Supporting protocol for use-case 1: Dimensionality reduction in "M2aia - Interactive, fast and memory efficient analysis of 2D and 3D multi-modal mass spectrometry imaging data"
Jonas Cordes
Abstract
M²aia is able to apply dimensionality reduction methods to 2D/3D MSI images. To demonstrate the DR methods we load a single peptide brain slice MS image, select multiple peaks from a peak list and start a PCA and t-SNE.
Before start
a) Download and install M²aia
Software
| Value | Label |
|---|---|
| M²aia | NAME |
| Windows/Linux | OS_NAME |
| https://github.com/jtfcordes/m2aia | REPOSITORY |
| Jonas Cordes | DEVELOPER |
| https://jtfcordes.github.io/M2aia | LINK |
| 2021.07.00 | VERSION |
b) Download the data set
Steps
Load Data into M²aia
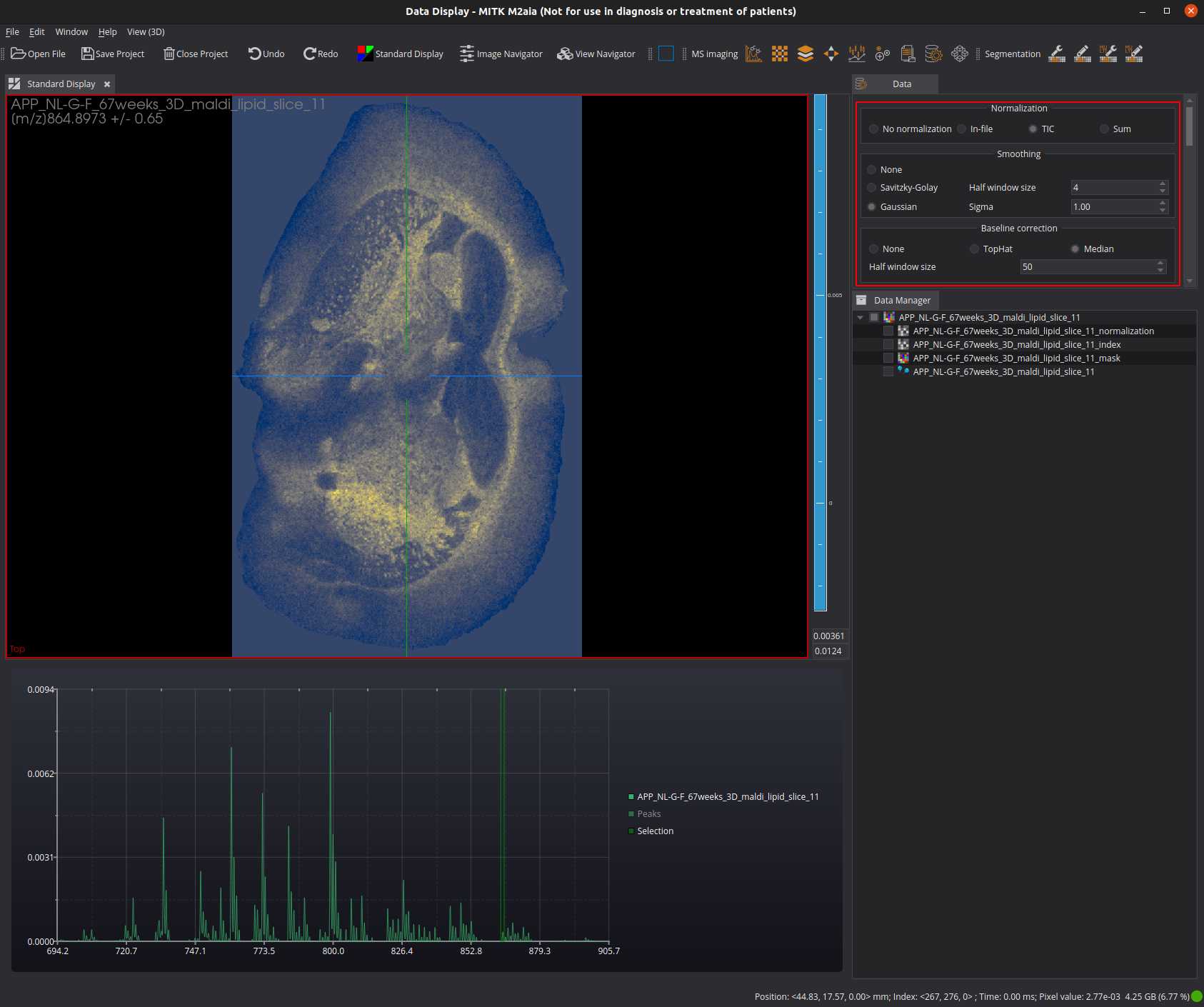
Open the Data view and define the signal processing:
- set your normalization strategy (e.g. TIC)
- set your smoothing strategy (e.g. Gaussian)
- set your baseline correction strategy (e.g. Median)
Load at least one continuous profile *.imzML image file into M²aia: File > Open File or Ctrl + O.
Ensure, that the corresponding *.ibd file is available at the same directory level.
Peak picking
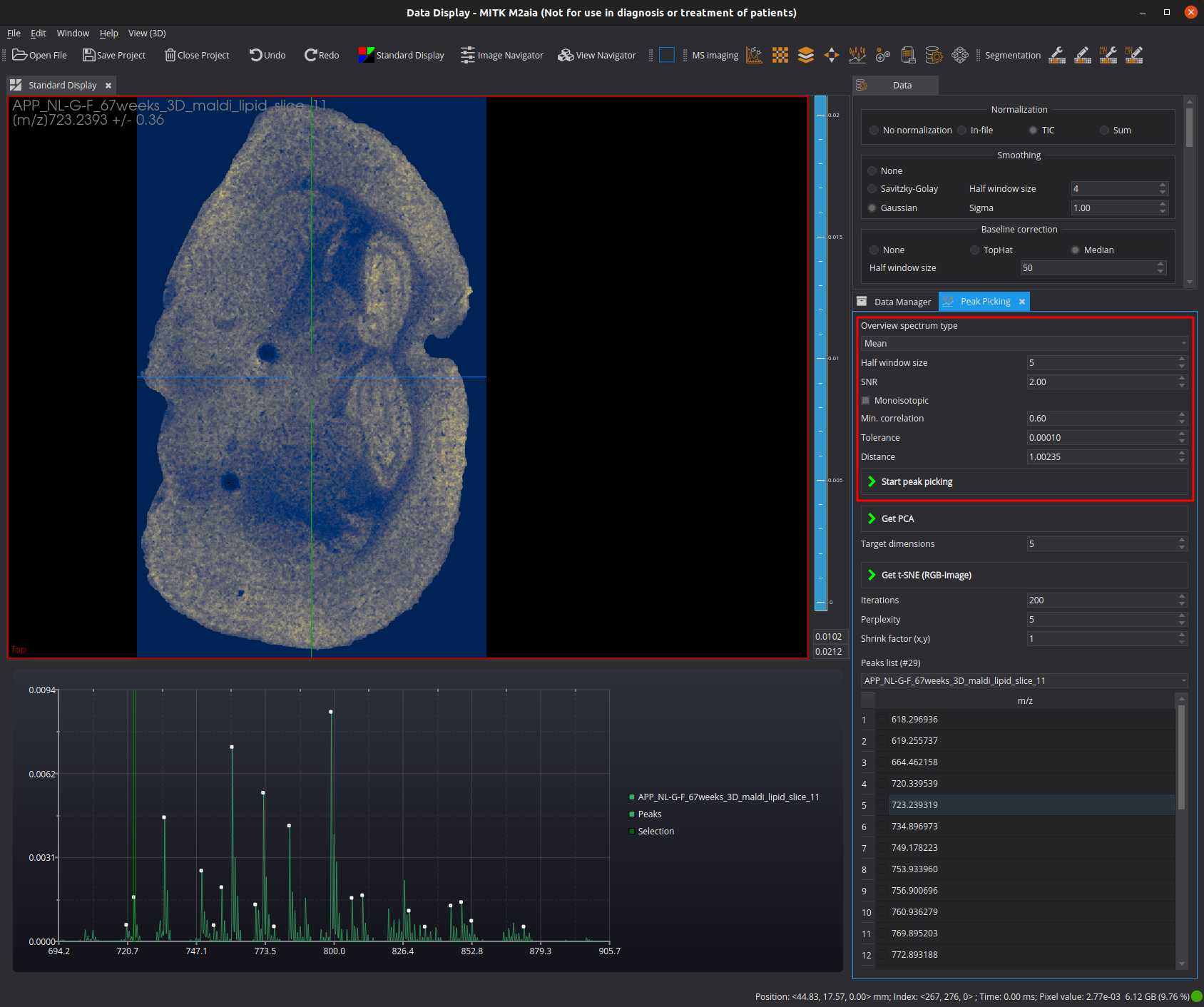
Open the Peak Picking view, e.g. from the menu: Window > Show View > Peak Picking .
Select an overview spectrum type in the drop-down menu (e.g. Mean).
Set the peak picking parameters (example):
- set the SNR to 2
- enable monoisotopic peak harvesting
- set the min correlation to 0.6
Start the peak picking.
Peak selection and PCA
Select peaks of interest (red annotations).
Select the target dimensionality and start the PCA (blue annotation).
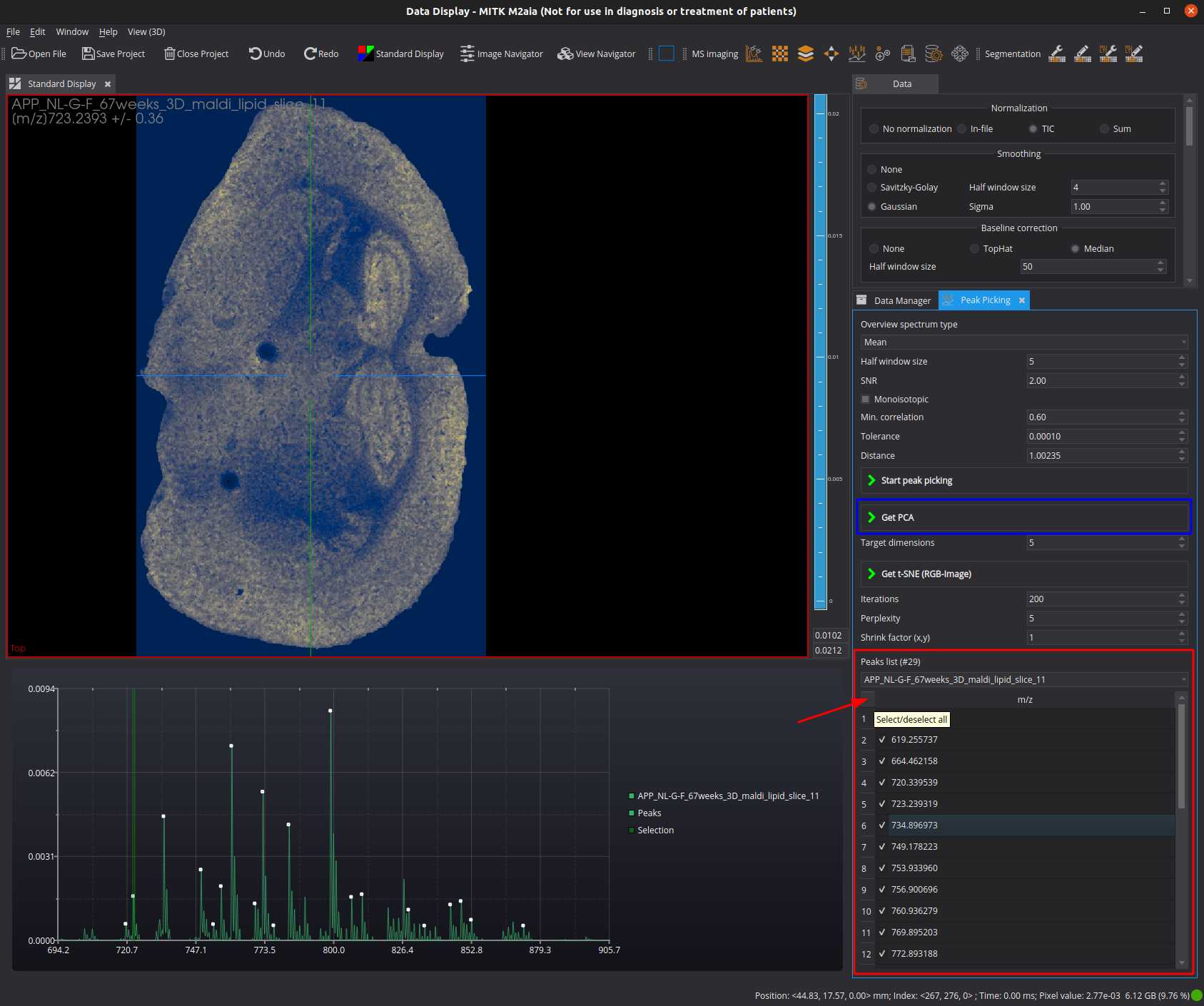
The result image is now available in the Data Manager view. You can change the currently displayed projected image by using the right click menu of the "PCA"-datanode and the Component slider.
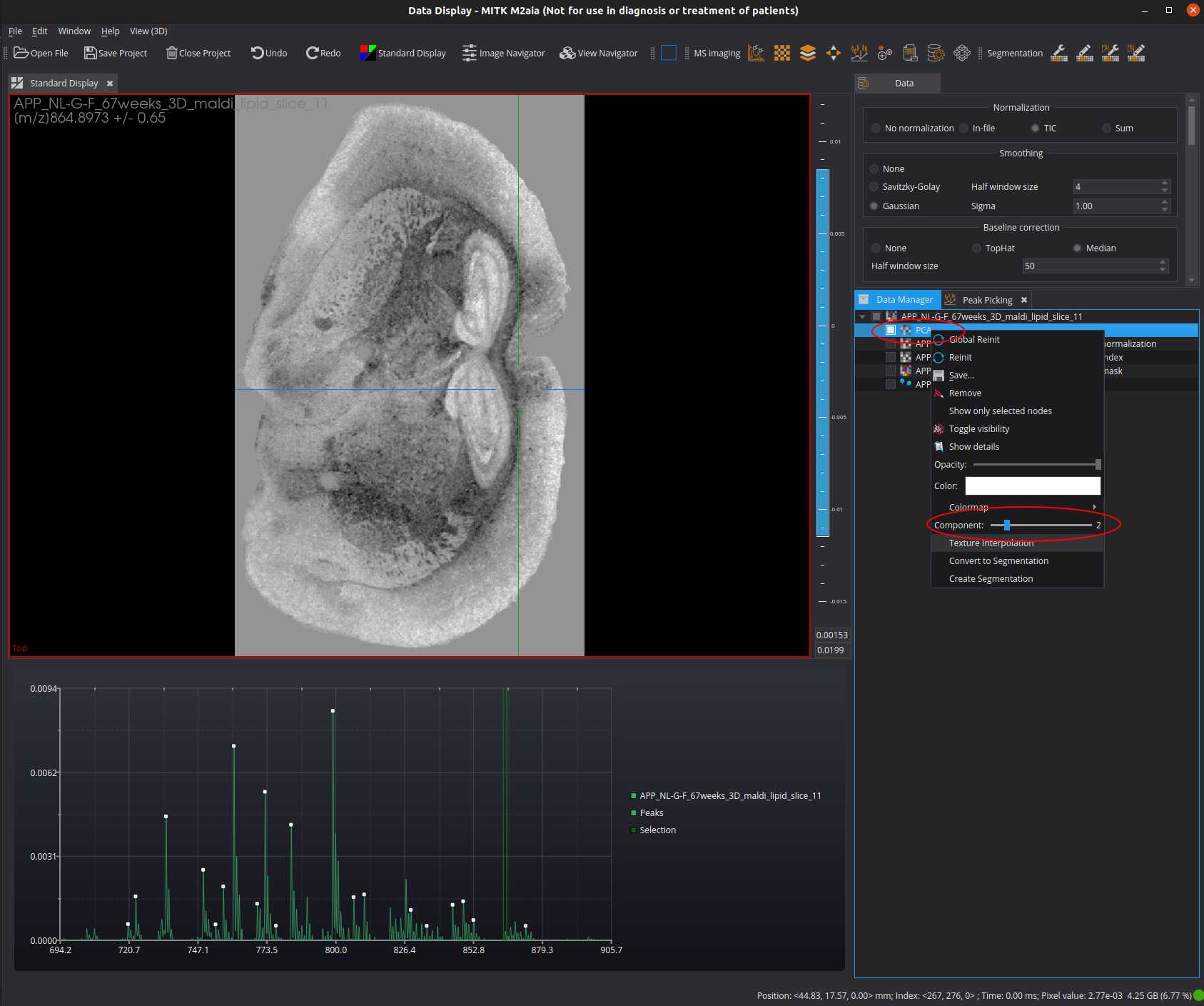
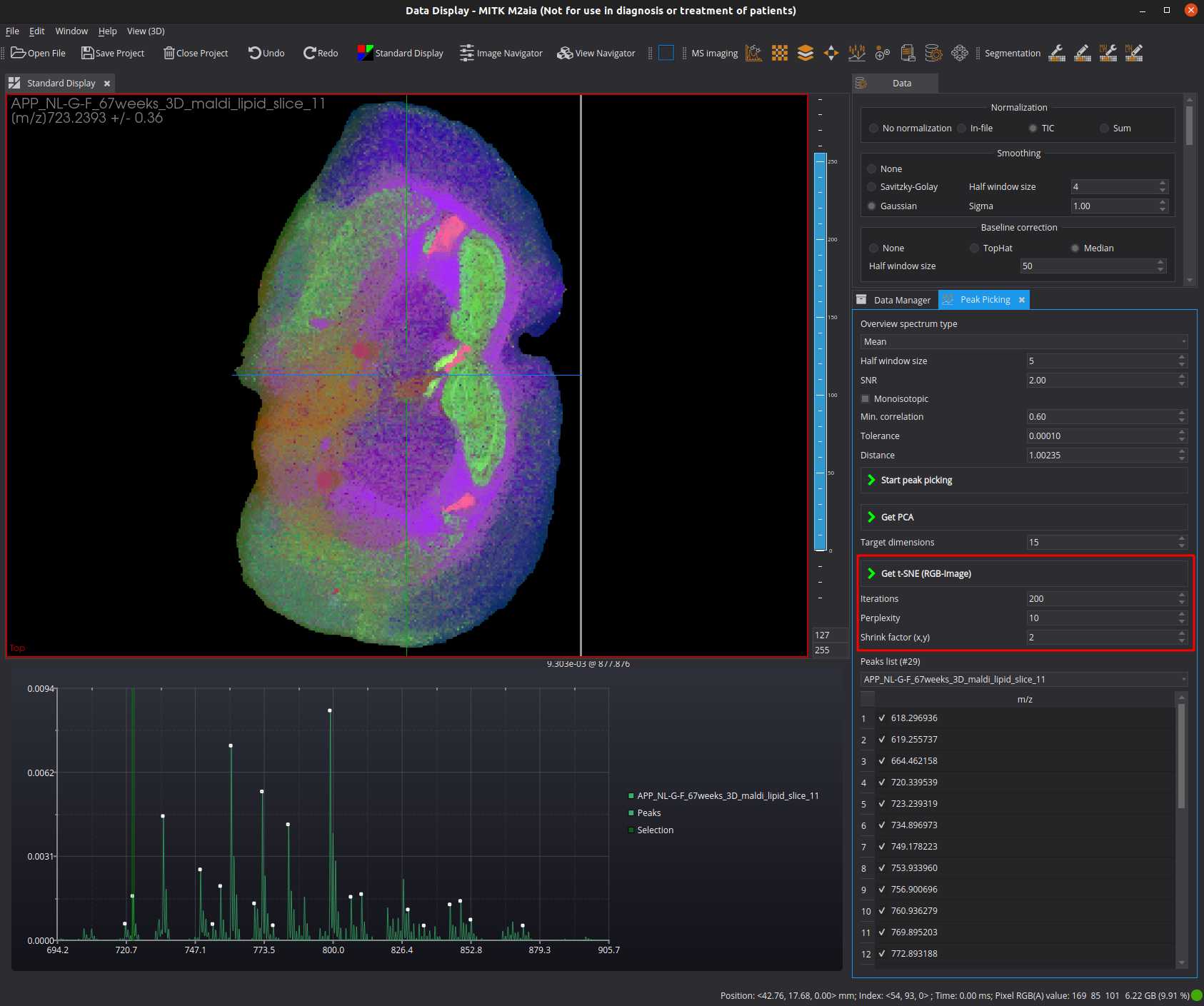
t-SNE
Set the t-SNE parameters (example):
- set iterations to 200
- set perplexity to 10
- set shrink factor to 2
Make sure that a "PCA" named datanode is available as derived node of the MSI datanode. Start the t-SNE.