Quantitative Neuroanatomical Phenotyping of the Embryonic Mouse Brain
Sylvie Nguyen, Sylvie Nguyen, Meghna Kannan, Meghna Kannan, Marion Gaborit, Marion Gaborit, Stephan C. Collins, Stephan C. Collins, Binnaz Yalcin, Binnaz Yalcin
brain morphology
coronal and sagittal planes
embryo development
neurodevelopmental disorders
standardized histology pipeline
Abstract
Congenital neurodevelopmental anomalies are present from birth and are characterized by an abnormal development of one or more structures of the brain. Brain structural anomalies are highly comorbid with neurodevelopmental and neuropsychiatric disorders such as intellectual disability, autism spectrum disorders, epilepsy, and schizophrenia, and 80% are of genetic origin. We aim to address an important neurobiological question: How many genes regulate the normal anatomy of the brain during development. To do so, we developed a quantitative approach for the assessment of a total of 106 neuroanatomical parameters in mouse mutant embryos at embryonic day 18.5 across two planes commonly used in brain anatomical studies, the coronal and sagittal planes. In this article we describe the techniques we developed and explain why ultrastandardized procedures involving embryonic mouse brains are even more of prime importance for morphological phenotyping than adult mouse brains. We focus our analysis on brain size anomalies and on the most frequently altered brain regions including the cortex, corpus callosum, hippocampus, ventricles, caudate putamen, and cerebellum. Our protocols allow a standardized histology pipeline from embryonic mouse brain preparation to sectioning, staining, and scanning and neuroanatomical analyses at well-defined positions on the coronal and sagittal planes. Together, our protocols will help scientists in deciphering congenital neurodevelopmental anomalies and anatomical changes between groups of mouse embryos in health and genetic diseases. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Fixation and preparation of embryonic mouse brain samples
Basic Protocol 2 : Sectioning, staining, and scanning of embryonic mouse brain sections
Basic Protocol 3 : Coronal neuroanatomical measurements of embryonic mouse brain structures
Basic Protocol 4 : Sagittal neuroanatomical measurements of embryonic mouse brain structures
INTRODUCTION
In vivo modeling of human congenital neurodevelopmental anomalies is important in basic and clinical research since the brain cannot be easily accessed in humans. We thus turned to the long-standing mouse model where both the environment and genetics can be controlled. As a primary screen with no phenotypic a priori , we assess neuroanatomical phenotypes (NAPs; defined as any phenotype pertaining to brain neuroanatomical changes) throughout the embryonic mouse brain and at many different levels, from cellularly to macroscopically evident changes. In contrast to conventional histopathological embryonic screens, we rely on a quantitative phenotyping approach where each brain section has to pass well-defined stereotaxic coordinates, defined according to the Atlas of the Developing Mouse Brain (Paxinos, Halliday, Watson, Koutcherov, & Wang, 2007), to detect any NAPs ranging from mild to moderate to severe effect in size.
To do so, we developed precision histological pipelines for phenotyping the embryonic mouse brain consisting of 106 quantifiable neuroanatomical brain parameters across multiple brain sections, down to cell-level resolution achieved using a digital slide scanner from Hamamatsu. Similar to our existing screens in adult mouse brain (see Current Protocols articles: Collins et al., 2018; Mikhaleva, Kannan, Wagner, & Yalcin, 2016), brain parameters encompass six main categories: brain size, commissures, ventricles, cortex, subcortex, and cerebellum. We previously described how the adult mouse brain can be studied for large-scale neuroanatomical profiling assays in over 1000 mouse models of neurodevelopmental disorders (Collins, Mikhaleva, et al., 2019) produced by the International Mouse Phenotyping Consortium, a large-scale mouse knockout program (Birling et al., 2021). This new protocol explores the benefits of combining our previous procedures in the adult with ones presented in the developing mouse brain at embryonic day 18.5 (E18.5).
Here, we describe a series of standard operating procedures to perform neuroanatomical phenotyping of the developing mouse brain at E18.5. More specifically, we first describe the fixation and preparation of embryonic brain tissue and trimming across the sagittal and coronal planes (Basic Protocol 1) and sectioning using well-defined anatomical features, staining of sections using Nissl, and imaging of slides using a digital slide scanner (Basic Protocol 2). Quality control steps and data collection of 66 and 40 quantitative neuroanatomical parameters across the coronal and sagittal sectioning planes are then described in Basic Protocols 3 and 4, respectively. Although each specific step is described for one embryonic mouse brain, the protocols are compatible for a larger number of embryonic brain samples.
Basic Protocol 1: FIXATION AND PREPARATION OF EMBRYONIC MOUSE BRAIN SAMPLES
Fixation is an important step to preserve the structural features of the developing brain. It is preferable to use Bouin's fixative for the whole embryo where preservation of morphology is important. After immersion fixation, brain samples are dissected and transferred to 70% ethanol before being embedded in paraffin wax. For standardization purposes, the number of days the sample is left in ethanol needs to be consistent between samples to limit unwanted variation due to tissue shrinkage. Although coronal and sagittal sections could be done using the same embryo, we recommend the use of one embryo per orientation (coronal or sagittal) should you need the two orientations to address more specific questions. There are a small number of specific steps should you opt for coronal or sagittal sections. For the sagittal plane, the brain is cut into two halves along the midline. For the coronal plane, the brain must be cut at the level of interest relative to Bregma. Coronal-specific instructions are indicated as cor-, whereas sagittal-specific instructions are indicated as sag-.
Materials
-
Gestant female mouse
-
Bouin's fixative (e.g., Fischer Scientific, cat. no. 001401)
-
70%, 95%, and 100% ethanol (e.g., Fischer Scientific, cat. no. 10041814)
-
Xylene
-
Paraffin wax (e.g., CliniSciences, cat. no. 4810025)
-
Dissection instruments:
- Surgical scissors (e.g., Dominique Dutscher, cat. no. 2492680)
- Extra-thin straight scissors (e.g., Dominique Dutscher, cat. no. 2522789)
- Short forceps (e.g., Dominique Dutscher, cat. no. 2522705)
- Long forceps (e.g., Dominique Dutscher, cat. no. 2522689)
- Surgical blade (e.g., Braun Medical AS, cat. no. 16600584)
- Forceps, #5 (e.g., Sigma-Aldrich, cat. no. F6521-1A)
- Scalpel blades (e.g., Sigma-Aldrich, cat. no. S2771-100EA)
-
Precision balance (e.g., Ohaus 092567)
-
15-ml conical tubes (e.g., Fisher Scientific, cat. no. 352097)
-
Cassette printer (e.g., Epredia PrintMate AS, model no. A83800001)
-
Small histological cassettes (e.g., VWR, cat. no. 720-2192)
-
Weigh boat (e.g., Fischer Scientific, cat. no. HS1420CC)
-
Brightfield microscope
-
Glass bottles
-
Tissue processor (e.g., Leica, model no. ASP300)
-
Tissue-embedding station (e.g., Leica, model no. EG1160)
-
Reusable metal base molds, 15 × 15 (e.g., Leica, cat. no. 3803081)
-
Additional reagents and equipment for mouse euthanasia (see Current Protocols article: Donovan & Brown, 2005)
NOTE : All protocols using live animals must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of laboratory animals (see Karp et al., 2015).
NOTE : The experimenter must have a good knowledge of embryonic brain anatomy to be able to identify whether a section is in the correct stereotaxic position and symmetrical both along the dorsoventral and rostrocaudal orientations.
Fix mouse embryos
1.Euthanize gestant female mouse at E18.5 (1 day before birth) using an approved method.
2.Remove fetus from the embryonic sac, and keep embryo cold on ice with a paper towel or other barriers between the embryo and the ice. If the head and rest of the body are required for independent experiments, harvest and process the head separately (Fig. 1A,B), instead of processing the full body, following the same steps below (except for steps 4 and 11, which are no longer required).
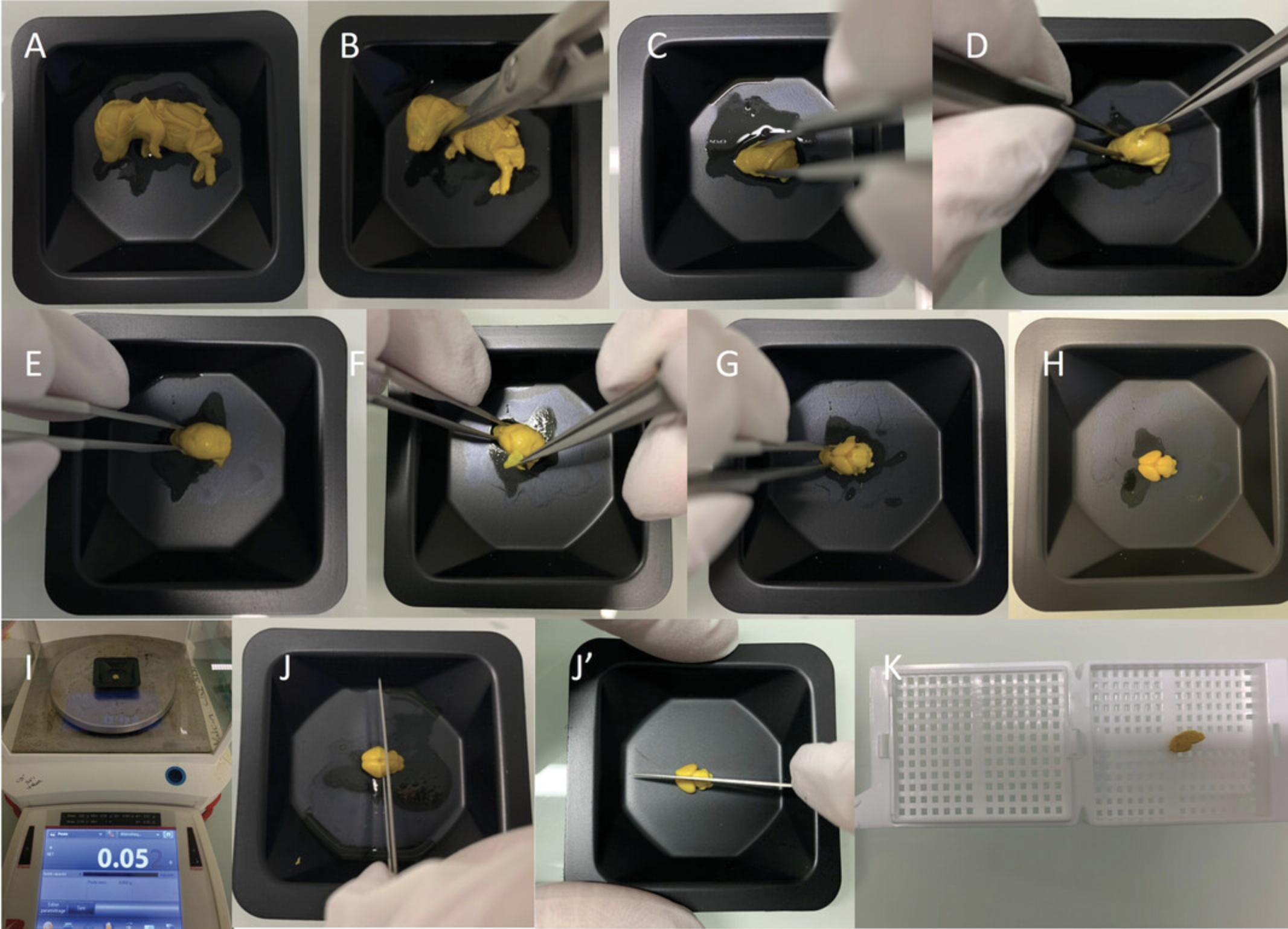
3.Weigh embryo and record weight.
4.Kill embryo by making a cut between the brain and spinal cord junction.
5.Repeat steps 2 to 4 for each embryo.
6.Cut a piece of embryonic tail at the tip for genotyping and sexing purposes.
7.Fix embryos in a 15-ml conical tube containing 10 ml Bouin's fixative for exactly 48 hr at room temperature away from direct sunlight. To allow better absorption of the fixative, pinch skin and make a lateral incision in the neck with surgical scissors.
8.Transfer embryos to a new 15-ml conical tube containing 10 ml of 70% ethanol for a few days.
Dissect embryonic mouse brain samples
9.Print a unique barcode on small histological cassettes using an Inkjet cassette printer to uniquely label individual brain samples.
10.Place fixed embryos into a weigh boat (Fig. 1A).
11.Cut neck close to the base using a pair of surgical scissors to harvest the head of the embryo (Fig. 1B).
12.Carefully stabilize head of the embryo using short forceps, pinching laterally at the tip of the head without touching the actual brain tissue (Fig. 1C).
13.Carefully peel away the calvaria, and gently pull the skin and meninges away from the brain using the tip of #5 forceps (Fig. 1D-G).
14.Recover intact embryonic brain sample (Fig. 1H), and take a high-quality picture for future records.
15.Weigh brain using a precision balance and record data (Fig. 1I).
16.Trim embryonic brain sample:
-
cor- Trim through Bregma first, which is just above the longitudinal cerebral fissure (Fig.1J), producing two chunks (anterior and posterior). Then cut the anterior chunk coronally into two parts of equal size.
-
sag- Trim along the median sagittal suture using a scalpel (Fig.1J') to obtain two halves of tissue (right and left hemispheres).
17.Put each trimmed brain tissue into its corresponding preprinted histological cassette prepared in step 9 (Fig. 1K).
18.Place cassettes in a glass bottle with lid filled with 70% ethanol.
Perform dehydration and paraffin embedding
19.Transfer histological cassettes containing the trimmed embryonic brain tissues to the basket of the tissue processor.
20.Set paraffin bath of the tissue processor to 59°C.
21.Place basket in the processor, and begin the paraffin infiltration process, including ethanol, xylene, and paraffin (see Table 1 for incubation steps).
| Solution | Time, min | Temperature, °C |
|---|---|---|
| 70% ethanol | 30 | 35 |
| 95% ethanol | 30 | 35 |
| 95% ethanol | 60 | 35 |
| 95% ethanol | 60 | 35 |
| 100% ethanol | 30 | 35 |
| 100% ethanol | 60 | 35 |
| 100% ethanol | 60 | 35 |
| Xylene | 30 | 35 |
| Xylene | 60 | 35 |
| Xylene | 60 | 35 |
| Paraffin | 120 | 59 |
| Paraffin | 120 | 59 |
| Paraffin | 180 | 59 |
22.Remove tissue basket when the program ends, and empty basket containing cassettes into the paraffin bath incorporated on the surface of the modular tissue-embedding system (Fig. 2A).

23.Remove brain tissue from each cassette, and place individual chunk in the center of a metal base mold.
24.Fill mold with hot paraffin (preheated to 59°C) up to the internal rounded corners of the mold (Fig. 2B).
25.Correct the orientation of the tissue in the mold using forceps:
-
cor- Place forebrain with the posterior side facing down, and place midbrain and hindbrain chunks with the anterior side facing down.
-
sag- Orient left and right halves so that the cerebellum faces toward the printed side of the cassette.
The correct orientation of the tissue in the cassette is important for minimization of tissue loss during sectioning and for maximization of storage for future experiments.
26.Place base (labeled half) of the cassette at the brim of the mold.
27.Place mold on the cold plate at −5°C, and let paraffin cool down (Fig. 2C).
28.Remove formed paraffin block from the mold, and store at room temperature until sectioning (Fig. 2D).
Basic Protocol 2: SECTIONING, STAINING, AND SCANNING OF EMBRYONIC MOUSE BRAIN SECTIONS
In this protocol we describe the steps involved in (1) sectioning embryonic brain tissue embedded in paraffin blocks using a sliding microtome; (2) staining of slides using Nissl stain; and (3) scanning of the histological slides using a NanoZoomer whole-slide scanner. We highly recommend using the NanoZoomer scanner to produce cell-resolution digital images in a high-throughput setting, although other whole-slide scanners or motorized XY microscopes capable of imaging could be used if the NanoZoomer is not available at your institution. The sectioning process relies on a set of well-defined anatomical landmarks in the embryonic mouse brain that correspond to previously defined positions in the adult mouse brain both on coronal and sagittal planes (see Current Protocols articles: Collins et al., 2018; Mikhaleva et al., 2016). The sagittal plane allows for simultaneously testing three main regions involved in cognition: cortex, hippocampus, and cerebellum. The coronal plane permits the study of the right and left hemispheres. The benefits of each plane are described in more detail in a Current Protocols article by Collins et al. (2018). The staining process relies on neuronal Nissl bodies of the endoplasmic reticulum, stained purple using cresyl violet, to identify neuronal structures in the embryonic brain. It is worthwhile to mention that the main development of myelination in the mouse brain starts during the first postnatal months. It is thus not necessary to stain myelin with Luxol as done in our previous protocols adapted for the adult mouse (Collins et al., 2018; Mikhaleva et al., 2016). The scanning process generates two formats—TIFF (Tagged Image File Format) and NDPI (NanoZoomer Digital Pathology Image). However, we recommend the use of the nonproprietary TIFF format throughout the protocol for data collection (Basic Protocols 3 and 4).
Materials
-
Peptone water (see recipe)
-
Paraffin-embedded embryonic brain tissue (see Basic Protocol 1)
-
0.1% (w/v) cresyl violet acetate (see recipe)
-
Xylene (e.g., VWR, cat. no. 28073.328)
-
70%, 90%, 95%, and 100% ethanol (e.g., Fischer Scientific, cat. no. 10041814)
-
Pertex mounting medium (e.g., Biosystems Switzerland AG)
-
Microtome (e.g., Epredia HM 450)
-
Microtome blades (e.g., Dura Edge, cat. no. 7223)
-
Stereomicroscope (e.g., Leica MZ6 or Zeiss Stemi 508) with vertical incident optic illumination (e.g., SCHOTT KL Series, model no. KL1500) and optional camera and HDMI screen
-
Hot plate (e.g., Slimline Slide Drying Hotplate)
-
Frosted glass slides (e.g., Thermo Fisher Scientific, cat. no. ISO 8037/1)
-
Staining racks (e.g., Electron Microscopy Sciences, cat. no. 70321-10)
-
37°C incubator (e.g., BODE Science Center, model no. 159905)
-
56°C water bath (e.g., Memmert, model no. WNB 29)
-
Glass staining dishes with lids (e.g., Electron Microscopy Sciences, cat. no. 71424-DL)
-
Automated slide staining system (e.g., Leica, model no. XL ST5010)
-
Filter paper (e.g., VWR, cat. no. 516 0838)
-
Glass coverslips (e.g., Leica, cat. no. 3800100)
-
Slide organizer for microscope slides (e.g., VWR, cat. no. 631-1069)
-
Permanent marker
-
Hamamatsu NanoZoomer digital slide scanner, 2.0HT, C9600 series, and associated software (NanoZoomer digital pathology, version 2.5.64)
Section tissue
1.Prepare for sectioning (Fig. 3A):
-
Switch on microtome, stereomicroscope camera, and monitor.
-
Bring peptone water to room temperature.
-
Set hot plate to 42°C.
-
Using a pencil, label frosted glass slide with the corresponding unique barcode.
-
Place a drop of peptone water on the slide.

2.Chill paraffin block on wet ice for a couple of minutes before sectioning.
3.Mount paraffin block into the microtome specimen holder (Fig. 3B):
-
cor- Sequentially mount the three coronal blocks.
-
sag- Mount right hemisphere block.
4.Orient paraffin block so the microtome blade will cut straight across the block.
5.Trim block before sectioning using the trimming function of the microtome, set to 20-µm slices.
6.Ensure positioning is correct, and fix any asymmetry detected while sectioning before reaching the section of interest.
7.Determine how close, in μm, the observed section is to the targeted section by comparing the image with the reference atlas of the developing mouse brain at E18.5 (Paxinos et al., 2007; also see https://developingmouse.brain-map.org/static/atlas).
8.Slow down about 100 µm before the section to be collected, cutting 5-µm thick slices (corresponding to one-cell thickness, a parameter allowing single-cell resolution) as the section of interest approaches (see Critical Parameters and Troubleshooting).
9.Place a drop of peptone water on a glass slide, and place slide on the heated part of the hot plate set at 42°C.
10.Retrieve section of interest, and place on top of the water drop on the glass slide.
11.Allow section to spread over the area of the glass containing the water for about 1 minute (Fig. 3C).
12.Repeat steps 7 to 11 so that two consecutive sections of interest are mounted on the slide.
13.Place slide in a slide holder, and place holder in an oven to let dry overnight at 37°C (Fig. 3D).
Stain with cresyl violet
14.Set water bath to 56°C, and place closed glass staining dish containing 0.1% cresyl violet acetate in the water bath.
15.Take dried slide out of the incubator (Fig. 3E).
16.Place slide in the automatic staining station (Fig. 3F) for paraffin removal following the incubation steps with xylene, ethanol, and water as indicated in Table 2.
| Solution | Time, min | Temperature |
|---|---|---|
| Xylene | 3 | Room temperature |
| Xylene | 3 | Room temperature |
| 100% ethanol | 3 | Room temperature |
| 90% ethanol | 3 | Room temperature |
| 70% ethanol | 3 | Room temperature |
| Distilled water | 1 | Room temperature |
17.Place slide in the dish containing warm 0.1% cresyl violet acetate for 1 min.
18.Wash slide in distilled water for a few seconds to remove excess cresyl violet.
19.Remove excess distilled water. Place slide on filter paper, and let dry at room temperature for 15 min.
20.Mount slide with a drop of Pertex mounting medium, and lower coverslip on the slide at an angle, working carefully to avoid the formation of air bubbles. Dry slide overnight at room temperature (Fig. 3G).
21.Place slide in a storage box, and proceed with scanning.
Scan and process images
22.Observe slide under the microscope, and mark the section of interest with a cross using a permanent marker.
23.Clean glass slide with 70% ethanol using tissue wipes.
24.Switch on NanoZoomer scanner (Fig. 3H) and associated computer, and load slide into the scanner rack.
25.Choose "Brightfield" scanning profile with 20× magnification, and configure output settings to save data as uncompressed TIFF files.
26.Choose folder where all digital files should be saved.
27.Start batch scanning.
28.Transfer images to a server with automatic backup, and conduct image analyses.
Basic Protocol 3: CORONAL NEUROANATOMICAL MEASUREMENTS OF EMBRYONIC MOUSE BRAIN STRUCTURES
Our histology pipeline on the coronal plane is designed based on three sections of interest at Bregma +2.19 mm, Bregma +3.51 mm, and Bregma +6.75 mm, spanning 28 developmentally distinct brain regions of E18.5 mouse embryos (Fig. 4). Some parameters can be taken twice per section, on the right and left hemispheres, totaling 66 neuroanatomical measurements including 19 at Bregma +2.19 mm (Table 3), 29 at Bregma +3.51 mm (Table 4), and 18 at Bregma +6.75 mm (Table 5). Some parameters are common across the three sections, and some are specific to the section of interest. Parameters involve measurements of area and length. For large-scale projects, we recommend a well-designed database to accommodate the large amount of E18.5 morphometric data as described in detail in our previous papers (see Current Protocols articles: Collins et al., 2018; Mikhaleva et al., 2016). Each image is checked for quality control before analysis. Staff should be trained and competent to perform the measurements. Once staff are fully trained, it takes an estimated 20 to 30 min per section to make the measurements and another 50 to 60 min to verify the measurements, assess the quality of the measures, and rectify any potential human errors.
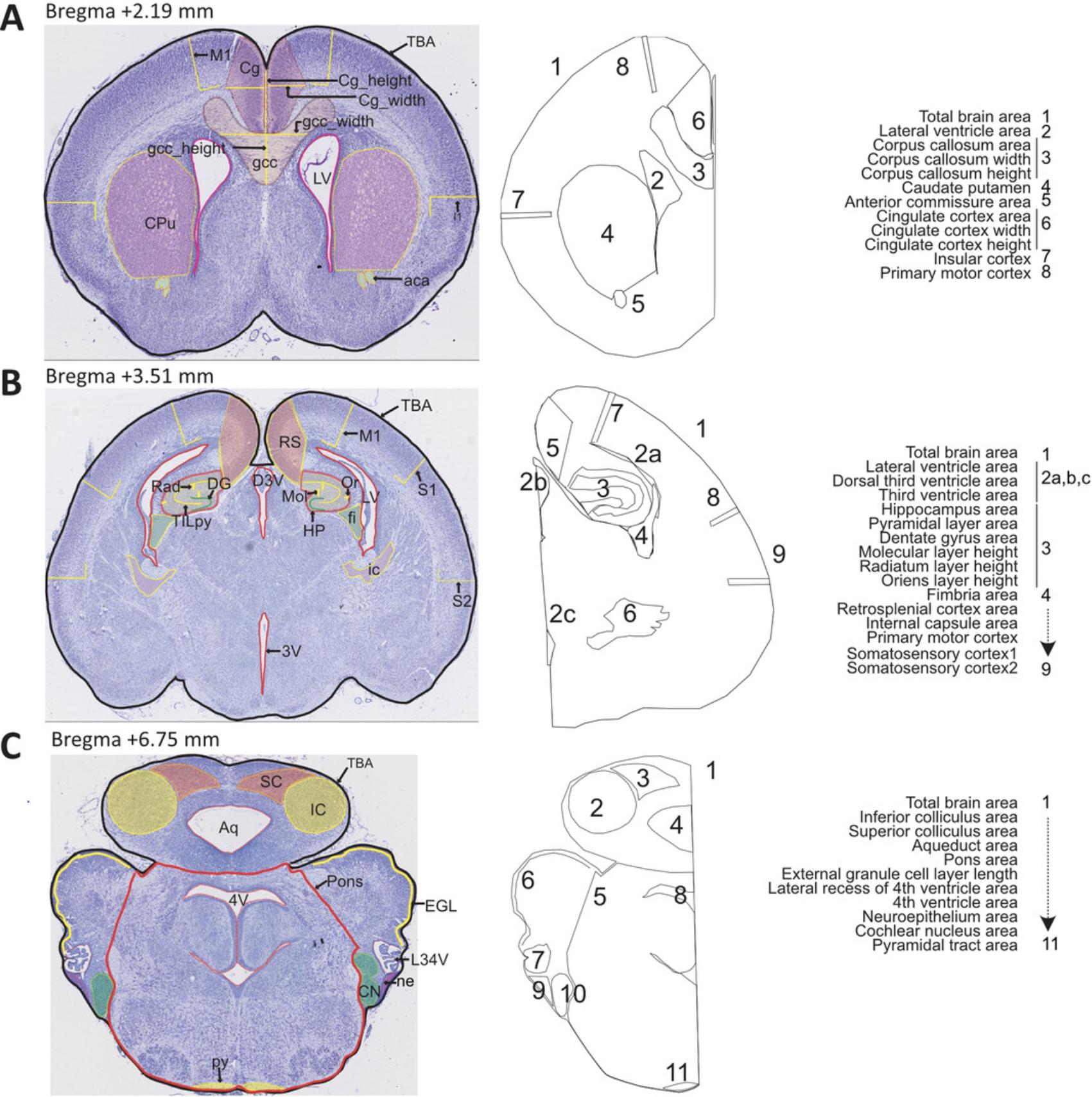
| Neuroanatomical parameter | Brief description | Measurement unit |
|---|---|---|
| TBA | Total brain area | cm² |
| LV_L | Area of the lateral ventricle, left hemisphere | cm² |
| LV_R | Area of the lateral ventricle, right hemisphere | cm² |
| Gcc | Area of the genu of the corpus callosum | cm² |
| gcc_width | Width of the genu of the corpus callosum | cm |
| gcc_height | Height of the genu of the corpus callosum | cm |
| CPu_L | Area of the caudate putamen, left hemisphere | cm² |
| CPu_R | Area of the caudate putamen, right hemisphere | cm² |
| aca_L | Area of the anterior commissure, left hemisphere | cm² |
| aca_R | Area of anterior commissure, right hemisphere | cm² |
| Cg_L | Area of the cingulate cortex, left hemisphere | cm² |
| Cg_R | Area of the cingulate cortex, right hemisphere | cm² |
| Cg_width_L | Width of the cingulate cortex, left hemisphere | cm |
| Cg_width_R | Width of the cingulate cortex, right hemisphere | cm |
| Cg_height | Height of the cingulate cortex | cm |
| I_L | Height of the insular cortex, left hemisphere | cm |
| I_R | Height of the insular cortex, right hemisphere | cm |
| M1_L | Height of the primary motor cortex, left hemisphere | cm |
| M1_R | Height of the primary motor cortex, right hemisphere | cm |
| Neuroanatomical parameter | Brief description | Measurement unit |
|---|---|---|
| TBA | Total brain area | cm² |
| LV_L | Area of the lateral ventricle, left hemisphere | cm² |
| LV_R | Area of the lateral ventricle, right hemisphere | cm² |
| D3V | Area of the dorsal third ventricle | cm² |
| 3V | Area of the third ventricle | cm² |
| HP_L | Area of the hippocampus, left hemisphere | cm² |
| HP_R | Area of the hippocampus, right hemisphere | cm² |
| TILpy_L | Total internal length of the pyramidal layer of the hippocampus, left hemisphere | cm |
| TILpy_R | Total internal length of the pyramidal layer of the hippocampus, right hemisphere | cm |
| DG_L | Length of the dentate gyrus, left hemisphere | cm |
| DG_R | Length of the dentate gyrus, right hemisphere | cm |
| Mol_L | Height of the molecular layer of the hippocampus, left hemisphere | cm |
| Mol_R | Height of the molecular layer of the hippocampus, right hemisphere | cm |
| Rad_L | Height of the stratum radiatum of the hippocampus, left hemisphere | cm |
| Rad_R | Height of the stratum radiatum of the hippocampus, right hemisphere | cm |
| Or_L | Height of the stratum oriens of the hippocampus, left hemisphere | cm |
| Or_R | Height of the stratum oriens of the hippocampus, right hemisphere | cm |
| fi_L | Area of the fimbria of the hippocampus, left hemisphere | cm² |
| fi_R | Area of the fimbria of the hippocampus, right hemisphere | cm² |
| RS_L | Area of the retrosplenial granular cortex, left hemisphere | cm² |
| RS_R | Area of the retrosplenial granular cortex, right hemisphere | cm² |
| ic_L | Area of internal capsule, left hemisphere | cm² |
| ic_R | Area of internal capsule, right hemisphere | cm² |
| M1_L | Height of the primary motor cortex, left hemisphere | cm |
| M1_R | Height of the primary motor cortex, right hemisphere | cm |
| S1_L | Height of the primary somatosensory cortex, left hemisphere | cm |
| S1_R | Height of the primary somatosensory cortex, right hemisphere | cm |
| S2_L | Height of the secondary somatosensory cortex, left hemisphere | cm |
| S2_R | Height of the secondary somatosensory cortex, right hemisphere | cm |
| Neuroanatomical parameter | Brief description | Measurement unit |
|---|---|---|
| TBA | Total brain area | cm² |
| IC_L | Area of the inferior colliculus, left hemisphere | cm² |
| IC_R | Area of the inferior colliculus, right hemisphere | cm² |
| SC_L | Area of the superior colliculus, left hemisphere | cm² |
| SC_R | Area of the superior colliculus, right hemisphere | cm² |
| Aq | Area of the aqueduct | cm² |
| Pons | Area of the pons | cm² |
| EGL_L | Length of the external granule cell layer, left hemisphere | cm |
| EGL_R | Length of the external granule cell layer, right hemisphere | cm |
| LR4V_L | Area of the lateral recess of the fourth ventricle, left hemisphere | cm² |
| LR4V_R | Area of the lateral recess of the fourth ventricle, right hemisphere | cm² |
| 4V | Area of the fourth ventricle | cm² |
| ne_L | Area of the neuroepithelium, left hemisphere | cm² |
| ne_R | Area of the neuroepithelium, right hemisphere | cm² |
| CN_L | Area of the cochlear nucleus, left hemisphere | cm² |
| CN_R | Area of the cochlear nucleus, right hemisphere | cm² |
| py_L | Area of the pyramidal tract, left hemisphere | cm² |
| py_R | Area of the pyramidal tract, right hemisphere | cm² |
Materials
- Computer running ImageJ software (version 1.53c) and the following specifications: Windows 11, 64 bits, Processor 8M, 16 GB RAM, and appropriate graphic card such as NVIDIA GeForce RTX 2060
- Images of tissue of interest (see Basic Protocol 2)
Prepare computer
1.If needed, download ImageJ software (http://imagej.nih.gov/ij/; freely available).
2.Set up ImageJ settings as follows: 9 decimal places, “cm” for unit of length.
Measure stepwise each of the 19 coronal parameters at Bregma +2.19 mm
See Figure 4A and Table 3 for regions of interest. Make area measurements using the Polygon selection tool, length measurements using the Segmented lines selection tool, and width or height measurements using the Straight lines selection tool with the Shift button pressed for vertical or horizontal lines.
Use a two-letter code to fail any measurements that cannot be taken: NC for not critical, when the measurement lies within a part of the section that is not located at the correct stereotaxic position; NM for not measurable, when the section is too badly damaged and prevents making the measurement; NS for not symmetrical, when the measurement lies within a part of the section that is not symmetrical; and NV for not visible, when the experimenter is unable to locate the brain structure due to, for example, poor staining.
3.Measure total brain area (TBA), delineating the entire section.
4.Delineate area of the lateral ventricle on the left (LV_L) and right (LV_R) hemispheres.
5.Measure area, width, and height of the genu of the corpus callosum (cc, gcc_width, and gcc_height, respectively) at the midline.
6.Measure area of the caudate putamen on the left (CPu_L) and right (CPu_R) hemispheres.
7.Measure area of the anterior commissure on the left (aca_L) and right (aca_R) hemispheres.
8.Measure area of the cingulate cortex on the left (Cg_L) and right (Cg_R) hemispheres. In addition, measure width of the cingulate cortex on the left (Cg_width_L) and right (Cg_width_R) hemispheres, as well as the height of the cingulate cortex (Cg_height).
9.Measure height of the insular cortex on the left (I_L) and right (I_R) hemispheres.
10.Measure height of the primary motor cortex on the left (M1_L) and right (M1_R) hemispheres.
Measure stepwise each of the 29 coronal parameters at Bregma +3.51 mm
See Figure 4B and Table 4 for regions of interest.
11.Delineate total brain area (TBA).
12.Delineate area of the lateral ventricle on the left (LV_L) and right (LV_R) hemispheres, as well as the area of the dorsal third ventricle (D3V) and third ventricle (3V) along the midline.
13.Measure area of the hippocampal formation on the left (HP_L) and right (HP_R) hemispheres, as well as total internal length of the pyramidal layer on the left (TILpy_L) and right (TILpy_R) hemispheres. In addition, measure length of the dentate gyrus on the left (DG_L) and right (DG_R) hemispheres, height of the molecular layer on the left (Mol_L) and right (Mol_R) hemispheres, height of the stratum radiatum on the left (Rad_L) and right (Rad_R) hemispheres, and height of the stratum oriens on the left (Or_L) and right (Or_R) hemispheres.
14.Measure area of the fimbria of the hippocampus on the left (fi_L) and right (fi_R) hemispheres.
15.Measure area of the retrosplenial granular cortex on the left (RS_L) and right (RS_R) hemispheres.
16.Measure area of the internal capsule on the left (ic_L) and right (ic_R) hemispheres.
17.Measure height of the primary motor cortex on the left (M1_L) and right (M1_R) hemispheres.
18.Measure height of the primary somatosensory cortex on the left (S1_L) and right (S1_R) hemispheres.
19.Measure height of the secondary somatosensory cortex on the left (S2_L) and right (S2_R) hemispheres.
Measure stepwise each of the 18 coronal parameters at Bregma +6.75 mm
See Figure 4C and Table 5 for regions of interest.
20.Delineate total brain area (TBA).
21.Measure area of the inferior colliculus on the left (IC_L) and right (IC_R) hemispheres.
22.Measure area of the superior colliculus on the left (SC_L) and right (SC_R) hemispheres.
23.Measure area of the aqueduct (Aq).
24.Delineate area of the pons (Pons).
25.Take length of the external granule cell layer of the cerebellum on the left (EGL_L) and right (EGL_R) hemispheres.
26.Measure area of the lateral recess of the fourth ventricle on the left (LR4V_L) and right (LR4V_R) hemispheres.
27.Measure area of the fourth ventricle (4V).
28.Measure area of the neuroepithelium on the left (ne_L) and right (ne_R) hemispheres.
29.Measure area of the cochlear nucleus on the left (CN_L) and right (CN_R) hemispheres.
30.Measure area of the pyramidal tract on the left (py_L) and right (py_R) hemispheres.
Assess quality of data collected and perform downstream analysis
31.Check data collected for the presence of outliers, typographical errors, and any other erroneous measurements, and correct measurement when necessary.
32.Ask project manager to reveal the genotype.
33.Check data distribution within each genotype group, and retake measurements if necessary.
34.Produce final set of verified raw data.
35.Proceed with statistical analysis (see Number and Statistical Analysis).
Basic Protocol 4: SAGITTAL NEUROANATOMICAL MEASUREMENTS OF EMBRYONIC MOUSE BRAIN STRUCTURES
The main benefit of using one single section (e.g., Lateral +0.60 mm) on the sagittal plane is to reduce time to determine whether there is a presence or absence of brain structural changes. Time being one important factor in large-scale neuroanatomical studies of embryonic brain samples, we recommend the sagittal plane for projects involving thousands of samples. For such projects, we recommend a relational database to accommodate large amount of morphometric data, multiple quality control steps, and statistical analysis, as described in detail in our previous papers (see Current Protocols articles: Collins et al., 2018; Mikhaleva et al., 2016). Each image is checked for quality control before analysis. Staff should be trained and competent to perform the measurements. Once staff are fully trained, it takes an estimated 20 to 30 min per image to make the measurements and another 50 to 60 min to verify the measurements, assess the quality of the measures, and rectify any potential human errors. The sagittal measurements are based on 40 well-defined neuroanatomical brain parameters on the sagittal plane spanning 21 developmentally distinct brain regions of E18.5 mouse embryos (Fig. 5). Overall, 15 of the 40 parameters correspond to length measurements (Fig. 5; indicated in red) and the remaining 25 to area measurements (Fig. 5; indicated in black). The full list of measurement abbreviations is provided in Table 6, together with a short description of the measurement and its associated unit in cm2 or cm.
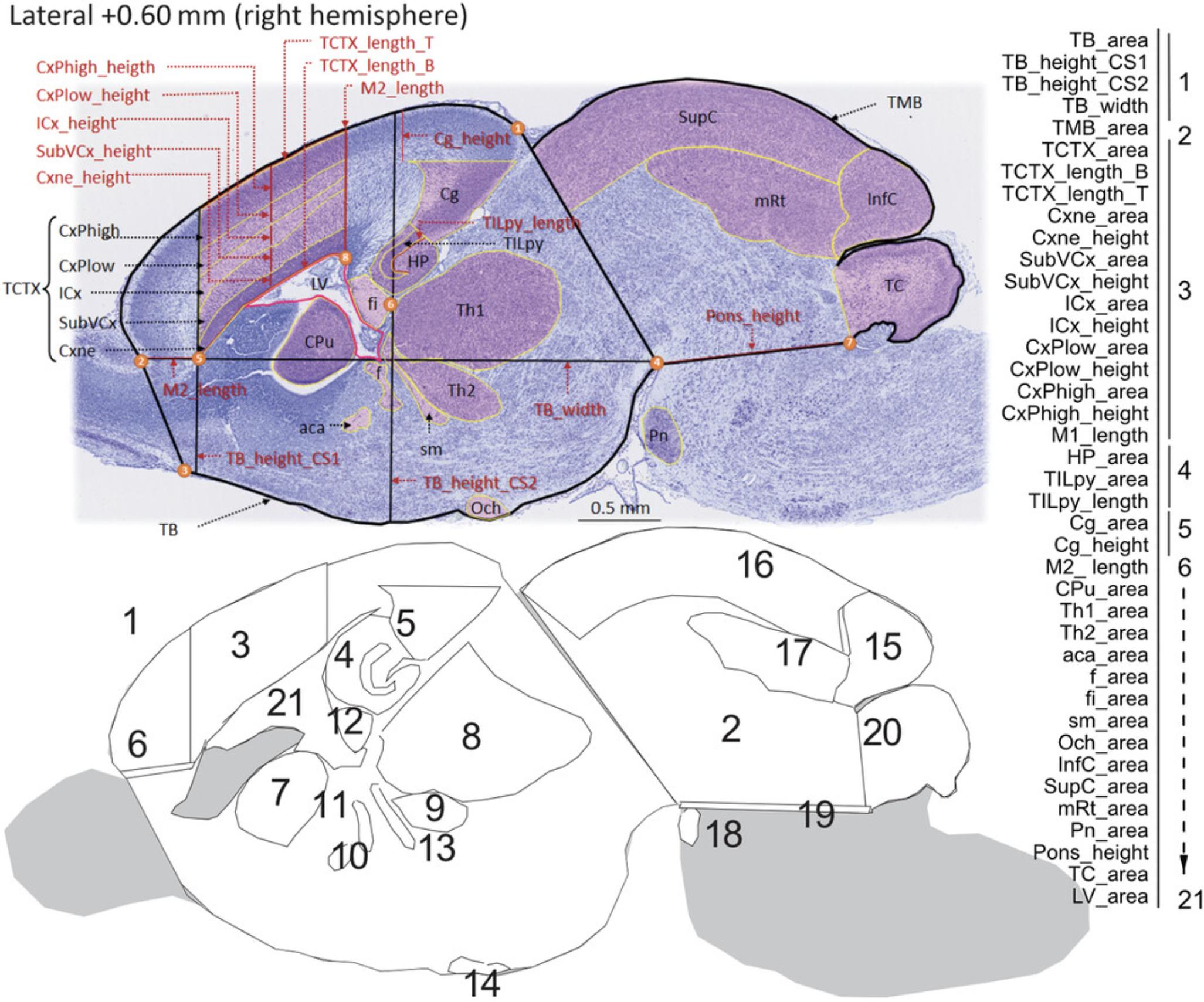
| Neuroanatomical parameter | Brief description | Measurement unit |
|---|---|---|
| TB_area | Total area of the brain | cm2 |
| TB_height_CS1 | Height of the total brain (matching section 1 at Bregma +2.19 mm) | cm |
| TB_height_CS2 | Height of the total brain (matching section 2 at Bregma +3.51 mm) | cm |
| TB_width | Width of the brain | cm |
| TMB_area | Total area of the midbrain | cm2 |
| TCTX_area | Total area of the cortical plate | cm2 |
| TCTX_length_B | Top length of the cortical area | cm |
| TCTX_length_T | Bottom length of the cortical area | cm |
| Cxne_area | Area of the neuroepithelial cortical layer | cm2 |
| Cxne_height | Height of the neuroepithelial cortical layer | cm |
| SubVCx_area | Area of the subventricular cortical layer | cm2 |
| SubVCx_height | Height of the subventricular cortical layer | cm |
| ICx_area | Area of the intermediate cortical layer | cm2 |
| ICx_height | Height of the intermediate cortical layer | cm |
| CxPlow_area | Area of the lower layer of the cortical plate | cm2 |
| CxPlow_height | Height of the lower layer of the cortical plate | cm |
| CxPhigh_area | Area of the upper layer of the cortical plate | cm2 |
| CxPhigh_height | Height of the upper layer of the cortical plate | cm |
| M1_length | Length of the primary motor cortex | cm |
| HP_area | Area of the hippocampal formation | cm2 |
| TILpy_area | Total internal area of pyramidal cells of the hippocampal formation | cm2 |
| TILpy_length | Total internal length of pyramidal cells of the hippocampal formation | cm |
| Cg_area | Area of the cingulate cortex | cm2 |
| Cg_height | Height of the cingulate cortex | cm |
| M2_length | Length of the secondary motor cortex | cm |
| CPu_area | Area of the caudate putamen | cm2 |
| Th1_area | Area of thalamic nucleus 1 | cm2 |
| Th2_area | Area of thalamic nucleus 2 | cm2 |
| aca_area | Area of the anterior commissure | cm2 |
| f_area | Area of the fornix | cm2 |
| fi_area | Area of the fimbria of the hippocampus | cm2 |
| sm_area | Area of the stria medullaris | cm2 |
| Och_area | Area of the optic chiasm | cm2 |
| InfC_area | Area of the inferior colliculus | cm2 |
| SupC_area | Area of the superior colliculus | cm2 |
| mRt_area | Area of the mesencephalic reticular formation | cm2 |
| Pn_area | Area of the pontine nuclei | cm2 |
| Pons_height | Height of the pons | cm |
| TC_area | Total area of the cerebellum | cm2 |
| LV_area | Area of the lateral ventricle | cm2 |
Materials
- Computer running ImageJ software (version 1.53c) and the following specifications: Windows 11, 64 bits, Processor 8M, 16 GB RAM, and appropriate graphic card such as NVIDIA GeForce RTX 2060
- Images of tissue of interest (see Basic Protocol 2)
Prepare computer
1.If needed, download ImageJ software (http://imagej.nih.gov/ij/; freely available).
2.Set up ImageJ settings as follows: 9 decimal places, “cm” for unit of length.
Measure stepwise each of the 40 sagittal parameters at Lateral +0.60 mm
See Figure 5 and Table 6 for regions of interest. Make area measurements using the Polygon selection tool, length measurements using the Segmented lines selection tool, and width or height measurements using the Straight lines selection tool with the Shift button pressed for vertical or horizontal lines.
Use a two-letter code to fail any measurements that cannot be taken: NC for not critical, when the measurement lies within a part of the section that is not located at the correct stereotaxic position; NM for not measurable, when the section is too badly damaged and prevents making the measurement; NP for not parallel, when the measurement lies within a part of the section that is not parallel to the sagittal suture; and NV for not visible, when the experimenter is unable to locate the brain structure due to, for example, poor staining.
3.Measure total brain area (TB_area) using the Polygon selection tool, starting the drawing from the apex of the cingulate cortex (landmark 1 in Fig. 5), moving along the whole cortical plate until reaching the first junction between the olfactory bulb and the rest of the brain (landmark 2 in Fig. 5). Continue drawing toward the point where the olfactory tubercle appears (landmark 3 in Fig. 5), and follow the ventral side of the brain including the area of the optic chiasm until reaching the junction between the hindbrain and the rest of the brain (landmark 4 in Fig. 5), more precisely at the apex of the pontine nuclei. End the drawing by rejoining the starting point (landmark 1 in Fig. 5).
4.Take height of the forebrain at a position corresponding to Bregma +2.19 mm (TB_height_CS1), drawing a straight vertical line using the left-hand-side sharp edge of the neuroepithelial zone area (Cxne) as a landmark (5 in Fig. 5) where the vertical line should pass over.
5.Take height of the midbrain at a position corresponding to Bregma +3.51 mm (TB_height_CS2), drawing a straight vertical line using the far-right edge of the fimbria (fi) of the hippocampus as a landmark (6 in Fig. 5) where the line should pass over.
6.Take width (TB_width) of the brain area starting from the junction between the olfactory bulb and the rest of the brain (landmark 1 in Fig. 5 used to draw TB_area) and ending at the point between the hindbrain and the rest of the brain at the apex of the pontine nuclei (landmark 4 in Fig. 5 used to draw TB_area).
7.Measure total midbrain area (TMB_area) superimposing a line over the same line drawn to delineate the total brain area on the right part, from just below landmark 1 to 4 in Figure 5.Continue drawing to reach the point corresponding to the lower base of the developing cerebellum (landmark 7 in Fig. 5), and delineate the entire cerebellum and midbrain until reaching the starting point just below landmark 1.
8.Measure total area of the cortical plate (TCTX_area) by drawing a straight vertical line using the apex of the lateral ventricle (LV) as a landmark (8 in Fig. 5) reaching the cortical plate upward and then moving along the whole cortical plate until reaching the line used to draw TB_height_CS1, following it downward until reaching the left-hand-side edge of the neuroepithelial zone area (Cxne) at landmark 5 before joining with the starting point at landmark 8 in Figure 5.
9.Take length of the bottom line that delineates the total area of the cortical plate (TCTX_length_B).
10.Take length of the top line that delineates the total area of the cortical plate (TCTX_length_T).
11.Measure area (Cxne_area) and height (Cxne_height) of the neuroepithelial zone of the cortex.
12.Measure area (SubVCx_area) and height (SubVCx_height) of the subventricular layer of the cortex, which is the layer located just above the neuroepithelial zone.
13.Measure area (ICx_area) and height (ICx_height) of the intermediate layer of the cortex, which is the layer adjacent to the subventricular layer.
14.Measure area (CxPlow_area) and height (CxPlow _height) of the lower layer of the cortical plate.
15.Measure area (CxPhigh_area) and height (CxPhigh _height) of the upper layer of the cortical plate.
16.Measure length of the primary motor cortex (M1_length), which we define as the sum of the height of the various layers of the cortex at the far right-hand-side of the TCTX_area.
17.Delineate total area of the hippocampus (HP_area) drawing a line around the hippocampal formation.
18.Measure total area occupied by the pyramidal cells of the hippocampal formation (TILpy_area), as well as the length of the lower line that delineates the area of the pyramidal cells (TILpy_length).
19.Measure area (Cg_area) and length (Cg_length) of the developing cingulate cortex by drawing a vertical line from the left-hand-side apex of the Cg_area measurement upward until reaching the TB_area lining.
20.Measure length of the motor cortex that partially corresponds to the secondary motor cortex (M2_length) by joining two points previously defined as the junction between the olfactory bulb and the rest of the brain and the point making the left-hand-side edge of the neuroepithelial zone area.
21.Delineate area of the caudate putamen (CPu_area) drawing a line around the developing striatum.
22.Delineate area of thalamic nucleus 1 (Th1_area) and nucleus 2 (Th2_area) drawing independently around each nucleus.
23.Delineate area of the anterior commissure (aca_area).
24.Delineate area of the fornix (f_area).
25.Delineate area of the fimbria (fi_area) of the hippocampus located immediately below the hippocampus.
26.Delineate area of the stria medullaris (sm_area) located below the thalamus.
27.Delineate area of the optic chiasm (Och_area).
28.Delineate area of the inferior colliculus (InfC_area).
29.Delineate area of the superior colliculus (SupC_area).
30.Delineate area of the mesencephalic reticular formation (mRt_area).
31.Delineate area of the pontine nuclei (Pn_area).
32.Measure height of the pons (Pons_height) by joining the reference point between the hindbrain and the rest of the brain and the point delineating the upper base of the developing cerebellum.
33.Delineate total area of the developing cerebellum (TC_area).
34.Delineate total area of the lateral ventricle (LV_area).
Assess quality of data collected and perform downstream analysis
35.Check data collected for the presence of outliers, typographical errors, and any other erroneous measurements, and correct measurement if necessary.
36.Ask project manager to reveal the genotype.
37.Check data distribution within each genotype group, and correct measurement if necessary.
38.Produce final set of verified raw data.
39.Proceed with statistical analysis (see Number and Statistical Analysis).
REAGENTS AND SOLUTIONS
Cresyl violet acetate, 0.1%
Dissolve 0.4 g cresyl violet acetate (e.g., Sigma-Aldrich, cat. no. C5042) in 400 ml deionized water. Add 4 ml of 100% glacial acetic acid (1% final; e.g., Merck, cat. no. 100063). Stir with a magnetic stirrer overnight, and filter through a 0.45-µm vacuum filter (e.g., Chromoptic, cat. no. 30162087) into a bottle covered with aluminum foil to protect from light. Store at room temperature for up to 2 years.
Peptone water
Add a few pellets (30 mg) of gelatin (e.g., Sigma-Aldrich, cat. no. G7041) to 1 L deionized water. Store at 4°C for up to 6 months.
COMMENTARY
Background Information
Congenital neurodevelopmental anomalies are a wide range of medical conditions present from birth and characterized by an abnormal development of one or more regions of the brain. Brain development is a complex process requiring coordinated development of multiscale structures and relying on a system of cues, which together contribute to the proper formation of circuits and cognitive functions. In humans, the process starts with the formation of the neural tube, and from the fourth week of embryonic growth, the brain undergoes abundant neuronal proliferation, migration, and differentiation. At birth, basic structures are roughly established and continue to develop up to the age of 20 to 25 years, at which point the brain reaches its mature form and has grown four times in size. Defects in any of these neurodevelopmental stages may lead to defects in synaptogenesis, neurogenesis, cell survival, neuronal migration, axonal guidance, and cerebrospinal fluid (CSF) dispersion. This diversity of causal pathophysiological mechanisms consequently results in an array of quantifiable neuroanatomical anomalies.
Different classifications of brain anomalies have been proposed including specific forms of defects such as microcephaly, hydrocephalus, or corpus callosum agenesis. Microcephaly is one of the most studied brain anomalies characterized by decreased brain size compared with an age-matched population. It has an incidence of 1 in 5000 births and is associated with growth and cognitive impairment often resulting from an abnormal regulation of the cell cycle in progenitor cells. Congenital hydrocephalus has an incidence of 1 in 1500 births and is characterized by distortion in the flow or reabsorption of CSF resulting in ventricular dilatation. Obstruction of CSF flow within the ventricles is classified as noncommunicating hydrocephalus, whereas obstruction of CSF flow in the subarachnoid spaces is communicating hydrocephalus. Congenital hydrocephalus is often associated with developmental defects and can be either isolated or part of a syndrome with other anomalies such as Joubert syndrome. However, there is still little knowledge about the underlying genetic mechanisms that cause hydrocephalus in humans. Anomalies of the corpus callosum are the most frequent viable human brain malformation, affecting 1 in 4000 newborns, and can range from thin to thick corpus callosum. Corpus callosum agenesis is the most severe anomaly, characterized by complete or partial absence of the corpus callosum as a result of the disruption of any of its developmental steps. Corpus callosum anomalies occurring in conjunction with other brain structural defects will almost always lead to intellectual disability (syndromic forms). In contrast, when occurring without any other visible brain defects (seen by ultrasound from the second trimester of pregnancy), corpus callosum anomalies can be associated with normal (70% of cases) or abnormal (30%) cognition. This constitutes a major clinical challenge as genetic counseling is based on these statistics. Other studies have also suggested that a small corpus callosum is associated with a higher risk for autism and schizophrenia.
In vivo modeling of a wide spectrum of congenital neurodevelopmental anomalies is therefore of prime importance in biomedical research. To address this, we adopt a quantitative method for the assessment of neuroanatomical parameters in mouse embryos at E18.5 using an ultrastandardized paraffin-based histology procedure. The main advantage of our approach is our ability to detect subtle changes in embryonic brain anatomy relying on well-defined sections with no asymmetry along both the anteroposterior and dorsoventral axis at single-cell resolution. It is worthwhile to mention that the protocols presented here can be adapted to cryosections using a cryostat for sectioning of thicker slices of about 30 to 40 µm. While these standard 2D histological practices can provide cost-effective and relatively straightforward answers when it comes to quantifying NAPs, high-resolution 3D techniques are becoming increasingly accessible for embryonic brain exploration (Wendling et al., 2021) but are typically low throughput and require important informatic resources to acquire, store, and analyze large datasets (Pan et al., 2016; Zhong et al., 2021). About 80% of congenital neurodevelopmental anomalies are of genetic etiology, and our increasingly sophisticated ability to phenotype the embryonic mouse brain will continue to help in gene discovery in mice, which remains crucial to progress in the postgenomic era (Collins, Mikhaleva, et al., 2019). It will also allow for determining the time scale, that is whether NAPs are present from the embryonic stage (primary) or are progressive (secondary), by combining the present standardized procedures of the embryonic mouse brain with those of the adult mouse brain (see Current Protocols articles: Collins et al., 2018; Mikhaleva et al., 2016).
Finally, the molecular and pathophysiological mechanisms underlying NAPs detected using procedures presented in this article can be further characterized using immunofluorescence techniques on the same paraffin blocks. For example, sections can be stained with Ki67 for neuronal proliferation; Pax6, Sox2, and Tbr2 for differentiation; cleaved caspase-3 for neuronal survival; and Robo3 for axonal guidance.
Critical Parameters and Troubleshooting
There are three main critical parameters. First, dissection of the mouse embryo needs to be done without any damage since the slightest puncture while dissecting the embryonic brain will have a strong impact on the final sectioning quality. To facilitate dissection, we recommend fixation of the whole embryos first, then harvest of the embryonic brain. This does not apply to the adult mouse brain, which is dissected before fixation. Second, incubation time in fixative and ethanol solutions must be ultrastandardized and identical for samples belonging to the same project to avoid any artifacts due to uneven shrinkage and swelling of the sample. The time each histological slide is left on the 42°C hot plate must also be homogeneous. Third, visual trimming of the embryonic mouse brain into chunks of tissue on the coronal or sagittal orientations must be carried out on a perfectly symmetrical plane. This will prevent significant time loss at the microtome, at which point the issue can often (but not always) be rescued by adjusting the angle of the specimen holder and thin sectioning.
There are two common problems encountered during the method. First, streaks can appear on tissue that can be resolved by regularly changing the microtome blade. Second, sections cannot be identified as easily as in the adult mouse brain. The embryonic mouse brain is about three times smaller than a mature adult brain, and anatomical features change more rapidly between two consecutive sections of the atlas. It is therefore necessary to slow down when approaching the section of interest, otherwise it will be missed. Other troubleshooting aspects have been previously described (see Current Protocols articles: Collins et al., 2018; Mikhaleva et al., 2016).
Finally, it is important to mention that, although the complexity of the brain regions and structures are similar between the embryonic and adult mouse brain, the size of the embryonic mouse brain is about three times smaller than the adult mouse brain. This means that the distance in µm between sections characterized with defined landmarks is also smaller and requires more attention to not be missed.
Number and statistical analysis
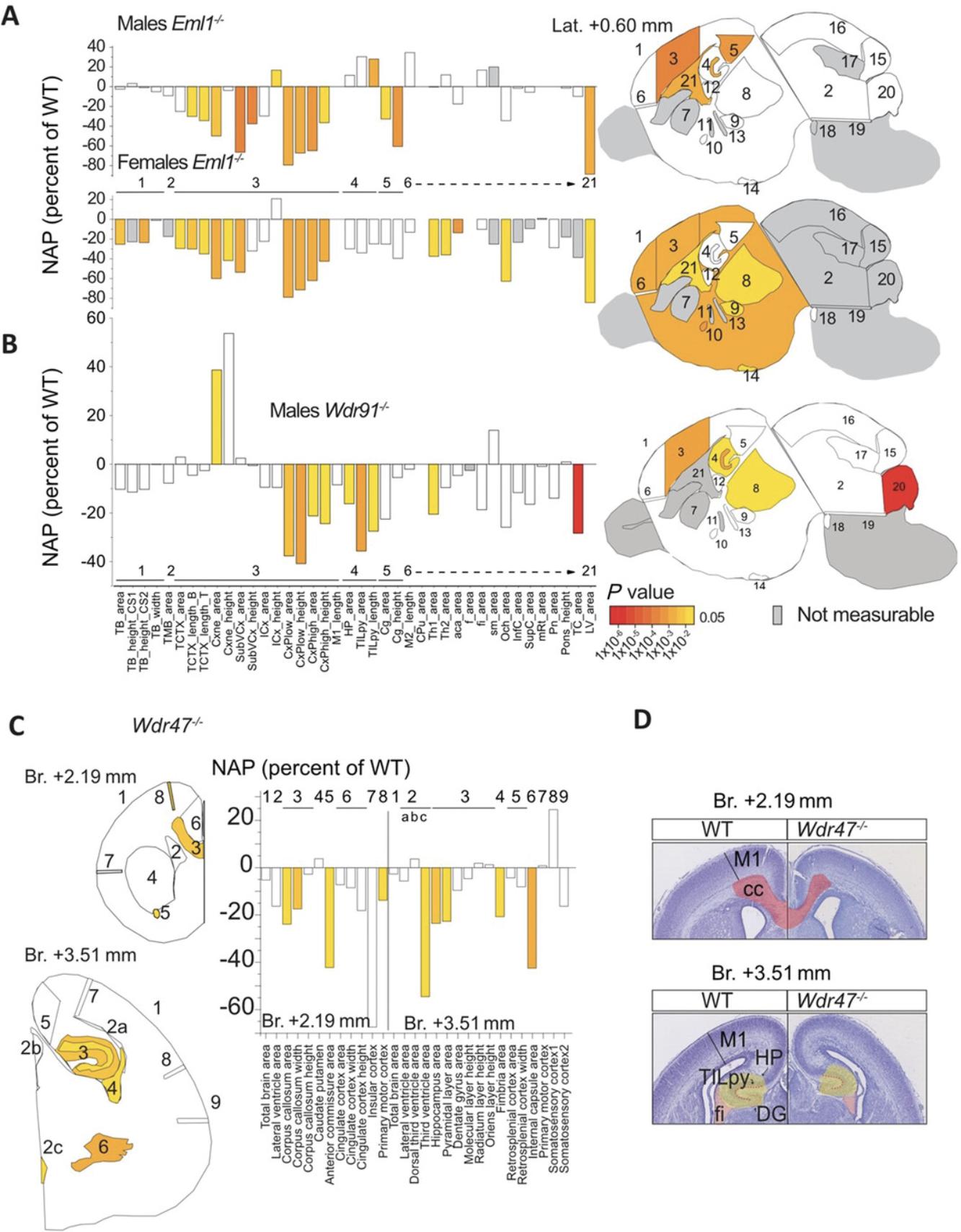
Embryonic brain samples from six male and six female mice per genotype should be collected, which was estimated based on power calculation with an 80% chance of detecting an effect of a size of about 5% at the 0.05 level, limited throughput of necropsy collection and histological workflows, and the 3Rs of research (replacement, reduction, refinement) with regard to animal use. In addition, up to six embryonic male and six embryonic female control brains should be collected to form a wild-type baseline population for analysis. For statistical analysis, to assess the relationship between genotype and phenotype, we provide a detailed description of the model framework elsewhere (Collins, Mikhaleva, et al., 2019). In brief, for large-scale projects, we recommend a linear mixed model and multiple testing corrections using the Benjamini-Hochberg method and a two-tailed Student's t -test with equal variance for smaller-size projects. Data generated consists of an array of neuroanatomical measurements, detailed size effects expressed as percent of wild-type, and directionality of brain areas (increased or decreased) that can be visualized using a standard bar graph where statistics are reported using the same color code as NAPs (Fig. 6). Considering the broad range of the measurement values, which are large or small (e.g., total brain area versus anterior commissure size), we chose to use bar graphs of percent of wild-type because it provides a chance to see changed neuroanatomical features.
Understanding Results
Using these protocols, we have processed 288 embryonic mouse brain samples to date, representing 144 mutant and 144 wild-type samples, all on C57BL/6N backgrounds for 12 unique gene mutations. Here, we illustrate three examples of genes (Eml1 , Wdr91 , and Wdr47) belonging to the WD40-repeat gene family, which is characterized by seven repetitive blades of 40 amino acids that end with a tryptophan-aspartic acid dipeptide at the C terminus, which is enriched in neurodevelopmental anomalies (Collins, Uzquiano, et al., 2019; Kannan et al., 2017). Key results are summarized, which were obtained for Eml1 , Wdr91 , and Wdr47 full knockout embryos, ordered here from extremely severe (Eml1) to very severe (Wdr91) to severe (Wdr47) brain anatomical perturbations.
EML1 is a microtubule-associated protein implicated in human ribbon-like subcortical heterotopia (Kielar et al., 2014). We identified extremely severe NAPs at E18.5 in both male and female Eml1 ‒/‒ embryos (Collins, Uzquiano, et al., 2019) studied on the sagittal plane (Fig. 6A). Most measurements pertaining to the cortical plate (cortical neuroepithelial zone, cortical subventricular zone, cortical intermediate layer, and cortical plate) and the height of the cingulate cortex were decreased in size compared with wild-type brains both in male and female embryos. Some NAPs were, however, gender specific. For example, the length of the pyramidal layer of the hippocampus was enlarged in male Eml1 ‒/‒ embryos only, whereas the size of the anterior commissure appeared smaller in the female Eml1 ‒/‒ embryos analyzed. This demonstrates that our protocol can be used efficiently to assess sexual dimorphism in brain structural defects at high resolution.
WDR91 protein was recently discovered in Caenorhabditis elegans in a complex with WDR81 acting as a negative regulator of phosphatidylinositol 3-phosphate levels in early-to-late endosome conversion (Liu et al., 2016) through a direct interaction with active GTP-bound Rab7 (Liu et al., 2017). Interestingly, we found very severe NAPs in the E18.5 homozygous knockout Wdr91 embryos (unpublished data) with 10 assessed embryonic brain regions reduced in size on the sagittal plane compared with wild-types (Fig. 6B). The most significantly affected brain parameter comprised the total area of the cerebellum, which was smaller by 28% (P = 9.74E‒06). The area of the thalamic nucleus was smaller by 20% (P = 0.025), and the hippocampal formation was reduced in size by 16% (P = 0.011).
Like EML1, WDR47 is a microtubule-associated protein that our laboratory identified for the first time as essential for brain development (Kannan et al., 2017). In Wdr47 −/− embryos, nine NAPs on the coronal plane emerged as decreased in size (Fig. 6C), including the size of the genu of the corpus callosum area at Bregma +2.19 mm, the thickness of the primary motor cortex at both Bregma +2.19 mm and Bregma +3.51 mm, and the size of the hippocampus and the fimbria of the hippocampus (Fig. 6D). There were no differences between male and female Wdr47 −/− embryos. Combining these developmental studies with similar morphological screens in 16-week-old Wdr47 −/− mice, we were able to point toward primary neurodevelopmental anomalies that worsen postnatally.
Time Considerations
Time considerations for each step of the protocols are summarized in Table 7. The maximum time required to complete the protocol for one sample is 14 days (±2 days depending on the experimenter). The minimum time required for one sample is 5 days when the time in 70% ethanol is reduced to the minimum (N.B., fixation time in Bouin's fixative cannot be reduced). Should the time required to generate embryos at E18.5 be considered, it is an additional 19 days to account for the average gestation length for C57BL/6N mice (up to 21 days depending on strain). Dissection of the brain should take no longer than 10 min once the experimenter is trained to the task. Brain sectioning on the microtome should take about 5 min; however, if the quality of dissection and trimming was poor, then this step can take much longer, up to 1 hr. Nissl staining takes about 30 min and can easily be done in a batch of up to 20 slides. Although most steps can be done simultaneously, steps involving image analysis and data control quality are carried out sequentially and can take a significant amount of time.
| Step | Time for one embryo |
|---|---|
| Embryo generation | 19 days |
| Embryo harvest | 30 min |
| Fixation with Bouin's fixation | 2 days |
| Ethanol transfer | 7 days |
| Brain dissection | 10 min |
| Paraffin embedding | 15 hr |
| Tissue sectioning | cor-, 15 min; sag-, 5 min |
| Slide drying | Overnight |
| Nissl staining | 30 min |
| Slide coverslipping and drying | Overnight |
| Image scanning | cor-, 45 min; sag-, 15 min |
| Image analysis | cor-, 60 min; sag-, 20 min |
| Data quality control | cor-, 3 hr; sag-, 1 hr |
| Data reporting | 1 day |
Acknowledgments
This work was supported by multiple grants to Dr. Yalcin from the French National Research Agency Young Team Investigator Award and the National Institute of Health and Medical Research Inserm First Step Program. Dr. Yalcin thanks all members of her laboratory who contributed over the years to the development of these standardized procedures for embryonic mouse brain phenotyping and discussions, in particular Charlotte Montillot, Perrine Kretz, Anna Mikhaleva, Léo Gagliardi, Christel Wagner, and Marie-Christine Fischer. The authors also thank Audrey Geissler, Laura Guenot, Rim Belkaid, and Amandine Bataille, members of the Histopathology Facility CellImap at Inserm Unit 1231 in Dijon (https://www.cellimap.fr/), for histological advice and discussions.
Author Contributions
Sylvie Nguyen : methodology; Meghna Kannan : methodology; Marion Gaborit : methodology; Stephan C. Collins : investigation, methodology, resources, supervision; Binnaz Yalcin : conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing—original draft, review, and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Literature Cited
- Birling, M. C., Yoshiki, A., Adams, D. J., Ayabe, S., Beaudet, A. L., Bottomley, J., … Murray, S. A. (2021). A resource of targeted mutant mouse lines for 5,061 genes. Nature Genetics , 53, 416–419. doi: 10.1038/s41588-021-00825-y
- Collins, S. C., Mikhaleva, A., Vrcelj, K., Vancollie, V. E., Wagner, C., Demeure, N., … Yalcin, B. (2019). Large-scale neuroanatomical study uncovers 198 gene associations in mouse brain morphogenesis. Nature Communication , 10, 3465. doi: 10.1038/s41467-019-11431-2
- Collins, S. C., Uzquiano, A., Selloum, M., Wendling, O., Gaborit, M., Osipenko, M., … Francis, F. (2019). The neuroanatomy of Eml1 knockout mice, a model of subcortical heterotopia. Journal of Anatomy , 235, 637–650. doi: 10.1111/joa.13013
- Collins, S. C., Wagner, C., Gagliardi, L., Kretz, P. F., Fischer, M. C., Kessler, P., … Yalcin, B. (2018). A method for parasagittal sectioning for neuroanatomical quantification of brain structures in the adult mouse. Current Protocols in Mouse Biology , 8, e48. doi: 10.1002/cpmo.48
- Donovan, J., & Brown, P. (2005). Euthanasia. Current Protocols in Neuroscience , 33, A.4H.1–A.4H.4. doi: 10.1002/0471142301.nsa04hs33
- Kannan, M., Bayam, E., Wagner, C., Rinaldi, B., Kretz, P. F., Tilly, P., … Yalcin, B. (2017). WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy. Proceedings of the National Academy of Sciences of the United States of America , 114, E9308–E9317. doi: 10.1073/pnas.1713625114
- Karp, N. A., Meehan, T. F., Morgan, H., Mason, J. C., Blake, A., Kurbatova, N., … Brown, S. D. (2015). Applying the ARRIVE guidelines to an in vivo database. PLoS Biology , 13, e1002151. doi: 10.1371/journal.pbio.1002151
- Kielar, M., Tuy, F. P., Bizzotto, S., Lebrand, C., de Juan Romero, C., Poirier, K., … Francis, F. (2014). Mutations in Eml1 lead to ectopic progenitors and neuronal heterotopia in mouse and human. Nature Neuroscience , 17, 923–933. doi: 10.1038/nn.3729
- Liu, K., Jian, Y., Sun, X., Yang, C., Gao, Z., Zhang, Z., … Yang, C. (2016). Negative regulation of phosphatidylinositol 3-phosphate levels in early-to-late endosome conversion. The Journal of Cell Biology , 212, 181–198. doi: 10.1083/jcb.201506081
- Liu, K., Xing, R., Jian, Y., Gao, Z., Ma, X., Sun, X., … Yang, C. (2017). WDR91 is a Rab7 effector required for neuronal development. The Journal of Cell Biology , 216, 3307–3321. doi: 10.1083/jcb.201705151
- Mikhaleva, A., Kannan, M., Wagner, C., & Yalcin, B. (2016). Histomorphological phenotyping of the adult mouse brain. Current Protocols in Mouse Biology , 6, 307–332. doi: 10.1002/cpmo.12
- Pan, C., Cai, R., Quacquarelli, F. P., Ghasemigharagoz, A., Lourbopoulos, A., Matryba, P., … Erturk, A. (2016). Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nature Methods , 13, 859–867. doi: 10.1038/nmeth.3964
- Paxinos, G., Halliday, G. M., Watson, C., Koutcherov, Y., & Wang, H. (2007). Atlas of the developing mouse brain. Elsevier.
- Wendling, O., Hentsch, D., Jacobs, H., Lemercier, N., Taubert, S., Pertuy, F., … Herault, Y. (2021). High resolution episcopic microscopy for qualitative and quantitative data in phenotyping altered embryos and adult mice using the new “Histo3D” system. Biomedicines , 9, 767. doi: 10.3390/biomedicines9070767
- Zhong, Q., Li, A., Jin, R., Zhang, D., Li, X., Jia, X., … Luo, Q. (2021). High-definition imaging using line-illumination modulation microscopy. Nature Methods , 18, 309–315. doi: 10.1038/s41592-021-01074-x
Key References
- Paxinos et al. (2007). See above.
Atlas of the developing mouse brain at E17.5, day of birth, and postnatal day 6. Represents an essential tool for researchers and students who investigate the development of the murine brain or for those who interpret findings from genetic manipulation.
Internet Resources
Allen Developing Mouse Brain Atlas, a high-resolution anatomical reference atlas at E18.5 accompanied by a systematic developmental taxonomy of mouse brain structures.
Citing Literature
Number of times cited according to CrossRef: 3
- Ailong Sha, Hongrun Chen, Xuewen Zhao, Exploration of the mechanisms of improving learning and memory in the offspring of aging pregnant mice by supplementation with Paris polyphylla polysaccharide based on the P19-P53-P21 and Wnt/β-catenin signaling pathways, Journal of Ethnopharmacology, 10.1016/j.jep.2023.116883, 318 , (116883), (2024).
- Perrine F. Kretz, Christel Wagner, Anna Mikhaleva, Charlotte Montillot, Sylvain Hugel, Ilaria Morella, Meghna Kannan, Marie-Christine Fischer, Maxence Milhau, Ipek Yalcin, Riccardo Brambilla, Mohammed Selloum, Yann Herault, Alexandre Reymond, Stephan C. Collins, Binnaz Yalcin, Dissecting the autism-associated 16p11.2 locus identifies multiple drivers in neuroanatomical phenotypes and unveils a male-specific role for the major vault protein, Genome Biology, 10.1186/s13059-023-03092-8, 24 , 1, (2023).
- Charlotte Montillot, Emilia Skutunova, undefined Ayushma, Morgane Dubied, Adam Lahmar, Sylvie Nguyen, Benazir Peerally, Fabrice Prin, Yannis Duffourd, Christel Thauvin-Robinet, Laurence Duplomb, Heng Wang, Muhammad Ansar, Laurence Faivre, Nicolas Navarro, Shilpi Minocha, Stephan C. Collins, Binnaz Yalcin, Characterization of Vps13b-mutant mice reveals neuroanatomical and behavioral phenotypes with females less affected, Neurobiology of Disease, 10.1016/j.nbd.2023.106259, 185 , (106259), (2023).

