One-step RT-PCR Ins214EPE assay for Omicron (B.1.1.529) variant detection
Nikita Yolshin, Andrey Komissarov, Kirill Varchenko, Kseniia Komissarova, Daria Danilenko, Dmitry Lioznov
Abstract
On 26 November 2021 WHO designated a new variant of concern B.1.1.529 named Omicron. This variant has a large number of mutations, some of which are concerning. Preliminary evidence suggests an increased risk of reinfection with this variant and reduced neutralization by convalescent and vaccinated sera, as compared to other VOCs. Implementation of the high-throughput rRT-PCR screening for Omicron is of great importance for monitoring the spread of this VOC in the population, especially in resource-limited countries lacking sufficient sequencing capacity.
Omicron lineage B.1.1.529 (BA.1) has some indels that turned out to be a good target for its detection. In the current protocol, we use ins214EPE for this purpose. Here we describe the 1-step quantitative multiplex RT-qPCR assay consisting of the newly developed Ins214EPE detection set and widely used Hong Kong University N gene assay for SARS-CoV-2 detection (Chu et al., 2020).

The assay was validated on the Omicron variant RNA kindly provided by the Pathogenic Microorganisms Variability Laboratory (Dr. Vladimir Guschin, Gamaleya Institute, Moscow, Russia) and RNA from the collection of Smorodintsev Research Institute of Influenza.
| A | B |
|---|---|
| hCoV-19/Russia/MOW-Moscow_PMVL-O11/2021 | EPI_ISL_7263932 |
| hCoV-19/Russia/MOW-Moscow_PMVL-O16/2021 | EPI_ISL_7263933 |
| hCoV-19/Russia/SPE-RII-MH44356S/2021 | EPI_ISL_7717296 |
Virus RNA specimens used for rRT-PCR ins214EPE assay validation
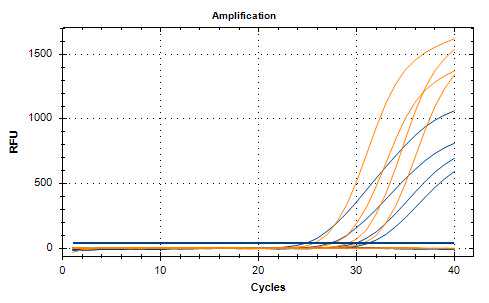
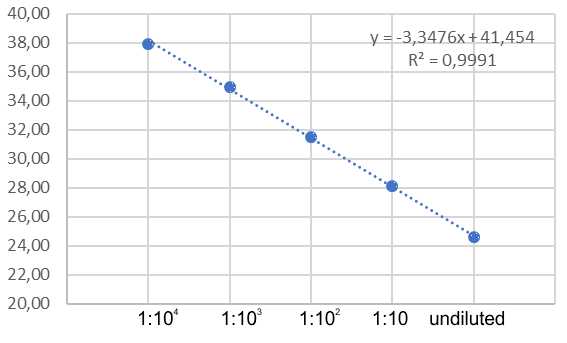
Omicron RNA specimens were positive in the assay as expected. Negative controls were found negative. 10-fold serial dilutions of Omicron RNA were used to assess ins214EPE assay amplification efficiency. The amplification efficiency was 98,9% (R2 = 0,99).

The developed rRT-PCR assay demonstrates high specificity. It was tested on 26 clinical samples (RNA extracted from oropharyngeal swabs) with previously characterized viruses belonging to 8 different SARS-CoV-2 lineages (including Delta B.1.617.2+AY.*) Specific signal was detected only in samples with SARS-CoV-2 Omicron lineage RNA (confirmed by whole-genome sequencing). Specificity was additionally tested on clinical samples positive for other respiratory viruses from the collection of Smorodintsev Research Institute of Influenza - influenza, parainfluenza, human seasonal coronaviruses (OC43, NL63, 229E, HKU1), hRSV, rhinoviruses, bocaviruses, metapneumovirus (33 in total) - with no false-positive results.
| A | B | C |
|---|---|---|
| B.1.1.7 | 15,30 | - |
| B.1.351 | 16,08 | - |
| P.1 | 16,15 | - |
| B.1.617.2 | 16,70 | - |
| B.1 | 17,47 | - |
| AT.1 | 18,36 | - |
| B.1.617.2 | 21,87 | - |
| B.1.1.7 | 22,49 | - |
| B.1.617.2 | 24,79 | - |
| B.1.617.2 | 26,16 | - |
| B.1.1.7 | 26,24 | - |
| B.1.1.529 | 26,24 | 26,29 |
| B.1.617.2 | 28,06 | - |
| B.1.1.7 | 28,54 | - |
| B.1.1.529 | 28,84 | 28,72 |
| AT.1 | 29,65 | - |
| AY.122 | 31,18 | - |
| AY.122 | 31,67 | - |
| AY.122 | 33,51 | - |
| B.1.1.7 | 33,63 | - |
| AT.1 | 33,80 | - |
| AT.1 | 34,01 | - |
| AT.1 | 35,02 | - |
| B.1.1.7 | 35,60 | - |
| B.1.351 | 36,33 | - |
| B.1.617.2 | 37,30 | - |
Analytical specificity of the assay was checked on 26 samples of 8 different SARS-CoV-2 lineages
| A | B | C | D |
|---|---|---|---|
| hRV | 32,85 | ||
| hRV | 33,76 | ||
| hRV | 27,75 | ||
| hRSV B | 31,49 | ||
| hRSV B | 30,98 | ||
| hRSV B | 32,33 | ||
| hRSV A | 28,76 | ||
| hRSV A | 30,56 | ||
| hRSV A | 27,70 | ||
| hPIV4 | 23,90 | ||
| hPIV3 | 27,01 | ||
| hPIV2 | 24,72 | ||
| hPIV1 | 28,63 | ||
| hCoV OC43 | 30,34 | ||
| hCoV OC43 | 30,69 | ||
| hCoV OC43 | 28,64 | ||
| hCoV NL63 | 32,20 | ||
| hCoV NL63 | 30,42 | ||
| hCoV NL63 | 24,95 | ||
| hMPV | 30,12 | ||
| hCoV HKU1 | 30,06 | ||
| hCoV HKU1 | 28,30 | ||
| hCoV HKU1 | 30,73 | ||
| Human influenza A (H3N2) | 28,16 | 39,06 | |
| Human influenza A (H3N2) | 29,13 | ||
| Human influenza A (H3N2) | 28,45 | ||
| SARS-CoV-2 B.1.1.529 | 34,15 | 26,61 | 28,44 |
| hBoV | 32,26 | ||
| hBoV | 30,75 | ||
| hBoV | 27,25 | ||
| hAdV | 29,47 | ||
| hCoV 229E | 29,11 | ||
| hCoV 229E | 32,52 | ||
| hCoV 229E | 29,37 |
Analytical specificity of the assay was checked on 33 clinical samples positive for other respiratory viruses. RP assay (human RNAse P assay) developed by US CDC was used to check the presence of human RNA in clinical samples
Analytical sensitivity determination is underway.
We consider developed assay to be useful in wide populational RT-PCR screening to assess the spread of Omicron variant.
Steps
Order oligonucleotides with following sequences 5'->3':
| A | B |
|---|---|
| Ins214EPE F | ATATTCTAAGCACACGCCTATT |
| Ins214EPE R | GGCAAATCTACCAATGGTTCTA |
| Ins214EPE P | ROX-TGCGTGAGCCAGAAGATCTCCCT-BHQ-2 |
| HKU-NF | TAATCAGACAAGGAACTGATTA |
| HKU-NR | CGAAGGTGTGACTTCCATG |
| HKU-NP | FAM-GCAAATTGTGCAATTTGCGG-BHQ |
Ins214EPE oligonucleotide set developed in Smorodintsev Research Institute of Influenza (St.Petersburg, Russia) tfor specific detection of B.1.1.529 (Omicron) lineageHKU oligolucleotide set was developed by the University of Hong Kong to detect SARS-CoV-2 in human clinical specimens (Chou et al., 2020)Please, change the dye if you use ROX as passive reference dye
Briefly vortex and centrifuge reagents before use.
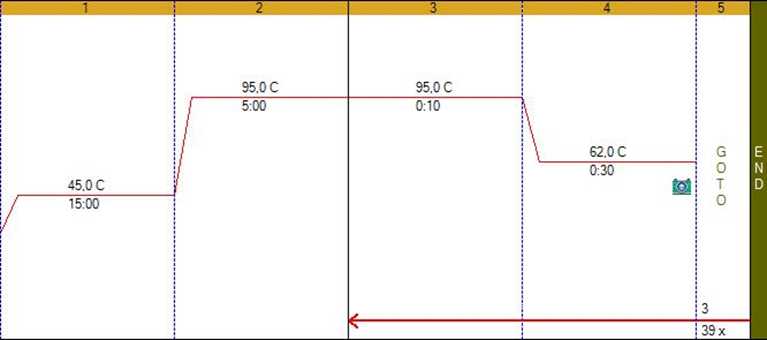
Prepare the PCR reaction mixture following the specifications below:
| A | B |
|---|---|
| 2X RT-qPCR Buffer | 12.5 μl |
| 25X RT-qPCR Enzyme Mix | 1 μl |
| Ins214EPE F | 400 nM |
| Ins214EPE P | 400 nM |
| Ins214EPE R | 400 nM |
| HKU-NF | 400 nM |
| HKU-NR | 400 nM |
| HKU-NP | 400 nM |
| RNA template | 5 μl |
| Nuclease-free water | to 25 μl |
We used Biomaster qRT-PCR mix (Biolabmix, Russia)
| A |
|---|
How to interpret the results: detection of fluorescence of FAM probe (HKU-NP) means thepresence of SARS-CoV-2 RNA in the sample, detection of fluorescence in ROX channel (Ins214EPE probe) means the presence of B.1.1.529 (BA.1)(omicron) RNA.