RNA-Seq Data Analysis: A Practical Guide for Model and Non-Model Organisms
Enrique Pola-Sánchez, Enrique Pola-Sánchez, Karen Magdalena Hernández-Martínez, Karen Magdalena Hernández-Martínez, Rafael Pérez-Estrada, Rafael Pérez-Estrada, Nelly Sélem-Mójica, Nelly Sélem-Mójica, June Simpson, June Simpson, María Jazmín Abraham-Juárez, María Jazmín Abraham-Juárez, Alfredo Herrera-Estrella, Alfredo Herrera-Estrella, José Manuel Villalobos-Escobedo, José Manuel Villalobos-Escobedo
Abstract
RNA sequencing (RNA-seq) has emerged as a powerful tool for assessing genome-wide gene expression, revolutionizing various fields of biology. However, analyzing large RNA-seq datasets can be challenging, especially for students or researchers lacking bioinformatics experience. To address these challenges, we present a comprehensive guide to provide step-by-step workflows for analyzing RNA-seq data, from raw reads to functional enrichment analysis, starting with considerations for experimental design. This is designed to aid students and researchers working with any organism, irrespective of whether an assembled genome is available. Within this guide, we employ various recognized bioinformatics tools to navigate the landscape of RNA-seq analysis and discuss the advantages and disadvantages of different tools for the same task. Our protocol focuses on clarity, reproducibility, and practicality to enable users to navigate the complexities of RNA-seq data analysis easily and gain valuable biological insights from the datasets. Additionally, all scripts and a sample dataset are available in a GitHub repository to facilitate the implementation of the analysis pipeline. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Analysis of data from a model plant with an available reference genome
Basic Protocol 2 : Gene ontology enrichment analysis
Basic Protocol 3 : De novo assembly of data from non-model plants
INTRODUCTION
The development of high-throughput sequencing technologies has made it possible to uncover many organisms' transcriptional profiles at the whole-genome level. The technology of RNA-seq, or messenger RNA (transcriptome) sequencing, has permitted researchers to explore gene expression patterns with great precision in hundreds of model and non-model organisms. Even without a reference genome, a wealth of understanding related to processes such as cellular development, gene function, and responses to environmental stimuli, among others, has been uncovered (Stark et al., 2019). The RNA-seq methodology has often set the basis for developing molecular genetic analysis in non-model organisms and has become an essential tool. Researchers focused mainly on wet lab and field work sometimes struggle to exploit the data available from “next-generation sequencing” because they lack experience in bioinformatics, which is perceived to require in-depth computational and programming skills. This guide is intended to help bridge this gap.
Conventionally, an RNA-seq experiment involves subjecting organisms or cells to at least two experimental conditions and subsequently freezing the tissue or cell group of interest to halt transcriptional activity and initiate the RNA extraction process. Once the RNA is purified, cDNA libraries are constructed for all transcripts present at a given moment in the cell. These transcripts commonly include different classes of RNAs, such as tRNAs, ribosomal RNA, noncoding RNA, and messenger RNAs (mRNAs; Stark et al., 2019). However, given their importance, researchers are mainly interested in mRNAs that give rise to proteins. In recent years, there has also been a growing interest in determining the differential expression of other types of RNAs, such as small interfering RNAs (siRNAs) and microRNAs (Kasschau et al., 2007; Liu et al., 2019; Mehdi et al., 2021), which also play important roles in the regulation of different cellular processes. Once the cDNA libraries are built, they are sequenced using any of various different possible sequencing technologies; some of the technologies most widely used currently are those available commercially from Illumina.
In general, these protocols present the quality analysis of the FastQ files, the trimming and mapping process, and the differential gene expression analysis. Furthermore, we have included global analyses to facilitate the interpretation of the analyzed data, such as functional category enrichment analysis and data visualization through easily interpretable graphics. We also present a processing approach for non-model organisms for which no assembled and annotated genome is available. We encourage users of these protocol to share their scripts and output data on freely accessible platforms such as GitHub, providing the scientific community with the steps to obtain the results presented in scientific reports such as theses or indexed papers. All scripts and flowcharts provided here are documented in a GitHub repository, making it easier for users to implement these protocols and ensure repeatability when analyzing this type of data.
STRATEGIC PLANNING
When planning a differential gene expression experiment using RNA-seq, it is essential to consider several factors. First, the experimental design must clearly define the conditions that will be contrasted to correspond to the biological question of interest: the more precisely defined these are, the better the results. Second, having at least three biological replicates per condition is advisable. It is crucial to strive for as much consistency as possible between these replicates during the experiments. Given that RNA-seq is highly susceptible to small fluctuations in experimental conditions, experimental noise can often be reflected in the results.
Additionally, it is essential to consider the number of reads obtained after sequencing. Typically, having around 10 million reads per library is sufficient for most organisms. However, in the case of plants with large genomes, such as maize, which is ∼2.4 Gb, it is advisable to aim for at least 20-30 million reads for a meaningful analysis of differential gene expression.
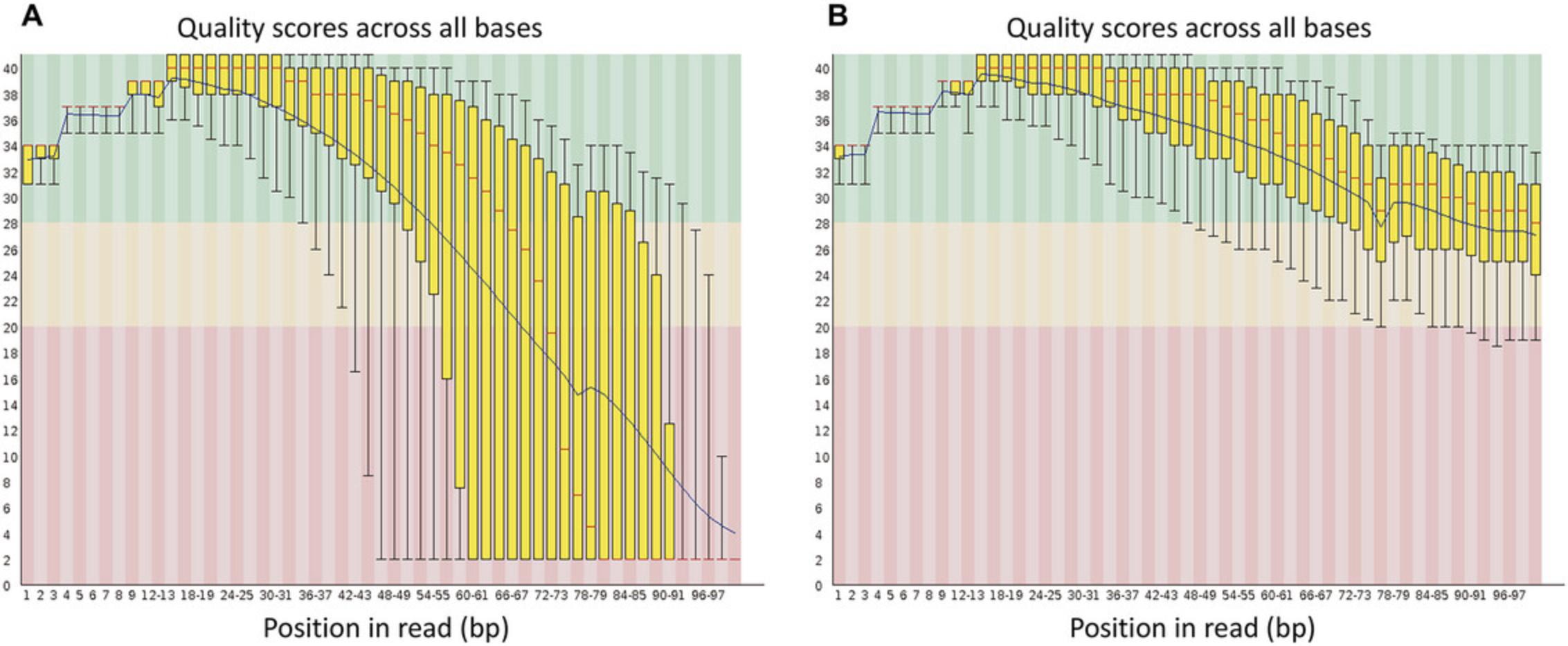
The sequencing files for each sample are recorded in FastQ format, which typically contains the read identifier, the sequence expressed in the four bases (CGAT; and in some cases “N” to indicate an undefined base), the read position (+ or −), and the quality with which each base was read. The quality is expressed in ASCII code and refers to Sacred values (Ewing & Green, 1998). These values are used to assess sequencing quality, and conventionally, RNA-seq analysis begins by examining read quality using the FastQC program. After this analysis, a trimming process to eliminate low-quality sequences and remove adapters is highly recommended. For this process, Trimmomatic has been one of the most versatile and valuable programs developed so far (Bolger et al., 2014). After data are trimmed and high-quality FASTQ files are obtained, the next stage is mapping the RNA-seq sequences to an assembled and annotated genome if this option is available. When these resources are not available, it is necessary to perform a de novo assembly.
Once the read counts per gene are obtained, the user can analyze differential expression levels to identify those genes whose expression changes (or differs) significantly between the two conditions being compared. These protocols present a differential expression analysis based on the DESeq2 program, which runs in the R environment (Love et al., 2014). As an alternative, we also recommend using edgeR (Robinson et al., 2010), which provides various examples for different cases in its manual and essentially performs the same processing as DESeq2. We have not extensively covered this program, for practical purposes, but the variations are minor. Figure 1 proposes a general workflow for model and non-model organisms.
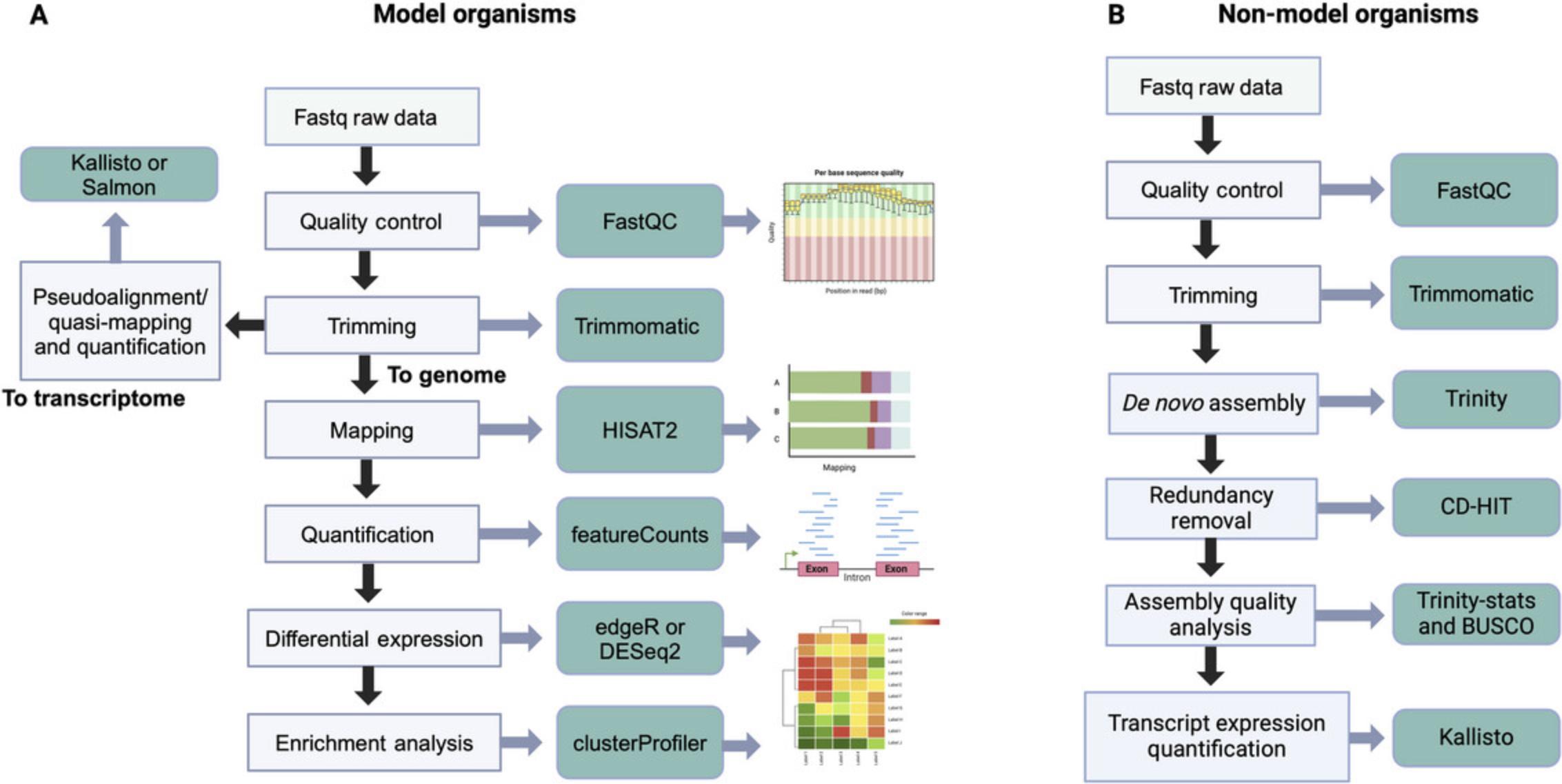
Basic Protocol 1: ANALYSIS OF DATA FROM A MODEL PLANT WITH AN AVAILABLE REFERENCE GENOME
This protocol meticulously outlines RNA-seq data analysis using the Arabidopsis thaliana reference genome. A typical RNA-seq analysis involves quality control and filtering of reads based on quality measures. Additionally, it includes alignment with the reference genome and, ultimately, the generation of the count matrix. These steps are executed using various programs, and their key characteristics and essential parameters are detailed below. Following this protocol as described will enable the analysis of differential expression in the dataset and the visualization of results through graphs such as principal component analysis (PCA) plots, M (log ratio)-A (mean average) (MA) plots, volcano plots, and heatmaps, though which differentially expressed genes can be identified.
Efficient progress in these tasks can be achieved by utilizing the command line within a Unix/Linux environment. We conducted our work on a 64-bit Ubuntu 23.04 operating system. For Windows users, a Windows system employing the Windows Subsystem for Linux (WSL; https://learn.microsoft.com/en-us/windows/wsl/install) is a viable option. For macOS users, it is feasible to execute identical commands in the terminal because of the shared Unix foundation of both operating systems.
Necessary Resources
Software
- FastQC: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Trimmomatic: http://www.usadellab.org/cms/?page=trimmomatic
- HISAT2: https://daehwankimlab.github.io/hisat2/manual/
- featureCounts: https://subread.sourceforge.net/featureCounts.html
- R: https://www.r-project.org/
Hardware
- At least 4 GB of RAM and 50 GB of storage space
- Scripts and Docker containers (detailed in the steps) available at the authors’ GitHub repository, https://github.com/jmvillalobos/RNA-seq-protocol
- Dataset(s) to be analyzed: the datasets used in the example here (detailed in the steps) are available in a public repository on Zenodo: https://zenodo.org/records/10537097
Part 1: Installing packages and creating directories
1.To install the bioinformatics tools that we will use in this protocol, go to the terminal and create a general folder in which the analysis will be carried out:
- user:∼$ mkdir project_2023
2.Move into the folder project_2023 using the cd command, and create subdirectories using the mkdir command:
- user:∼$ cd project_2023/
- user:∼/project_2023$ mkdir bioinformatics_tools
- user:∼/project_2023$ mkdir raw_data
- user:∼/project_2023$ mkdir trimming_data
- user:∼/project_2023$ mkdir alignment_hisat2
- user:∼/project_2023$ mkdir quantification_featureCounts
- user:∼/project_2023$ mkdir genome_arabidopsis
- user:∼/project_2023$ mkdir index_hisat2
3.To look at the folders created, use the ls command:
- user:∼/project_2023$ ls
4.The sudo command enables users to execute programs with the security privileges of another user, typically the root user, granting temporary superuser access. To execute this action, enter each of the following commands separately in the terminal:
- user:∼/project_2023/bioinformatics_tools$ sudo apt update
- user:∼/project_2023/bioinformatics_tools$ sudo apt install zip unzip
- user:∼/project_2023/bioinformatics_tools$ sudo apt install fastqc
- user:∼/project_2023/bioinformatics_tools$ sudo apt install r-base
- user:∼/project_2023/bioinformatics_tools$ sudo apt install hisat2
- user:∼/project_2023/bioinformatics_tools$ sudo apt install subread
5.To check the version or request help, you can execute the following command:
- user:∼/project_2023/bioinformatics_tools$ fastqc --version
- user:∼/project_2023/bioinformatics_tools$ fastqc --help
6.Trimmomatic installation : To install Trimmomatic, download the available zip file from http://www.usadellab.org/cms/?page=trimmomatic using the wget command:
- user:∼/project_2023$ cd bioinformatics_tools/
- user:∼/project_2023/bioinformatics_tools$ wget
- http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic/Trimmomatic-0.39.zip
7.Decompress:
- user:∼/project_2023/bioinformatics_tools$ unzip Trimmomatic-0.39.zip
8.To install the complete R system, use:
- user:∼/project_2023/bioinformatics_tools$ sudo apt-get install r-base
Part 2: Data download
The data we will use as our example here pertain to a study published in The Plant Journal (Villalobos-Escobedo et al., 2020) characterizing the roles of the Nox genes of the fungus Trichoderma atroviride during its interaction with A. thaliana. The raw RNA-seq data are publicly available in the NCBI SRA database at www.ncbi.nlm.nih.gov/sra/ with the accession number PRJNA575031.To simplify the process, we will use the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home) to find compressed versions of the FASTQ files.
9a. To download the files, copy each file link address by right-clicking on each file and, using the wget command, proceed to download into the raw_data folder (once per file):
- user:∼/project_2023/raw_data$ wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR102/004/SRR10207204/SRR10207204_1.fastq.gz
9b. Another way to download sequencing files is using the SRA Toolkit. To utilize this method, you must first install the SRA Toolkit by following the instructions on GitHub (https://github.com/ncbi/sra-tools/wiki/02.-Installing-SRA-Toolkit). Once that is installed, you can download the FASTQ files as follows:
- user:∼/project_2023/raw_data$ fasterq-dump --split-files SRR10207204
10.In the end, all 12 available library files should have been downloaded:
- SRR10207204_1.fastq.gz
- SRR10207204_2.fastq.gz
- SRR10207206_1.fastq.gz
- SRR10207206_2.fastq.gz
- SRR10207210_1.fastq.gz
- SRR10207210_2.fastq.gz
- SRR10207212_1.fastq.gz
- SRR10207212_2.fastq.gz
- SRR10207216_1.fastq.gz
- SRR10207216_2.fastq.gz
- SRR10207218_1.fastq.gz
- SRR10207218_2.fastq.gz
Part 3: Quality check of the raw reads with FastQC
One initial stage in an RNA-seq data analysis is assessing data quality. To accomplish this, we use FastQC v0.11.9, a widely recognized tool enabling quality checks on sequencing data (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
11.To evaluate the quality of the raw data, navigate to the raw_data directory and execute the following command:
- user:∼/project_2023/raw_data$ fastqc *.gz
12.If you need to determine the quality of a particular file, simply run fastqc followed by the file name:
- user:∼/project_2023/raw_data$ fastqc SRR10207204_1.fastq.gz
13.Maintaining order in your working directories is essential, so we suggest creating a folder for the raw data quality into which the files generated from the quality analysis can be moved:
- user:∼/project_2023/raw_data$ mkdir quality_raw
- user:∼/project_2023/raw_data$ mv *.zip quality_raw/
- user:∼/project_2023/raw_data$ mv *.html quality_raw/
Part 4: Read trimming with Trimmomatic
Now that we have assessed the quality of the raw data, a common next task is read trimming. For this purpose, various bioinformatics tools have been developed, including Trimmomatic, Trim Galore (available at https://github.com/FelixKrueger/TrimGalore), Cutadapt, Skewer, SOAPnuke, and fastp. These tools are designed to remove low-quality bases, adapter sequences, and bases that exceed a predefined threshold (Bolger et al., 2014; Chen, Chen, et al., 2018; Chen, Zhou, et al., 2018; Jiang et al., 2014; Martin, 2011). We carry out the read-trimming process for this protocol using Trimmomatic, which can handle both paired-end and single-end Illumina sequencing data.
14.Trimmomatic: With Trimmomatic, perform smooth read trimming into the trimming_data folder. The command is the following:
- user:∼/project_2023/trimming_data$ java-jar /home/enriquepola/project_2023/bioinformatics_tools/Trimmomatic-0.39/trimmomatic-0.39.jar PE -phred33 ../raw_data/SRR10207204_1.fastq.gz ../raw_data/SRR10207204_2.fastq.gz SRR10207204_P_1.fastq.gz SRR10207204_U_1.fastq.gz SRR10207204_P_2.fastq.gz SRR10207204_U_2.fastq.gz ILLUMINACLIP:/home/enriquepola/project_2023/bioinformatics_tools/Trimmomatic-0.39/adapters/TruSeq3-PE.fa:2:30:10 SLIDINGWINDOW:4:15 MINLEN:36
15.The Trimmomatic output should look like this:
TrimmomaticPE: Started with arguments:
- -phred33 ../raw_data/SRR10207204_1.fastq.gz ../raw_data/SRR10207204_2.fastq.gz SRR10207204_P_1.fastq.gz SRR10207204_U_1.fastq.gz SRR10207204_P_2.fastq.gz SRR10207204_U_2.fastq.gz ILLUMINACLIP:/home/enriquepola/project_2023/bioinformatics_tools/Trimmomatic-0.39/adapters/TruSeq3-PE.fa:2:30:10 SLIDINGWINDOW:4:15 MINLEN:36
Multiple cores found: Using 4 threads
- Using PrefixPair: ‘TACACTCTTTCCCTACACGACGCTCTTCCGATCT’ and ‘GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT’
- ILLUMINACLIP: Using 1 prefix pair, 0 forward/reverse sequences, 0 forward-only sequences, 0 reverse-only sequences
- Input Read Pairs: 11645096 Both Surviving: 11117409 (95.47%) Forward Only Surviving: 178980 (1.54%) Reverse Only Surviving:265455 (2.28%) Dropped: 83252 (0.71%)
TrimmomaticPE: Completed successfully
16.The last part of the Trimmomatic output shows the percentage of reads that survived the process, which is reasonably satisfactory. To optimize the execution, we can implement a “for” loop, in which we set the last three numbers of the sequencing data as a sample because they are the only ones that vary between each file:
- user:∼/project_2023/trimming_data
{sample}_1.fastq.gz ../raw_data/SRR10207 {sample}_P_1.fastq.gz SRR10207 {sample}_P_2.fastq.gz SRR10207${sample}_U_2.fastq.gz ILLUMINACLIP:/home/enriquepola/project_2023/bioinformatics_tools/Trimmomatic-0.39/adapters/TruSeq3-PE.fa:2:30:10 SLIDINGWINDOW:4:15 MINLEN:36; done|sh
Part 5: Alignment of reads
Once the read-trimming process is completed, the next stage in RNA-seq analysis is to align reads to a reference. When RNA-seq data come from an organism for which a reference genome is available, it is possible to infer the identity of the expressed transcripts by mapping the reads to the genome or transcriptome (Conesa et al., 2016). To align reads to a reference genome, tools such as HISAT2, STAR, TopHat2, Bowtie 2, and Rsubread can be used (Dobin et al., 2013; Kim et al., 2013; Kim et al., 2019; Liao et al., 2019; Langmead & Salzberg, 2012). Conversely, Kallisto or Salmon can be employed for pseudoalignment/quasi-mapping toward a reference transcriptome (Bray et al., 2016; Patro et al., 2017). In this protocol, we will only use HISAT2 (version 2.2.1), but we have provided an alternative protocol for the use of Kallisto and Salmon on GitHub (https://github.com/jmvillalobos/RNA-seq-protocol).
Part 5.1: Alignment to a reference genome using HISAT2
The aligner we will use is HISAT2, which employs a graph-based data structure and alignment algorithm for rapid and sensitive alignment of sequencing reads to a genome and a comprehensive collection of minor variants (Kim et al., 2019). Before commencing read mapping, it is essential to have an index of the reference genome. For this purpose, the specific reference genome is necessary; for the example data, we use the TAIR10 version of the A. thaliana genome, which can be obtained from EnsemblPlants (https://plants.ensembl.org/info/data/ftp/index.html). Subsequently, proceed to decompress the genome using the gunzip command and create the index into the genome_arabidopsis folder.
17.Download genome data:
- user:∼/project_2023/genome_arabidopsis$ wget
- https://ftp.ebi.ac.uk/ensemblgenomes/pub/release-57/plants/fasta/arabidopsis_thaliana/dna/Arabidopsis_thaliana.TAIR10.dna.toplevel.fa.gz
18.Decompress:
- user:∼/project_2023/genome_arabidopsis$ gunzip Arabidopsis_thaliana.TAIR10.dna.toplevel.fa.gz
19.Create the index in the index_hisat2 folder with HISAT2:
- user:∼/project_2023/index_hisat2$ hisat2-build -p 4 ../genome_arabidopsis/Arabidopsis_thaliana.TAIR10.dna.toplevel.fa genome
Where:
- hisat2-build is the HISAT2 command used to build the index;
- -p specifies the number of computer cores to be used—in this case, four are used;
- ../genome_arabidopsis indicates the genome path, including the specific file (Arabidopsis_thaliana.TAIR10.dna.toplevel.fa);
- genome is the name assigned to the files resulting from the index.
20.The process should generate a directory containing eight files:
- user:∼/project_2023/index_hisat2$ ls:
- genome.1.ht2
- genome.2.ht2
- genome.3.ht2
- genome.4.ht2
- genome.5.ht2
- genome.6.ht2
- genome.7.ht2
- genome.8.ht2
21.Once the index has been successfully created, it is possible to align the clean reads to the reference genome (per each sample) in the alignment_hisat2 folder:
- user:∼/project_2023/alignment_hisat2$ hisat2 -p 4 -x ../index_hisat2/genome -1 ../trimming_data/SRR10207204_P_1.fastq.gz -2 ../trimming_data/SRR10207204_P_2.fastq.gz -S SRR10207204.sam
Where:
- -x is the index path and base name of the reference genome index;
- -1 y -2 are paired-end files to be aligned with their respective paths;
- -S is the output of the SAM alignment file.
22.Generate a loop to speed up the analysis:
- user:∼/project_2023/alignment_hisat2
{sample}_P_1.fastq.gz -2 ../trimming_data/SRR10207 {sample}.sam; done|sh
23.Each alignment will generate an alignment output, which is essential as it indicates the percentage of reads that are aligned to the genome:
11117409 reads; of these:
- 11117409 (100.00%) were paired; of these:
- 415513 (3.74%) aligned concordantly 0 times
- 10136746 (91.18%) aligned concordantly exactly 1 time
- 565150 (5.08%) aligned concordantly >1 time
- —-
- 415513 pairs aligned concordantly 0 times; of these:
- 59514 (14.32%) aligned discordantly 1 time
- —-
355999 pairs aligned 0 times concordantly or discordantly; of these:
- 711998 mates make up the pairs; of these:
- 556751 (78.20%) aligned 0 times
- 141225 (19.84%) aligned exactly 1 time
- 14022 (1.97%) aligned >1 times
- 97.50% overall alignment rate
Part 5.2: Convert SAM to BAM file and sort by coordinates
The alignment result produces SAM (sequence alignment map) files, which are text files containing sequence alignment information in the reference genome (Li et al., 2009). BAM (binary alignment map) files are compressed versions of SAM files containing the same information but in a non-human-readable format. BAM files are more efficient and smaller, making them easier to process. After generating a BAM file, it is essential to sort and index it because reads are randomly assigned in the original file. The choice to sort by sequence identifier or genomic coordinates depends on the application, with sorting by coordinates being common for genomic data. Samtools is a commonly used tool to perform these tasks (Danecek et al., 2021).
24.To convert SAM to BAM format and sort it, execute the following command in the folder containing the SAM alignment files (per sample):
- user:∼/project_2023/alignment_hisat2$ samtools sort SRR10207204.sam -o SRR10207204_sorted.bam
This should generate a sorted file:
- user:∼/project_2023/alignment_hisat2$ ls
- SRR10207204.sam SRR10207204_sorted.bam
25.Generate a loop to speed up the analysis.
- user:∼/project_2023/alignment_hisat2
{sample}.sam -o SRR10207${sample}_sorted.bam; done|sh
Part 6: Quantification of the number of mapped reads
Read quantification is a crucial task in RNA-seq analysis. The central operation in this context involves counting the number of reads overlapping specific genomic features. Specialized tools such as featureCounts and HTSeq have emerged as critical elements for this task, enabling efficient counting of reads mapped to particular genomic features (Anders et al., 2015; Liao et al., 2014).
26.Before starting the quantification, it is essential to download the EnsemblPlants GTF annotation file and unzip it in the genome_arabidopsis directory as follows:
- user:∼/project_2023/genome_arabidopsis$ wget
- https://ftp.ebi.ac.uk/ensemblgenomes/pub/release-57/plants/gtf/arabidopsis_thaliana/Arabidopsis_thaliana.TAIR10.57.gtf.gz
27.Decompress:
- user:∼/project_2023/genome_arabidopsis$ gunzip Arabidopsis_thaliana.TAIR10.57.gtf.gz
Part 7: Quantifying with featureCounts
We will use the files sorted by coordinates we obtained previously to quantify the number of mapped reads.
28.Navigate to the quantification_featureCounts folder and execute the following command:
- user:∼/project_2023/quantification_featureCounts$ featureCounts -p --count ReadPairs -t exon -g gene_id -a ../genome_arabidopsis/Arabidopsis_thaliana.TAIR10.57.gtf -o SRR10207204.txt ../alignment_hisat2/SRR10207204_sorted.bam
29.We can use a “for” loop to automate the quantification of all samples, following a similar approach to that used with Trimmomatic, HISAT2, and Samtools. To simplify the process further, we propose creating a shell script that dynamically operates on the sorted.bam files in the alignment_hisat2 directory. This script can be inserted into a text editor such as Nano or Vim and saved with the name featureCounts.sh. Subsequently, necessary permissions are granted using the command chmod +x featureCounts.sh, and finally, the script is executed in the quantification_featureCounts directory.
-
#!/bin/bash
-
Input folder containing sorted BAM files
-
input_folder="../alignment_hisat2/"
-
GTF file
-
gtf_file="../genome_arabidopsis/Arabidopsis_thaliana.TAIR10.57.gtf"
-
Output folder for featureCounts results
-
output_folder="featureCounts_output/"
-
Create the output folder if it does not exist
-
mkdir -p $output_folder
-
Loop over each BAM file in the input folder
-
for bam_file in $input_folder/*.bam; do
-
Get the base name of the file without the .bam extension
-
base_name=
bam_file .bam) -
Run featureCounts for the current file
-
featureCounts -p --count ReadPairs -t exon -g gene_id -a
output_folder/ bam_file -
done
30.The user can save this script from the command line:
- vim featureCounts.sh
31.Run the script:
- user:∼/project_2023/quantification_featureCounts$./featureCounts.sh
32.To achieve this, run a series of commands that generate a count matrix for all samples with their respective identifiers. For this purpose, install two data manipulation packages:
- user:∼/project_2023/quantification_featureCounts$ sudo apt install moreutils
- user:∼/project_2023/quantification_featureCounts$ sudo apt install parallel
33.Extract counts (column 7):
- user:∼/project_2023/quantification_featureCounts$ ls -1 *.txt | parallel ‘cat {} | sed ‘1d’ | cut -f7 {} > {/.}_clean.txt’
34.Extract id (column 1):
- user:∼/project_2023/quantification_featureCounts$ ls -1 *.txt | head -1 | xargs cut -f1 > genes.txt
35.Arrange data in a matrix:
- user:∼/project_2023/quantification_featureCounts$ paste genes.txt *clean.txt > matriz_arabidopsis_2023.txt
| Accession number | Library name | Replicates | Treatment |
|---|---|---|---|
| SRR10207204 | Only Arabidopsis growing 5 dpi (without Trichoderma) | Biological replicate 1 | Control_1 |
| SRR10207210 | Only Arabidopsis growing 5 dpi (without Trichoderma) | Biological replicate 2 | Control_2 |
| SRR10207216 | Only Arabidopsis growing 5 dpi (without Trichoderma) | Biological replicate 3 | Control_3 |
| SRR10207206 | Arabidopsis with wild-type strain 5 dpi | Biological replicate 1 | Treatment_1 |
| SRR10207212 | Arabidopsis with wild-type strain 5 dpi | Biological replicate 2 | Treatment_2 |
| SRR10207218 | Arabidopsis with wild-type strain 5 dpi | Biological replicate 3 | Treatment_3 |
| Geneid | Control_1 | Control_2 | Control_3 | Treatment_1 | Treatment_2 | Treatment_3 |
|---|---|---|---|---|---|---|
| AT1G30814 | 0 | 1 | 0 | 0 | 1 | 0 |
| AT1G78930 | 54 | 88 | 25 | 71 | 44 | 7 |
| AT1G71695 | 339 | 297 | 90 | 372 | 157 | 37 |
| AT1G58983 | 0 | 0 | 0 | 0 | 0 | 0 |
| AT1G12980 | 0 | 0 | 0 | 0 | 0 | 0 |
| AT1G45223 | 0 | 0 | 0 | 0 | 0 | 0 |
| AT1G56250 | 3 | 4 | 0 | 149 | 164 | 30 |
| AT1G66852 | 0 | 0 | 0 | 0 | 0 | 0 |
| AT1G69810 | 756 | 911 | 319 | 893 | 434 | 74 |
| AT1G72450 | 159 | 186 | 63 | 510 | 273 | 52 |
Part 8: Differential gene expression analysis
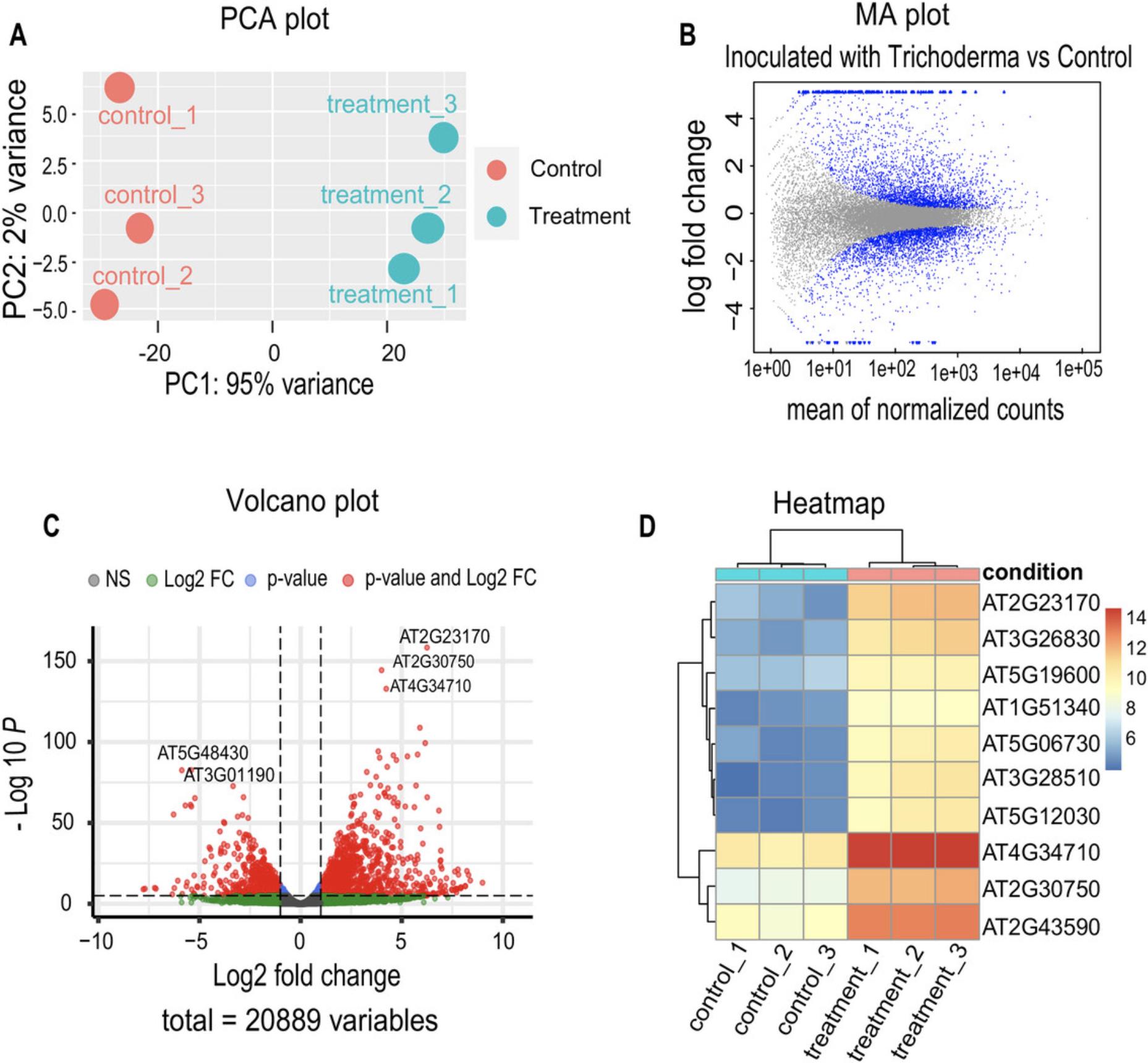
Differential expression analysis is essential in interpreting RNA-seq data, allowing the determination of quantitative changes in gene expression levels between experimental groups (Fig. 3). Specialized tools such as edgeR and DESeq2, based on negative binomial distributions, are prominent in differential expression analysis (Love et al., 2014; Robinson et al., 2010). In this specific protocol, we will focus on implementing DESeq2 to explore changes in gene expression of A. thaliana plants inoculated with T. atroviride (Treatment) versus non-inoculated plants (Control).
To perform the differential expression analysis, we will use R and the packages indicated in the script described here, provided on GitHub, as a supplementary script (https://github.com/jmvillalobos/RNA-seq-protocol).
36.Install packages to use in the R session:
-
Library Installation
-
Installing DESeq2
-
if (!require("BiocManager", quietly = TRUE))
-
install.packages("BiocManager")
-
BiocManager::install("DESeq2")
-
Installing ggplot2
-
install.packages("ggplot2")
-
Installing EnhancedVolcano
-
BiocManager::install("EnhancedVolcano")
-
Installing pheatmap
-
install.packages("pheatmap")
37.Load the library for differential gene expression (DGE) analysis and plotting:
-
Loading the library for DGE
-
library(DESeq2)
-
Loading the library for plots
-
library("ggplot2")
-
Loading the library for the volcano plot
-
library("EnhancedVolcano")
-
Loading the library for heatmap
-
library("pheatmap")
38.Load the contingency table of the accounts from featureCounts into the R session:
-
Setting the path to the featureCounts count matrix
-
setwd("C:/project_2023/quantification_featureCounts")
-
Viewing Files in the Directory
-
list.files()
-
Reading the count matrix
-
countData <- read.delim("./matriz_arabidopsis_2023.txt", header = TRUE, row.names = 1)
-
head(countData)
-
Define the new order of the replicates
-
column_order <- c("control_1", "control_2", "control_3", "treatment_1", "treatment_2", "treatment_3")
-
Rearrange the columns
-
countData <- countData[, column_order]
-
Displays the header of the new table
-
head(countData)
39.Create the “data.frame” object to perform differential expression analysis:
-
Description of samples
-
condition <- factor(c("Control", "Control", "Control", "Treatment", "Treatment", "Treatment"))
-
colData <- data.frame(row.names = colnames(countData), condition)
-
head(colData)
-
Creating a DESeqDataSet
-
dds <- DESeqDataSetFromMatrix(countData = countData, colData = colData, design = ∼ condition)
-
dds
40.Generate a PCA plot to verify consistency between replicates.
-
Generating a PCA Plot
-
rld <- rlog(dds, blind = F)
-
plotPCA(rld, intgroup = "condition") + geom_text(aes(label=name),
- vjust=0.2)
41.Filter the genes to be considered in the analysis by the number of reads.
-
Filtering genes with very low expression
-
dds <- dds[rowSums(counts(dds)) > 10,]
-
dds
-
Performing DGE analysis
-
dds <- DESeq(dds)
-
dds
-
Extracting results
-
res <- results(dds)
-
res
-
Summary of DGE
-
summary(res)
-
Sorting the summary list by p-adj
-
res <- res[order(res$padj),]
-
head(res)
42.Set the comparison of interest.
-
Extracting contrasts between conditions
-
Treatment_vs_Control <- results(dds, contrast = c("condition", "Treatment", "Control"))
-
summary(Treatment_vs_Control)
-
Obtaining a list of differentially expressed genes (DEGs) with stricter filtering
-
deg <- subset(Treatment_vs_Control, padj < 0.05 & abs(log2FoldChange) > 1)
-
print(deg)
-
summary(deg)
43.Export results:
-
Exporting the DEGs table
-
write.csv(deg, file = "DEG_Treatment_vs_Control_strict.csv")
-
Printing and exporting up-DEGs
-
up <- subset(deg, log2FoldChange > 1)
-
print(up)
-
summary(up)
-
write.csv(up, file = "Up_Treatment_vs_Control_strict.csv")
-
Printing and exporting down-DEGs
-
down <- subset(deg, log2FoldChange < (-1))
-
print(down)
-
summary(down)
-
write.csv(down, file = "Down_Treatment_vs_Control_strict.csv")
| baseMean | log2FoldChange | lfcSE | stat | p-value | padj | |
|---|---|---|---|---|---|---|
| AT1G56250 | 54.351119 | 5.87533399 | 0.65010368 | 9.03753377 | 1.60E-19 | 7.14E-18 |
| AT1G72450 | 163.108104 | 1.02177379 | 0.20647886 | 4.94856367 | 7.48E-07 | 8.23E-06 |
| AT1G79700 | 53.0739385 | −1.7029217 | 0.34330325 | −4.9604009 | 7.03E-07 | 7.79E-06 |
| AT1G52200 | 1506.68968 | −1.3297206 | 0.16382535 | −8.116696 | 4.79E-16 | 1.61E-14 |
| AT1G80870 | 310.280349 | −1.1105098 | 0.21694316 | −5.1188976 | 3.07E-07 | 3.61E-06 |
| AT1G24575 | 68.0515638 | 2.75544694 | 0.35408813 | 7.78181113 | 7.15E-15 | 2.15E-13 |
| AT1G53625 | 128.236024 | 2.25057089 | 0.28583177 | 7.87376033 | 3.44E-15 | 1.07E-13 |
| AT1G51440 | 9.75718656 | 1.98154125 | 0.70965146 | 2.7922739 | .0052339 | 0.02253924 |
| AT1G80840 | 91.2133689 | 3.4596998 | 0.3633223 | 9.52239873 | 1.69E-21 | 8.81E-20 |
| AT1G68470 | 54.7916789 | 1.87428097 | 0.32386298 | 5.78726522 | 7.15E-09 | 1.08E-07 |
44.To generate a basic volcano plot:
-
Generating a volcano plot with EnhancedVolcano
-
EnhancedVolcano(res, lab = rownames(res), x = ‘log2FoldChange’, y = ‘pvalue’)
45.To generate an MA plot:
-
Setting the contrast of interest
-
plotMA(Treatment_vs_Control, alpha = 0.05, main = "Inoculated with Trichoderma vs Control", xlab = "mean of normalized counts")
46.To generate a heatmap:
-
Selecting the top 10 DEGs in the Treatment_vs_Control comparison
-
res_ordered <- Treatment_vs_Control[order(Treatment_vs_Control$padj),]
-
top_genes <- row.names(res_ordered)[1:10]
-
Extracting and normalizing counts
-
counts <- counts(dds, normalized = TRUE)
-
counts_top <- counts[top_genes,]
-
Applying a logarithmic transformation to counts
-
log_counts_top <- log2(counts_top + 1)
-
Creating an annotation data frame based on condition information (colData)
-
df <- colData
-
Displaying the annotation data frame
-
df
-
Generating a heatmap using the pheatmap library
-
heatmap_20 <- pheatmap(log_counts_top, annotation = df)
Basic Protocol 2: GENE ONTOLOGY ENRICHMENT ANALYSIS
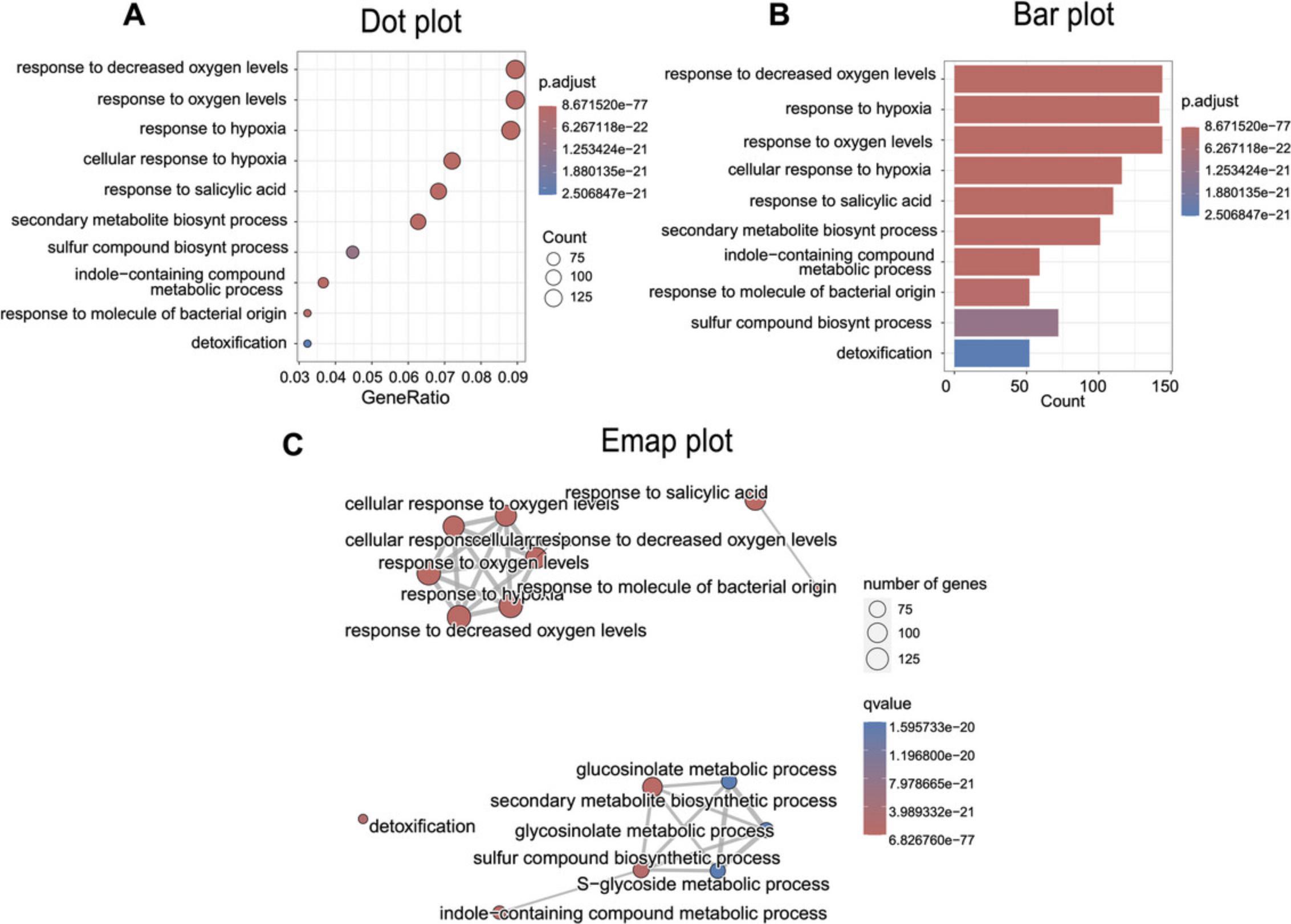
In Basic Protocol 2, we will perform a functional enrichment analysis to comprehensively understand the underlying biological processes (Gene Ontology or GO terms; Fig. 4). Various tools are available for this type of analysis, such as CAMERA, GOseq, topGO, clusterProfiler, and gprofiler2 (Alexa & Rahnenfuhrer, 2023; Robertson, H., & Robertson, N., 2024; Kolberg et al., 2020; Wu & Smyth, 2012; Wu et al., 2021; Young et al., 2010). In this protocol, we focus on using clusterProfiler because of its user-friendly interface and efficient integration with the differential expression output generated by DESeq2.clusterProfiler performs over-representation analysis (ORA; Boyle et al., 2004), a widely used approach to determine whether known biological functions or processes are over-represented (enriched) in an experimentally derived list of genes, such as a list of differentially expressed genes. To perform this analysis, follow the R script described here and on GitHub (https://github.com/jmvillalobos/RNA-seq-protocol).
In terms of systems resources, efficient progress in this task can be achieved by utilizing the command line within a Unix/Linux environment. We conducted this work on a 64-bit Ubuntu 23.04 operating system. For Windows users, a Windows system employing the Windows Subsystem for Linux (WSL; https://learn.microsoft.com/en-us/windows/wsl/install) is a viable option. For macOS users, it is feasible to execute identical commands in the terminal because of the shared Unix foundation of both operating systems. This protocol is compatible with the R desktop application, contingent on your operating system.
Necessary Resources
Software
Hardware
- A minimum of 4 GB of RAM and 1 GB of storage space for optimal performance
Part 1: Installation of the package to use in the R session
1.Run R script:
-
Installing clusterProfiler
-
if (!require("BiocManager", quietly = TRUE))
- install.packages("BiocManager")
-
BiocManager::install("clusterProfiler")
-
Installing biomaRt
-
BiocManager::install("biomaRt")
-
Installing enrichplot
-
BiocManager::install("enrichplot")
-
Installing org.At.tair.db
-
BiocManager::install("org.At.tair.db")
-
Installing tidyverse
-
install.packages("tidyverse")
2.Load required libraries:
-
library("clusterProfiler")
-
library("biomaRt")
-
library("enrichplot")
-
library("org.At.tair.db")
-
library("tidyverse")
3.Load the DESeq2 table into the R session:
-
Setting the path to the DESeq2 table.
-
setwd("C:/project_2023/quantification_featureCounts")
-
list.files()
-
Read the DESeq2 table (up or down).
-
diff_genes <- read_delim(file = "Up_Treatment_vs_Control_strict.csv", delim = ",")
-
Assign names to the first column.
-
colnames(diff_genes)[1] <- "genes"
-
Create a new table with the columns of interest.
-
diff_genes <- diff_genes[, c("genes", "log2FoldChange")]
-
Save the new table to a file.
-
write.table(diff_genes, file = "diff_genes_up.tsv", sep = "\t", row.names = FALSE, quote = FALSE)
4.Perform DEGs annotation with Ensembl and biomaRt:
- #Loading Arabidopsis database from EnsemblPlants with biomaRt
-
arabidopsis_mart <- useMart(biomart = ‘plants_mart’,
- host = ‘plants.ensembl.org’, dataset = ‘athaliana_eg_gene’)
-
head(listAttributes(arabidopsis_mart))
- #Retrieve the entrezgene_id and description for the DEGs
-
annot_diff <- getBM(
- values = diff_genes$genes,
- mart = arabidopsis_mart,
- attributes = c(‘ensembl_gene_id’, ‘entrezgene_id’,
- ‘description’),filters = ‘ensembl_gene_id’)
Part 2: Over-representation analysis (ORA) with clusterProfiler
5.Change the working directory:
-
setwd("C:/project_2023/quantification_featureCounts")
6.Read all Arabidopsis genes from a file (universe):
-
universe_arabidopsis <- read.delim("matriz_arabidopsis_2023.txt", header = TRUE, stringsAsFactors = FALSE)[,1]
- #Retrieve the entrezgene_id and description for the universe
-
annot_universe <- getBM(
- values = universe_arabidopsis,
- mart = arabidopsis_mart,
- attributes = c(‘ensembl_gene_id’, ‘entrezgene_id’,
- ‘description’),filters = ‘ensembl_gene_id’)
7.For compatibility with the enrichGO function, annot_universe genes must be characters, not integers, so it is necessary to convert them:
-
annot_universe
entrezgene_id)
8.Perform ORA for the Gene Ontology Biological Process class:
-
ora_analysis_BP <- enrichGO(
- gene = annot_diff$entrezgene_id,
- universe = annot_universe$entrezgene_id,
- OrgDb = org.At.tair.db,
- keyType = "ENTREZID",
- ont = "BP",
- pAdjustMethod = "BH",
- qvalueCutoff = 0.05,
- readable = TRUE,
- pool = FALSE
- )
9.Simplify the ORA results:
-
ora_analysis_BP_final <- clusterProfiler::simplify(ora_analysis_BP)
10.Export results and plot figures:
-
Write simplified results in a CSV file.
-
write_delim(
- x = as.data.frame(ora_analysis_BP@result),
- path = "go_results_up.csv",
- delim = ","
- )
-
Plot 1 - Dotplot.
-
dotplot(ora_analysis_BP_final, showCategory = 10)
-
Plot 2 - Barplot.
-
barplot(ora_analysis_BP_final, showCategory = 10)
-
Plot 3 - Enrichment Map (Emap Plot).
-
ora_analysis_BP <- pairwise_termsim(ora_analysis_BP, method = "JC")
-
emapplot(ora_analysis_BP, color = "qvalue", showCategory = 15)
Basic Protocol 3: DE NOVO ASSEMBLY OF DATA FROM NON-MODEL PLANTS
In Basic Protocol 1, we analyzed the transcriptome using a reference genome; in the absence of a reference genome, it is necessary to construct the transcriptome de novo. For this purpose, in Basic Protocol 3, we use a dataset from Sarwar et al. (2019), who generated it while exploring drought tolerance in Agave sisalana. Employing Trinity (Grabherr et al., 2011), we perform de novo assembly on a curated dataset of 10,000 reads for efficient execution on personal computers. Part of the workflow used in this protocol is shown in Figure 1.The dataset is available on GitHub (https://github.com/jmvillalobos/RNA-Seq-protocol).
In terms of systems, efficient progress in this task can be achieved utilizing the command line within a Unix/Linux environment. We conducted our work on a 64-bit Ubuntu 23.04 operating system. For Windows users, a Windows system, employing the Windows Subsystem for Linux (WSL; https://learn.microsoft.com/en-us/windows/wsl/install) is a viable option. For macOS users, it is feasible to execute identical commands in the terminal because of the shared Unix foundation of both operating systems. It is important to note that accurate data analysis demands substantial computational resources to execute effectively (see below).
Necessary Resources
Software
- Trinity: https://github.com/trinityrnaseq/trinityrnaseq/wiki
- CD-HIT: https://sites.google.com/view/cd-hit
- Kallisto: https://pachterlab.github.io/kallisto/
Hardware
- A minimum of 4 GB RAM and 5 GB storage space (we recommend ∼1 GB of RAM for every 1 million Illumina read pairs)
Part 1: Assembling with RNA-seq data using Trinity
1.Establish directories and install Trinity:
- user:∼/project_2023$ mkdir novo_assembly
- user:∼/project_2023/novo_assembly$ mkdir trimming_data_agave
- user:∼/project_2023/novo_assembly$ mkdir trinity_analysis
2.Install Trinity and CD-HIT:
- user:∼/project_2023/bioinformatics_tools$ sudo apt install trinityrnaseq
- user:∼/project_2023/bioinformatics_tools$ sudo apt install cd-hit
3.Organize data in preparation for de novo assembly using Trinity: Before initiating the de novo assembly, we consolidate sequencing data from the treatment and control conditions into a unified input file for Trinity (--samples_file). To accomplish this, furnish Trinity with a list of FASTQ files organized by treatment/control and replicate name, adhering to the structure outlined in a file named samples.txt, as explained below.
- user:∼/project_2023/novo_assembly/trinity_analysis$ head samples.txt
- Tra Tra_SRR5137658 ../trimming_data_agave/Tra_SRR5137658_1_P.fastq.gz ../trimming_data_agave/Tra_SRR5137658_2_P.fastq.gz
- Tra Tra_SRR5137660 ../trimming_data_agave/Tra_SRR5137660_1_P.fastq.gz ../trimming_data_agave/Tra_SRR5137660_2_P.fastq.gz
- Tra Tra_SRR5137663 ../trimming_data_agave/Tra_SRR5137663_1_P.fastq.gz ../trimming_data_agave/Tra_SRR5137663_2_P.fastq.gz
- Con Con_SRR5137659 ../trimming_data_agave/Con_SRR5137659_1_P.fastq.gz ../trimming_data_agave/Con_SRR5137659_2_P.fastq.gz
- Con Con_SRR5137661 ../trimming_data_agave/Con_SRR5137661_1_P.fastq.gz ../trimming_data_agave/Con_SRR5137661_2_P.fastq.gz
- Con Con_SRR5137662 ../trimming_data_agave/Con_SRR5137662_1_P.fastq.gz ../trimming_data_agave/Con_SRR5137662_2_P.fastq.gz
4.Using the samples.txt file, perform de novo transcriptome assembly of the reads with Trinity as follows:
- user:∼/project_2023/novo_assembly/trinity_analysis$ Trinity --seqType fq --samples_file samples.txt --CPU 4 --max_memory 12G
5.After the assembly, you will obtain two files and a directory; the file trinity_out_dir.Trinity.fasta represents the assembled transcripts:
- user:∼/project_2023/novo_assembly/trinity_analysis$ ls -l
- trinity_out_dir
- trinity_out_dir.Trinity.fasta
- trinity_out_dir.Trinity.fasta.gene_trans_map
6.Proceeding with the process, rename the files for enhanced manageability:
- user:∼/project_2023/novo_assembly/trinity_analysis$ mv trinity_out_dir.Trinity.fasta Trinity.fasta
- user:∼/project_2023/novo_assembly/trinity_analysis$ mv trinity_out_dir.Trinity.fasta.gene_trans_map Trinity.fasta.gene_trans_map
7.The assembly header should look like this:
- user:∼/project_2023/novo_assembly/trinity_analysis$ head Trinity.fasta
-
TRINITY_DN992_c0_g1_i1 len=223 path=[0:0-222]
- CTGGGGTCAGAGAAGCTGTAAAGTTATGCACTACTGCTGGCGTGAAGGTGCGGATGGTCACTGGAGATAACCTTAAGACTGCTAAA GCCATTGCTCTGGAGTGTGGGATACTTGATTCGGAAGCAGAAGCAACAGAGCCCACATTGATACACGGACAAACATTCCGTGCGAT GCGTGAAAAAGATAGAGACTCAGTTGCTGACAGGATCTCTGTGATGGGAAG
-
TRINITY_DN967_c1_g1_i1 len=217 path=[0:0-216]
- CAACCAAGACCTGTACTTCAAGAAGACCGTGAAATATGTCGGAGAGCCAATGACTCATTTGGAGTCAATTGCTTCATCTGCTGTAC GTGCTGCAATTAAAGTTAAAGCTTCTGTCATCGTTGTCTTTACTTCATCTGGAAGGGCAGCTAGATTAATTGCAAAGTATAGGCCT ACAATGCCTGTATTATCAGTTGTCATTCCTCGGCTCAAAACAAAC
Part 2: Evaluating the assembly
After completing the de novo assembly, it is crucial to evaluate its quality, meticulously considering potential errors. Before proceeding to further analyses, it is vital to perform a comprehensive assessment of the de novo transcriptome assembly, as a subpar assembly may lead to misinterpretation of gene identification and differential expression analysis (Raghavan et al., 2022). Key quality metrics include read the alignment rate, ExN50 statistic, and the count of universal genes with matches according to the BUSCO values (Simão et al., 2015). Notably, in this protocol, we exclusively visualize the basic statistics of the assembly. In a high-quality assembly, one would expect the reads used to construct such assembly to exhibit a high alignment percentage. An alignment rate of 70% could indicate satisfactory transcriptome quality. For BUSCO, however, there is no consensus about the optimal value, as it is a relative metric that can vary between different assemblers or assembly configurations. Nevertheless, the general expectation is to find the most complete genes. In this context, the N50 value is another important metric indicating that at least half of the assembled bases are contained within contigs equal to or longer than that value. However, this metric is not suitable for transcriptome assemblies because there the goal is the recovery of many assembled sequences and not just the construction of a few long contigs. A more appropriate statistic for evaluating transcriptome assemblies is the ExN50 statistic, which excludes contigs with low expression that tend to be very short because of insufficient read coverage. This statistic often yields higher values than the conventional N50 statistic and is more representative in this context. The ExN50 value in an RNA-seq assembly conducted with Trinity is crucial and can vary significantly depending on several factors, such as the species under study, the quality of the sequencing data, and the transcriptome complexity. In general terms, a high ExN50 value suggests a more complete and contiguous assembly, which is highly desirable. However, it is important to note that there is no universally “good” threshold for ExN50, as its assessment depends heavily on the aforementioned factors. Consequently, it is advisable to interpret the ExN50 value in the specific context of each project and to consider other assembly quality parameters for a more comprehensive and accurate evaluation. In general, all of these metrics depend on several factors, such as the species studied, the quality of the sequencing data, and the assembly tools used.
8.To determine the count of assembled transcripts, employ the following command:
- user:∼/project_2023/novo_assembly/trinity_analysis$ grep ‘>’ Trinity.fasta | wc -l
- 2116
9.If desired, a more comprehensive assembly statistics can be obtained using a Trinity-loaded script called TrinityStats.pl. To locate the script's path, execute the following command:
- user:∼/project_2023/novo_assembly/trinity_analysis$ dpkg-query -L trinityrnaseq | grep TrinityStats.pl
10.After identifying the path, execute it on the file Trinity.fasta:
-
user:∼/project_2023/novo_assembly/trinity_analysis$ perl /usr/lib/trinityrnaseq/util/TrinityStats.pl Trinity.fasta
-
################################
-
Counts of transcripts, etc.
-
################################
-
Total trinity ‘genes’: 1884
-
Total Trinity transcripts: 2116
-
Percent GC: 49.26
-
########################################
-
Stats based on ALL transcript contigs:
-
########################################
-
Contig N10: 1268
-
Contig N20: 981
-
Contig N30: 736
-
Contig N40: 585
-
Contig N50: 462
-
Median contig length: 315
-
Average contig: 431.78
-
Total assembled bases: 913641
-
#####################################################
-
Stats based on ONLY LONGEST ISOFORM per ‘GENE’:
-
#####################################################
-
Contig N10: 1227
-
Contig N20: 845
-
Contig N30: 632
-
Contig N40: 504
-
Contig N50: 413
-
Median contig length: 303
-
Average contig: 402.23
-
Total assembled bases: 757802
Part 3: Redundancy removal
De novo transcriptome assemblers often generate many sequences, exceeding the gene count in the genome. This complexity results from transcriptional artifacts, pre-mRNA, noncoding RNA (ncRNA), and isoforms generated through alternative splicing processes (Freedman et al., 2021). Assembly reduction is commonly employed to simplify and obtain a manageable, nonredundant set of sequences (Raghavan et al., 2022). Practical tools for this purpose include CD-HIT, Corset, Grouper, and Compacta (Davidson & Oshlack, 2014; Fu et al., 2012; Li & Godzik, 2006; Malik et al., 2018; Razo-Mendivil et al., 2020).
11.To eliminate redundancy from the transcriptome, employ CD-HIT by executing the following command:
- user:∼/project_2023/novo_assembly/trinity_analysis$ cd-hit-est -i Trinity.fasta -o Trinity_90.fasta -c 0.9 -n 9 -T 4 -M 3000
12.After this clustering process, the user can revisit the statistics, as mentioned previously, and observe a reduction in the number of contigs.
-
user:∼/project_2023/novo_assembly/trinity_analysis$ perl /usr/lib/trinityrnaseq/util/TrinityStats.pl Trinity_90.fasta
-
################################
-
Counts of transcripts, etc.
-
################################
-
Total trinity ‘genes’: 1869
-
Total trinity transcripts: 1904
-
Percent GC: 49.19
-
########################################
-
Stats based on ALL transcript contigs:
-
########################################
-
Contig N10: 1198
-
Contig N20: 857
-
Contig N30: 646
-
Contig N40: 514
-
Contig N50: 419
-
Median contig length: 305
-
Average contig: 405.38
-
Total assembled bases: 771847
-
#####################################################
-
Stats based on ONLY LONGEST ISOFORM per ‘GENE’:
-
#####################################################
-
Contig N10: 1227
-
Contig N20: 843
-
Contig N30: 630
-
Contig N40: 505
-
Contig N50: 413
-
Median contig length: 303
-
Average contig: 402.57
-
Total assembled bases: 752,400
Part 4: Transcript expression quantification with Kallisto
After eliminating redundancy from the assembly, the subsequent step entails estimating the expression values of the transcripts. This task will be performed using Kallisto, executing a script seamlessly integrated with Trinity.
13.The quantification process can be carried out according to the following details for each replicate within a particular condition:
- user:∼/project_2023/novo_assembly/trinity_analysis$ perl /usr/lib/trinityrnaseq/util/align_and_estimate_abundance.pl --seqType fq -- samples_file samples.txt --transcripts Trinity_90.fasta --est_method kallisto -- trinity_mode --prep_reference
14.This procedure generates six new directories, each corresponding to the quantification conducted by Kallisto for individual replicates:
-
user:∼/project_2023/novo_assembly/trinity_analysis$ ls -l
-
drwxr-xr-x 2 user 4096 Jan 17 15:54 Con_SRR5137659
-
drwxr-xr-x 2 user 4096 Jan 17 15:54 Con_SRR5137661
-
drwxr-xr-x 2 user 4096 Jan 17 15:54 Con_SRR5137662
-
drwxr-xr-x 2 user 4096 Jan 17 15:54 Tra_SRR5137658
-
drwxr-xr-x 2 user 4096 Jan 17 15:54 Tra_SRR5137660
-
drwxr-xr-x 2 user 4096 Jan 17 15:54 Tra_SRR5137663
15.You can now inspect the contents of each directory as follows:
- user:∼/project_2023/novo_assembly/trinity_analysis$ ls -l Tra_SRR5137660/
- -rw-r--r-- 1 user 82266 Jan 17 15:54 abundance.tsv
- -rw-r--r-- 1 user 85282 Jan 17 15:54 abundance.tsv.genes
- -rw-r--r-- 1 user 614 Jan 17 15:54 run_info.json
16.You can further examine one of the files containing the quantification in transcripts per million (TPM) for a specific replicate:
- user:∼/project_2023/novo_assembly/trinity_analysis$ head Tra_SRR5137660/abundance.tsv |column -t
| target_id | length | eff_length | est_counts | tpm |
|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Part 5: Counting and gene expression matrices
Using the quant.sf files, a matrix with estimated counts and another with TPM expression values can be generated and normalized across samples using the TMM method.
17.Initially, compile a list of quant.sf files for each replicate:
-
user:∼/project_2023/novo_assembly/trinity_analysis$ find Tra_* Con_* -name "abundance.tsv" | tee quant_files.list
-
Tra_SRR5137658/abundance.tsv
-
Tra_SRR5137660/abundance.tsv
-
Tra_SRR5137663/abundance.tsv
-
Con_SRR5137659/abundance.tsv
-
Con_SRR5137661/abundance.tsv
-
Con_SRR5137662/abundance.tsv
18.Using the quant_files.list file, employ another Trinity script to generate two matrices:
- user:∼/project_2023/novo_assembly/trinity_analysis$ perl /usr/lib/trinityrnaseq/util/abundance_estimates_to_matrix.pl --est_method kallisto--out_prefix kallisto --name_sample_by_basedir --quant_files quant_files.list --gene_trans_map Trinity_90.fasta.gene_trans_map
You have now produced count matrices at both the gene and isoform levels. You can proceed with the differential expression analysis using the scripts integrated into Trinity or opt to perform Basic Protocol 1 manual with minimal adjustments to the conditions.
COMMENTARY
Background Information
This article offers a comprehensive guide for performing differential gene expression analysis using two distinct approaches. In the first approach (presented in Basic Protocols 1 and 2), HISAT2 is employed to align with the reference genome; in contrast, in the second approach (detailed in an alternative protocol on GitHub at https://github.com/jmvillalobos/RNA-seq-protocol), Kallisto is utilized, relying on pseudoalignment to predict transcripts of the organism. Given the nature of the analysis, we strongly recommend using HISAT2, as it tends to yield more precise results. However, our comparison of results at the level of differentially expressed genes highlights the remarkable similarity between the results obtained with the two strategies (Fig. 5). Therefore, for users with limited computational resources, opting for Kallisto can be an excellent strategy to reduce time and computational load significantly.
![Details are in the caption following the image Differentially expressed genes (DEGs) obtained by HISAT2 and Kallisto. (A) Bar graph illustrating the number of induced and repressed genes for each bioinformatics tool. (B and C) Venn diagrams illustrating upregulated (B) and downregulated (C) genes shared and unique between HISAT2 and Kallisto. Those meeting the criteria of p<sub>adj</sub> <0.05 and abs[log<sub>2</sub>(fold change)] >1, according to DESeq2 analysis, were considered. We generated this Venn diagram using the online tool available at https://bioinformatics.psb.ugent.be/webtools/Venn/.](https://static.yanyin.tech/literature_test/cpz11054-fig-0005-m.jpg)
Additionally, Basic Protocol 3 addresses the application of the Trinity software in a scenario without a reference genome, conducting de novo assembly for a dataset from a non-model organism. Although the process we describe here was expedited by limiting the reads to only 10,000, we must emphasize the substantial demand for computational resources when working with more realistic datasets. For this reason, in real-world scenarios, we recommend performing the assembly on servers with higher RAM resources. We encourage the use of services such as Amazon Web Services.
This protocol is a practical tool for experienced researchers and a valuable resource for students seeking to delve into RNA-seq analysis. Discussing differences between mappers, models for differential expression analysis, and considerations when working with or without a reference genome provides a deeper and more contextual understanding for new users. This inclusive approach facilitates the entry and participation of students in this research field and contributes to the continuous development of skills in genomic data analysis.
Critical Parameters
A crucial step in executing this series of protocols is carefully selecting a suitable version of a Linux-based system. When opting for Ubuntu, using at least Ubuntu version 20.04 or higher is strongly recommended. Another vital consideration is to run command lines with appropriate computational resources, adjusting them to the user's computer capabilities and avoiding exceeding the established limits for RAM and threads.
Although many of the commands in these protocols are executed with default parameters, it is highly advisable to review the list of parameters to be used thoroughly. These may vary in some cases depending on the organism or project objectives. Nevertheless, these protocols are entirely functional and highly interpretable for various organisms.
Troubleshooting Table
For a list of possible problems, causes, and solutions, see Table 4.
| Problem | Possible cause | Solution |
|---|---|---|
| Killed process | Limited memory; limited space | Use high-performance server |
| Unsuccessful program installation | Versions available for your device | Install via conda |
Understanding Results
Basic Protocol 1
In this protocol, we tested the differential gene expression analysis of A. thaliana inoculated with spores of the fungus T. atroviride (Treatment) as compared to non-inoculated plants (Control condition). The principal component analysis (PCA) plot reveals consistent clustering between biological replicates for each condition (Fig. 3A). This type of analysis is of fundamental importance in RNA-seq experiments because it provides valuable insights from the initial stages of the investigation. PCA visualizes sample variability and alerts us of possible discrepancies between the replicates.
Next, an MA plot showing the log base 2 (log2) value of fold changes is obtained (Fig. 3B). In this graph, we set a significance value cutoff (alpha) of 0.05, so points below this threshold were colored blue. On the other hand, the volcano plot in Figure 3C represents differentially expressed genes in the Treatment versus Control contrast, including those induced and those repressed. This graph is informative because it highlights the genes with the most significant changes in expression levels based on the log2(fold change) and adjusted p -value criteria. The visual representation of this information provides a precise and rapid perspective on the magnitude of changes in gene expression, allowing the efficient identification of the most relevant genes in the Treatment versus Control contrast. Finally, Figure 3D shows the top ten differentially expressed genes of this contrast, in which a well-defined group of negatively regulated genes is observed in the control condition, whereas in the treatment condition, they are overexpressed. These differential expression results are consistent with those reported in the article from which these data are taken (Villalobos-Escobedo et al., 2020).
Basic Protocol 2
After identifying the differentially expressed genes, a quick method to grasp the overall landarabidopscape of the biological phenomenon in experimental comparisons is to conduct a functional category enrichment analysis. This analysis relies heavily on the annotation level of the genome used. In the case of Arabidosis, the genomic annotation is quite comprehensive, making the use of GO terms highly convenient. Our protocol employs the clusterProfiler tool due to its versatility in enrichment analysis and in generating graphs that aid in result comprehension. We present three useful types of graphs (Fig. 4). Firstly, a dot plot allows visualization of the overall change quantity between comparisons (GeneRatio), the significance of enrichment (p -adjusted, p adj), and the count number. A similar graph is the bar plot, though the GeneRatio is not shown here to simplify the figure and enhance comprehension. Lastly, we introduced the emap plot, which displays the relationship between GO terms and indicates how related these terms are, potentially revealing linked and enriched processes. These graphs are valuable tools to demonstrate result consistency and comprehensively highlight significant processes.
Basic Protocol 3
This protocol involves the analysis of data derived from an organism that lacks a reference genome (a non-model organism). It shares with Basic Protocol 1 the quality analysis phase with FastQC and the trimming of reads with Trimmomatic. Subsequently, an assembly is carried out using Trinity with a curated data set. Examining assembly statistics such as alignment rate, ExN50, and BUSCO statistics is essential. However, this protocol does not address these statistics because of the low quality of the assembly obtained, which we attribute to the use a minimal data set to run the process on a personal computer in this demonstration. Eliminating redundancy in the assembly is highly recommended; here, CD-HIT was used successfully to reduce redundant transcripts. Subsequently, an abundance quantification analysis was performed using Kallisto, generating count matrices at the isoform level (Kallisto.isoform.counts.matrix) and at the gene level (Kallisto.gene.counts.matrix). Because of space limitations, Basic Protocol 3 does not include details of the differential expression analysis and transcriptome annotation steps. In general, however, these are very similar to those discussed in Basic Protocols 1 and 2.
Time Considerations
The time required for the analyses presented varies depending on the protocol used and the amount of data to be analyzed. Basic Protocols 1 and 2 will take ∼1 day of work, depending mainly on the computational power of the equipment used for analysis. Execution of Basic Protocol 3 is estimated to take ∼4 hr.
Acknowledgments
E.P.-S. and K.M.H.-M. received grants no. 989265 and 827463 for their M.Sc. theses from CONAHCyT Mexico. The authors thank Tania Quintana and Laura Gálvez for executing this protocol series and giving us their valuable comments and suggestions for improvement.
Author Contributions
Enrique Pola-Sánchez : Conceptualization; methodology; writing—original draft; writing—review and editing. Karen Magdalena Hernández-Martínez : Methodology; visualization; writing—review and editing. Rafael Pérez-Estrada : Visualization. Nelly Selem-Mojica : Writing—review and editing. June Simpson : Writing—review and editing. María Jazmín Abraham-Juárez : Writing—review and editing. Alfredo Herrera-Estrella : Writing—review and editing. José Manuel Villalobos-Escobedo : Conceptualization; methodology, writing—original draft; writing—review and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data are openly available in the public repository Zenodo at https://zenodo.org/records/10537097.
Literature Cited
- Alexa, A., & Rahnenfuhrer, J. (2023). topGO: Enrichment analysis for gene ontology. GO R package version 2.54.0. https://bioconductor.org/packages/topGO
- Anders, S., Pyl, P. T., & Huber, W. (2015). HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics , 31(2), 166–169. https://doi.org/10.1093/bioinformatics/btu638
- Blighe, K., Rana, S., & Lewis, M. (2019). EnhancedVolcano: Publication-ready volcano plots with enhanced coloring and labeling. R package version 1(0), https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html
- Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics , 30(15), 2114–2120. https://doi.org/10.1093/bioinformatics/btu170
- Boyle, E. I., Weng, S., Gollub, J., Jin, H., Botstein, D., Cherry, J. M., & Sherlock, G. (2004). GO::TermFinder—open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics , 20(18), 3710–3715. https://doi.org/10.1093/bioinformatics/bth456
- Bray, N. L., Pimentel, H., Melsted, P., & Pachter, L. (2016). Near-optimal probabilistic RNA-Seq quantification. Nature Biotechnology , 34(5), 525–527. https://doi.org/10.1038/nbt.3519
- Carlson, M. (2019). org.At.tair.db: Genome wide annotation for Arabidopsis. R package version 3.8.2. https://bioconductor.org/packages/org.At.tair.db/
- Chen, Y., Chen, Y., Shi, C., Huang, Z., Zhang, Y., Li, S., Li, Y., Ye, J., Yu, C., Li, Z., Zhang, X., Wang, J., Yang, H., Fang, L., & Chen, Q. (2018). SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience , 7(1), 1–6. https://doi.org/10.1093/gigascience/gix120
- Chen, S., Zhou, Y., Chen, Y., & Gu, J. (2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics , 34(17), i884–i890. https://doi.org/10.1093/bioinformatics/bty560
- Conesa, A., Madrigal, P., Tarazona, S., Gomez-Cabrero, D., Cervera, A., McPherson, A., Szcześniak, M. W., Gaffney, D. J., Elo, L. L., Zhang, X., & Mortazavi, A. (2016). A survey of best practices for RNA-Seq data analysis. Genome Biology , 17, 13. https://doi.org/10.1186/s13059-016-0881-8
- Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., Whitwham, A., Keane, T., McCarthy, S. A., Davies, R. M., & Li, H. (2021). Twelve years of SAMtools and BCFtools. GigaScience , 10(2), giab008. https://doi.org/10.1093/gigascience/giab008
- Davidson, N. M., & Oshlack, A. (2014). Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biology , 15(7), 410. https://doi.org/10.1186/s13059-014-0410-6
- Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M., & Gingeras, T. R. (2013). STAR: Ultrafast universal RNA-Seq aligner. Bioinformatics , 29(1), 15–21. https://doi.org/10.1093/bioinformatics/bts635
- Durinck, S., Moreau, Y., Kasprzyk, A., Davis, S., de Moor, B., Brazma, A., & Huber, W. (2005). BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics , 21(16), 3439–3440. https://doi.org/10.1093/bioinformatics/bti525
- Durinck, S., Spellman, P., Birney, E., & Huber, W. (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature Protocols , 4, 1184–1191. https://doi.org/10.1038/nprot.2009.97
- Ewing, B., & Green, P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research , 8(3), 186–194. https://doi.org/10.1101/gr.8.3.186
- Freedman, A. H., Clamp, M., & Sackton, T. B. (2021). Error, noise and bias in de novo transcriptome assemblies. Molecular Ecology Resources , 21(1), 18–29. https://doi.org/10.1111/1755-0998.13156
- Fu, L., Niu, B., Zhu, Z., Wu, S., & Li, W. (2012). CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics , 28(23), 3150–3152. https://doi.org/10.1093/bioinformatics/bts565
- Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., Adiconis, X., Fan, L., Raychowdhury, R., Zeng, Q., Chen, Z., Mauceli, E., Hacohen, N., Gnirke, A., Rhind, N., di Palma, F., Birren, B. W., Nusbaum, C., Lindblad-Toh, K., … Regev, A. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology , 29(7), 644–652. https://doi.org/10.1038/nbt.1883
- Jiang, H., Lei, R., Ding, S. W., & Zhu, S. (2014). Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics , 15, 182. https://doi.org/10.1186/1471-2105-15-182
- Kasschau, K. D., Fahlgren, N., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., Givan, S. A., & Carrington, J. C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biology , 5(3), e57. https://doi.org/10.1371/journal.pbio.0050057
- Kim, D., Paggi, J. M., Park, C., Bennett, C., & Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology , 37(8), 907–915. https://doi.org/10.1038/s41587-019-0201-4
- Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., & Salzberg, S. L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology , 14(4), R36. https://doi.org/10.1186/gb-2013-14-4-r36
- Kolberg, L., Raudvere, U., Kuzmin, I., Vilo, J., & Peterson, H. (2020). gprofiler2—an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Research , 9, ELIXIR–709. https://doi.org/10.12688/f1000research.24956.2
- Langmead, B., & Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods , 9(4), 357–359. https://doi.org/10.1038/nmeth.1923
- Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R., 1000 Genome Project Data Processing Subgroup. (2009). The sequence alignment/map format and SAMtools. Bioinformatics , 25(16), 2078–2079. https://doi.org/10.1093/bioinformatics/btp352
- Li, W., & Godzik, A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics , 22(13), 1658–1659. https://doi.org/10.1093/bioinformatics/btl158
- Liao, Y., Smyth, G. K., & Shi, W. (2014). featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics , 30(7), 923–930. https://doi.org/10.1093/bioinformatics/btt656
- Liao, Y., Smyth, G. K., & Shi, W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Research , 47(8), e47. https://doi.org/10.1093/nar/gkz114
- Liu, T., Tang, J., Chen, L., Zeng, J., Wen, J., Yi, B., Ma, C., Tu, J., Fu, T., & Shen, J. (2019). Differential expression of miRNAs and their targets in wax-deficient rapeseed. Scientific Reports , 9(1), 12201. https://doi.org/10.1038/s41598-019-48439-z
- Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biology , 15(12), 1–21. https://doi.org/10.1186/s13059-014-0550-8
- Malik, L., Almodaresi, F., & Patro, R. (2018). Grouper: Graph-based clustering and annotation for improved de novo transcriptome analysis. Bioinformatics , 34(19), 3265–3272. https://doi.org/10.1093/bioinformatics/bty378
- Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal , 17(1), 10–12. https://doi.org/10.14806/ej.17.1.200
- Mehdi, S. M. M., Krishnamoorthy, S., Szczesniak, M. W., & Ludwików, A. (2021). Identification of novel miRNAs and their target genes in the response to abscisic acid in Arabidopsis. International Journal of Molecular Sciences , 22(13), 7153. https://doi.org/10.3390/ijms22137153
- Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., & Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature Methods , 14(4), 417–419. https://pubmed.ncbi.nlm.nih.gov/26813401/
- Raghavan, V., Kraft, L., Mesny, F., & Rigerte, L. (2022). A simple guide to de novo transcriptome assembly and annotation. Briefings in Bioinformatics , 23(2), bbab563. https://doi.org/10.1093/bib/bbab563
- Razo-Mendivil, F. G., Martínez, O., & Hayano-Kanashiro, C. (2020). Compacta: A fast contig clustering tool for de novo assembled transcriptomes. BMC Genomics , 21(1), 148. https://doi.org/10.1186/s12864-020-6528-x
- Robertson, H., & Robertson, N. (2024). TOP: TOP constructs transferable model across gene expression platforms. R package version 1.4.0. https://doi.org/10.18129/B9.bioc.top
- Robinson, M. D., McCarthy, D. J., & Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics , 26(1), 139–140. https://doi.org/10.1093/bioinformatics/btp616
- Sarwar, M. B., Ahmad, Z., Rashid, B., Hassan, S., Gregersen, P. L., Leyva, M. O., Nagy, I., Asp, T., & Husnain, T. (2019). De novo assembly of Agave sisalana transcriptome in response to drought stress provides insight into the tolerance mechanisms. Scientific Reports , 9(1), 396. https://doi.org/10.1038/s41598-018-35891-6
- Simão, F. A., Waterhouse, R. M, Ioannidis, P., Kriventseva, E. V., & Zdobnov, E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics , 31(19), 3210–3212. https://doi.org/10.1093/bioinformatics/btv351
- Stark, R., Grzelak, M., & Hadfield, J. (2019). RNA sequencing: The teenage years. Nature Reviews Genetics , 20(11), 631–656. https://doi.org/10.1038/s41576-019-0150-2
- Villalobos-Escobedo, J. M., Esparza-Reynoso, S., Pelagio-Flores, R., López-Ramírez, F., Ruiz-Herrera, L. F., López-Bucio, J., & Herrera-Estrella, A. (2020). The fungal NADPH oxidase is an essential element for the molecular dialog between Trichoderma and Arabidopsis. The Plant Journal , 103(6), 2178–2192. https://doi.org/10.1111/tpj.14891
- Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., Feng, T., Zhou, L., Tang, W., Zhan, L., Fu, X., Liu, S., Bo, X., & Yu, G. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation , 2(3), 100141. https://doi.org/10.1016/j.xinn.2021.100141
- Wu, D., & Smyth, G. K. (2012). Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Research , 40(17), e133. https://doi.org/10.1093/nar/gks461
- Young, M. D., Wakefield, M. J., Smyth, G. K., & Oshlack, A. (2010). Gene ontology analysis for RNA-Seq: Accounting for selection bias. Genome Biology , 11(2), R14. https://doi.org/10.1186/gb-2010-11-2-r14
- Yu, G. (2023). enrichplot: Visualization of Functional Enrichment Result. R package version 1.22.0. https://bioconductor.org/packages/enrichplot

