Initiative added over 1100 other genome sequences for different natural accessions of A. thaliana from all around the world, which enabled groundbreaking work on natural plant variation (Alonso-Blanco et al., 2016; Weigel, 2012). None of these breakthroughs would have been possible without the adoption of A. thaliana as a universal plant model.
The development of plant transformation
The adoption of A. thaliana as a model plant coincided with another major technological advance that propelled molecular plant research forward: the development of plant transformation as a method to create transgenic plants (1983) (Somssich, 2019). Plant transformation has its roots in the study of A. tumefaciens -induced crown galls, which are tumorous outgrowths on plant tissue (Smith and Townsend, 1907; Somssich, 2019). In an important breakthrough paper in 1941, White and Braun laid the foundation for future explorations of A. tumefaciens -mediated plant transformation when they demonstrated the ability to culture crown gall tumors in vitro and that while such explants retained their tumorous character, they no longer contained any Agrobacteria (White and Braun, 1941). Thus, these explants appeared to be permanently transformed, leading Braun to conclude that there must be a “tumor-inducing principle,” a substance that is transferred from the bacterium to the plant (White and Braun, 1941). Speculating, in 1947, about the identity of this “principle,” he already considered DNA a possible candidate, based on contemporary findings that Pneumococci could exchange certain traits by transferring their DNA (Avery, MacLeod, and McCarty, 1944; Braun, 1947). However, without molecular biology tools, Braun and his colleagues were unable to identify the “tumor-inducing principle.” The second major breakthrough happened in 1967 when Rob Schilperoort et al. showed that a short strand of agrobacterial RNA could hybridize with DNA isolated from cultured tumors (Schilperoort, Veldstra, Warnaar, Mulder, and Cohen, 1967). This short RNA strand would otherwise only hybridize with A. tumefaciens DNA, but not plant DNA, indicating that bacterial DNA was indeed transferred into the plant cell and somehow induced the tumors (Schilperoort et al., 1967). This publication reignited interest in how A. tumefaciens induces plant tumors and specifically inspired Mary-Dell Chilton, Jeff Schell, and Marc van Montagu, which would prove key (Angenon, Van Lijsebettens, and van Montagu, 2013; Chilton, 2001).
None of the three investigators were plant biologists or particularly interested in A. tumefaciens. Chilton was a chemist interested in microbiology, while Schell and van Montagu were bacterial and phage geneticists, respectively. All three, however, had a great sense of “ the next big thing ,” and this observation of a cross-species DNA transfer held the potential to be just that (Angenon et al., 2013; Heimann, 2018). So, all three of them set out to study how A. tumefaciens could perform such a feat of bioengineering. In her previous position, Chilton had pioneered a new DNA-detection technique based on changes in double-strand renaturation kinetics in the presence or absence of additional complementary molecules (Chilton, 2018). Thus, she first set out to confirm the findings from Schilperoort et al. (1967). Curiously though, no matter which bit of the A. tumefaciens genome she tested, she could not detect any complementary sequences in tumor tissue (Chilton et al., 1974). That same year, the now integrated Schell/van Montagu lab reported that they had identified an extrachromosomal plasmid in all tumor-inducing A. tumefaciens strains but in none of the non-oncogenic strains and that the presence of this plasmid is indeed essential for the bacterium's ability to cause plant tumors (Larebeke et al., 1974; Zaenen, Van Larebeke, van Montagu, and Schell, 1974). They christened it the tumor-inducing (Ti)-plasmid (Engler et al., 1975). For Chilton, this meant that she had tested the wrong DNA when she screened the DNA from plant tumors for sequences complementary to the A. tumefaciens genome. She and her team repeated the work using the Ti-plasmid as a template and were able to identify the specific fragment within the Ti-plasmid that is transferred into the plant cell: the transferred (T)-DNA (Chilton et al., 1977). The implications of this finding were immediately clear. If A. tumefaciens could transfer the T-DNA into plant cells, it must be possible to use this bacterium as a shuttle to transfer specific genes of interest into plants. And so, a scientific race toward generating the first transgenic line began.
To start the race, the Schell/van Montagu lab found that the sequences immediately flanking the T-DNA seemed to be highly conserved among Ti-plasmids from different A. tumefaciens strains, suggesting the importance of these sequences in determining the region that would be transferred (Depicker, van Montagu, and Schell, 1978). By analyzing T-DNAs transferred into plants, they confirmed that these borders—now labeled the left and right border—indeed always flanked the inserted T-DNA. Intriguingly, they also found that in plants, these borders always flank the T-DNA on one side but genomic plant DNA on the other side, providing the first real evidence that the T-DNA was not just shuttled into the plant cell but stably integrated into the plant's genome (Zambryski et al., 1980). In 1980, the Schell/van Montagu lab demonstrated that it was possible to introduce foreign DNA into a Ti-plasmid between the left and right border. This insert would then be shuttled into the plant when the bacterium infected a plant (Hernalsteens et al., 1980). The group inserted Transposon 7 of E. coli into the T-DNA and demonstrated its presence in the transformed plant tissue. However, since the Ti-plasmid still carried cancer-inducing genes, the transformed tissue was still tumorous, and it was not possible to regenerate a healthy plant from it (Hernalsteens et al., 1980). The following year, the Nester, Schilperoort, and Schell/van Montagu labs all published on Ti-plasmid mutants, resulting in the first map of the plasmid and a first clue on which regions are responsible for the tumor-inducing properties of the T-DNA (Garfinkel et al., 1981; Greve, Decraemer, Seurinck, van Montagu, and Schell, 1981; Ooms, Hooykaas, Moolenaar, and Schilperoort, 1981). Based on these results, the Schell/van Montagu lab used a “partially disarmed” (less oncogenic) Ti-plasmid to again transfer the E. coli Transposon 7 into plants. This time, they successfully regenerated a healthy, transformed plant that passed the transgene on to the next generation in Mendelian fashion (Otten et al., 1981). This may be regarded as the first-ever lab-generated transgenic plant line; however, the plant still had some tumorous tissue and expressed unwanted agrobacterial octopine and nopaline synthase genes. These genes are transferred to the plant during natural infection by A. tumefaciens , and their gene products direct the synthesis of opines in the plant cell, which the bacterium can use as a carbon and nitrogen source (Bomhoff et al., 1976; Koncz et al., 1983). However, these opines also interfere with the plant's metabolism, so to obtain healthy transgenic plants, it was necessary to avoid the formation of tumorous tissue and eliminate the transfer of opine synthesis genes (Zambryski et al., 1983). Furthermore, the transgene was not expressed and did not confer a new trait to the plant, which was the ultimate goal. The finish line was now in sight, however, and at the Miami Winter Symposium in January 1983, Jeff Schell, Mary-Dell Chilton, and Bob Horsch (from Monsanto) all gave talks presenting their own transgenic plant lines, all expressing an antibiotic resistance gene and conferring this resistance to the plant cells (O'Brien, 1983). The critical publications came in the following months: first, the Chilton lab published on the regeneration of a healthy transgenic N. tabacum plant line carrying the yeast ALCOHOL DEHYDROGENASE I gene (Barton, Binns, Matzke, and Chilton, 1983). However, this transgene was not expressed due to the lack of plant-active regulatory sequences. Next, the Schilperoort lab published the first binary vector set for plant transformation, separating the T-DNA and the virulence genes necessary to facilitate T-DNA transport onto two different plasmids (the T-DNA-carrying binary vector and the virulence -providing helper-plasmid) (Hoekema, Hirsch, Hooykaas, and Schilperoort, 1983). This system makes maintenance and modification of the now smaller T-DNA plasmid easier and is used to this day. Just a week later, the Schell/van Montagu lab published their transgenic N. tabacum plant line expressing a chloramphenicol acetyltransferase gene from E. coli , making the plant cells resistant to this antibiotic (Herrera-Estrella, Depicker, van Montagu, and Schell, 1983). The Chilton lab followed 2 months later with their own transgenic N. tabacum plant line expressing a gene for resistance to the G418 antibiotic (Bevan, Flavell, and Chilton, 1983). And two weeks after that, the Monsanto lab published their transgenic P. hybrida lines carrying the aminoglycoside-3'-phosphotransferase (npt) gene, providing resistance to kanamycin (Fraley et al., 1983). In all three cases, the researchers had inserted the transgene into the Ti-plasmid at the position of the bacterial nopaline or octopine synthase genes, therefore indirectly exploiting endogenous regulatory sequences and circumventing the problem that no plant-active promoters were known at the time. In another 1983 paper, the Schell/van Montagu lab characterized these regulatory sequences in detail, providing information on the first two promoters suitable to express transgenes in plants (Koncz et al., 1983). Finally, at the end of this important year for plant molecular biology, the lab of Timothy Hall published their transgenic sunflower cells, expressing the bean phaseolin gene using the octopine synthase promoter or the gene's endogenous promoter, which they achieved by including about 1000 bp of sequences just upstream of the phaseolin coding region (Murai et al., 1983). The race toward the first transgenic plant thus ended with four independent plant lines published in 1983. However, none of the four papers presented healthy, regenerated, transgenic plants; all were published when the transgenic cell lines were still in the regeneration phase. So, to formalize the result, the Schell/van Montagu lab published, in 1984, on the regenerated, healthy plants and their transgenic offspring, which inherited the antibiotic resistance gene in a Mendelian fashion (De Block, Herrera-Estrella, van Montagu, Schell, and Zambryski, 1984).
Michael Bevan, a former member of the Chilton lab, followed their 1983 paper with a new binary vector, pBIN19 (Bevan, 1984). This became the most widely used plant transformation vector for years, often combined with the pMP90 helper-plasmid that the Schell lab published together with the GV3101 A. tumefaciens strain (Koncz and Schell, 1986). The labs of van Montagu and Patricia Zambryski followed up with two papers describing the gene transfer process. They demonstrated that the Ti-plasmid is cleaved near the right border of the T-DNA, where it contains a highly conserved 25-bp sequence (Wang, Herrera-Estrella, van Montagu, and Zambryski, 1984). Subsequently, a complementary DNA strand is synthesized from this position to the left border, and this copy is then shuttled into the plant cell, starting with the conserved 25-bp sequence. Because of this insert orientation, resistance genes are now typically inserted into transformation vectors at the left border so that incomplete transformation events, where the plant cell only receives part of the T-DNA, will not result in resistant cell lines. Another paper from the lab demonstrated that wounded plant cells excrete the chemical acetosyringone into the surrounding soil, which A. tumefaciens uses as a chemotactic signal, activating the expression of the bacterial virulence genes, thereby initiating the DNA transfer process (Stachel, Messens, van Montagu, and Zambryski, 1985). Thus, acetosyringone is typically added to plant transformation media today.
In the absence of a plant model, research teams tended to use N. tabacum or P. hybrida cell cultures, as these plants are well suited to cell and tissue culture methods, with subsequent plant regeneration—a requirement for these early transformation protocols. But once A. thaliana was adopted as a model plant, and plant transformation developed, the Monsanto lab was also quick to provide a transformation procedure for A. thaliana (Lloyd et al., 1986). They achieved this by adopting the cumbersome tissue culture and plant regeneration procedure used for their N. tabacum cell lines (Lloyd et al., 1986). However, the choice of A. thaliana immediately proved to be an advantage for plant transformation. In 1987, Kenneth Feldmann and David Marks showed that they could produce transgenic A. thaliana lines by simply co-culturing freshly germinated seeds with A. tumefaciens , thereby eliminating the laborious cell culturing and regeneration steps (Feldmann and Marks, 1987). Then, in 1993, Bechtold et al. demonstrated that they could obtain transgenic plant lines by uprooting adult flowering A. thaliana plants, immersing them in an infiltration buffer containing A. tumefaciens , applying a vacuum, re-planting them in soil, and simply harvesting the ripe seeds (Bechtold, Ellis, and Pelletier, 1993). While this process already reduced the workload enormously compared to the cell culturing and plant regeneration process, it was then shown that it was possible to simplify the process even further. In 1998, Steven Clough and Andrew Bent published their “floral dip” method. Here, a potted adult flowering A. thaliana plant is simply dipped upside down for a couple of seconds into a beaker containing the A. tumefaciens solution before returning it to the growth room in a humid environment (Clough and Bent, 1998). This dip is repeated once after 5 days. The seeds are collected, and transgenic lines are selected in the next generation. The ease of creating transgenic lines would have been unthinkable without adopting a simple and small model plant, such as A. thaliana. Since then, subsequent modifications have further simplified the method (Logemann, Birkenbihl, Ülker, and Somssich, 2006; Narusaka, Shiraishi, Iwabuchi, and Narusaka, 2010).
Finally, in the early 2000s, A. tumefaciens -mediated plant transformation, along with the availability of a complete and high-quality A. thaliana genome sequence, formed the basis for another major development: the creation of ready-to-order T-DNA insertion mutant lines for nearly every gene in the A. thaliana genome, which were organized in three major collections, the SALK, SAIL, and GABI-Kat lines (Alonso et al., 2003; Arabidopsis Genome Initiative, 2000; Rosso et al., 2003; Sessions et al., 2002). In all three cases, A. tumefaciens was used in a massive experimental setup to randomly insert T-DNAs throughout the A. thaliana genome, presumably knocking out the function of the gene into which the T-DNA was inserted. Tens of thousands of insertions were produced and mapped to the A. thaliana genome, enabling researchers to simply order the plant lines that carry insertions in their genes of interest for functional studies.
To this day, Agrobacterium-mediated transformation is the standard for creating transgenic lines. Its use, however, is not only limited to the plant field. Agrobacterium-mediated transformation has also been successfully applied to yeast, fungi, and even cultured human cells (Lacroix, Tzfira, Vainstein, and Citovsky, 2006). Secondary advances such as the use of T-DNA insertions for mutagenesis may, at one point, become obsolete thanks to modern genome editing techniques. Still, even those techniques rely on transformation procedures to introduce the relevant machinery (e.g., CRISPR) into the cell (Zhu, Li, and Gao, 2020).
Furthermore, plant transformation is not only important for researchers. Creating transgenic plants led to the development of resilient, high-performing, and biofortified crop plants, which are already essential to ensuring food security today (Schulman, 2020). Accordingly, Marc van Montagu, Mary-Dell Chilton, and Robert T. Fraley were awarded the World Food Prize in 2013 for the development of the technique (Angenon et al., 2013).
The description of the cauliflower mosaic virus 35S promoter
With the development of plant transformation, it was possible to transfer a transgene into a plant, but as mentioned above, when the van Montagu/Schell and Chilton labs first managed to regenerate a transgenic plant, the inserted transgene was not expressed due to the lack of plant-active regulatory sequences (Barton et al., 1983; Otten et al., 1981). The researchers circumvented the problem by using the A. tumefaciens nopaline or octopine synthase promoters. These, however, were not active in all plant tissues and are under strong developmental and environmental regulation, resulting in high variability in their activity and limiting their usefulness in transgenesis (An, Costa, Mitra, Ha, and Márton, 1988). This situation changed in 1985 with the description of the cauliflower mosaic virus (CaMV) 35S promoter (Odell, Nagy, and Chua, 1985; Somssich, 2018b).
CaMV is a pathogenic plant virus that infects several cruciferous crop plants, such as broccoli, cabbage, kale, turnip, and its namesake, cauliflower, from which C. M. Tompkins first isolated it in 1937 (Tompkins, 1937). Infection results in the mosaic disease, named after the mosaic-shaped necrotic lesions visible on the leaves of infected plants (Schultz, 1921). It was later shown that aphids serve as the viral vector, transporting it from plant to plant, and that, unlike the then better-studied tobacco mosaic virus, it is not an RNA but a DNA virus, representing the first-ever DNA virus described for plants (Day and Venables, 1961; Shepherd, Wakeman, and Romanko, 1968; Siegel and Wildman, 1960). Furthermore, in contrast to the second identified plant DNA virus family, the geminiviruses, the CaMV DNA is double-, not single-stranded (Goodman, 1977; Shepherd, Bruening, and Wakeman, 1970). In the absence of plant transformation techniques, this observation made scientists consider the CaMV as a vector to shuttle DNA into plant cells to produce transgenic crops (Hull, 1978). After all, infection of a plant with the virus requires the replication of viral DNA in the plant cell.
By 1980, the 8024-bp genome of CaMV had been mapped and annotated (Franck, Guilley, Jonard, Richards, and Hirth, 1980; Hohn, Hohn, Lesot, and Lebeurier, 1980). It contains six open reading frames transcribed as just two RNAs: the short 19s and the whole-genome covering 35S RNA (Franck et al., 1980). The 35S RNA is spliced into four individual, protein-coding mRNAs (Covey and Hull, 1981). The first major breakthrough came when both Covey and Hull (1981) and Guilley, Dudley, Jonard, Balàzs, and Richards (1982) found that four CaMV transcripts could be isolated from infected P. hybrida leaves, indicating that these viral genes were indeed transcribed in the plant cells and that the viral DNA contained all elements necessary to activate transcription in plants (Covey and Hull, 1981; Guilley et al., 1982). Teams working with Thomas and Barbara Hohn and Ingo Potrykus, as well as Jean-François Laliberté, subsequently managed to transform plant cells with engineered CaMV carrying a mammalian or bacterial gene in the position of the 35S gene (Brisson et al., 1984; Lefebvre, Miki, and Laliberté, 1987; Paszkowski et al., 1986). Unfortunately, however, CaMV would only tolerate the insertion of short stretches of foreign DNA (∼250 bp), and with the successful development of A. tumefaciens -mediated plant transformation at around the same time, research into CaMV-mediated plant transformation was abandoned (Gronenborn, Gardner, Schaefer, and Shepherd, 1981; Haas, Bureau, Geldreich, Yot, and Keller, 2002). Nevertheless, this left the 35S promoter as an interesting candidate for transgene expression in plants, and in 1985, Joan Odell and Ferenc Nagy from Nam-Hai Chua's lab published a detailed promoter analysis (Odell et al., 1985).
Odell and colleagues fragmented the roughly 1000-bp upstream region of the 35S open reading frame and fused the fragments to the human growth hormone (hgh) gene to identify the sequences needed for gene expression in plant cells (Odell et al., 1985). They then transformed N. tabacum cells with these constructs, using A. tumefaciens -mediated transformation, and detected hgh mRNA in the cultured plant cells via northern blotting. They discovered that a fragment containing 46 bp upstream of the 35S open reading frame resulted in minimal transgene expression, while an extended 343-bp fragment conferred strong expression across all plant tissues tested. Hence, they defined the 46 bp as the “ minimal promoter ” and the 343 bp as the “ CaMV 35S promoter ” (Odell et al., 1985). The Chua lab followed up these studies with two more publications, further subdividing the 343 bp promoter into individual regions that could control expression strength and pattern across different plant tissues in a combinatorial and/or additive fashion (Benfey and Chua, 1990; Fang, Nagy, Sivasubramaniam, and Chua, 1989). Robert Kay, Chan, Daly, and McPherson (1987) showed that placing two CaMV 35S promoters in tandem further enhanced expression. It was not, however, just this full CaMV 35S promoter that was valuable to the research community; the minimal promoter proved to be a useful tool. The 46-bp segment carried all the DNA binding sites necessary to initiate basal transcription but lacked activating elements. Researchers, therefore, could combine the minimal promoter with various transcriptional activator sites of interest, such as binding sites for auxin response factors, to create auxin-inducible transgenes, or WRKY transcription factor binding sites, to produce transgenes that would be activated in response to a pathogenic attack (Rushton, Reinstädler, Lipka, Lippok, and Somssich, 2002; Ulmasov, Murfett, Hagen, and Guilfoyle, 1997). The minimal promoter could also be combined with activator sites responsive to substances foreign to plants, such as human estrogen or alcohol, thereby giving researchers complete control over when the transgene is expressed (Caddick et al., 1998; Zuo, Niu, and Chua, 2000).
Throughout the following 20 to 30 years, the CaMV 35S promoter became the most widely used promoter in plant research and plant biotechnology. Nearly every genetically modified crop plant in the field carried a version of this promoter (Hull, Covey, and Dale, 2000). Next to the development of plant transformation, the CaMV 35S promoter was the second catalyst that got the field of plant biotechnology started. In 1986, the Monsanto lab combined these two scientific breakthroughs to create a transgenic P. hybrida cell line expressing the 5-enolpyruvylshikimate-3-phosphate synthase gene from the CaMV 35S promoter (Shah, Horsch, and Klee, 1986). This transgene rendered the petunia tolerant to the herbicide glyphosate.
THE PLANT BIOTECH INDUSTRY
By 1980, when it became evident that plant transformation was within reach, the usefulness of this technique to create herbicide- and pest-tolerant plants was obvious. For Monsanto, the agrochemical giant producing and selling herbicides, this was the initial motivation to get involved in the race toward the first transgenic plant. In 1982, this prospect led Marc van Montagu and Jeff Schell to form their own biotech company, Plant Genetic Systems, the first plant biotech company in Europe (Heimann, 2018). While Monsanto developed the herbicide-tolerant petunia, Plant Genetic Systems started by creating a pest-tolerant N. tabacum (Shah et al., 1986; Vaeck et al., 1987). The year before, Plant Genetic Systems had cloned the Bacillus thuringiensis (Bt) berliner 1715 Bt2-gene , encoding the active ingredient in bt toxin (Höfte et al., 1986). The bt-toxin is typically sprayed on conventional and organic-certified crop fields to kill off larvae of the hornworm, a major agricultural pest, while being completely safe for humans. In 1987, Plant Genetic Systems used the gene to create an N. tabacum plant expressing the Bt2 gene at low levels directly in the plant (Vaeck et al., 1987). With the transgenic plants now directly producing the toxin, only larvae feeding on the plant will be killed off, while insects that merely inhabit the field are spared. Fields growing these plants do not require spraying, saving labor, time, and money. Furthermore, the toxin can no longer reach and contaminate the environment and groundwater as it does when being sprayed, limiting the ecological damage done by modern agricultural practices. While the glyphosate-tolerant petunia and the Bt2 -expressing N. tabacum plants were just proof-of-concept plants, a Bt potato was the first Bt crop approved for the food market in the USA in 1995, followed by a glyphosate-resistant soybean in 1996 (Duke and Powles, 2008; Peferoen, 1997).
Both systems, glyphosate resistance and Bt expression, have succeeded and are still widely used in agriculture (Schulman, 2020). Despite what might be expected, however, most crops in the field today have not been created using A. tumefaciens -mediated transformation methods because once this plant transformation technique was established, the researchers immediately found themselves in a patent-induced gridlock (Nottenburg and Rodríguez, 2008; Somssich, 2019). In 1983, right after the successful development of the A. tumefaciens -mediated plant transformation technique, Monsanto filed the first patent for the invention of the method (Heimann, 2018; Nottenburg and Rodríguez, 2008). This was immediately countered by Jeff Schell and the Max Planck Society, as well as Mary-Dell Chilton and Washington University. Countless patents on specific variants of A. tumefaciens -mediated plant transformation followed. As a result, a patent interference was issued, meaning that no patent would be granted due to the legal uncertainty as to who should rightfully own it (Nottenburg and Rodríguez, 2008). This interference was only resolved in 2005, when Monsanto, Bayer CropScience, and the Max Planck Society worked out a licensing agreement (the Max Planck Society subsequently used the back-royalties to fund the Jeff Schell Professorship at the University of Cologne). Consequently, this meant that everybody who used the A. tumefaciens -mediated plant transformation technique in the meantime was infringing on the patent, including researchers in academic, not-for-profit environments. Scientists at public research institutes, such as universities, generally believe that there is an “ experimental use exception ” due to the non-commercial nature of their work. This, however, is not the case (Nottenburg and Rodríguez, 2008). To circumvent this problem, most commercial transgenic crops were created with alternative transformation methods, such as biolistic transformation with a particle gun (McCabe, Swain, Martinell, and Christou, 1988). Similarly, the use of the CaMV 35S promoter to control marker gene expression in crop plants had been patented by Monsanto, prompting other companies to use related promoters, such as the figwort mosaic virus 34S promoter (Chi-Ham et al., 2012; Graff, Cullen, Bradford, Zilberman, and Bennett, 2003).
While these patent issues made things unnecessarily complicated, the plant biotech sector had been firmly established in the 1980s, and since the first genetically engineered crops were released in the 1990s, the success was overwhelming. The pest- or herbicide-resistant crop varieties that have been introduced since the early 1990s allow farmers to grow more food with less financial and labor input and maximize productivity on limited farmland. In addition, they allow for the cultivation of food crops with a lower environmental impact due to a reduced need for fertilizers, pesticides, and herbicides (Schulman, 2020). In fact, it would be difficult to feed the world without the transgenic food crops used to feed livestock or consumed directly by humans (Schulman, 2020).
CONCLUDING REMARKS
The major developments described here, which collectively kicked off the plant molecular biology revolution of the 1980s, catapulted the plant sciences forward and quickly advanced plant research to a level comparable with that of leading molecular biology research in other fields (Fig. 3). When the genomics era dawned in the 1990s, the A. thaliana genome was the first plant genome to be sequenced, the fourth eukaryotic organism, and the first one to include DNA regions difficult to sequence, such as centromere regions, thereby raising the bar for genome sequence quality (Arabidopsis Genome Initiative, 2000).
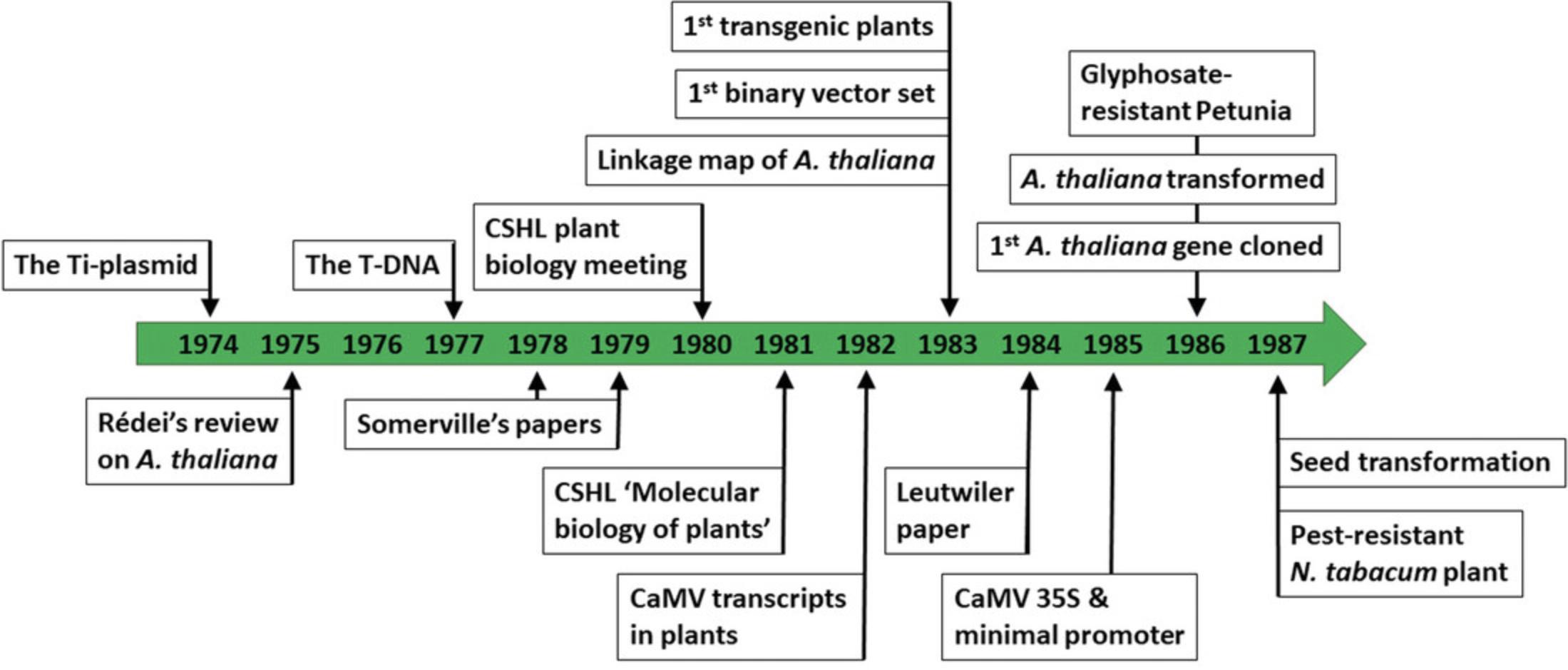
Since 2000, several secondary plant models have been established to study more specific aspects of plant life, including models for grasses, trees, the water-to-land transition of early plants, or plant parasitism (Cesarino et al., 2020). This was made possible because the methods and techniques developed for A. thaliana often merely had to be adapted rather than completely re-invented. A. thaliana remains at the forefront of plant sciences, allowing researchers to continually pioneer new research fields, facilitating the rapid adoption of new methodologies and by providing mechanistic insights into all aspects of plant biology at ever-increasing detail. A. tumefaciens -mediated plant transformation protocols exist for almost all plants that researchers are currently working on, and the CaMV 35S promoter has been the standard for over 30 years.
It is not just that these techniques are still in use today that makes these developments so important. For instance, all the large-scale “omic” tools developed in the plant genomic era depend directly on the high-quality genome sequence that could only be obtained thanks to the small and simple genome of A. thaliana and the associated pioneering work. Further, the fact that today we have inducible, or tissue- and developmental stage-specific promoters is the result of the knowledge gained when first dissecting and studying the full-length and minimal 35S promoter. And while several secondary plant models have been established since 2000, using Agrobacterium-mediated transformation to express a transgene from the CaMV 35S promoter in this plant is still the standard for establishing a new model. Similarly, first candidate genes for functional studies in secondary model or crop plants are usually selected based on sequence homology to well-studied genes in A. thaliana. The dawn of plant molecular biology in the early 1980s, brought on by these key developments, still directly impacts plant research today.
AUTHOR CONTRIBUTION
Marc Somssich : Conceptualization, Funding acquisition, Project administration, Visualization, Writing–original draft, Writing–review and editing.
CONFLICT OF INTEREST
The author declares no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
LITERATURE CITED
- Adler, S. P., & Nathans, D. (1973). Studies of simian virus 40 DNA. V. conversion of circular to linear SV 40 DNA by restriction endonuclease from Escherichia coli B. Biochimica et Biophysica Acta (BBA), Nucleic Acids and Protein Synthesis , 299, 177–188. doi: 10.1016/0005-2787(73)90340-7.
- Agarwal, K. L., Büchi, H., Caruthers, M. H., Gupta, N., Khorana, H. G., Kleppe, K., … Yamada, T. (1970). Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast. Nature , 227, 27–34. doi: 10.1038/227027a0.
- Alonso-Blanco, C., Andrade, J., Becker, C., Bemm, F., Bergelson, F., Borgwardt, K. M., … Zhou, X. (2016). 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell , 166, 481–491. doi: 10.1016/j.cell.2016.05.063.
- Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., … Ecker, J. R. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science , 301, 653–657. doi: 10.1126/science.1086391.
- Alwine, J. C., Kemp, D. J., & Stark, G. R. (1977). Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proceedings of the National Academy of Sciences of the United States of America , 74, 5350–5354. doi: 10.1073/pnas.74.12.5350.
- An, G., Costa, M. A., Mitra, A., Ha, S.-B., & Márton, L. (1988). Organ-specific and developmental regulation of the nopaline synthase promoter in transgenic tobacco plants. Plant Physiology , 88, 547–552. doi: 10.1104/pp.88.3.547.
- Angenon, G., Van Lijsebettens, M., & van Montagu, M. C. E. (2013). From the tumor-inducing principle to plant biotechnology and its importance for society. The International Journal of Developmental Biology , 57, 453–460. doi: 10.1387/ijdb.130295ga.
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature , 408, 796–815. doi: 10.1038/35048692.
- Arber, W. (1979). Promotion and limitation of genetic exchange. Science , 205, 361–365. doi: 10.1126/science.377489.
- Arber, W., & Roulland-Dussoix, D. (1962). Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage λ. Journal of Molecular Biology , 5, 18–36. doi: 10.1016/S0022-2836(62)80058-8.
- Astbury, W. T. (1961). Molecular biology or ultrastructural biology? Nature , 190, 1124–1124. doi: 10.1038/1901124a0.
- Astbury, W. T., Ephrussi, B., Cournand, A., Mirsky, A. E., Runnström, J., Ochoa, S., … Kabat, E. A. (1952). The Harvey Lectures: Delivered Under the Auspices of the Harvey Society of New York, 1950-1951. Springfield, Illinois, U.S.A: Charles C. Thomas.
- Ausubel, F. M. (2018). Tracing my roots: How I became a plant biologist. Annual Review of Genetics , 52, 1–20. doi: 10.1146/annurev-genet-120417-031722.
- Avery, O. T., MacLeod, C. M., & McCarty, M. (1944). Studies on the chemical nature of the substance inducing transformation of Pneumococcal types. Journal of Experimental Medicine , 79, 137–158. doi: 10.1084/jem.79.2.137.
- Barton, K. A., Binns, A. N., Matzke, A. J. M., & Chilton, M - D. (1983). Regeneration of intact tobacco plants containing full length copies of genetically engineered T-DNA, and transmission of T-DNA to R1 progeny. Cell , 32, 1033–1043. doi: 10.1016/0092-8674(83)90288-X.
- Bechtold, N., Ellis, J., & Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Comptes Rendus de l'Academie des Sciences, Serie 3: Sciences de la Vie , 316, 1194–1199. doi: 10.1007/978-3-642-79247-2_3.
- Benfey, P. N., & Chua, N.-H. (1990). The Cauliflower Mosaic Virus 35S Promoter: Combinatorial regulation of transcription in plants. Science , 250, 959–966. doi: 10.1126/science.250.4983.959.
- Berg, P. (1981). Dissections and reconstructions of genes and chromosomes. Science , 213, 296–303. doi: 10.1126/science.6264595.
- Berg, P. (2008). Asilomar 1975: DNA modification secured. Nature , 455, 290–291. doi: 10.1038/455290a.
- Berg, P., Baltimore, D., Boyer, H. W., Cohen, S. N., Davis, R. W., Hogness, D. S., … Zinder, N. D. (1974a). NAS ban on plasmid engineering. Nature , 250, 175–175. doi: 10.1038/250175a0.
- Berg, P., Baltimore, D., Boyer, H. W., Cohen, S. N., Davis, R. W., Hogness, D. S., … Zinder, N. D. (1974b). Potential biohazards of recombinant DNA molecules. Proceedings of the National Academy of Sciences of the United States of America , 71, 2593–2594. doi: 10.1073/pnas.71.7.2593.
- Berg, P., Baltimore, D., Boyer, H. W. et al. (1974c). Potential biohazards of recombinant DNA molecules. Science , 185, 303–303. doi: 10.1126/science.185.4148.303.
- Berg, P., Baltimore, D., Brenner, S., Roblin, III R. O., & Singer, M. F. (1975). Summary statement of the Asilomar conference on recombinant DNA molecules. Proceedings of the National Academy of Sciences of the United States of America , 72, 1981–1984. doi: 10.1073/pnas.72.6.1981.
- Bevan, M. W. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Research , 12, 8711–8721. doi: 10.1093/nar/12.22.8711.
- Bevan, M. W., Flavell, R. B., & Chilton, M.-D. (1983). A chimaeric antibiotic resistance gene as a selectable marker for plant cell transformation. Nature , 304, 184–187. doi: 10.1038/304184a0.
- De Block, M., Herrera-Estrella, L., van Montagu, M. C. E., Schell, J., & Zambryski, P. C. (1984). Expression of foreign genes in regenerated plants and in their progeny. The EMBO Journal , 3, 1681–1689. doi: 10.1002/j.1460-2075.1984.tb02032.x.
- Bomhoff, G., Klapwijk, P. M., Kester, H. C. M., Schilperoort, R. A., Hernalsteens, J.-P., & Schell, J. (1976). Octopine and nopaline synthesis and breakdown genetically controlled by a plasmid of Agrobacterium tumefaciens. Molecular and General Genetics , 145, 177–181.
- Braun, A. C. (1947). Thermal studies on the factors responsible for tumor initiation in crown gall. American Journal of Botany , 34, 234–240. doi: 10.1002/j.1537-2197.1947.tb12981.x.
- Brisson, N., Paszkowski, J., Penswick, J. R., Gronenborn, B., Potrykus, I., & Hohn, T. (1984). Expression of a bacterial gene in plants by using a viral vector. Nature , 310, 511–514. doi: 10.1038/310511a0.
- Caddick, M. X., Greenland, A. J., Jepson, L., Krause, K.-P., Qu, N., Riddell, K. V., … Tomsett, A. B. (1998). An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nature Biotechnology , 16, 177–180. doi: 10.1038/nbt0298-177.
- Cesarino, I., Dello Ioio, R., Kirschner, G. K., Ogden, M. S., Picard, K. L., Rast-Somssich, M. I., & Somssich, M. (2020). Plant science's next top models. Annals of Botany , 126, 1–23. doi: 10.1093/aob/mcaa063.
- Chang, A. C. Y., & Cohen, S. N. (1974). Genome construction between bacterial species in vitro: Replication and expression of Staphylococcus plasmid genes in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America , 71, 1030–1034. doi: 10.1073/pnas.71.4.1030.
- Chang, C., & Meyerowitz, E. M. (1986). Molecular cloning and DNA sequence of the Arabidopsis thaliana alcohol dehydrogenase gene. Proceedings of the National Academy of Sciences of the United States of America , 83, 1408–1412. doi: 10.1073/pnas.83.5.1408.
- Chi-Ham, C. L., Boettiger, S., Figueroa-Balderas, R., Bird, S., Geoola, J. N., Zamora, P., … Bennett, A. B. (2012). An intellectual property sharing initiative in agricultural biotechnology: Development of broadly accessible technologies for plant transformation. Plant Biotechnology Journal , 10, 501–510. doi: 10.1111/j.1467-7652.2011.00674.x.
- Chien, A., Edgar, D. B., & Trela, J. M. (1976). Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. Journal of Bacteriology , 127, 1550–1557. doi: 10.1128/jb.127.3.1550-1557.1976.
- Chilton, M.-D. (2001). Agrobacterium. A Memoir. Plant Physiology , 125, 9–14. doi: 10.1104/pp.125.1.9.
- Chilton, M.-D. (2018). My secret life. Annual Review of Plant Biology , 69, 1–20. doi: 10.1146/annurev-arplant-032717-090606.
- Chilton, M.-D., Currier, T. C., Farrand, S. K., Bendich, A. J., Gordon, M. P., & Nester, E. W. (1974). Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proceedings of the National Academy of Sciences of the United States of America , 71, 3672–3676. doi: 10.1073/pnas.71.9.3672.
- Chilton, M.-D., Drummond, M. H., Merlo, D. J., Sciaky, D., Montoya, A. L., Gordon, M. P., & Nester, E. W. (1977). Stable incorporation of plasmid DNA into higher plant cells: The molecular basis of crown gall tumorigenesis. Cell , 11, 263–271. doi: 10.1016/0092-8674(77)90043-5.
- Church, G. M., & Gilbert, W. (1984). Genomic sequencing. Proceedings of the National Academy of Sciences of the United States of America , 81, 1991–1995. doi: 10.1073/pnas.81.7.1991.
- Clough, S. J., & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal , 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x.
- Cohen, S. N., Chang, A. C. Y., Boyer, H. W., & Helling, R. B. (1973). Construction of biologically functional bacterial plasmids in vitro. Proceedings of the National Academy of Sciences of the United States of America , 70, 3240–3244. doi: 10.1073/pnas.70.11.3240.
- Covey, S. N., & Hull, R. (1981). Transcription of cauliflower mosaic virus DNA. Detection of transcripts, properties, and location of the gene encoding the virus inclusion body protein. Virology , 111, 463–474. doi: 10.1016/0042-6822(81)90349-4.
- Danna, K. J., Sack, G. H., & Nathans, D. (1973). Studies of Simian Virus 40 DNA. VII. A Cleavage Map of the SV40 Genome. Journal of Molecular Biology , 78, 363–376. doi: 10.1016/0022-2836(73)90122-8.
- Day, M. F., & Venables, D. G. (1961). The transmission of cauliflower mosaic virus by Aphids. Australian Journal of Biological Sciences , 14, 187. doi: 10.1071/BI9610187.
- Depicker, A. G., van Montagu, M. C. E., & Schell, J. (1978). Homologous DNA sequences in different Ti-plasmids are essential for oncogenicity. Nature , 275, 150–153. doi: 10.1038/275150a0.
- Duke, S. O., & Powles, S. B. (2008). Glyphosate: A once-in-a-century herbicide. Pest Management Science , 64, 319–325. doi: 10.1002/ps.1518.
- Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D., Bombarely, A., … Mueller, L. A. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics , 18, 448. doi: 10.1186/s12864-017-3791-6.
- Engler, G., Holsters, M., van Montagu, M. C. E., Schell, J., Hernalsteens, J.-P., & Schilperoort, R. A. (1975). Agrocin 84 sensitivity: A plasmid determined property in Agrobacterium tumefaciens. Molecular and General Genetics , 138, 345–349. doi: 10.1007/BF00264804.
- Fang, R.-X., Nagy, F., Sivasubramaniam, S., & Chua, N. (1989). Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. The Plant Cell , 1, 141–150. doi: 10.1105/tpc.1.1.141.
- Feenstra, W. J. (1964). Isolation of nutritional mutants in Arabidopsis thaliana. Genetica , 35, 259–269. doi: 10.1007/BF01804894.
- Feldmann, K. A., & Marks, M. D. (1987). Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana : A non-tissue culture approach. Molecular and General Genetics , 208, 1–9. doi: 10.1007/BF00330414.
- Fraley, R. T., Rogers, S. G., Horsch, R. B., Sanders, P. R., Flick, J. S., Adams, S. P., … Woo, S. C. (1983). Expression of bacterial genes in plant cells. Proceedings of the National Academy of Sciences of the United States of America , 80, 4803–4807. doi: 10.1073/pnas.80.15.4803.
- Franck, A., Guilley, H., Jonard, G., Richards, K. E., & Hirth, L. (1980). Nucleotide sequence of cauliflower mosaic virus DNA. Cell , 21, 285–294. doi: 10.1016/0092-8674(80)90136-1.
- Garfinkel, D. J., Simpson, R. B., Ream, L. W., White, F. F., Gordon, M. P., & Nester, E. W. (1981). Genetic analysis of crown gall: Fine structure map of the T-DNA by site-directed mutagenesis. Cell , 27, 143–153. doi: 10.1016/0092-8674(81)90368-8.
- Gerats, T., & Vandenbussche, M. (2005). A model system for comparative research: Petunia. Trends in Plant Science , 10, 251–256. doi: 10.1016/j.tplants.2005.03.005.
- Gilbert, W. (1981). DNA sequencing and gene structure. Science , 214, 1305–1312. doi: 10.1126/science.7313687.
- Goodman, R. M. (1977). Single-stranded DNA genome in a whitefly-transmitted plant virus. Virology , 83, 171–179. doi: 10.1016/0042-6822(77)90220-3.
- Graff, G. D., Cullen, S. E., Bradford, K. J., Zilberman, D., & Bennett, A. B. (2003). The public-private structure of intellectual property ownership in agricultural biotechnology. Nature Biotechnology , 21, 989–995. doi: 10.1038/nbt0903-989.
- Greve de, H., Decraemer, H., Seurinck, J., van Montagu, M. C. E., & Schell, J. (1981). The functional organization of the octopine Agrobacterium tumefaciens plasmid pTiB6s3. Plasmid , 6 ( 2 ) , 235–248. doi: 10.1016/0147-619x(81)90069-x.
- Gronenborn, B., Gardner, R. C., Schaefer, S., & Shepherd, R. J. (1981). Propagation of foreign DNA in plants using cauliflower mosaic virus as vector. Nature , 294, 773–776. doi: 10.1038/294773a0.
- Guilley, H., Dudley, R. K., Jonard, G., Balàzs, E., & Richards, K. E. (1982). Transcription of cauliflower mosaic virus DNA: Detection of promoter sequences, and characterization of transcripts. Cell , 30, 763–773. doi: 10.1016/0092-8674(82)90281-1.
- Haas, M., Bureau, M., Geldreich, A., Yot, P., & Keller, M. (2002). Cauliflower mosaic virus: Still in the news. Molecular Plant Pathology , 3, 419–429. doi: 10.1046/j.1364-3703.2002.00136.x.
- Heimann, J. M. (2018). Using Nature's Shuttle: The making of the first genetically modified plants and the people who did it. The Netherlands: Wageningen Academic Publishers.
- Hernalsteens, J.-P., Vliet, F. V., Beuckeleer, M. De, Depicker, A., Engler, G., Lemmers, M., … Schell, J. (1980). The Agrobacterium tumefaciens Ti plasmid as a host vector system for introducing foreign DNA in plant cells. Nature , 287, 654–656. doi: 10.1038/287654a0.
- Herrera-Estrella, L., Depicker, A., van Montagu, M. C. E., & Schell, J. (1983). Expression of chimaeric genes transferred into plant cells using a Ti-plasmid-derived vector. Nature , 303, 209–213. doi: 10.1038/303209a0.
- Hoekema, A., Hirsch, P. R., Hooykaas, P. J. J., & Schilperoort, R. A. (1983). A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature , 303, 179–180. doi: 10.1038/303179a0.
- Höfte, H., Greve de, H., Seurinck, J., Jansens, S., Mahillon, J., Ampe, C., … Zabeau, M. (1986). Structural and functional analysis of a cloned delta endotoxin of Bacillus thuringiensis Berliner 1715. European Journal of Biochemistry , 161, 273–280. doi: 10.1111/j.1432-1033.1986.tb10443.x.
- Hohn, T., Hohn, B., Lesot, A., & Lebeurier, G. (1980). Restriction map of native and cloned cauliflower mosaic virus DNA. Gene , 11, 21–31. doi: 10.1016/0378-1119(80)90083-9.
- Hull, R. (1978). The possible use of plant viral DNAs in genetic manipulation in plants. Trends in Biochemical Sciences , 3, 254–256. doi: 10.1016/S0968-0004(78)95435-X.
- Hull, R., Covey, S. N., and Dale, P. (2000). Genetically modified plants and the 35S promoter: Assessing the risks and enhancing the debate. Microbial Ecology in Health and Disease , 12, 1–5. doi: 10.3402/mehd.v12i1.8034.
- Hutchison, C. A., Phillips, S., Edgell, M. H., Gillam, S., Jahnke, P., & Smith, M. (1978). Mutagenesis at a specific position in a DNA sequence. Journal of Biological Chemistry , 253, 6551–6560. doi: 10.1016/S0021-9258(19)46967-6.
- Jackson, D. A., Symons, R. H., & Berg, P. (1972). Biochemical method for inserting new genetic information into DNA of simian virus 40: Circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America , 69, 2904–2909. doi: 10.1073/pnas.69.10.2904.
- Jay, E., Bambara, R., Padmanabhan, R., & Wu, R. (1974). DNA sequence analysis: A general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Research , 1, 331–354. doi: 10.1093/nar/1.3.331.
- Johansson, B. G. (1972). Agarose gel electrophoresis. Scandinavian Journal of Clinical and Laboratory Investigation , 29, 7–19. doi: 10.3109/00365517209102747.
- Kay, R., Chan, A., Daly, M., & McPherson, J. (1987). Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science , 236, 1299–1302. doi: 10.1126/science.236.4806.1299.
- Khorana, H. G. (1979). Total synthesis of a gene. Science , 203, 614–625. doi: 10.1126/science.366749.
- Kleppe, K., Ohtsuka, E., Kleppe, R., Molineux, I., & Khorana, H. G. (1971). Studies on polynucleotides: XCVI. Repair replication of short synthetic DNAs as catalyzed by DNA polymerases. Journal of Molecular Biology , 56, 341–361. doi: 10.1016/0022-2836(71)90469-4.
- Koncz, C., De Greve, H., André, D., Deboeck, F., van Montagu, M. C. E., & Schell, J. (1983). The opine synthase genes carried by Ti plasmids contain all signals necessary for expression in plants. The EMBO Journal , 2, 1597–1603. doi: 10.1002/j.1460-2075.1983.tb01630.x.
- Koncz, C., & Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics , 204, 383–396. doi: 10.1007/BF00331014.
- Koornneef, M. (1978). Gibberellin-sensitive mutants in Arabidopsis thaliana. Arabidopsis Information Service , 15, 17–20.
- Koornneef, M. (1983). The use of telotrisomics for centromere mapping in Arabidopsis thaliana (L.) Heynh. Genetica , 62, 33–40. doi: 10.1007/BF00123307.
- Koornneef, M. (2021). A central role for genetics in plant biology. Annual Review of Plant Biology , 72, 1–16. doi: 10.1146/annurev-arplant-071720-111039.
- Koornneef, M., Barbaro, A., & van der Veen, J. H. (1977). Nongerminating, gibberellic acid responsive mutants in Arabidopsis thaliana. Arabidopsis Information Service , 14, 14–17.
- Koornneef, M., van Eden, J., Hanhart, C. J., Stam, P., Braaksma, F. J., & Feenstra, W. J. (1983). Linkage map of Arabidopsis thaliana. Journal of Heredity , 74, 265–272. doi: 10.1093/oxfordjournals.jhered.a109781.
- Koornneef, M., & van der Veen, J. H. (1983). Trisomics in Arabidopsis thaliana and the location of linkage groups. Genetica , 61, 41–46. doi: 10.1007/BF00563230.
- Lacroix, B., Tzfira, T., Vainstein, A., & Citovsky, V. (2006). A case of promiscuity: Agrobacterium's endless hunt for new partners. Trends in Genetics , 22, 29–37. doi: 10.1016/j.tig.2005.10.004.
- Laibach, F. (1943). Arabidopsis thaliana (L.) Heynh. als Objekt für genetische und entwicklungsphysiologische Untersuchungen. Botanisches Archiv , 44, 439–455.
- Laibach, F. (1965). 60 Jahre Arabidopsis-Forschung, 1905-1965. Arabidopsis Information Service , 01, 16.
- Larebeke, N., Engler, G., Holsters, M., Elsacker, S., Zaenen, I., Schilperoort, R. A., & Schell, J. (1974). Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature , 252, 169–170. doi: 10.1038/252169a0.
- Lazarides, E., & Weber, K. (1974). Actin antibody: The specific visualization of actin filaments in non-muscle cells. Proceedings of the National Academy of Sciences of the United States of America , 71, 2268–2272. doi: 10.1073/pnas.71.6.2268.
- Lefebvre, D. D., Miki, B. L., & Laliberté, J.-F. (1987). Mammalian metallothionein functions in plants. Nature Biotechnology , 5, 1053–1056. doi: 10.1038/nbt1087-1053.
- Leutwiler, L. S., Hough-Evans, B. R., & Meyerowitz, E. M. (1984). The DNA of Arabidopsis thaliana. Molecular and General Genetics , 194, 15–23. doi: 10.1007/BF00383491.
- Lloyd, A. M., Barnason, A. R., Rogers, S. G., Byrne, M. C., Fraley, R. T., & Horsch, R. B. (1986). Transformation of Arabidopsis thaliana with Agrobacterium tumefaciens. Science , 234, 464–466. doi: 10.1126/science.234.4775.464.
- Logemann, E., Birkenbihl, R. P., Ülker, B., & Somssich, I. E. (2006). An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods , 2, 16. doi: 10.1186/1746-4811-2-16.
- McCabe, D. E., Swain, W. F., Martinell, B. J., and Christou, P. (1988). Stable transformation of soybean (Glycine Max) by particle acceleration. Nature Biotechnology , 6, 923–926. doi: 10.1038/nbt0888-923.
- Mertz, J. E., & Davis, R. W. (1972). Cleavage of DNA by R1 Restriction Endonuclease Generates Cohesive Ends. Proceedings of the National Academy of Sciences of the United States of America , 69, 3370–3374. doi: 10.1073/pnas.69.11.3370.
- Meselson, M., & Yuan, R. (1968). DNA restriction enzyme from E. coli. Nature , 217, 1110–1114. doi: 10.1038/2171110a0.
- Meyerowitz, E. M. (1987). Arabidopsis thaliana. Annual Review of Genetics , 21, 93–111. doi: 10.1146/annurev.ge.21.120187.000521.
- Mullis, K. B. (1994). The Polymerase Chain Reaction (Nobel Lecture). Angewandte Chemie International Edition in English , 33, 1209–1213. doi: 10.1002/anie.199412091.
- Mullis, K. B., Faloona, F., Scharf, S. J., Saiki, R. K., Horn, G. T., & Erlich, H. A. (1986). Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. Cold Spring Harbor Symposia on Quantitative Biology , 51(Pt 1), 263–273. doi: 10.1101/SQB.1986.051.01.032.
- Murai, N., Sutton, D. W., Murray, M. G., Murray, M. G., Slightom, J. L., Merlo, D. J., … Hall, T. C. (1983). Phaseolin gene from bean is expressed after transfer to sunflower via tumor-inducing plasmid vectors. Science , 222, 476–482. doi: 10.1126/science.222.4623.476.
- Narusaka, M., Shiraishi, T., Iwabuchi, M., & Narusaka, Y. (2010). The floral inoculating protocol: A simplified Arabidopsis thaliana transformation method modified from floral dipping. Plant Biotechnology , 27, 349–351. doi: 10.5511/plantbiotechnology.27.349.
- Nathans, D. (1979). Restriction endonucleases, Simian Virus 40, and the new genetics. Science , 206, 903–909. doi: 10.1126/science.228393.
- Nottenburg, C., & Rodríguez, C. R. (2008). Agrobacterium-mediated gene transfer: A Lawyer's Perspective. Agrobacterium: From Biology to Biotechnology (pp. 699–735). New York, NY: Springer New York. doi: 10.1007/978-0-387-72290-0_20.
- O'Brien, S. J. (1983). Miami winter symposium. Nature Biotechnology , 1, 36–38. doi: 10.1038/nbt0383-36.
- Odell, J. T., Nagy, F., & Chua, N.-H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature , 313, 810–812. doi: 10.1038/313810a0.
- Ooms, G., Hooykaas, P. J. J., Moolenaar, G., & Schilperoort, R. A. (1981). Grown gall plant tumors of abnormal morphology, induced by Agrobacterium tumefaciens carrying mutated octopine Ti plasmids; analysis of T-DNA functions. Gene , 14, 33–50. doi: 10.1016/0378-1119(81)90146-3.
- Otten, L., Greve, H., Hernalsteens, J.-P., van Montagu, M. C. E., Schieder, O., Straub, J., & Schell, J. (1981). Mendelian transmission of genes introduced into plants by the Ti plasmids of Agrobacterium tumefaciens. Molecular and General Genetics , 183, 209–213. doi: 10.1007/BF00270619.
- Paszkowski, J., Pisan, B., Shillito, R. D., Hohn, T., Hohn, B., & Potrykus, I. (1986). Genetic transformation of Brassica campestris var. rapa protoplasts with an engineered cauliflower mosaic virus genome. Plant Molecular Biology , 6, 303–312. doi: 10.1007/BF00034937.
- Peferoen, M. (1997). Progress and prospects for field use of bt genes in crops. Trends in Biotechnology , 15, 173–177. doi: 10.1016/S0167-7799(97)01018-4.
- Pennisi, E. (2000). Arabidopsis comes of age. Science , 290, 32–35. doi: 10.1126/science.290.5489.32.
- Pruitt, R. E., & Meyerowitz, E. M. (1986). Characterization of the genome of Arabidopsis thaliana. Journal of Molecular Biology , 187, 169–183. doi: 10.1016/0022-2836(86)90226-3.
- Rédei, G. P. (1962). Single locus heterosis. Zeitschrift für Vererbungslehre , 93, 164–170.
- Rédei, G. P. (1975). Arabidopsis as a genetic tool. Annual Review of Genetics , 9, 111–127. doi: 10.1146/annurev.ge.09.120175.000551.
- Rédei, G. P. (1992). A heuristic glance at the past of Arabidopsis genetics. Methods in Arabidopsis Research , 1–15.
- Rosso, M. G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., & Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology , 53, 247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a.
- Roulland-Dussoix, D., & Arber, W. (1962). Host specificity of DNA produced by Escherichia coli II, Control over acceptance of DNA from infecting phage λ. Journal of Molecular Biology , 5, 37–49. doi: 10.1016/S0022-2836(62)80059-X.
- Rushton, P. J., Reinstädler, A., Lipka, V., Lippok, B., & Somssich, I. E. (2002). Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. The Plant cell , 14, 749–762. doi: 10.1105/tpc.010412.
- Sack, G. H., & Nathans, D. (1973). Studies of Simian Virus 40 DNA. VI. Cleavage of SV40 DNA by restriction endonuclease from Hemophilus parainfluenzae. Virology , 51, 517–520. doi: 10.1016/0042-6822(73)90455-8.
- Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn G. T., Mullis, K. B., & Erlich, H. A. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science , 239, 487–491. doi: 10.1126/science.2448875.
- Sanger, F. (1981). Determination of nucleotide sequences in DNA. Science , 214, 1205–1210. doi: 10.1126/science.7302589.
- Sanger, F., Nicklen, S., & Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America , 74, 5463–5467. doi: 10.1073/pnas.74.12.5463.
- Schilperoort, R. A., Veldstra, H., Warnaar, S. O., Mulder, G., & Cohen, J. A. (1967). Formation of complexes between DNA isolated from tobacco crown gall tumours and RNA complementary to Agrobacterium tumefaciens DNA. Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis , 145, 523–525. doi: 10.1016/0005-2787(67)90075-5.
- Schulman, A. H. (2020). The impact of GM crops on agriculture. In V. Andersen (Ed.), Genetically Modified and Irradiated Food: Controversial Issues: Facts Versus Perceptions (pp. 195-213). Academic Press Inc Elsevier Science. https://doi.org/10.1016/B978-0-12-817240-7.00012-7.
- Schultz, E. S. (1921). A transmissible mosaic disease of Chinese cabbage, mustard and turnip. Journal of Agricultural Research , 22, 173–177.
- Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., & Goff, S. A. (2002). A high-throughput Arabidopsis reverse genetics system. The Plant Cell , 14, 2985–2994. doi: 10.1105/tpc.004630.
- Shah, D. M., Horsch, R. B., Klee, H. J., Kishore, G. M., Winter, J. A., Tumer, N. E., … Fraley, R. T. (1986). Engineering herbicide tolerance in transgenic plants. Science , 233, 478–481. doi: 10.1126/science.233.4762.478.
- Shepherd, R. J., Bruening, G. E., & Wakeman, R. J. (1970). Double-stranded DNA from cauliflower mosaic virus. Virology , 41, 339–347. doi: 10.1016/0042-6822(70)90086-3.
- Shepherd, R. J., Wakeman, R. J., & Romanko, R. R. (1968). DNA in cauliflower mosaic virus. Virology , 36, 150–152. doi: 10.1016/0042-6822(68)90127-X.
- Siegel, A., & Wildman, S. G. (1960). Some aspects of the structure and behavior of tobacco mosaic virus. Annual Review of Plant Physiology , 11, 277–298. doi: 10.1146/annurev.pp.11.060160.001425.
- Smith, H. O. (1979). Nucleotide sequence specificity of restriction endonucleases. Science , 205, 455–462. doi: 10.1126/science.377492.
- Smith, M. (1994). Synthetic DNA and Biology (Nobel Lecture). Angewandte Chemie International Edition in English , 33, 1214–1221. doi: 10.1002/anie.199412141.
- Smith, E. F., & Townsend, C. O. (1907). A plant-tumor of bacterial origin. Science , 25, 671–673. doi: 10.1126/science.25.643.671.
- Smith, H. O., & Wilcox, K. W. (1970). A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. Journal of Molecular Biology , 51, 379–391. doi: 10.1016/0022-2836(70)90149-X.
- Somerville, C. R. (1989). Arabidopsis blooms. The Plant Cell , 1, 1131–1135. doi: 10.1105/tpc.1.12.1131.
- Somerville, C. R., & Koornneef, M. (2002). A fortunate choice: The history of Arabidopsis as a model plant. Nature reviews. Genetics , 3, 883–889. doi: 10.1038/nrg927.
- Somerville, C. R., & Ogren, W. L. (1979). A phosphoglycolate phosphatase-deficient mutant of Arabidopsis. Nature , 280, 833–836. doi: 10.1038/280833a0.
- Somerville, C. R., & Ogren, W. L. (1980). Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature , 286, 257–259. doi: 10.1038/286257a0.
- Somssich, M. (2018a). A short history of Arabidopsis thaliana (L.) Heynh. Columbia-0. PeerJ Preprints , e26931v3, 1–7. doi: 10.7287/peerj.preprints.26931v5.
- Somssich, M. (2018b). A short history of the CaMV 35S promoter. PeerJ Preprints , 6, e27096v2, 1–16. doi: 10.7287/peerj.preprints.27096v3.
- Somssich, M. (2019). A short history of plant transformation. PeerJ Preprints , 1–28. doi: 10.7287/peerj.preprints.27556v2.
- Somssich, M. (2021). A short history of plant light microscopy. Zenodo , 4682572. doi: 10.5281/zenodo.4682572.
- Southern, E. M. (1975). Detection of specific sequences among DNA fragments separated by gel electrophoresis. Journal of Molecular Biology , 98, 503–517. doi: 10.1016/S0022-2836(75)80083-0.
- Stachel, S. E., Messens, E., van Montagu, M. C. E., & Zambryski, P. (1985). Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature , 318, 624–629. doi: 10.1038/318624a0.
- Tompkins, C. M. (1937). A transmissible mosaic disease of cauliflower. Journal of Agricultural Research , 55, 33–46.
- Towbin, H., Staehelin, T., & Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proceedings of the National Academy of Sciences of the United States of America , 76, 4350–4354. doi: 10.1073/pnas.76.9.4350.
- Ulmasov, T., Murfett, J., Hagen, G., & Guilfoyle, T. J. (1997). Aux/lAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell , 9, 1963–1971. doi: 10.1105/tpc.9.11.1963.
- Vaeck, M., Reynaerts, A., Höfte, H., Jansens, S., De Beuckeleer, M., Dean, C., … Leemans, J. (1987). Transgenic plants protected from insect attack. Nature , 328, 33–37. doi: 10.1038/328033a0.
- Wang, K., Herrera-Estrella, L., van Montagu, M. C. E., & Zambryski, P. (1984). Right 25 by terminus sequence of the nopaline t-DNA is essential for and determines direction of DNA transfer from Agrobacterium to the plant genome. Cell , 38, 455–462. doi: 10.1016/0092-8674(84)90500-2.
- Weber, K., & Osborn, M. (1969). The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. Journal of Biological Chemistry , 244, 4406–4412. doi: 10.1016/S0021-9258(18)94333-4.
- Weigel, D. (2012). Natural variation in Arabidopsis: From molecular genetics to ecological genomics. Plant Physiology , 158, 2–22. doi: 10.1104/pp.111.189845.
- Wells, R. D., Jacob, T. M., Narang, S. A., & Khorana, H. G. (1967). Studies on polynucleotides: LXIX. Synthetic deoxyribopolynucleotides as templates for the DNA polymerase of Escherichia coli : DNA-like polymers containing repeating trinucleotide sequences. Journal of Molecular Biology , 27, 265–272. doi: 10.1016/0022-2836(67)90019-8.
- White, P. R., & Braun, A. C. (1941). Crown gall production by bacteria-free tumor tissues. Science , 94, 239–241. doi: 10.1126/science.94.2436.239.
- Zaenen, I., Van Larebeke, N., van Montagu, M. C. E., & Schell, J. (1974). Supercoiled circular DNA in crown-gall inducing Agrobacterium strains. Journal of Molecular Biology , 86, 109–127. doi: 10.1016/S0022-2836(74)80011-2.
- Zambryski, P. C., Holsters, M., Kruger, K., Depicker, A., Schell, J., van Montagu, M. C. E., & Goodman, H. M. (1980). Tumor DNA structure in plant cells transformed by A. tumefaciens. Science , 209, 1385–1391. doi: 10.1126/science.6251546.
- Zambryski, P. C., Joos, H., Genetello, C., Leemans, J., van Montagu, M. C. E., & Schell, J. (1983). Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. The EMBO Journal , 2, 2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x.
- Zhu, H., Li, C., & Gao, C. (2020). Applications of CRISPR–Cas in agriculture and plant biotechnology. Nature Reviews Molecular Cell Biology , 21, 661–677. doi: 10.1038/s41580-020-00288-9.
- Zuo, J., Niu, Q.-W., & Chua, N.-H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. The Plant Journal , 24, 265–273. doi: 10.1046/j.1365-313x.2000.00868.x.
Citing Literature
Number of times cited according to CrossRef: 4
- Camille Soulhat, Houssein Wehbi, Yannick Fierlej, Patrick Berquin, Thomas Girin, Pierre Hilson, Oumaya Bouchabké-Coussa, Fast-track transformation and genome editing in Brachypodium distachyon, Plant Methods, 10.1186/s13007-023-01005-1, 19 , 1, (2023).
- Geoffrey Thomson, Lauren Dickinson, Yannick Jacob, Genomic consequences associated with Agrobacterium‐mediated transformation of plants, The Plant Journal, 10.1111/tpj.16496, 117 , 2, (342-363), (2023).
- Jiyue Huang, Xiang Li, Cong Wang, Yingxiang Wang, Evaluation of Crossover Number, Distribution, and Interference Using Cytological Assays in Arabidopsis, Current Protocols, 10.1002/cpz1.599, 2 , 12, (2022).
- Marc Somssich, A Short History of Plant Light Microscopy, Current Protocols, 10.1002/cpz1.577, 2 , 10, (2022).

