SARS-CoV-2 McGill Nextera Flex sequencing protocol_Lunascript_ARTIC.V3_5uLRT
Anne-Marie Roy, Shu-Huang Chen, Ioannis Ragoussis, Sarah J Reiling, Marie-Michelle Simon
Abstract
How the Nextera DNA Flex Assay Works
The Nextera DNA Flex library prep kit uses a bead-based transposome complex to tagment genomic DNA, which is a process that fragments DNA and then tags the DNA with adapter sequences in one step. After it is saturated with input DNA, the bead-based transposome complex fragments a set number of DNA molecules. This fragmentation provides flexibility to use a wide DNA input range to generate normalized libraries of consistent tight fragment size distribution. Following tagmentation, a limited-cycle PCR adds Nextera DNA Flex-specific index adapter sequences to the ends of a DNA fragment. This step enables capability across all Illumina sequencing platforms. A subsequent Sample Purification Beads (SPB) cleanup step then purifies libraries for use on an Illumina sequencer. The double-stranded DNA library is denatured before hybridization of the biotin probe oligonucleotide pool.
PCR Amplicons for Nextera Flex
When starting with PCR amplicons, the PCR amplicon must be > 150 bp. The standard clean up protocol depletes libraries < 500 bp. Therefore, Illumina recommends that amplicons < 500 bp undergo a 1.8 x sample purification bead volume ratio to supernatant during Clean Up Libraries on page 11. Shorter amplicons can otherwise be lost during the library cleanup step. Tagmentation cannot add an adapter directly to the distal end of a fragment, so a drop in sequencing coverage of ~50 bp from each distal end is expected. To ensure sufficient coverage of the amplicon target region, design primers to extend beyond the target region by 50 bp per end.
More information can be found here: https://emea.support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera_dna_flex/nextera-dna-flex-library-prep-reference-guide-1000000025416-07.pdf
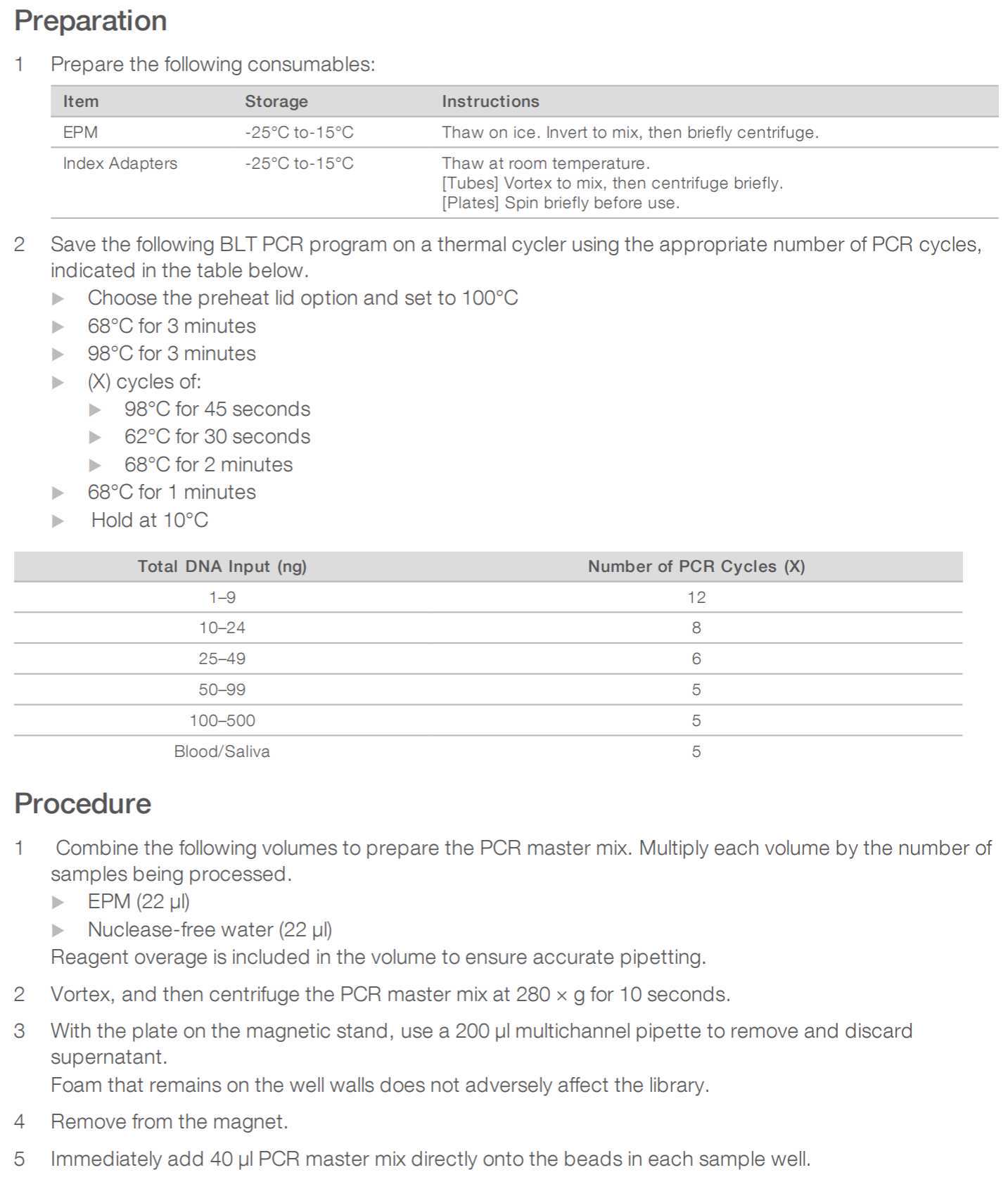
Steps
cDNA preparation
cDNA PREPARATION
Mix the following components in a 0.2 mL 8-strip tube or in a 96-well plate:
Component Volume
LunaScript RT SuperMix (5x) 4µL
Nuclease-free water 5µL
Template RNA 11µL
Total 20µL
Gently mix by pipetting and pulse spin the tube to collect liquid at the bottom of the tube.
Incubate the reaction as follows:
25°C for 0h 2m 0s
55°C for 0h 20m 0s
95°C for 0h 1m 0s
Place on ice for 0h 1m 0s or store cDNA at -20C
Primer pool preparation
PRIMER POOL PREPARATION
If required resuspend lyophilised primers at a concentration of 100 µM each
ARTIC nCov-2019 only primers for this protocol were designed using Primal Scheme and generate overlapping 400 nt amplicons. Primer names and dilutions are listed in the table below. https://github.com/sarahreiling/artic-ncov2019/blob/master/primer_schemes/nCoV-2019/V3/nCoV-2019_V3only.scheme.bed
Generate primer pool stocks by adding 5µL of each primer pair to a 1.5mL Eppendorf labelled either “Pool 1 (100 µM)” or “Pool 2 (100 µM)”. Total volume should be 490µL for Pool 1 (100 µM) and 490µL for Pool 2 (100 µM). These are your 100 µM stocks of each primer pool.
Dilute this primer pool 1:10 in molecular grade water, to generate 10 µM primer stocks. It is recommend that multiple aliquots of each primer pool are made to in case of degradation or contamination.
Multiplex PCR
MULTIPLEX PCR
In the extraction and sample addition cabinet add 5µL RT product to each tube and mix well by pipetting.
In the mastermix hood set up the multiplex PCR reactions as follows in 0.2mL 8-strip PCR tubes:
Component Pool 1 [10 uM primer] Pool 2 [10 uM]
Q5 Hot Start High-Fidelity 2X Master Mix 12.5µL 12.5µL
Primer Pool 1 or 2 3.7µL 3.7µL
Nuclease-free water 3.8µL 3.8µL
Total 20µL 20µL
Add 5 ul RT product as mentioned in step 10.
Pulse centrifuge the tubes to collect the contents at the bottom of the tube.
Set-up the following program on the thermal cycler:
Step Temperature Time Cycles
Heat Activation 98°C 0h 0m 30s 1
Denaturation 98°C 0h 0m 15s 36
Annealing 63°C 0h 5m 0s 36
Hold 4°C Indefinite 1
PCR clean-up
PCR CLEANUP
Combine the entire contents of “Pool 1” and “Pool 2” PCR reactions for each biological sample into to a single 1.5mLEppendorf tube.
Clean-up the amplicons using the following protocol:
Add a volume ratio of 0.8x of SPRI beads to the sample tube and mix gently by either flicking or pipetting.
Incubate for 5 min at room temperature.
Pellet on magnet for 5 min. Remove supernatant.
Add 200 ul of 80% ethanol to the pellet and wash twice.
Elute in 30 ul elution buffer.
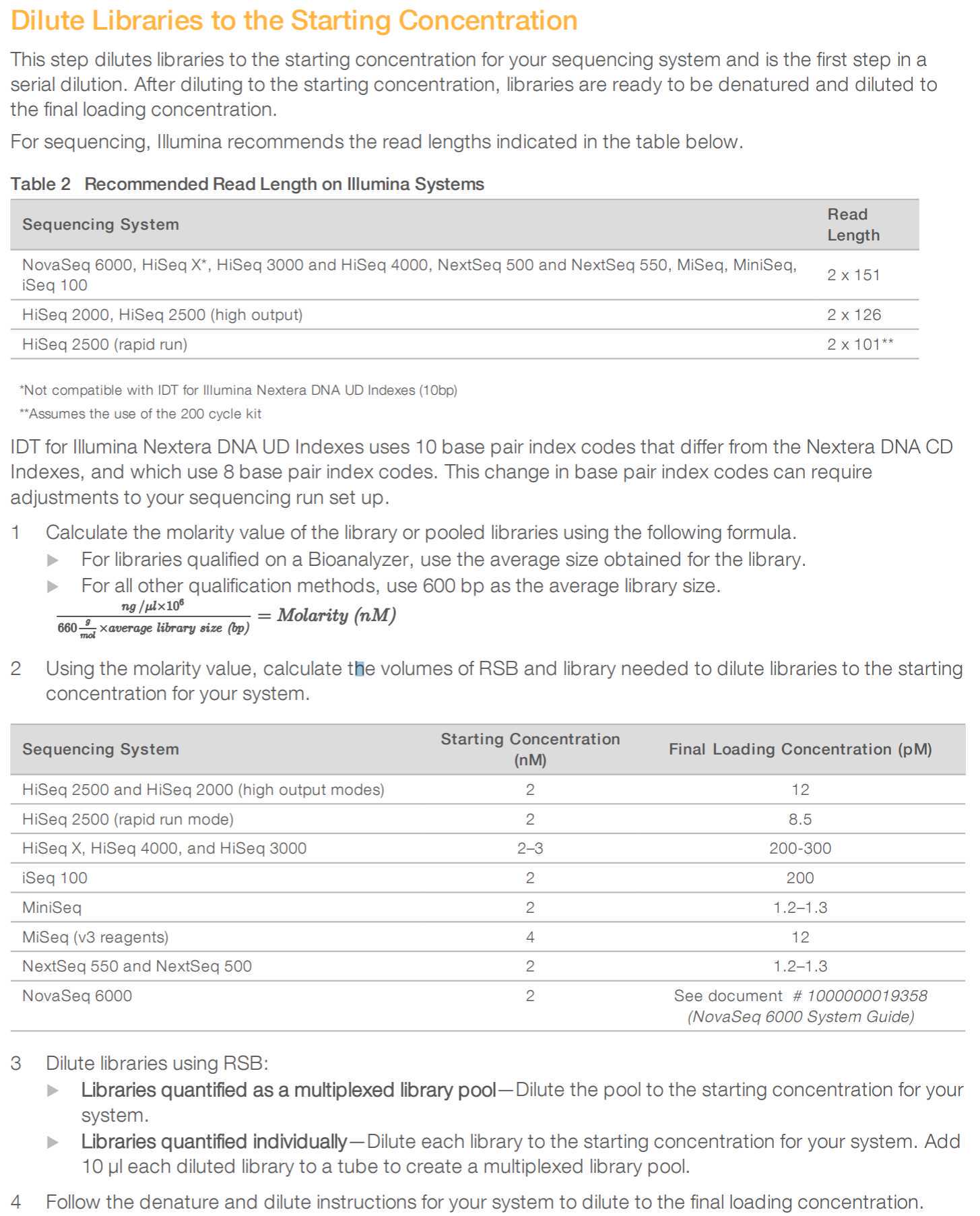
Amplicon Quantification and normalisation
AMPLICON QUANTIFICATION AND NORMALIZATION
Quantify the amplicon pools using a fluorimetric dsDNA assay.
We expect following concentrations:
Pool 1+2 combined:
100-150 ng/ul for Ct 14-24
30-80 ng/ul for Ct 25-29
10-30 ng/ul for Ct 30-36
Use the 30 ul with 150 ng DNA for Nextera Flex Tagmentation.
Protocol can be found here:
Start on page 7 with the Tagmentation.