Processing human frontal cortex brain tissue for population-scale Oxford Nanopore long-read DNA sequencing SOP
Kimberley J Billingsley, Ramita Dewan, Laksh Malik, Pilar Alvarez Jerez, Stith Kiley, Cornelis Blauwendraat, on behalf of the CARD Long-read Team
Disclaimer
In development
We are still developing and optimizing this protocol.
Abstract
Processing human frontal cortex brain tissue for population-scale Oxford Nanopore long-read DNA sequencing SOP
At the NIH's Center for Alzheimer's and Related Dementias (CARD) https://card.nih.gov/research-programs/long-read-sequencing we will generate long-read sequencing data from roughly 4000 patients with Alzheimer's disease, frontotemporal dementia, Lewy body dementia, and healthy subjects. With this research, we will build a public resource consisting of long-read genome sequencing data from a large number of confirmed people with Alzheimer's disease and related dementias and healthy individuals. To generate this large-scale nanopore sequencing data we have developed a protocol for processing and long-read sequencing human frontal cortex brain tissue, targeting an N50 of ~30kb and ~30X coverage.
†Correspondence to: Kimberley Billingsley billingsleykj@nih.gov and Cornelis Blauwendraat cornelis.blauwendraat@nih.gov
Acknowledgements:
We would like to thank the Nanopore team (Androo Markham &Hannah Lucio), Circulomics Inc team (Jeffrey Burke, Michelle Kim, Duncan Kilburn & Kelvin Liu) and the whole CARD long-read team listed below => UCSC : Benedict Paten, Mikhail Kolmogorov, Miten Jain, Kishwar Shafin, Trevor Pesout; NHGRI : Adam Phillippy, Arang Rhie; Baylor: Fritz Sedlazeck; JHU : Winston Timp; NINDS: Sonja Scholz; NIA : Cornelis Blauwendraat, Kimberley Billingsley, Frank Grenn, Pilar Alvarez Jerez, Bryan Traynor, Shannon Ballard, Caroline Pantazis; CZI : Paolo Carnevali.

Attachments
Steps
Part 1: Brain Tissue Cutting (~2.5 hours for 16 samples) )
Add dry ice to a ice bucket
Place supplies (sterile weigh boat, razor blade, labeled empty 2mL protein LoBind microcentrifuge tubes and cooling block) in dry ice and allow to chill for ~ 0h 5m 0s
Obtain tissue samples from -80°Cfreezer and place in dry ice
Wear all necessary protective equipment (lab coat, face shield, double gloved) and complete the following steps within a chemical fume hood
Weigh labeled empty tube to tare scale, ensuring tube is centered
Working in the chemical fume hood, place the chilled weighing boat on top of the cooling tissue (use right hand to grip blade and cut tissue), use left hand just above, in between tissue and in front of hood, to shield any flying pieces)
Using the chilled razor blade, gently lift the cut tissue piece and transfer to the chilled labeled empty tube and weigh immediately, place back in dry ice immediately to avoid tissue thawing
Add or remove tissue using the method outlined above as required by input specifications ~40mg for frontal cortex, DNA recovery varies based on amount of gray matter vs. white matter)
Dispose of used weighing boat in a burn box and razor blade in the sharps waste container between each sample to prevent inter-sample contamination and keep all tissue samples on dry ice when not in use
Part 2: TissueRuptor Brain Tissue Disruption (~3 hours)
Place 15mL round tubes and cold Buffer CT on ice, chill centrifuge to4°C, and warm ThermoMixer to 55°C
Add 20µL of Proteinase K to the previous pellet
Add 50µL1X TE pH 8
Add 60µL Buffer CS
Add 100µL Buffer CLE3 and pipette mix 15X with a wide bore P200 pipette
Incubate for 1h 0m 0s on a ThermoMixer at 55°Cand 900 rpm
Spin on a mini-centrifuge for 0h 0m 2s to remove liquid from the cap
Add 20µL of RNaseA and pipette mix 3X with a wide bore P200 pipette
Incubate for 0h 30m 0son a ThermoMixer at 55°C and 900 rpm
Spin the tube on a mini-centrifuge for0h 0m 2s to remove liquid from the cap
Add 50µL Buffer SB and vortex for0h 0m 10s at maximum speed (transfer any foam that appears)
Transfer brain tissue from previous steps to 15mL round tubes (keep on ice during the entire disruption process)
Add 750µL of cold Buffer CT
Submerge TissueRuptor probe tip in buffer and blend at max speed for 0h 0m 10s(place probe tip off to side to be cleaned later)
Transfer homogenate to a 2 mL Protein LoBind microcentrifuge tube including all undisrupted tissue chunks and any foam that forms
Pellet homogenate by centrifuging at 6,000 x g and 4°C for 0h 5m 0s. Discard supernatant (pellet may not be visible, so pipette carefully and avoid pipetting from the bottom of tube)
Add 1mLof cold Buffer CT and pipette mix 10X with a wide bore P1000 pipette to resuspend tissue
Pellet homogenate by centrifuging at 6,000 x g and 4°C for 0h 2m 0s. Discard supernatant (pellet may not be visible, so pipette carefully and avoid pipetting from the bottom of tube)
Pulse vortex for 0h 0m 1sx 5 times (max setting) to dislodge pellet
Part 3: KingFisher Apex Nanobind Tissue Big DNA protocol (~2 hours)
Prepare KingFisher Apex plates as follows:
Plate 1 - Lysis Binding: Sample + 50µL BL3
Plate 2 - Nanobind Storage: one 3mm Nanobind disk
Plate 3 - CW1 Wash 1: 600µLBuffer CW1
Plate 4 - CW1 Wash 2: 600µL Buffer CW1
Plate 5 - CW2 Wash 1: 600µL Buffer CW2
Plate 6 - CW2 Wash 2: 600µL Buffer CW2
Plate 7 - Elution: 100µL Buffer EB
Plate 8 - Tip: KingFisher Flex 96-Tip Combo
Run KingFisher Apex program “210804_nanobind_tissue_kf_apex_v2.kfx” (KF script available by request from Circulomics Inc)
After 0h 12m 0s when the program pauses, add 300µLIPA
Collect DNA by transferring eluate to a new 1.5 mL microcentrifuge tube OR if sample is to be sheared, the sample can be transferred to a DNA Fluid+ tube.
Let the sample rest at room temperature overnight to allow DNA to solubilize
Part 4: DNA Shearing (8 hours per 48 samples)
Hand shear DNA
Hand-shear 10X with 1mL Luer-Lock syringes and 1.5” needles (bringing sample up into needle and depressing plunger counts as 1 cycle)
Quantify:
- Quantify the samples with the nanodrop and Qubit and size using the Agilent Tapestation 4200
- Upload the tapestation reports
- Add the quantifications
- Only take forward samples that are > 4.5ug. If a sample does not reach this requirement then repeat DNA extraction from Part 1.
Megaruptor 3 Shear with DNAFluid+ kit:
- Transfer all of sample to DNA Fluid+ tubes if not in already
- Make sample up to
150µLwith nuclease free water - Attach the DNA Fluid+ needle onto the tube and push the entire item into the Megaruptor 3 slots until it fits snugly. If running fewer than 8 samples, put the tubes in the 1st and/or 8th slots, working your way in. Samples should always be balanced, if running an odd number of samples, samples can be balanced with an empty corresponding tube. Shear at speed 45 (takes around ~
1h 0m 0s) - Once the MR3 shearing is finished, repeat the run by navigating back to the main menu and select speed 45 again (takes around ~
1h 0m 0s) - Avoid any vortexing of DNA from this point on to avoid any unnecessary further shearing, instead mix by gently flicking the tube and spin down
Part 5: DNA Quantification
Quantify the samples with the nanodrop and Qubit and size using the Agilent Tapestation 4200
Upload the tapestation reports and and quantifications
For Step 6 the starting material must be > 4.5ug. If a sample does not reach this requirement then repeat DNA extraction from Part 1.
DNA can be stored at 4° C for up to four weeks, or -80°Cindefinitely
Part 6: Library Prep (~6 hours, not including flushing and returning cells) )
A. DNA Repair and End-Prep
-
Put all the necessary reagents on ice to thaw and the Agencourt AMPure XP beads out at room temperature
-
Prepare the following in a 0.2 mL thin-walled PCR tube:
48µLDNA (load 4.5 ug, this will likely be over 48 μL but that is fine, just adjust the amount of beads to match the total volume of this mixture)
-
3.5µLNEBNext FFPE DNA Repair Buffer (vortex)-
3.5µLUltra II End-prep reaction buffer (vortex) -
3µLUltra II End-prep enzyme mix (do not vortex) -
2µLNEBNext FFPE DNA Repair Mix (do not vortex)
-
Mix thoroughly by gently flicking tube or pipetting up and down 10X, and then spin down
-
Using a thermal cycler, incubate samples at
20°Cfor0h 5m 0sand65°C
for0h 5m 0s
- Start and pause thermal cycler to allow lid to come t
85°Cbefore putting samples in
-
Allow Thermocycler to cool to
4°Cand then remove your samples. -
Resuspend the AMPure XP beads by vortexing
-
Transfer DNA samples to clean 1.5 mL Eppendorf DNA LoBind tube
-
Add
60µL(or equivalent volume, see step 2) of resuspended beads to the reaction and mix by pipetting up and down 10X -
Incubate for
0h 5m 0sat room temperature -
Prepare
500µLper sample of fresh 75% ethanol in Nuclease-free water -
Pellet sample on magnet until eluate is clear and colorless, about
0h 2m 0s -
Pipette off the supernatant and retain, just in case the following quant is uncharacteristically or surprisingly low
-
With the samples remaining on the magnet, wash the beads with
200µLof the ethanol, pipetting on the opposite wall (the goal here is to make sure the beads are fully covered) and making sure not to disturb the pellet, count to 3 and remove and discard ethanol -
Repeat previous step
-
Spin down and place the tube back on magnet, pipetting off any residual ethanol
-
Allow to dry for ~
0h 0m 30sbut do not over-dry -
Remove the tube from the magnetic rack and resuspend the pellet in
61µLNuclease-free water, incubate for0h 3m 0sat room temperature (hold in hands and gently flick every so often) -
Pellet the samples on a magnet until eluate is clear and colorless
-
Remove and retain
61µLof eluate into a clean 1.5 mL Eppendorf DNA LoBind tube -
The sample concentration must be > 40ng/ul. If the sample does not reach this requirement restart from Part 6.
-
It is possible to store samples at
4°Covernight at this step if needed
B. Adapter Ligation and Clean-Up
- Spin down the AMX-F, Quick T4 ligase, and LNB, then return to ice
- Do not allow AMX-F to remain at room temperature for too long
-
Thaw LNB at RT and mix by pipetting up and down (vortexing is ineffective due to viscosity)
-
Thaw EB at RT, mix by vortexing, spin down, and place on ice
-
Thaw SFB at RT, mix by vortexing, spin down, and keep at RT
-
In a 1.5 mL Eppendorf DNA LoBind tube, mix the following in order:
-
60µLDNA sample (if not 60, add water until it is) -
25µLLNB -
10µLQuick T4
-
5µLAMX-F -
-
Mix gently by flicking the tube and spin down
-
Incubate the reaction for
0h 10m 0sat RT -
During this time, put flow cells out at RT
-
Resuspend beads by vortexing
-
Add
45µLof resuspended beads to the reaction and mix by flicking -
Incubate for
0h 5m 0sat RT -
Spin down sample and pellet on magnet
-
Pipette off the supernatant and retain, just in case the final elution quant is uncharacteristically or surprisingly low
-
Wash the beads with
250µLSFB, flick to resuspend and repellent, remove and discard supernatant -
Repeat previous step
-
Spin down and place the tube back on magnet, pipetting off any residual SFB
-
Allow to dry for ~
0h 0m 30s, but do not over-dry -
Remove the tube from magnet and resuspend pellet in 25 μL EB, spin down, and incubate for
0h 20m 0sat37°C -
During this time, QC the flow cells (only use flow cells with >6000 pores)
-
Pellet the beads on magnet until eluate is clear and colorless
-
Remove and retain
25µLof eluate into a clean 1.5 mL Eppendorf DNA LoBind tube (this is the DNA library) -
Quantify samples on Qubit
-
Reprep library from Part 6 if < 1200 ng.
-
Keep libraries on ice until ready to load on flow cell
C. Priming and Loading Flow Cell
-
Thaw SBII, FLT, and FB, vortex, and spin down
-
Thaw LBII
-
Add
30µLof thawed and mixed FLT directly to tube of FB and vortex -
Expose inlet port on flow cell and draw back a small volume to remove any air bubbles (usually about 20-30 μL, just until a small volume of buffer enters the pipette tip)
-
Flush
500µLof Priming Mix into the inlet port of the flow cell, being extremely careful to avoid the introduction of air bubbles at the end -
Wait
0h 5m 0s -
During this time, separate the DNA library into three equal aliquots (ideally with 400 ng of DNA each). Bring each aliquot up to
24µLwith EB. (i.e, if your final elution is exactly 1200 ng in 24 ul, move8µLto three separate tubes and add16µLof EB to each.) -
Prepare the first library mix for loading:
-
75µLSBII (vortex) -
51µLLB (pipette up and down immediately before use) -
24µLDNA library in EB (400 ng)
-
-
Immediately load all
150µLof the library mix -
Close valve to seal inlet port and close PromethION lid
-
Wait
0h 10m 0sand then initiate sequencing -
Ideally, the library quants yielded at least 1200 ng to allow for 3X 400 ng loads, the latter 2 loaded approximately after 24 and 48 hours. However this will vary slightly depending on pore usage, data generated, as well as other factors i.e. if after 24 hours there are still +3000 pores then the sample does not need to be reloaded until 48 hours.
- To wash and reload a flow cell, begin by thawing Wash Mix (WMX) on ice and Wash Diluent (DIL) at RT (DIL should be vortexed, WMX should NOT be vortexed, only spun) - Add`2µL`WMX into `398µL`DIL and pipette mix -Pause the PromethION runs and export .pdf reports -Rotate the inlet port cover to reveal inlet port 1 -Using a P1000, insert tip into inlet port and draw back a small volume using the wheel to remove any air (usually around 20-30 μL) -Load `400µL`Flow Cell Wash Mix into the inlet port, avoiding any introduction of air -Wait `1h 0m 0s` -Repeat priming steps and reload samples (steps 1 - 12)
D. Flushing and Recycling Flow Cells (~15 minutes per set of 4 flow cells)
-
Following the completion of the sequencing, flow cells may be removed from the sequencer
-
Place enough absorbent material to take up approximately 4 mL of flush waste
-
Rotate valve to reveal inlet port 1
-
Place flow cell at a 45° angle on the absorbent material and, using a P1000, flush
1mLof DI water into the inlet port -
Repeat 3 more times for a total of
4mL -
Once complete, close the input port cover and remove all liquid from the waste port
-
Dispose of absorbent material as local biological waste guidelines dictate
-
Return flow cells to clear plastic tray in which it was shipped, making sure to record the flow cell IDs
-
Seal the tray with the sticker provided in the packaging
-
Put the clear plastic lid back on the tray
-
Place the tray back in the packaging
-
Place packaged cells in the returns box (large box can hold 80)
-
Once returns box is filled, follow the instructions here and follow the prompts to request the box to be sent back to Nanopore
Sequencing results:
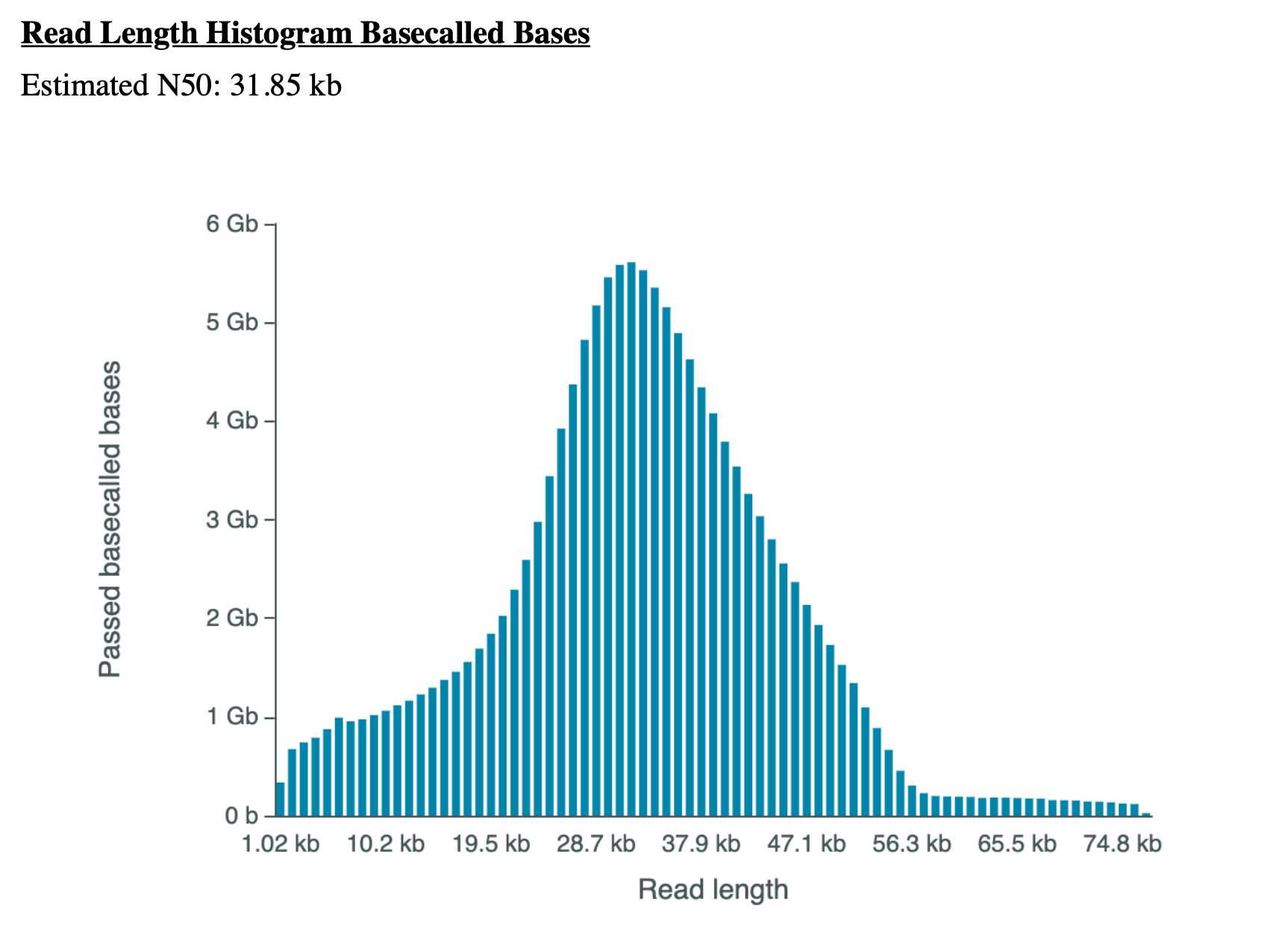
- Following 72 hours of sequencing the sample should yield an N50 ≥ 30kb with a data output ~ 100-160GB.
#尊敬的用户,由于网络监管政策的限制,部分内容暂时无法在本网站直接浏览。我们已经为您准备了相关原始数据和链接,感谢您的理解与支持。
https://lh6.googleusercontent.com/0xryslPCmkGicbTEKEzYakeiLrt4TpN5_BbIQE-zCnz3LowEsyyo29nlu0Eb48eGsg5jTkDxkmNKfNY4IEXwv2hss9GhK4HR9mx3hIX81yJ7xQjEcaFEZ6lhJe6E0BrCDqJjAWav