SNP-SVant: A Computational Workflow to Predict and Annotate Genomic Variants in Organisms Lacking Benchmarked Variants
Clarissa J. Nobile, Clarissa J. Nobile, Deepika Gunasekaran, Deepika Gunasekaran, David H. Ardell, David H. Ardell
computational pipeline
short-read paired-end sequencing
single nucleotide polymorphisms
structural variations
variant calling
Abstract
Whole-genome sequencing is widely used to investigate population genomic variation in organisms of interest. Assorted tools have been independently developed to call variants from short-read sequencing data aligned to a reference genome, including single nucleotide polymorphisms (SNPs) and structural variations (SVs). We developed SNP-SVant, an integrated, flexible, and computationally efficient bioinformatic workflow that predicts high-confidence SNPs and SVs in organisms without benchmarked variants, which are traditionally used for distinguishing sequencing errors from real variants. In the absence of these benchmarked datasets, we leverage multiple rounds of statistical recalibration to increase the precision of variant prediction. The SNP-SVant workflow is flexible, with user options to tradeoff accuracy for sensitivity. The workflow predicts SNPs and small insertions and deletions using the Genome Analysis ToolKit (GATK) and predicts SVs using the Genome Rearrangement IDentification Software Suite (GRIDSS), and it culminates in variant annotation using custom scripts. A key utility of SNP-SVant is its scalability. Variant calling is a computationally expensive procedure, and thus, SNP-SVant uses a workflow management system with intermediary checkpoint steps to ensure efficient use of resources by minimizing redundant computations and omitting steps where dependent files are available. SNP-SVant also provides metrics to assess the quality of called variants and converts between VCF and aligned FASTA format outputs to ensure compatibility with downstream tools to calculate selection statistics, which are commonplace in population genomics studies. By accounting for both small and large structural variants, users of this workflow can obtain a wide-ranging view of genomic alterations in an organism of interest. Overall, this workflow advances our capabilities in assessing the functional consequences of different types of genomic alterations, ultimately improving our ability to associate genotypes with phenotypes. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Predicting single nucleotide polymorphisms and structural variations
Support Protocol 1 : Downloading publicly available sequencing data
Support Protocol 2 : Visualizing variant loci using Integrated Genome Viewer
Support Protocol 3 : Converting between VCF and aligned FASTA formats
INTRODUCTION
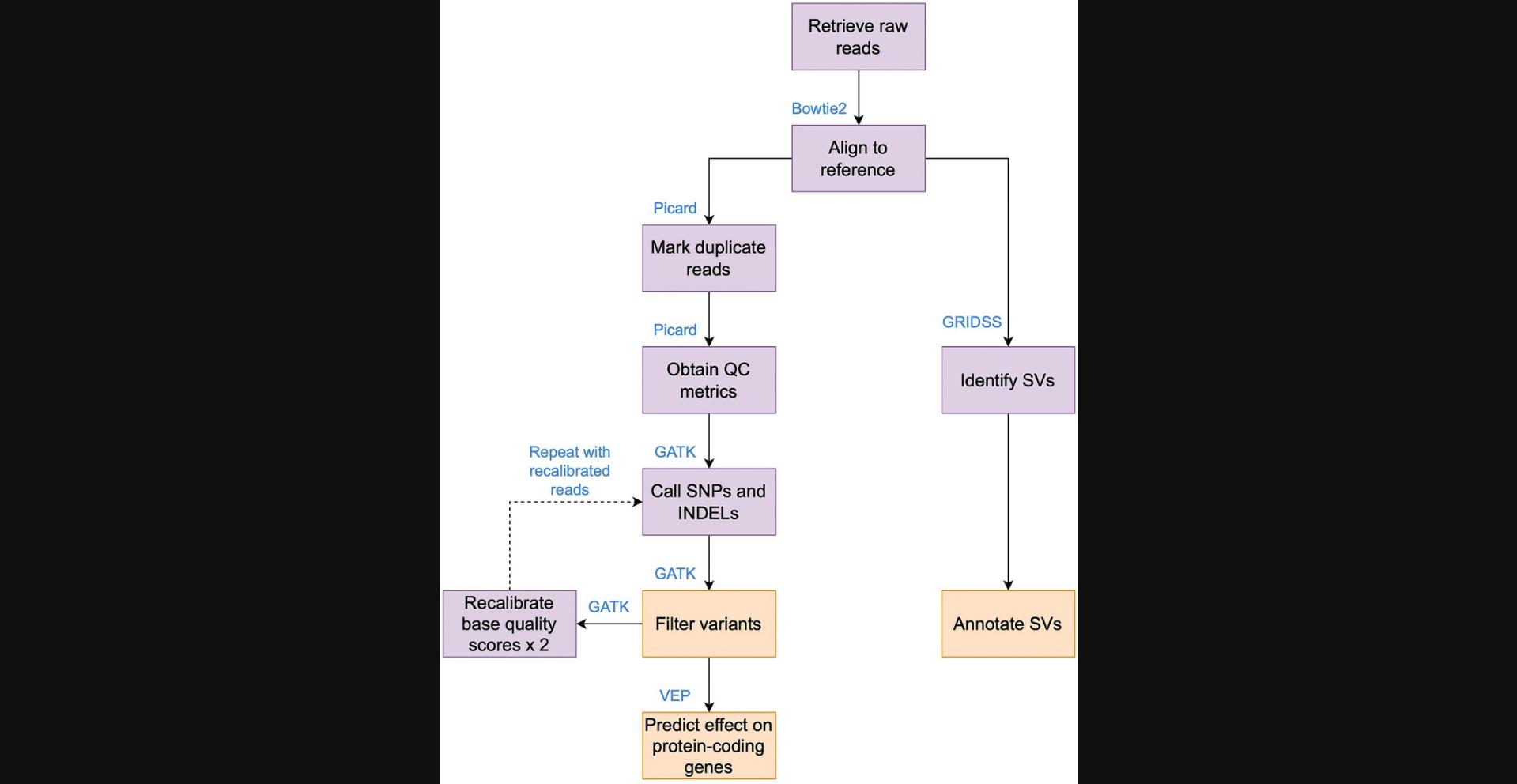
Variant calling refers to the process of identifying differences or “variants” in DNA sequences of individuals or samples compared to a reference genome. These variants can be changes in single nucleotides, known as single nucleotide polymorphisms (SNPs); small insertions and deletions (INDELs); or large genomic alterations, such as insertions, deletions, duplications, or inversions (Mahmoud et al., 2019; Olson et al., 2015). These large genomic alterations, encompassing at least 50 base pairs, are known as structural variations (SVs) (Mahmoud et al., 2019). Variant calling is a critical step in genomic analyses and plays a fundamental role in several applications, including population genetics and disease association studies. Studies aiming to associate genotypes with phenotypes have focused largely on SNPs (Uffelmann et al., 2021). Genome-wide studies in humans, however, have shown that SVs account for more variation within species compared to SNPs (Sudmant et al., 2015) and are also more frequently implicated in disease states (Weischenfeldt et al., 2013). Although identification of SVs is challenging due to low sequencing depth, especially in short-read sequencing data, recent advances in next-generation sequencing technologies (de Coster & van Broeckhoven, 2019) and algorithmic development (Kosugi et al., 2019) for detection of SVs have made it possible to predict SVs more accurately. Existing tools to predict SNPs and SVs currently do so independently. Here, we developed a computational workflow called SNP-SVant to streamline the comprehensive prediction of genomic variants, predicting both SNPs and SVs together. This workflow uses the Genome Analysis ToolKit (GATK) to predict SNPs and INDELs (McKenna et al., 2010) and the Genome Rearrangement IDentification Software Suite (GRIDSS) to predict SVs (Cameron et al., 2017, 2021) from short-read paired-end data. SNP-SVant was designed to be user friendly, such that users without extensive computational expertise can utilize the workflow. Additionally, SNP-SVant is tailored for variant prediction in organisms lacking benchmarked, known variants, which are traditionally used to distinguish sequencing errors from real variants (see Current Protocols article: Van der Auwera et al., 2013). Benchmarked variant datasets are used to recalibrate base quality scores obtained from sequencing machines (see Current Protocols article: Van der Auwera et al., 2013). In the absence of these benchmarked datasets, recalibration of base quality scores cannot be performed, which can result in biases stemming from genome context or read base position. To facilitate base quality score recalibration in the absence of benchmarked datasets, we follow the best practices recommended by developers of GATK (McKenna et al., 2010; see Current Protocols article: Van der Auwera et al., 2013) and leverage multiple rounds of statistical recalibration and variant calling to increase the precision of variant prediction.
SNP-SVant uses the Snakemake workflow management system to create reproducible data analyses (Köster et al., 2021). Variant calling is a computationally expensive process, especially when working with multiple samples. SNP-SVant runtime is optimized by leveraging parallelization and multicore processors when available, thus resolving dependencies for intermediate steps and checkpointing. Here, we demonstrate the use of SNP-SVant to predict variants in a strain of Candida albicans , a common human fungal pathogen. We use C. albicans as a test case for this pipeline because it is a medically relevant fungal pathogen that does not yet have experimentally validated and benchmarked variant data. However, SNP-SVant was developed to predict SNPs and SVs in any species lacking benchmarked, experimentally verified genomic variant data. SNP-SVant uses paired-end and short-read genomic DNA sequences as inputs to predict variants. The quality of the raw data is verified using FastQC (Andrews, 2010), reads are mapped to the indexed reference genome using Bowtie2 (Langmead & Salzberg, 2012), and aligned reads are sorted by genomic loci using samtools (Danecek et al., 2021). Duplicate reads in the alignment are then denoted using the MarkDuplicates command in the Picard toolkit (https://broadinstitute.github.io/picard/). In this latter step, duplicate reads that occur during the PCR amplification step of sequencing are marked to ensure that they are excluded from the statistical calculation to assess variant quality. Summaries of read alignment statistics, insert size distributions, and read depths are also generated to verify the quality of the sequenced data and alignment using the Picard toolkit and samtools. The first round of variant calling to predict SNPs and small INDELs is performed using the HaplotypeCaller function in GATK (McKenna et al., 2010), and variants with low mapping quality, strand biases, and low variant confidence scores accounting for coverage are removed.
The GATK thresholds of the workflow are set according to best practices described in the GATK documentation and can be modified by users based on the distributions of these metrics in the data. Base quality scores of the aligned reads are then recalibrated using filtered, high-quality variants to account for non-random errors due to genome context, especially homopolymer errors that can arise during sequencing (Stoler & Nekrutenko, 2021). This step is repeated twice, and the aligned reads with the recalibrated base scores are then used to perform another round of variant calling using HaplotypeCaller and are filtered again using the same criteria. These final filtered variants are then retained as the high-confidence variants. The effects of these variants on protein-coding regions are then predicted using the Variant Effect Predictor (VEP) software suite (McLaren et al., 2016).
SVs can be identified from short-read paired-end sequencing data based on the patterns of distribution of read pair distances and orientations at specific genomic regions, and different types of SVs result in different distributional patterns (Mahmoud et al., 2019). GRIDSS uses short-read paired-end sequencing data to predict SVs (Cameron et al., 2017, 2021). GRIDSS retains reads that are partially aligned, reads with incorrect orientation, reads with only a single mapped end, and reads with unusual insert sizes and filters out reads with low mapping quality and reads mapping to low-complexity genomic regions. The remaining high-quality reads are then used to construct a positional de Bruijn graph (Cameron et al., 2017), which is used to identify single break-end contigs. Break-end contigs are defined as contigs that extend out and flank a single breakpoint of the SV from each side and are generated by assembling reads that span a breakpoint region (Cameron et al., 2017). The break-end contigs are subsequently realigned to the reference to precisely identify breakpoints. Given that GRIDSS reports the break-ends but does not annotate SVs, we wrote a custom R script to annotate the SVs. During the annotation step, we annotate SVs if the break-ends reported by GRIDSS are paired. This results in an annotation file comprising simple SVs such as duplications, insertions, deletions, and inversions.
Our workflow is especially useful for users without extensive computational knowledge because it not only integrates SNP and SV prediction and functional annotation but also includes custom scripts to visualize quality scores of predicted variants. These custom scripts simplify the process of parameter tuning to filter out low-quality variants. Additionally, we modify default SNP-calling strategies in our workflow by adapting this step for organisms that do not have benchmarked, experimentally validated variant data. As a result, users can predict genomic variants, including SNPs and SVs, and their functional consequences in organisms using only a reference genome, annotated protein-coding regions, and paired-end short-read sequencing data. By accounting for both small and large structural variants, users can obtain a wide-ranging view of genetic diversity in their organism of interest. Overall, this workflow advances our capabilities in assessing the functional consequences of different types of genomic alterations, ultimately improving our abilities to associate genotypes with phenotypes.
Basic Protocol: PREDICTING SINGLE NUCLEOTIDE POLYMORPHISMS AND STRUCTURAL VARIATIONS
In this protocol, we describe in detail the usage of our variant-calling workflow named SNP-SVant to predict SNPs and SVs in non-benchmarked organisms. Figure 1 depicts a summary of the computational workflow. The SNP-SVant workflow requires the input files described in Table 1. Briefly, the workflow relies on a contiguous and complete reference genome and short-read paired-end sequencing data. The outputs of SNP-SVant include a Variant Call Format (VCF) file with SNPs and small INDELs obtained using GATK (version 4.4.0.0), a VCF file with structural variants obtained using GRIDSS (version 2.12.0), and annotated SVs listed in BED format. Additionally, the scores used to assess the quality of the variant calls can be visualized and used to adjust the variant-filtering parameters listed in Table 2. A list of all available parameters for this workflow is given in Table 3.
| File | Description | Parameter |
|---|---|---|
| sample_metadata.tsv | Tab-delimited file with the first column containing the base name of the FASTQ file | samples='{metadata: /path/to/sample_metadata.tsv}' |
| reference.fasta | Reference FASTA file | reference='{genome: /path/to/reference.fasta}' |
| reference.gff | Reference GFF file with annotations of protein-coding genes in the reference genome | reference='{genome_gff: /path/to/reference.gff}' |
| Parameter | Description | Default value |
|---|---|---|
| QD_filter | Quality score of the variant normalized by depth | QD < 2.0 (for SNPs and INDELs) |
| FS_filter | Phred-scaled probability that the variant site has a strand bias | FS > 60.0 (for SNPs) FS > 200.0 (for INDELs) |
| SOR_filter | Strand odds ratio to estimate strand bias | SOR > 4 (for SNPs) SOR > 10 (for INDELs) |
| MQ_filter | Root mean square mapping quality over all reads | MQ < 40.0 (for SNPs) |
| MQRankSum_filter | Approximation from the rank sum test for mapping qualities | MQRankSum < −12.5 (for SNPs) |
| ReadPosRankSum_filter | Approximation from the rank sum test for the variant site position within the reads | ReadPosRankSum < −8.0 (for SNPs) |
| Parameter | Description | Default value | Required |
|---|---|---|---|
| samples=’{metadata: <path>}’ | Tab-delimited metadata file with the first column as sample IDs | test/metadata/test_samples.tsv | Yes |
| samples=’{header: <boolean>}’ | Indicates if the sample metadata contain a header | False | No |
| threads=<int> | Number of threads to be used for the workflow | 8 | No |
| output=<path> | Output directory | test | No |
| input_dir=<path> | Input directory containing FASTQ files | test/raw | No |
| logs=<path> | Directory to store log files | test/logs | No |
| import=<boolean> | Indicates if sample IDs in the metadata file are SRA IDs and if FASTQ files for these publicly available samples should be imported | False | No |
| trimming=’{trim: <boolean>}’ | Indicates if adapter trimming should be performed | False | No |
| trimming=’{adapters: <path>}’ | FASTA file with adapter sequences | resources/adapters/NexteraPE-PE.fa | No |
|
trimming=’{seed Mismatches: <int>}’ |
The maximum number of mismatches to be allowed in the seed | 2 | No |
| trimming=’{palindromeClipThreshold: <int>}’ | The minimum score for the full alignment of the reads in palindrome mode | 30 | No |
| trimming=’{simpleClipThreshold: <int>}’ | The minimum score threshold for the adapter alignment to the read for clipping to take place | 10 | No |
| trimming=’{leading: <int>}’ | The minimum quality required to retain the base at the beginning of the read | 3 | No |
| trimming=’{trailing: <int>}’ | The minimum quality required to retain the base at the end of the read | 3 | No |
| trimming=’{windowSize: <int>}’ | Number of bases to average scores across | 4 | No |
|
trimming=’{required Quality: <int>}’ |
Average quality required | 15 | No |
| trimming=’{minlength: <int>}’ | The minimum length of reads to be retained | 36 | No |
|
snp_filters=’{QD_ filter: <string>}’ |
Quality score of the variant normalized by depth for SNPs | “QD < 2.0” | No |
|
snp_filters=’{FS_ filter: <string>}’ |
Phred-scaled probability that the variant site has a strand bias for SNPs | “FS > 60.0” | No |
|
snp_filters=’{SOR_ filter: <string>}’ |
Strand odds ratio to estimate the strand bias for SNPs | ”SOR > 4” | No |
|
snp_filters=’{MQ_ filter: <string>}’ |
Root mean square mapping quality over all reads for SNPs | ”MQ < 40.0” | No |
|
snp_filters=’{MQRank Sum_ filter: <string>}’ |
Approximation from the rank sum test for mapping qualities for SNPs | ”MQRankSum < -12.5” | No |
|
snp_filters=’{ReadPosRankSum_ filter: <string>}’ |
Approximation from the rank sum test for the variant site position within the reads for SNPs | ”ReadPosRankSum < -8.0” | No |
|
indel_filters=’{QD_ filter: <string>}’ |
Quality score of the variant normalized by depth for INDELs | “QD < 2.0” | No |
|
indel_filters=’{FS_ filter: <string>}’ |
Phred-scaled probability that the variant site has a strand bias for INDELs | “FS > 200.0” | No |
|
indel_filters=’{SOR_ filter: <string>}’ |
Strand odds ratio to estimate the strand bias for INDELs | ”SOR > 10” | No |
| reference=’{genome: <path>}’ | Path for reference FASTA |
reference/genome/ reference.fasta |
Yes |
| reference=’{gff: <path>}’ | Path for annotated reference genes in GFF format |
reference/genome/ reference.gff |
Yes |
Support Protocol 1: DOWNLOADING PUBLICLY AVAILABLE SEQUENCING DATA
Previously published sequencing data are publicly available at the Sequencing Read Archive (SRA) on NCBI (https://www.ncbi.nlm.nih.gov/sra). FASTQ files for these samples can be downloaded directly from the NCBI database or using the SRA toolkit (https://github.com/ncbi/sra-tools). This support protocol uses the command line to download the sequenced reads using the SRA accession number.
Necessary Resources
Software
- The SRA toolkit provides tools for accessing and downloading high-throughput sequencing data files generated from sequencing platforms such as Illumina. Details on downloading the SRA toolkit (version 3.0.6 or higher) are available at https://github.com/ncbi/sra-tools.
1.Verify if the publicly available data are sequenced in paired-end mode. Navigate to the directory to store the input FASTQ files. Download the FASTQ files for the SRA accession using the command below:
- fasterq-dump <sra_accession> -e <number_of_threads>
Support Protocol 2: VISUALIZING VARIANT LOCI USING INTEGRATED GENOME VIEWER
The intermediary files generated during the alignment step and the variant-calling step can be visualized using IGV. The annotation of protein-coding genes for the reference strain can also be used to compare the variant loci across genomic elements. Additional information such as transcriptomic or epigenomic data, if available, can also be integrated into the visualization to facilitate interpretation of the variants.
Necessary Resources
Software
- IGV is available as a desktop or web application. The IGV desktop application can be downloaded at https://software.broadinstitute.org/software/igv/.
1.To load the reference genome and annotations, in the IGV desktop application, click on Genomes > Load Genome from file and select the reference FASTA file from the local device. To include the annotations of protein-coding genes, click on File > Load from File and select the annotation file in GFF format from the local device.
2.Ensure that the reference FASTA file is indexed.
3.To load the SNP and SV variant files, in the IGV desktop application, click on File > Load from File and select the variant files *_filtered_snps_final.vcf, *_filtered_indels_final.vcf, and *_gridss.vcf from the local device.
4.To save a snapshot of a region of interest, in the IGV desktop application, navigate to the region of interest using the search tab to enter the genomic position or gene of interest. Use the “+” and “−” symbols at the top right of the application to zoom in and out of the genomic range. Click on File > Save PNG image and navigate to the folder of interest to save the IGV snapshot.
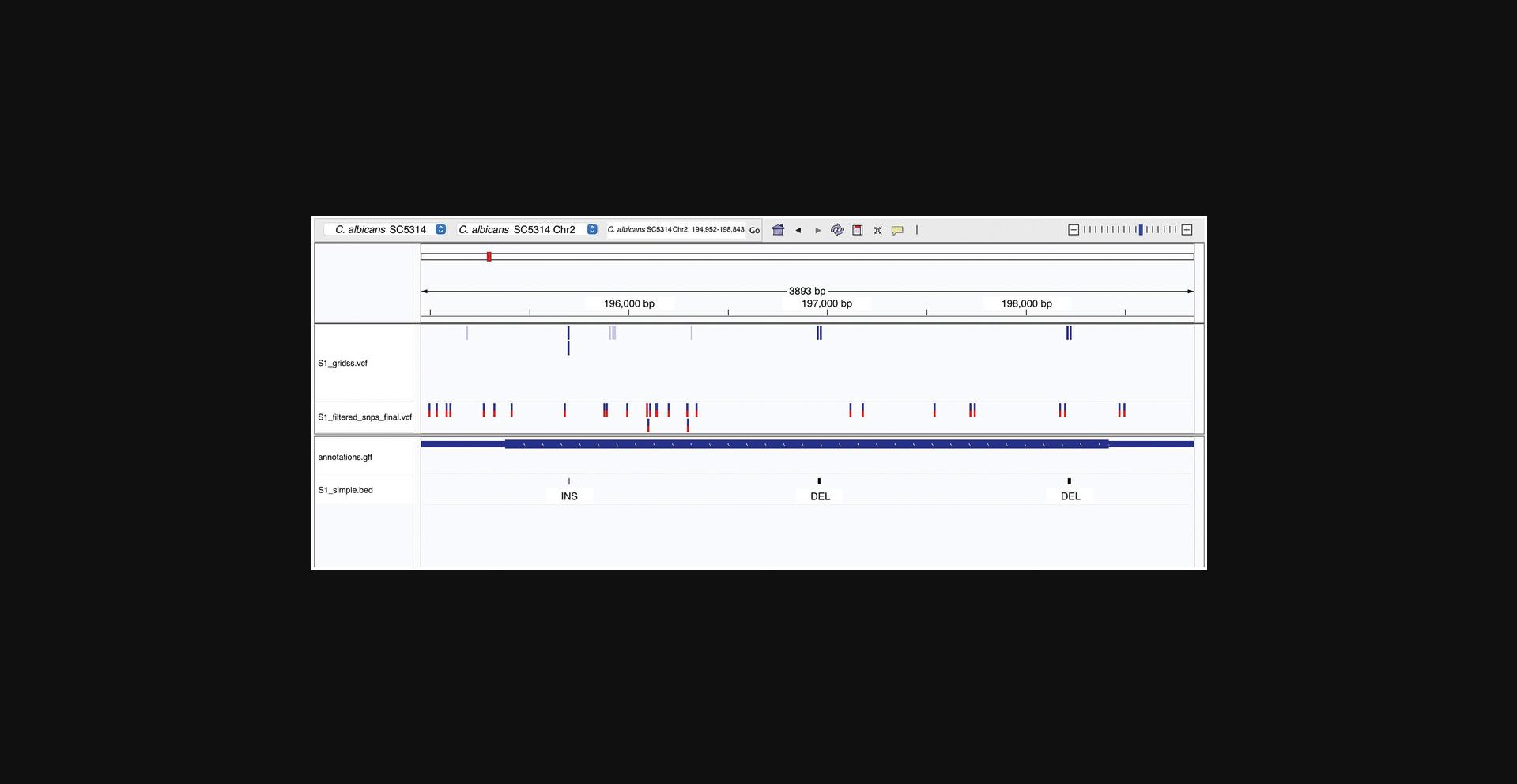
Support Protocol 3: CONVERTING BETWEEN VCF AND ALIGNED FASTA FORMATS
Genomic variants can be used to compute population genomics statistics. Some packages, such as alnpi, a module in the FAST toolkit, takes a multiFASTA file as input to compute population genomics statistics (Lawrence et al., 2015). In this support protocol, we describe the use of a custom python script to convert SNPs in VCF format to multiFASTA format along with the reference sequence. This script is available as a part of the snp_svant GitHub repository.
Necessary Resources
- This support protocol requires the same resources as those listed in the Basic Protocol.
1.Create and activate a new Conda environment to download packages required for the python script to run:
- conda create -n parse_vcf python pyvcf gffutils argparse biopython
- activate parse_vcf
2.Run the python script:
- python workflow/scripts/parse_vcf.py -i <*_filtered_snps_final.vcf> -r <reference.fasta> -lchromosome:start:end -s
-o <output_fasta>
Understanding Results
The primary outputs of this pipeline are variants predicted in the sample compared to the reference strain documented in VCF format. VCF is a standard file format used to represent information about genetic variants. This file contains information on the chromosome and position of the variant location, the reference allele, alternate alleles, and allele frequencies. Additionally, the VCF file contains information on the variant quality, including the metrics listed in Table 2.The VCF file also includes information about the sequencing depth, such as read depth at a variant location and allelic depth. A detailed description of all fields in VCF format can be found at http://samtools.github.io/hts-specs/.
The output for the SV annotations is a BED file. This BED file is a tab-delimited file with eight columns. The information represented in these columns is detailed below:
- 1.Chromosome location of the predicted SV.
- 2.Start coordinate of the SV.
- 3.End coordinate of the SV.
- 4.Type of SV, where INS denotes insertion, DEL denotes deletion, DUP denotes duplication, and INV denotes inversion.
- 5.Quality score of the SV, obtained from GRIDSS.
- 6.Length of the SV.
- 7.If the SV is an insertion, this column will show the nucleotide sequence.
- 8.Strand of the SV.
If the output files listed in Table 4 are not generated, then the execution of the SNP-SVant workflow has failed. Possible causes and solutions to resolve this execution failure are listed in Table 5. If the VCF files are generated but only headers are listed without any identified variants, this could indicate poor-quality data, either due to low sequencing depth or due to a failed sequencing run, both of which would necessitate resequencing the sample to improve variant prediction.
| Problem | Possible cause | Solution |
|---|---|---|
| Error importing packages (ImportError) | Mamba was not installed correctly or is not up to date |
Update Mamba in the Conda environment or re-install Mamba using the following command: conda install mamba -c conda-forge |
| Files are incomplete (IncompleteFilesException) | Previous run crashed unexpectedly, and some files were not completely generated | Re-run the pipeline or use additional parameter --rerun-incomplete to regenerate incomplete files |
COMMENTARY
Background Information
In this article, we present a user-friendly and computationally efficient workflow called SNP-SVant for identifying genomic variants. Users with minimal computational expertise should be able to run the workflow by following the protocols provided here. Because variant calling is complex and highly dependent on parameter choices, we calibrated SNP-SVant to ensure that default values are concordant with recommended, benchmarked standards. We also report quality metrics from variant callers visually, enabling the comparison of the comprehensive variant calls to these critical quality thresholds and allowing for the easy identification of low-quality samples.
SNP-SVant relies on a contiguous, complete reference genome. Thus, variant calling for organisms where the genome is fragmented or where the reference genome is incomplete could result in lower quality scores for the predicted variants, higher false-positive variant calls, and higher rates of undetected true genetic variations or false negatives. Another limitation of SNP-SVant is that only relatively simple SVs can be annotated. More complex SVs involving multiple genomic breakpoints are challenging to identify and annotate using short-read sequencing methods. The accurate identification of such complex SVs would thus require long-read sequencing data. Additionally, SVs with breakpoints where the break-end on one side cannot be unambiguously resolved are reported by GRIDSS and available in VCF format. These unpaired single break-ends contain information on SVs with parts of the sequence missing from the reference, likely due to the presence of foreign DNA or repetitive regions (Cameron et al., 2021). As a result, such SVs are not annotated due to the challenges in identifying the source and type of SV, but rather are retained in the GRIDSS output file in VCF format.
Variant calling is a computationally expensive and complex procedure. Existing methods for variant calling predict SNPs and SVs independently and typically require computational expertise to implement. Additionally, the accurate identification of SNPs relies on experimentally validated and benchmarked variant data, which are available primarily for humans (Karczewski et al., 2020) and model organisms, such as mice (Ringwald et al., 2022), fruit flies (Wang et al., 2015), and baker's yeast (Cherry et al., 2012). In the absence of these benchmarked datasets, it is difficult to distinguish between sequencing artifacts and true genomic variants. To predict SNPs, small INDELs, and SVs in the absence of benchmarked datasets, SNP-SVant uses multiple rounds of statistical recalibration to calibrate quality scores for the variants based on high-confidence variant calls. Calibration of base scores is an important step in SNP calling, and one major advantage of SNP-SVant is its ability to perform this step without benchmarked datasets. Although recalibration of base scores is known to reduce the false-positive rate in SNP prediction for early next-generation sequencing data (Depristo et al., 2011), it is also reported to compromise sensitivity in low-coverage data and when there is high divergence between the strain and reference genomes (Li & Wren, 2014; Tian et al., 2016). If users are working with low-coverage data or with strains that have diverged significantly from the reference, we recommend using VCF files for GATK SNP calls without base score recalibration, listed in Table 4. Further work is needed to identify sources of biases to refine best practices in SNP prediction for organisms without benchmarked variants. Overall, we integrate the prediction of SNPs, small INDELs, and SVs in the SNP-SVant workflow, which can be used to comprehensively identify genomic variants at different scales.
Troubleshooting
Troubleshooting steps for some common problems are listed in Table 5.
Acknowledgments
We thank all past and present members of the Nobile and Ardell laboratories for feedback on this manuscript. This work was supported by the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) award no. R35GM124594 and by the Kamangar family in the form of an endowed chair to C.J.N. The content is the sole responsibility of the authors and does not represent the views of the funders. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Author Contributions
Deepika Gunasekaran : Conceptualization; data curation; formal analysis; investigation; methodology; software; validation; visualization; writing—original draft; writing—review and editing. David H. Ardell : Investigation; project administration; resources; supervision; writing—review and editing. Clarissa J. Nobile : Funding acquisition; investigation; project administration; resources; supervision; writing—review and editing.
Conflict of Interest
C.J.N. is a cofounder of BioSynesis, Inc., a company developing diagnostics and therapeutics for biofilm infections. BioSynesis, Inc. was not involved in the content of this manuscript.
Open Research
Data Availability Statement
The source code describing the SNP-SVant workflow is freely available as a GitHub repository (https://github.com/dgunasekaran/snp_svant). The example paired-end sequencing data used to illustrate the usage of this workflow are publicly available and can be downloaded from SRA using the accession no. SRR7801919 (Sitterlé et al., 2019).
Literature Cited
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics , 30(15), 2114. https://doi.org/10.1093/BIOINFORMATICS/BTU170
- Cameron, D. L., Baber, J., Shale, C., Valle-Inclan, J. E., Besselink, N., van Hoeck, A., Janssen, R., Cuppen, E., Priestley, P., & Papenfuss, A. T. (2021). GRIDSS2: Comprehensive characterisation of somatic structural variation using single breakend variants and structural variant phasing. Genome Biology , 22(1), 1–25. https://doi.org/10.1186/S13059-021-02423-X
- Cameron, D. L., Schröder, J., Penington, J. S., Do, H., Molania, R., Dobrovic, A., Speed, T. P., & Papenfuss, A. T. (2017). GRIDSS: Sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Research , 27(12), 2050–2060. https://doi.org/10.1101/GR.222109.117
- Cherry, J. M., Hong, E. L., Amundsen, C., Balakrishnan, R., Binkley, G., Chan, E. T., Christie, K. R., Costanzo, M. C., Dwight, S. S., Engel, S. R., Fisk, D. G., Hirschman, J. E., Hitz, B. C., Karra, K., Krieger, C. J., Miyasato, S. R., Nash, R. S., Park, J., Skrzypek, M. S., … Wong, E. D. (2012). Saccharomyces genome database: The genomics resource of budding yeast. Nucleic Acids Research , 40(Database issue), D700. https://doi.org/10.1093/NAR/GKR1029
- Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., Whitwham, A., Keane, T., McCarthy, S. A., & Davies, R. M. (2021). Twelve years of SAMtools and BCFtools. GigaScience , 10(2), 1–4. https://doi.org/10.1093/GIGASCIENCE/GIAB008
- de Coster, W., & van Broeckhoven, C. (2019). Newest methods for detecting structural variations. Trends in Biotechnology , 37(9), 973–982. https://doi.org/10.1016/J.TIBTECH.2019.02.003
- Depristo, M. A., Banks, E., Poplin, R., Garimella, K. V., Maguire, J. R., Hartl, C., Philippakis, A. A., Del Angel, G., Rivas, M. A., Hanna, M., McKenna, A., Fennell, T. J., Kernytsky, A. M., Sivachenko, A. Y., Cibulskis, K., Gabriel, S. B., Altshuler, D., & Daly, M. J. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics , 43(5), 491–498. https://doi.org/10.1038/ng.806
- Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings, B. B., Alföldi, J., Wang, Q., Collins, R. L., Laricchia, K. M., Ganna, A., Birnbaum, D. P., Gauthier, L. D., Brand, H., Solomonson, M., Watts, N. A., Rhodes, D., Singer-Berk, M., England, E. M., Seaby, E. G., Kosmicki, J. A., … Xavier, R. J. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature , 581(7809), 434–443. https://doi.org/10.1038/s41586-020-2308-7
- Köster, J., Mölder, F., Jablonski, K. P., Letcher, B., Hall, M. B., Tomkins-Tinch, C. H., Sochat, V., Forster, J., Lee, S., Twardziok, S. O., Kanitz, A., Wilm, A., Holtgrewe, M., Rahmann, S., & Nahnsen, S. (2021). Sustainable data analysis with snakemake. F1000Research , 10, 33. https://doi.org/10.12688/F1000RESEARCH.29032.2
- Kosugi, S., Momozawa, Y., Liu, X., Terao, C., Kubo, M., & Kamatani, Y. (2019). Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biology , 20(1), 1–18. https://doi.org/10.1186/S13059-019-1720-5
- Langmead, B., & Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods , 9(4), 357. https://doi.org/10.1038/NMETH.1923
- Lawrence, T. J., Kauffman, K. T., Amrine, K. C. H., Carper, D. L., Lee, R. S., Becich, P. J., Canales, C. J., & Ardell, D. H. (2015). FAST: Fast analysis of sequences toolbox. Frontiers in Genetics , 6(MAY), 136911. https://doi.org/10.3389/FGENE.2015.00172
- Li, H., & Wren, J. (2014). Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics , 30(20), 2843. https://doi.org/10.1093/BIOINFORMATICS/BTU356
- Mahmoud, M., Gobet, N., Cruz-Dávalos, D. I., Mounier, N., Dessimoz, C., & Sedlazeck, F. J. (2019). Structural variant calling: The long and the short of it. Genome Biology , 20(1), 1–14. https://doi.org/10.1186/S13059-019-1828-7
- McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., Garimella, K., Altshuler, D., Gabriel, S., Daly, M., & DePristo, M. A. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research , 20(9), 1297–1303. https://doi.org/10.1101/GR.107524.110
- McLaren, W., Gil, L., Hunt, S. E., Riat, H. S., Ritchie, G. R. S., Thormann, A., Flicek, P., & Cunningham, F. (2016). The ensembl variant effect predictor. Genome Biology , 17(1), 1–14. https://doi.org/10.1186/S13059-016-0974–4
- Olson, N. D., Lund, S. P., Colman, R. E., Foster, J. T., Sahl, J. W., Schupp, J. M., Keim, P., Morrow, J. B., Salit, M. L., & Zook, J. M. (2015). Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Frontiers in Genetics , 6(JUL), 150309. https://doi.org/10.3389/FGENE.2015.00235
- Ringwald, M., Richardson, J. E., Baldarelli, R. M., Blake, J. A., Kadin, J. A., Smith, C., & Bult, C. J. (2022). Mouse Genome Informatics (MGI): Latest news from MGD and GXD. Mammalian Genome , 33(1), 4–18. https://doi.org/10.1007/S00335-021-09921-0
- Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., & Mesirov, J. P. (2011). Integrative genomics viewer. Nature Biotechnology , 29(1), 24–26. https://doi.org/10.1038/nbt.1754
- Sitterlé, E., Maufrais, C., Sertour, N., Palayret, M., d'Enfert, C., & Bougnoux, M. E. (2019). Within-host genomic diversity of candida albicans in healthy carriers. Scientific Reports , 9(1), 1–12. https://doi.org/10.1038/s41598-019-38768-4
- Stoler, N., & Nekrutenko, A. (2021). Sequencing error profiles of Illumina sequencing instruments. NAR Genomics and Bioinformatics , 3(1), lqab019. https://doi.org/10.1093/NARGAB/LQAB019
- Sudmant, P. H., Rausch, T., Gardner, E. J., Handsaker, R. E., Abyzov, A., Huddleston, J., Zhang, Y., Ye, K., Jun, G., Fritz, M. H. Y., Konkel, M. K., Malhotra, A., Stütz, A. M., Shi, X., Casale, F. P., Chen, J., Hormozdiari, F., Dayama, G., Chen, K., … Korbel, J. O. (2015). An integrated map of structural variation in 2,504 human genomes. Nature , 526(7571), 75. https://doi.org/10.1038/NATURE15394
- Tian, S., Yan, H., Kalmbach, M., & Slager, S. L. (2016). Impact of post-alignment processing in variant discovery from whole exome data. BMC Bioinformatics , 17(1), 403. https://doi.org/10.1186/S12859-016-1279-Z
- Uffelmann, E., Huang, Q. Q., Munung, N. S., de Vries, J., Okada, Y., Martin, A. R., Martin, H. C., Lappalainen, T., & Posthuma, D. (2021). Genome-wide association studies. Nature Reviews Methods Primers , 1(1), 1–21. https://doi.org/10.1038/s43586-021-00056-9
- van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., del Angel, G., Levy-Moonshine, A., Jordan, T., Shakir, K., Roazen, D., Thibault, J., Banks, E., Garimella, K. V., Altshuler, D., Gabriel, S., & DePristo, M. A. (2013). From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Current Protocols in Bioinformatics , 11(1110), 11.10.1–11.10.33. https://doi.org/10.1002/0471250953.bi1110s43
- Wang, F., Jiang, L., Chen, Y., Haelterman, N. A., Bellen, H. J., & Chen, R. (2015). FlyVar: A database for genetic variation in Drosophila melanogaster. Database: The Journal of Biological Databases and Curation , 2015, bav079. https://doi.org/10.1093/DATABASE/BAV079
- Weischenfeldt, J., Symmons, O., Spitz, F., & Korbel, J. O. (2013). Phenotypic impact of genomic structural variation: Insights from and for human disease. Nature Reviews Genetics , 14(2), 125–138. https://doi.org/10.1038/nrg3373
Internet Resources
GATK best practices, which can be used to understand quality thresholds.

