ONT Basecalling, Demultiplexing, and Analysis for Fungal Barcodes
Stephen Douglas Russell
Abstract
This protocol assumes that your MinION run has been completed and the data from the run has been saved. It should take you from raw data to useable FASTA files for each of your fungal barcodes.
Steps
This protocol assumes the experiment name is "FirstRun."
Create a new working folder on the desktop. Ex - FirstRun. Within that create a new folder called "fast5," another called "Programs," and a final one called "NGSpeciesID."
I will start by copying all of the fast5 files from:
/var/lib/minknow/data/./FirstRun/CellName/long_unique_name/fast5 to the newly created fast5 folder on the desktop
Create an index file from your extraction template papers. This will allow you to link all of your reads with the individual specimens. A template for 7 plates (672 specimens) can be found here:
NANOPORE TEMPLATE THIRD RUN.xlsx
This .xlsx is formatted to utilize the Lab Code and iNaturalist # columns as the only inputs. It will combine these and all of the other columns into a single cell - concatenating them all into the final file name. For the Lab Code, I will typically put these into the iNaturalist "Voucher Number(s)" Observational Field, and then export them all at once into a .csv from iNat. This allows me to simply copy and paste many iNat numbers at once, without ever needing to input any of the numbers manually.
After editing, save as a tab-delimited text file in the NGSpeciesID folder. You will need to remove most of the final columns from the template. The final output should be saved like this:
Copy these Python scripts into the Programs folder you just created.
#Run Guppy Basecalling (Pop!_OS 22.04)
guppy_basecaller -x "cuda:all" -i ~/Desktop/FirstRun/fast5 -s ~/Desktop/FirstRun/basecalling --flowcell FLO-FLG001 --kit SQK-LSK110 --records_per_fastq 0 --trim_adapters --trim_strategy dna
For a Flongle cell with 1.15Gb of bases and 1.18M reads, this command takes about 37 minutes to run. Example output:
Sometimes after the run I need to restart the CPU before this command runs successfully.
#Combine all FASTQ files into a single file (Pop!_OS 22.04)
cat ~/Desktop/FirstRun/basecalling/pass/*runid*.fastq > ~/Desktop/FirstRun/basecalling/pass/basecall.fastq
#Validate the number of reads in your file (Pop!_OS 22.04)
cat ~/Desktop/FirstRun/basecalling/pass/basecall.fastq | wc -l | awk '{print $1/4}'
#Remove the uncombined FASTQ files (Pop!_OS 22.04)
rm ~/Desktop/FirstRun/basecalling/pass/*runid*.fastq
#Move your fastq file and demultiplexer to a second location (Pop!_OS 22.04)
cp ~/Desktop/FirstRun/basecalling/pass/basecall.fastq ~/Desktop/FirstRun/NGSpeciesID/basecall.fastq
cp ~/Desktop/FirstRun/Programs/minibar.py ~/Desktop/FirstRun/NGSpeciesID/minibar.py
cp ~/Desktop/FirstRun/Programs/summarize.py ~/Desktop/FirstRun/NGSpeciesID/summarize.py
cp ~/Desktop/FirstRun/Programs/primers.txt ~/Desktop/FirstRun/NGSpeciesID/primers.txt
#MinIONQC.R (Pop!_OS 22.04)
cd ~/Desktop/FirstRun/Programs
Rscript MinIONQC.R -i ~/Desktop/SecondRun/basecalling/sequencing_summary.txt -o ~/Desktop/SecondRun/basecalling/pass/summary/
Review the images that are generated. Ensure the quality scores of your run are in an appropriate range. For a 9.4.1 Flongle with Q20+ (V12) K12 chemistry, I typically get a peak in the 12-13 range.
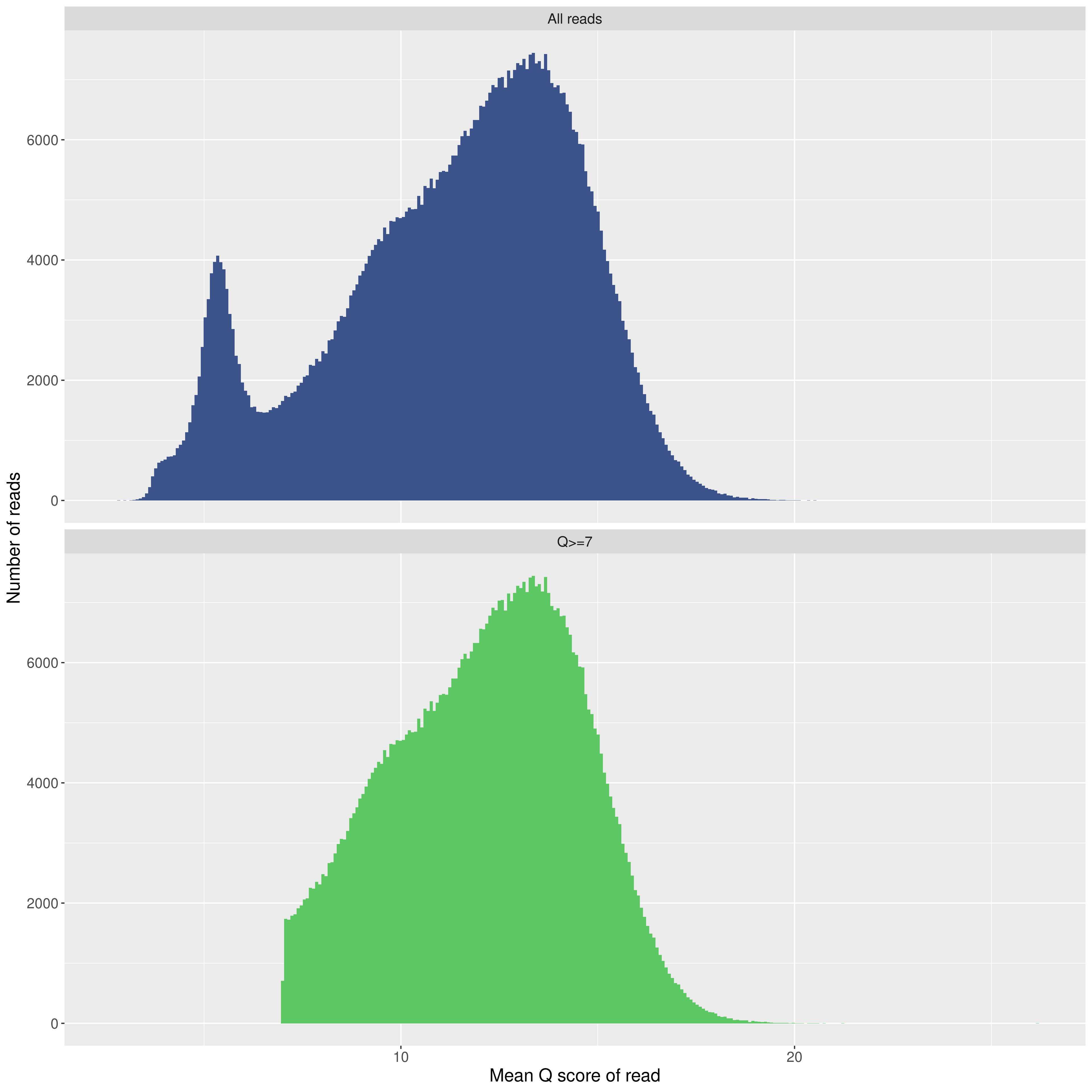
Example of all outputs from this command: MinIONQC.zip
#Go to your Programs folder and use MiniBar for Demultiplexing (Pop!_OS 22.04)
cd ~/Desktop/FirstRun/NGSpeciesID
./minibar.py -F Index.txt basecall.fastq
```This should take about 3-4 minutes to run.
<Note title="Citation" type="success" ><span>850800 seqs: H 734497 HH 581546 Hh 72882 hh 53453 IDs 707881 Mult_IDs 105306 (193.4800s)</span></Note>
#Run NGSpeciesID for read filtering, clustering, consensus generation and polishing (Pop!_OS 22.04)
conda activate NGSpeciesID
for file in *.fastq; do
bn=`basename $file .fastq`
NGSpeciesID --ont --consensus --sample_size 500 --m 800 --s 400 --medaka --primer_file primers.txt --fastq $file --outfolder ${bn}
done
```This program will take about 3-6 hours to complete.
#Create a summary of your NGSpeciesID consensus data (Pop!_OS 22.04)
python summarize.py ~/Desktop/FirstRun/NGSpeciesID

