Analysis of Genetic Ancestry from NGS Data Using EthSEQ
Davide Dalfovo, Davide Dalfovo, Alessandro Romanel, Alessandro Romanel
Abstract
Next-generation sequencing (NGS) is widely utilized both in translational cancer genomics studies and in the setting of precision medicine. Identification and stratification of an individual's ancestry is fundamental for the correct interpretation of genetic and genomic profiling. EthSEQ provides an easy and effective computational workflow to determine the ancestry of individuals, exploiting single nucleotide polymorphism genotypes computed from NGS data of whole-exome and targeted sequencing experiments. Genotypes are determined by EthSEQ from sequencing alignment files (BAM files) or can be provided as input in Variant Call Format (VCF) or CoreArray Genomic Data Structure (GDS) format. Ancestry is determined and assigned to individuals by EthSEQ exploiting a reference model and a standard or multi-step refinement approach based on Principal Component Analysis (PCA). A complete and detailed set of textual and graphical output files are generated by EthSEQ as result. EthSEQ is easy to use and can be integrated into any NGS-based processing pipeline also exploiting multi-core capabilities. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Perform ancestry analysis using a pre-computed reference model
Alternate Protocol : Perform ancestry analysis using a user-specified GDS file as reference model
Basic Protocol 2 : Perform ancestry analysis using multi-step refinement
Support Protocol 1 : Create a reference model from multiple VCF genotype data files
Support Protocol 2 : Create VCF genotype data files from a BAM file
INTRODUCTION
Next-generation sequencing (NGS) is widely utilized both in translational cancer genomics studies and in the setting of precision medicine. In particular, whole-exome sequencing (WES) and targeted sequencing (TS) are the preferred approaches for the exploration of the genomic characteristics of large-scale cohorts. Identification and stratification of individuals’ ancestry is fundamental for the correct interpretation of association studies and the investigation of the impact of personal genomic variations (Beltran et al., 2015). Of note, recent large-scale studies (Carrot-Zhang et al., 2020; Yuan et al., 2018) have shown the role of ancestry in mutation rates, DNA methylation, and mRNA expression, and have highlighted the importance of evaluating ancestry information when considering routes to disease and response to immunotherapies.
These considerations call for the development of computational tools that are able to effectively characterize ancestry from NGS data. Several model-based tools and tools based on Principal Component Analysis (PCA; Alexander, Novembre, & Lange, 2009; Li et al., 2016; Privé, Luu, Blum, McGrath, & Vilhjálmsson, 2020; Raj, Stephens, & Pritchard, 2014; Romanel, Zhang, Elemento, & Demichelis, 2017; Wang, Zhan, Liang, Abecasis, & Lin, 2015; Zhang et al., 2020) have been developed and proposed so far, but only few of them enable an easy NGS-based analysis.
In this space, we developed EthSEQ (Romanel et al., 2017), a tool that provides a fast and automated computational workflow to annotate ancestry information from WES and TS data, inspecting differential genotype profiles of SNPs while exploiting variants covered by the specific sequencing assays. As input, EthSEQ requires genotype data at single nucleotide polymorphism (SNPs) positions for a set of individuals with known ancestry (the reference model) and either a list of BAM files or genotype data of individuals with unknown ancestry (the target model). EthSEQ then uses a PCA-based approach applied on aggregated target and reference model data, and annotates the ancestry of each individual using an automated procedure. EthSEQ returns detailed information about individuals’ inferred ancestry, including visual reports. A multi-step refinement procedure is also available to better discern the annotation of ancestrally closed groups of individuals.
Here we present a documented version of EthSEQ version 3 to highlight all its features and hence make the tool available to a broader audience. Specifically, we describe detailed steps to perform ancestry analysis using different input file formats. Basic Protocol 1 describes how to perform ancestry analysis using a pre-computed reference model and genotype data of individuals with unknown ancestry in VCF format (Basic Protocol 1, a steps) or GDS format (Basic Protocol 1, b steps), or using a list of WES or TS BAM files (Basic Protocol 1, c steps). The Alternate Protocol describes how to perform ancestry analysis as described in Basic Protocol 1, but instead using a user-specified GDS file as reference model. Basic Protocol 2 describes how to perform a multi-step refinement procedure to better discern ancestry annotation across ancestrally close groups. Finally, Support Protocol 1 shows the instructions to automatically generate a reference model given a set of genomic regions of interest and genotype data of individuals with known ancestry and Support Protocol 2 shows how to externally compute the genotype of reference model's SNPs across a list of BAM files to use them as input for the Basic Protocol 1 a steps, or the Alternate Protocol.
A schematic representation of the EthSEQ version 3 computational workflow is shown in Figure 1A, together with an example of a visual report generated as results of the ancestry analysis (Fig. 1B).
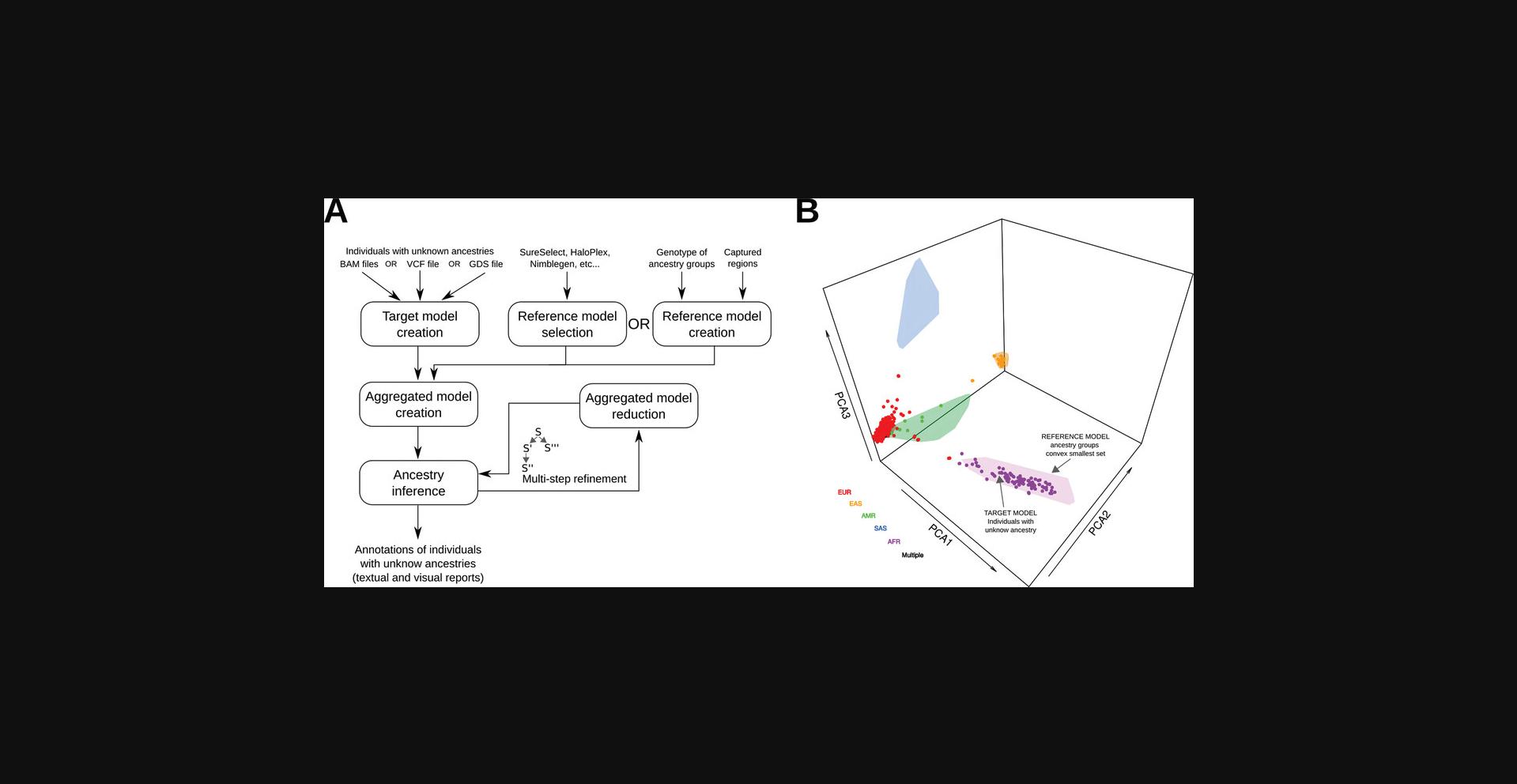
STRATEGIC PLANNING
To perform a genetic ancestry analysis, the user should provide, as input to EthSEQ, genotype data for a set of individuals of interest with unknown ancestry (the target model) and for a set of individuals with known ancestry (the reference model). Both models are required by EthSEQ to be in GDS (CoreArray Genomic Data Structures) format (Zheng et al., 2012, 2017); details about GDS file format can be found at https://bioconductor.org/packages/release/bioc/html/gdsfmt.html.
EthSEQ can handle different target model input file formats, automatically generating the proper GDS file when needed. Specifically, the user can provide a GDS data file that will be directly used as a target model to perform the analysis. Alternatively, the user can provide a list of BAM files from which genotype data are automatically computed and extracted, or a genotype data file in VCF format; details about BAM and VCF file formats can be found, respectively, at https://samtools.github.io/hts-specs/SAMv1.pdf and https://samtools.github.io/hts-specs/VCFv4.3.pdf. In both cases, EthSEQ will, as part of the processing, generate an intermediate GDS file that will be automatically used to perform the ancestry analysis.
On the other hand, the reference model must be obligatorily provided in the input as a GDS file. A set of reference models are already pre-computed and available for the user, to be automatically downloaded and used by EthSEQ. Alternatively, the user can exploit a specific functionality of EthSEQ to generate a reference model in GDS format from genotype data provided in VCF format.
Overall, to perform an ancestry analysis with EthSEQ on a set of individuals of interest, the user should necessarily have next-generation sequencing data or processed genotype data for those individuals. Data to build the reference model are instead optional and needed only when the pre-computed EthSEQ models are not suitable.
Reference Model
The set of pre-computed reference models provided by EthSEQ are built from genotype data of individuals with known ancestry. Specifically, 1000 Genomes Project genotype data were used to build whole-exome reference models considering overlapping exonic regions of SNPs as annotated by GENCODE (Frankish et al., 2021), or SNPs captured by common WES kits, including Agilent SureSelect, Twist Bioscience, Roche MedExome, and Roche KAPA. A specific function, getModelsList(), is provided by EthSEQ to retrieve the list of available reference models. The output of this function is a table (Table 1) showing all the pre-computed models that are available and that can be directly retrieved by the main analysis function, ethseq.Analysis. The first column of the table reports the name of the reference model (model.available parameter of the ethseq.Analysis function), the second column reports a detailed description of the model, and the third and fourth columns report, respectively, the list of genome assemblies (model.assembly parameter) and populations (model.pop parameter) used to build the models.
| Name | Description | Assembly | Pop |
|---|---|---|---|
| Gencode.Exome | Exon regions of protein coding genes retrieved from Gencode | hg19,hg38 | All,AFR,AMR,EAS,EUR,SAS |
| SS5.Regions | Agilent SureSelect DNA - SureSelect Human All Exon V5 | hg19,hg38 | All,AFR,AMR,EAS,EUR,SAS |
| SS6r2.Regions | Agilent SureSelect DNA - SureSelect Human All Exon V6 r2 | hg19,hg38 | All,AFR,AMR,EAS,EUR,SAS |
| SS7.Regions | Agilent SureSelect DNA - SureSelect Human All Exon V7 | hg19,hg38 | All,AFR,AMR,EAS,EUR,SAS |
| SS7.MergedProbes | Agilent SureSelect DNA - SureSelect Human All Exon V7 | hg19,hg38 | All,AFR,AMR,EAS,EUR,SAS |
- a The first and second columns report respectively the name (model.available parameter) and a description of the reference model. The third and fourth columns report, respectively, the list of genome assemblies (model.assembly parameter) and populations (model.pop parameter) available for each model.
Pre-computed EthSEQ reference models are typically used when WES data or TS data covering exonic SNPs are available and when the user is interested in characterizing the ancestry of a set of individuals against the populations described by the 1000 Genomes Project data. Of note, users who have NGS WES data or processed genotype data obtained from NGS WES data should choose the pre-computed reference model of the corresponding WES kit, if available; otherwise, we suggest using the model built with all the exonic regions defined by Gencode (model.available parameter). Moreover, the user should choose the reference model based on the human genome assembly used to align the sequence reads and to generate the genotype calls (model.assembly parameter). Finally, the user could select all or a single reference population to perform the ancestry analysis (model.pop parameter). More specifically, the user can set this parameter to ‘All’ to perform the ancestry analysis using all the superpopulation defined by the 1000 Genomes Project, or set the parameter to a specific available superpopulation to perform the analysis across the corresponding subpopulation groups. The function getSamplesInfo() could be used to list all available superpopulations and their related subpopulations.
In any other case, a more specific reference model should be used. Specifically, EthSEQ provides a function ethseq.RM to generate a reference model in GDS format from one or more VCF files (see Support Protocol 1) corresponding to genotype data of individuals with known ancestry. Once created, the reference model can be provided in input to the ethseq.Analysis function.
More generally, a reference model can be also provided to the main ethseq.Analysis function directly as a GDS file. In this case, the GDS file should provide the following minimum set of variables:
- sample.id
A unique identifier for each sample.
- snp.id
A unique identifier for each SNP.
- snp.rs.id
A character string for reference SNP ID that may not be unique.
- snp.position
The base position of each SNP on the chromosome, and 0 for unknown position; it does not allow NA.
- snp.chromosome
An integer or character mapping for each chromosome. Integer: numeric values 1-26, mapped in order from 1-22, 23=X, 24=XY (the pseudoautosomal region), 25=Y, 26=M (the mitochondrial probes), and 0 for probes with unknown positions; it does not allow NA. Character: “X”, “XY”, “Y” and “M” can be used here, and a blank string indicating unknown position. The integer/character coding must be coherent between target and reference models.
- genotype
An SNP genotypic numeric matrix. Stored in individual-major mode (SNP is the first dimension): nSNP × nsample. “0” indicates the homozygous minor allele, “1” indicates the heterozygous, “2” indicates the homozygous major allele, “3” indicates a missing genotype.
- snp.allele
A character string in the format A/B or B/A indicating if the minor allele is the reference allele (A/B) or the alternative allele (B/A).
- sample.annot
A two-column data.frame. It contains a character column ‘sex’ containing sample's sex and a character column ‘pop.group’ containing the sample's ancestry in the same order as the sample.id variable.
- snp.ref
A character array representing the reference alleles of all SNPs. This variable is mandatory together with the snp.alt variable only when EthSEQ is run using a list of BAM file as input (Basic Protocol Basic Protocol 1, c steps).
- snp.alt
A character array representing the alternative alleles of all SNPs. This variable is mandatory together with snp.ref variable, as explained above.
Target Model
To create the target model, whole-exome or targeted sequencing data or processed genotype data should be available to the user. Specifically, the target model is created by EthSEQ either from genotype data provided in VCF format (Basic Protocol 1, a steps, Alternate Protocol and Basic Protocol 2), in GDS format (Basic Protocol 1, b steps), or from an input list of individuals’ control (non-tumor) sequencing BAM files that are automatically genotyped at all reference model's positions by exploiting the genotyping module of the ASEQ tool (Romanel, Lago, Prandi, Sboner, & Demichelis, 2015; Basic Protocol 1, c steps).
Basic Protocol 1: PERFORM ANCESTRY ANALYSIS USING A PRE-COMPUTED REFERENCE MODEL
This protocol provides complete instructions for performing the ancestry analysis using a pre-computed reference model and three alternative file formats as input files. First, we show how to perform the analysis using a target model .vcf (VCF) file as input (alternative a steps). A VCF file contains sequence variations, and can be obtained from NGS data using Support Protocol 2 or any other of several available genotyping tools (FreeBayes (Garrison & Marth, 2012), GATK HaplotypeCaller (Poplin et al., 2017), VarScan (Koboldt et al., 2012), or SAMtools/BCFtools (Danecek et al., 2021)). Then, we show how to perform ancestry analysis using a GDS file as target model input (alternative b steps). To manage GDS files, EthSEQ uses a set of functions provided by the SNPRelate and gdsfmt R packages. Also, in this case, a minimum required set of variables should be stored in the GDS file (see Strategic Planning). Last, we show how to perform the ancestry analysis using a list of .bam (BAM) files as input (alternative c steps). A BAM format file is the compressed version of a SAM format file, and contains NGS aligned reads. EthSEQ expects a list of BAM files representing control (non-tumor) WES or TS NGS DNA data files obtained from a set of individuals of interest. EthSEQ determines the genotype calls for all available reference models’ SNPs for each individual from the corresponding BAM file, then merges the genotype calls of all individuals, and finally performs the ancestry analysis. Genotype information is extracted and computed by EthSEQ, exploiting the tool ASEQ. More specifically, EthSEQ downloads ASEQ and runs it automatically using its genotype mode to compute the genotype of all available reference model's SNPs across all input BAM files. Details about ASEQ are available at http://bcglab.cibio.unitn.it/aseq.
Necessary Resources
Hardware
- A 64-bit computer with ≥8 GB RAM
- An internet connection to download the reference model
- Note that the RAM required for using the tool is proportional to the size of the input VCF file (only alternative a steps). Usually, the maximum RAM required for processing a file with approximately 250,000 variants and 1000 individuals will not exceed 4.5 GB when performing the file conversion from VCF to GDS format. Using GDS or a list of BAM files as input files (alternative b and c steps, respectively) the minimum RAM required is 1 GB.
Software
- The library has been tested with R version 3.6.3+. The R software is free and can be downloaded from the official website https://cran.r-project.org/, where installation instructions are also available. Although EthSEQ is an R package and can be run across different operating systems (e.g., Windows, MacOS and Linux), the code provided in this protocol is designed to run under Linux systems.
Input files
- Target model: one of the following file formats must be provided as target model: VCF file format, GDS file format, or a list of BAM files. VCF files provided in input to EthSEQ should respect the following constraints: 1.Genotype field “GT” is used for the analysis and must be present; 2.Only positions with single reference and single alternative base are admitted; 3.No duplicated sample names are admitted; 4. No duplicated positions are admitted.
- GDS files must instead be formatted as explained in the Strategic Planning section.
Sample data
- Input data for the example analyses reported here are included and installed along with the EthSEQ package. The user can manually download and explore these data from https://github.com/cibiobcg/EthSEQ/tree/master/inst/extdata.
- Output results of the example analyses reported here are available at https://github.com/cibiobcg/EthSEQ_Data/tree/master/example_outputs/. The user can explore and compare the expected results available on the GitHub page with the results obtained by running the protocol.
R installation
- To download and install R, refer to the documentation available at the official website https://cran.r-project.org/. R is provided there as a precompiled binary for multiple operating systems.
Install and load EthSEQ
1.Run R from the command line:
- $ R
2.Install EthSEQ for the first time:
-
install.packages("EthSEQ")
3.Load the library:
-
library(EthSEQ)
- The user performs the ancestry analysis using the function ethseq.Analysis and, as previously described, must provide a reference model and a target model.
- The user defines the reference model using the parameters model.available, model.assembly, and model.pop (see Strategic Planning). The model is automatically downloaded and used by EthSEQ. Then, the user must follow one of the alternative steps based on the target model format available. The user can provide a genotype data file in VCF format (alternative a steps), a GDS data file (alternative b steps), or a list of BAM files (alternative c steps). A different set of parameters must be used when running the ethseq.Analysis function, depending on the target model file format used.
a. Ancestry analysis using a VCF file as input
4a. Run the analysis:
-
ethseq.Analysis(target.vcf = system.file("extdata", "Samples.HGDP.10000SNPs.vcf",
- package = "EthSEQ"),
- model.available = "Gencode.Exome",
- model.assembly = "hg38",
- model.pop = "All",
- out.dir = file.path(tempdir(),"EthSEQ_Analysis/"),
- verbose=TRUE,
- cores = 1,
- composite.model.call.rate = 1,
- space = "3D")
In this mode, the function ethseq.Analysis takes as input the following parameters:
- target.vcf: Path to the samples’ genotype data in VCF format;
- model.available: String specifying the pre-computed reference model to use (use getModelsList() function to retrieve the list of all available reference models);
- model.assembly (default = ‘hg38’): Version of the human assembly used to build the reference model (‘hg19’ or ‘hg38’);
- model.pop (default = ‘All’): Population of the samples to be included in the reference model; use “All” to perform the ancestry analysis using all the superpopulation defined by the 1000 Genomes Project or set the parameter to a specific available superpopulation (e.g., “EUR”) to perform the analysis across the corresponding subpopulation groups (use getSamplesInfo() function to retrieve all available populations);
- out.dir (default = tempdir()): Path to the folder where the output of the analysis is saved;
- model.folder (default = tempdir()): Path to the folder where reference models are already present or downloaded when needed;
- cores (default = 1): Number of parallel cores used for the analysis;
- verbose (default = TRUE): Print detailed execution information;
- composite.model.call.rate (default = 1): Minimum SNP call rate to include a SNP in the Principal Component Analysis (PCA);
- space (default = ‘2D’): Dimensions of PCA space used to infer ancestry (2D or 3D). 2D will use the first two principal components to infer the ancestry and generate the plot. 3D will use the first three principal components to infer the ancestry and generate the plots.
Of note, the function:
-
system.file("extdata", "Samples.HGDP.10000SNPs.vcf, package="EthSEQ")
retrieves the path to the sample VCF data file included in the EthSEQ package. Importantly, the file can also be manually downloaded from https://github.com/cibiobcg/EthSEQ/tree/master/inst/extdata and provided to the ethseq.Analysis function specifying the corresponding file system path.
The output of the analysis is written into the out.dir folder. EthSEQ produces a collection of textual and visual files reporting the inferred ancestries for all the target model's individuals. See Guidelines for Understanding Results for a detailed description of all EthSEQ outputs and their interpretation.
b. Ancestry analysis using a GDS file as input
4b. Run the analysis:
-
ethseq.Analysis(target.gds = system.file("extdata", "Samples.HGDP.10000SNPs.gds",
- package="EthSEQ"),
- model.available = "Gencode.Exome",
- model.assembly = "hg38",
- model.pop = "All",
- out.dir = file.path(tempdir(),"EthSEQ_Analysis/"),
- verbose= TRUE,
- cores = 1,
- composite.model.call.rate = 1,
- space = "3D")
In this mode, the function ethseq.Analysis takes as input the following parameters:
- target.gds: Path to the samples’ genotype data in GDS format;
- model.available: String specifying the pre-computed reference model to use (use getModelsList() function to retrieve the list of all available reference models);
- model.assembly (default = ‘hg38’): Version of the human assembly used to build the reference model model (‘hg19’ or ‘hg38’);
- model.pop (default = ‘All’): Population of the samples to be included in the reference model; use “All” to perform the ancestry analysis with all the superpopulation defined by the 1000 Genomes Project or set the parameter to a specific available superpopulation (e.g., “EUR”) to perform the analysis across the corresponding subpopulation groups (use getSamplesInfo() function to retrieve all available populations);
- out.dir (default = tempdir()): Path to the folder where the output of the analysis is saved;
- model.folder (default = tempdir()): Path to the folder where reference models are already present or downloaded when needed;
- cores (default = 1): Number of parallel cores used for the analysis;
- verbose (default = TRUE): Print detailed execution information;
- composite.model.call.rate (default = 1): Minimum SNP call rate to include a SNP in the Principal Component Analysis (PCA);
- space (default = ‘2D’): Dimensions of PCA space used to infer ancestry (2D or 3D). 2D will use the first two principal components to infer the ancestry and generate the plot. 3D will use the first three principal components to infer the ancestry and generate the plots.
As before, the function:
-
system.file("extdata", "Samples.HGDP.10000SNPs.gds, package="EthSEQ")
retrieves the path to the sample GDS data file included in the EthSEQ package. Importantly, the file can be also manually downloaded from https://github.com/cibiobcg/EthSEQ/tree/master/inst/extdata and provided to the ethseq.Analysis function specifying the corresponding file system path.
Also, in this case the output of the analysis is written into the out.dir folder, producing the same collection of textual and visual files reporting the inferred ancestries for all the target model's individuals. See Guidelines for Understanding Results for a detailed description of all EthSEQ outputs formats and their interpretation.
c. Ancestry analysis using a list of BAM files as input
4c. Create a text file containing the list of paths to the BAM files to use in the analysis:
-
write(c(file.path(tempdir(),"HGDP00228.sub_GRCh38.bam"),
- file.path(tempdir(),"HGDP01200.sub_GRCh38.bam"),
- file.path(tempdir(),"HGDP01201.sub_GRCh38.bam")),
- file.path(tempdir(),"BAMs_List.txt"))
Of note, in this example we rely on BAM files and their corresponding index files available in a temporary folder created with the function tempdir(). To download and save the files in the aforementioned temporary folder the user has to run the following R code:
-
download.file("https://github.com/cibiobcg/EthSEQ_Data/raw/master/BAM/HGDP00228.sub_GRCh38.bam",destfile = file.path(tempdir(),"HGDP00228.sub_GRCh38.bam"))
-
download.file("https://github.com/cibiobcg/EthSEQ_Data/raw/master/BAM/HGDP00228.sub_GRCh38.bam.bai",destfile = file.path(tempdir(),"HGDP00228.sub_GRCh38.bam.bai"))
-
download.file("https://github.com/cibiobcg/EthSEQ_Data/raw/master/BAM/HGDP01200.sub_GRCh38.bam",destfile = file.path(tempdir(),"HGDP01200.sub_GRCh38.bam"))
-
download.file("https://github.com/cibiobcg/EthSEQ_Data/raw/master/BAM/HGDP01200.sub_GRCh38.bam.bai",destfile = file.path(tempdir(),"HGDP01200.sub_GRCh38.bam.bai"))
-
download.file("https://github.com/cibiobcg/EthSEQ_Data/raw/master/BAM/HGDP01201.sub_GRCh38.bam",destfile = file.path(tempdir(),"HGDP01201.sub_GRCh38.bam"))
-
download.file("https://github.com/cibiobcg/EthSEQ_Data/raw/master/BAM/HGDP01201.sub_GRCh38.bam.bai",destfile = file.path(tempdir(),"HGDP01201.sub_GRCh38.bam.bai"))
5c. Run the analysis:
-
ethseq.Analysis(bam.list = file.path(tempdir(),"BAMs_List.txt"),
- model.available = "Gencode.Exome",
- out.dir = file.path(tempdir(),"EthSEQ_Analysis/"),
- verbose = TRUE,
- cores = 1,
- aseq.path = file.path(tempdir(),"EthSEQ_Analysis/"),
- run.genotype = TRUE,
- mbq = 20,
- mrq = 20,
- mdc = 10,
- composite.model.call.rate = 1,
- space = "3D",
- bam.chr.encoding = TRUE)
In this mode, the function ethseq.Analysis takes as input the following parameters:
- bam.list: Path to a file containing a list of BAM files paths;
- model.available: String specifying the pre-computed reference model to use (use getModels() function to retrieve the list of all available reference models);
- model.assembly (default = ‘hg38’): Version of the human assembly used to build the reference model (‘hg19’ or ‘hg38’);
- model.pop (default = ‘All’): Population of the samples to be included in the reference model; use “All” to perform the ancestry analysis using all the superpopulation defined by the 1000 Genomes Project or set the parameter to a specific available superpopulation (e.g., “EUR”) to perform the analysis across the corresponding subpopulation groups (use getSamplesInfo() function to retrieve all available populations);
- out.dir (default = tempdir()): Path to the folder where the output of the analysis is saved;
- model.folder (default = tempdir()): Path to the folder where reference models are already present or downloaded when needed;
- run.genotype (default = FALSE): Logical values indicating whether the ASEQ genotype should be run;
- aseq.path (default = tempdir()): Path to the folder where ASEQ binary is available or is downloaded when needed;
- mbq (default = 20): Minimum base quality used in the pileup by ASEQ;
- mrq (default = 20): Minimum read quality used in the pileup by ASEQ;
- mdc (default = 10): Minimum read count acceptable for genotype inference by ASEQ;
- cores (default = 1): Number of parallel cores used for the analysis;
- verbose (default = TRUE): Print detailed execution information;
- composite.model.call.rate (default = 1): Minimum SNP call rate to include a SNP in the Principal Component Analysis (PCA);
- space (default = ‘2D’): Dimensions of PCA space used to infer ancestry (2D or 3D). 2D will use the first two principal components to infer the ancestry and generate the plot. 3D will use the first three principal components to infer the ancestry and generate the plots.
- bam.chr.encoding (default = FALSE): Logical value indicating whether input BAM files have chromosomes encoded with "chr" prefix.
As previously mentioned, when NGS is used as input, EthSEQ invokes ASEQ to produce for each input BAM file a VCF file reporting the genotype calls for the reference model's SNPs. Specifically, ASEQ calls a heterozygous genotype for an SNP if the proportion of the coverage for the alternative base with respect to the total coverage is in the range [0.2,0.8]; otherwise, ASEQ calls a homozygous genotype, either for the reference or the alternative base. The precision of this approach is shown and discussed in Romanel et al. (2015). Although the execution of ASEQ and the processing of its output is managed automatically and transparently to the user by EthSEQ, when several BAM files are analyzed, the ASEQ processing step can take some time. In those cases, ASEQ could also be run externally (Support Protocol 2) and its output could be passed in input to EthSEQ in a secondary moment, setting the run.genotype parameter to FALSE. Importantly, in this case the ASEQ VCF output files should be provided to EthSEQ in the same format and folder as EthSEQ would have done automatically.
Setting the parameter run.genotype to FALSE is also useful when the user wants to re-run only the ancestry analysis (e.g., with different parameters) without running the entire ASEQ genotyping again.
Also in this case, the output of the analysis is written into the out.dir folder, producing the same collection of textual and visual files reporting the inferred ancestries for all the target model's individuals. See Guidelines for Understanding Results for a detailed description of all EthSEQ outputs formats and their interpretation.
Alternate Protocol: PERFORM ANCESTRY ANALYSIS USING A USER-SPECIFIED GDS FILE AS REFERENCE MODEL
This protocol provides complete instructions to perform ancestry analysis with a user-specified GDS file as reference model. In this case, a minimum set of variables should be stored in the GDS file, as described in Strategic Planning. In this protocol we show how to perform the analysis using a target model VCF file as in Basic Protocol 1, a steps.
Necessary Resources
Hardware
- 64-bit computer with ≥1 GB RAM
Software
- As described in Basic Protocol 1
Input files
- As described in Basic Protocol 1
Sample data
- Input data for the example analyses reported here are included and installed along with the EthSEQ package. The user can manually download and explore these data from https://github.com/cibiobcg/EthSEQ/tree/master/inst/extdata.
- Output results of the example analyses reported here are available at https://github.com/cibiobcg/EthSEQ_Data/tree/master/example_outputs/. The user can explore and compare the expected results available on the GitHub page with the results obtained by running the protocol.
The user should start by running step 1 to 3 as described in Basic Protocol 1 and then:
4.Run the analysis:
-
ethseq.Analysis(target.vcf = system.file("extdata", "Samples.HGDP.10000SNPs.vcf",
- package = "EthSEQ"),
- model.gds = system.file("extdata", "Reference.Gencode.Exome.10000SNPs.gds",
- package = "EthSEQ"),
- out.dir = file.path(tempdir(),"EthSEQ_Analysis/"),
- verbose = TRUE,
- cores = 1,
- composite.model.call.rate = 1,
- space = "3D")
In this mode, the function ethseq.Analysis takes as input the following parameters:
- target.vcf: path to the samples’ genotype data file in VCF format. Only one file is allowed as input in the analysis;
- model.gds: path to a GDS file specifying the reference model;
- out.dir (default = tempdir()): path to the folder where the output of the analysis is saved; 4.verbose (default = TRUE): print detailed execution information;
- cores (default = 1): number of parallel cores used for the analysis;
- composite.model.call.rate (default = 1): SNP call rate used to run the Principal Component Analysis (PCA). The SNPs with a call rate lower than this value are not included in the analysis;
- space (default = ‘2D’): Dimensions of PCA space used to infer ancestry (2D or 3D). 2D will use the first two principal components to infer the ancestry and generate the plot. 3D will use the first three principal components to infer the ancestry and generate the plots.
Of note, the functions:
-
system.file("extdata", "Samples.HGDP.10000SNPs.vcf, package="EthSEQ")
-
system.file("extdata","Reference.Gencode.Exome.10000SNPs.gds,package="EthSEQ")
retrieve the paths to the sample data files included in the EthSEQ package. Importantly, these files can be also manually downloaded from https://github.com/cibiobcg/EthSEQ/tree/master/inst/extdata and provided to the ethseq.Analysis function specifying the corresponding file system paths.
Also, in this case the output of the analysis is written into the out.dir folder, producing the same collection of textual and visual files reporting the inferred ancestries for all the target model's individuals. See Guidelines for Understanding Results for a detailed description of all EthSEQ outputs formats and their interpretation.
The same analysis can be performed using as target input a GDS file (similar to Basic Protocol 1, b steps) or a list of BAM files (similar to Basic Protocol 1, c steps). Specifically, using the procedure described in Basic Protocol 1, b steps and Basic Protocol 1, c steps, it is enough to replace the parameters model.available, model.assembly, and model.pop with the parameter model.gds to perform the analysis using a user-specified GDS reference model, as described above.
Basic Protocol 2: PERFORM ANCESTRY ANALYSIS USING MULTI-STEP REFINEMENT
This protocol provides complete instructions to perform an ancestry analysis using a VCF file as input (as Basic Protocol 1, a steps), but exploiting a multi-step refinement procedure to better discern ancestry annotations across ancestrally close groups. This approach relies on a tree structure (Fig. 2A), provided in input to EthSEQ as a matrix encoding (Fig. 2B), that represents recursively the annotation of close ancestry groups that we want to refine. Specifically, given a tree of ancestry group sets such that sibling nodes have non-intersecting ancestry groups and child nodes have ancestry groups included in the parent node ancestry groups, ancestry of individuals is inferred following a pre-order traversal of the tree (Fig. 2C). At each node with ancestry groups S , annotations resulting from the analysis of the parent node are refined by reducing both reference and target models on individuals with annotations in S only, and performing again the PCA analysis on these individuals. Global annotation of all individuals is updated throughout the tree traversal.
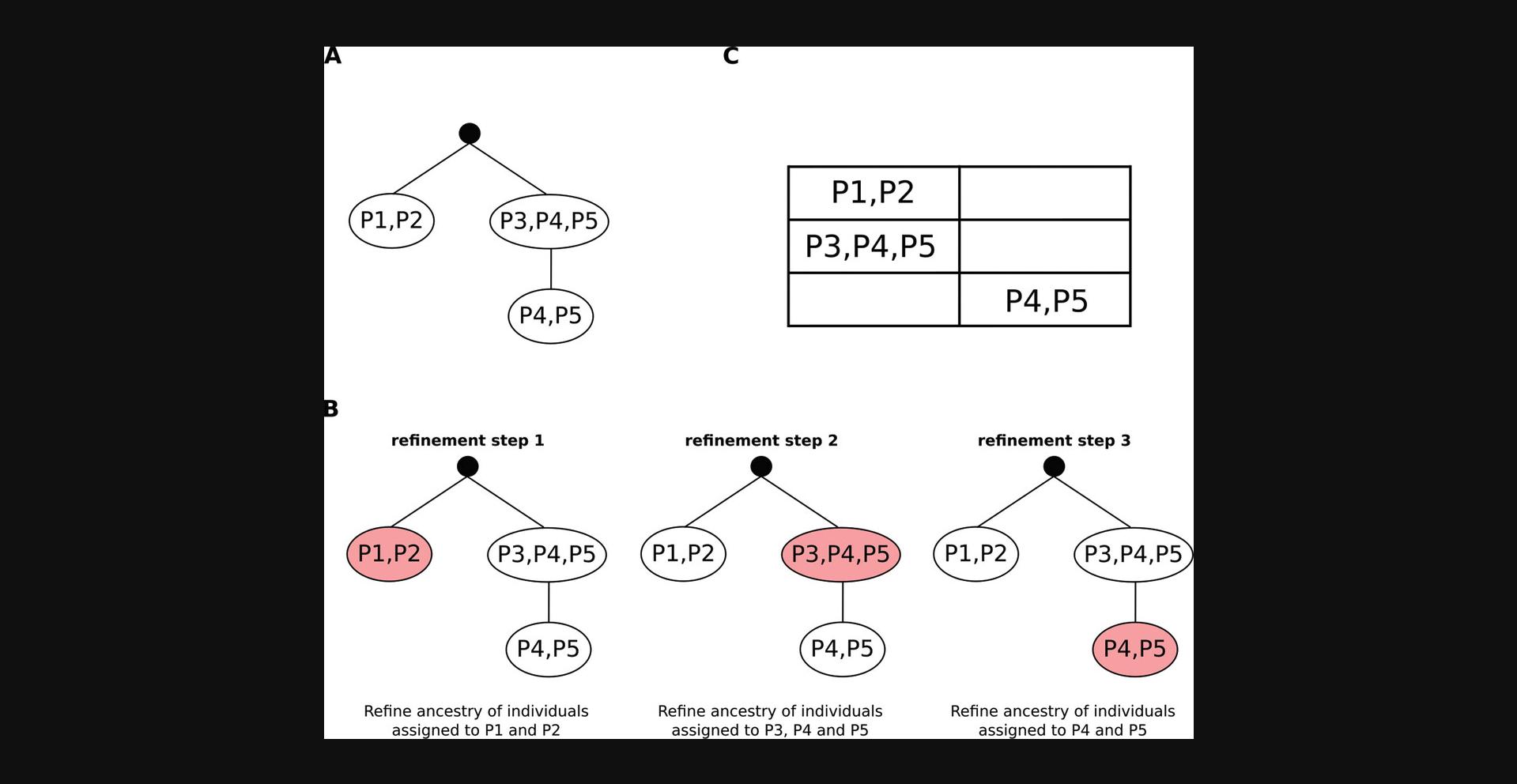
Necessary Resources
Hardware
- As described in Basic Protocol 1
Software
- As described in Basic Protocol 1
Input files
- As described in Basic Protocol 1
Sample data
- Input data for the example analyses reported here are included and installed along with the EthSEQ package. The user can manually download and explore these data from https://github.com/cibiobcg/EthSEQ/tree/master/inst/extdata.
- Output results of the example analyses reported here are available at https://github.com/cibiobcg/EthSEQ_Data/tree/master/example_outputs/. The user can explore and compare the expected results available on the GitHub page with the results obtained by running the protocol.
The user should start by running step 1 to 3 as described in Basic Protocol 1 and then:
4.Prepare the multi-step refinement tree encoding:
-
m = matrix("",ncol=2,nrow=2)
-
m[1,1] = "EUR|AFR|AMR"
-
m[2,2] = "EUR|AMR"
A 2 × 2 matrix is created representing a tree with a root node containing three populations and a child node containing two populations.
5.Run the ancestry analysis:
-
ethseq.Analysis(target.vcf = system.file("extdata",
- "Samples.HGDP.10000SNPs.vcf", package="EthSEQ"),
- out.dir = file.path(tempdir(),"EthSEQ_Analysis/"),
- model.available = "Gencode.Exome",
- verbose = TRUE,
- refinement.analysis = m,
- composite.model.call.rate = 1,
- space = "3D")
The same analysis can be performed using as target input: a GDS file (similar to Basic Protocol 1, b steps) or a list of BAM files (similar to Basic Protocol 1, c steps). Briefly, using the same procedure described in the Basic Protocol 1, b steps, and Basic Protocol 1, c steps, it is enough to add, in the “Run the analysis” step, the refinement.analysis parameter to perform the multi-step refinement procedure, as described above.
Support Protocol 1: CREATE A REFERENCE MODEL FROM MULTIPLE VCF GENOTYPE DATA FILES
This protocol provides complete information to generate a reference model given genotype data for a set of individuals already annotated for ancestry. This function takes as input a list of paths to VCF files to build the reference model. Optionally, the user can provide a path to a Browser Extensible Data (BED) file describing a set of genomic regions of interest to subset the VCF files. This is particularly useful when reference models for TS panels are to be created.
Necessary Resources
Hardware
- A 64-bit computer with ≥8 GB RAM. Note that the RAM required for performing this step is proportional to the size of each input VCF file. In most cases, less than 8 GB are enough. The maximum RAM required for processing a file with approximately 250,000 variants and 1000 individuals will not exceed 4.5 GB when performing the file conversion from VCF to GDS format.
Software
- As described in Basic Protocol 1
Input files
- As described in Basic Protocol 1
Sample data
- Input data for the example analyses reported here are included and installed along with the EthSEQ package. The user can manually download and explore these data from https://github.com/cibiobcg/EthSEQ/tree/master/inst/extdata.
- Output results of the example analyses reported here are available at https://github.com/cibiobcg/EthSEQ_Data/tree/master/example_outputs/. The user can explore and compare the expected results available on the GitHub page with the results obtained by running the protocol.
The user should start by running step 1 to 3 as described in Basic Protocol 1 and then:
4.Prepare the input data:
-
vcf.files = c(system.file("extdata","RefSample1.vcf", package="EthSEQ"),
- system.file("extdata","RefSample2.vcf", package="EthSEQ"))
-
annot.samples = read.delim(system.file("extdata", "Annotations_Test_v3.txt",
- package="EthSEQ"))
5.Run the analysis:
-
ethseq.RM(vcf.fn = vcf.files,
- annotations = annot.samples,
- out.dir = file.path(tempdir(),"EthSEQ_Analysis/"),
- model.name = "Reference.Model")
The function ethseq.RM takes as input the following parameters:
- vcf.fn: Vector of paths to genotype files in VCF format;
- annotations: data.frame with mapping of all samples names, known ancestries and sex;
- out.dir (default = tempdir()): Path to output folder; 4.model.name (default = “Reference.Model”): Name of the output model; 5.bed.fn (default = NA): Path to a BED file with a list of genomic regions of interest;
- call.rate (default = 1): SNP call rate cutoff for inclusion in the final reference model;
- cores (default = 1): Number of parallel cores to be use in the generation of the reference model.
Of note, the functions:
-
system.file("extdata","RefSample1.vcf", package="EthSEQ")
-
system.file("extdata","RefSample2.vcf", package="EthSEQ")
-
system.file("extdata", "Annotations_Test_v3.txt",package="EthSEQ")
retrieve the paths to the sample data files included in the EthSEQ package. Importantly, these files can be also manually downloaded from https://github.com/cibiobcg/EthSEQ/tree/master/inst/extdata and provided to the ethseq.Analysis function specifying the corresponding file system paths.
Table 2 shows the header and the first 10 rows of the annotation data.frame. Three columns (sample, pop, sex) must be present and are used to perform the analysis. The sample column reports the sample name—all the sample id's in the genotype files must be present in the sample column of the annotation data.frame. The pop column reports the known ancestry for each sample. The sex column reports the sex of each individual. If more columns are present in the data.frame, they will be ignored by EthSEQ and not included in the reference model.
The output of the analysis will be written to the out.dir folder. EthSEQ produces the file Reference.Model.gds that can be used as input reference model, as described in the Alternate Protocol. See Guidelines for Understanding Results for details on all EthSEQ outputs and their interpretation.
| Sample | pop | sex |
|---|---|---|
| HG00096 | EUR | M |
| HG00100 | EUR | F |
| HG00101 | EUR | M |
| HG00103 | EUR | M |
| HG00106 | EUR | F |
| HG00108 | EUR | M |
| HG00111 | EUR | F |
| HG00112 | EUR | M |
| HG00116 | EUR | M |
| HG00117 | EUR | M |
- a
First column represents individuals’ names, the second column contains the associated known ancestry and column three contains the individuals’ sex.
Support Protocol 2: CREATE VCF GENOTYPE DATA FILE FROM A BAM FILE USING ASEQ
This protocol provides complete information to generate the genotype calls of a set of SNPs positions from a BAM file using ASEQ. Here we describe also how to use the output VCF genotype data file as input to perform ancestry analysis as in Basic Protocols 1, a and c steps, Alternate Protocol and Basic Protocol 2.
Necessary Resources
Hardware
- 64-bit computer with ≥8 GB RAM
Software
- ASEQ and wget. Although ASEQ can be run across different operating systems (e.g., Windows, MacOS and Linux), the code provided in this protocol is designed to run under Linux systems.
Input files
- A BAM file containing aligned reads
- A VCF file containing a list of SNP positions. Of note, only positions with single reference and single alternative base are admitted in the file.
Sample data
- Input data are already included and installed along with the EthSEQ package. The user can manually download these data from https://github.com/cibiobcg/EthSEQ/tree/master/inst/extdata.
- Output results are available from (https://github.com/cibiobcg/EthSEQ_Data/tree/master/example_outputs/). The user can explore and compare the expected results on the GitHub page with the results obtained from the protocol.
1.From the command line, download ASEQ:
2.Add execute permission to ASEQ:
- $ chmod u+x ASEQ
3.Run the analysis:
- $./ASEQ vcf=ModelPositions.vcf bam=HGDP00228.sub_GRCh38.bam mode=GENOTYPE threads=1 htperc=0.2 mbq=20 mrq=20 mdc=20 out=./
ASEQ takes as input the following parameters:
1.bam: path to the sequence alignment data in BAM format; 2.vcf: path to the SNP positions in VCF format; 3.out: Path to the folder where the output of the analysis is saved; 4. mode (default = PILEUP): execution mode; 5. threads (default = 1): number of threads to be use for ASEQ computation; 6. htperc (default = 0.2): this value specifies the allelic fraction range [htperc,1-htperc] to call a SNP as heterozygous; 7. mbq (default = 1): Minimum base quality used in the pileup; 8. mrq (default = 1): Minimum read quality used in the pileup; 9. mdc (default = 1): Minimum read count acceptable for genotype calculation.
This example relies on the data that can be downloaded from the command line using the following commands:
- $ wget https://github.com/cibiobcg/EthSEQ_Data/raw/master/BAM/HGDP00228.sub_GRCh38.bam
- $ wget https://github.com/cibiobcg/EthSEQ_Data/raw/master/ModelPositions.vcf
The output of the analysis is written to the out folder. In this example, ASEQ produces the file HGDP00228.sub_GRCh38.genotype.vcf, containing the genotype calls for all SNPs position listed in the ModelPositions.vcf file. Having obtained a VCF file for each target individual of interest, the user can either run Basic Protocol 1, c steps, moving the files into the expected EthSEQ folder (see Guidelines for Understanding Results for details), or aggregate all VCF files into a single VCF file and run Basic Protocol 1, a steps, Alternate Protocol and Basic Protocol 2.
GUIDELINES FOR UNDERSTANDING RESULTS
Both ethseq.Analysis and ethseq.RM functions produce an output folder named according to the value specified in the out.dir parameter. This folder contains several files including intermediate results. These files are kept to examine the results of intermediate steps and, if needed, to be used as input to other downstream analyses.
If EthSEQ is run successfully using the ethseq.Analysis function to annotate the ancestry of the input individuals (Basic Protocol 1, a, b, and c steps), then the output folder will always contain the following files:
├── Aggregated.gds ├── Report.PCAcoord ├── Report.pdf ├── Report.txt └── Target.gds
- Target.gds:
EthSEQ builds the GDS corresponding to target model from the VCF or BAM files provided in input. This file is not generated when EthSEQ is run using a GDS file as target model in input (Basic Protocol Basic Protocol 1, b steps).
- Aggregated.gds
This GDS file contains the genotype data of the combined target and reference models. EthSEQ creates this file and uses it to perform the PCA.
- Report.pdf
This file (Fig. 3) contains the plot(s) of the 2- or 3-dimensional space built with the first two or three principal components results of the PCA. If the parameter space is set to ‘3D’, the file contains a 3-dimensional (Fig. 3A) and all the 2-dimensional (one for each pair of components) representations of the first 3 principal components (Fig. 3B-D). Otherwise, if the parameter space is set to ‘2D’, the file contains only a 2-dimensional representation of the first 2 principal components (Fig. 3B). In the plots, the polygons represent the smallest convex sets (convex hull) identifying the ancestry groups described in the reference model. The points instead represent the individuals contained in the target model (i.e., the individuals of which we want to perform the ancestry analysis). The color of each target individual point is set according to the estimated ancestry and refers to the corresponding reference ancestry color; individuals positioned inside two or more ancestry groups are colored in black.
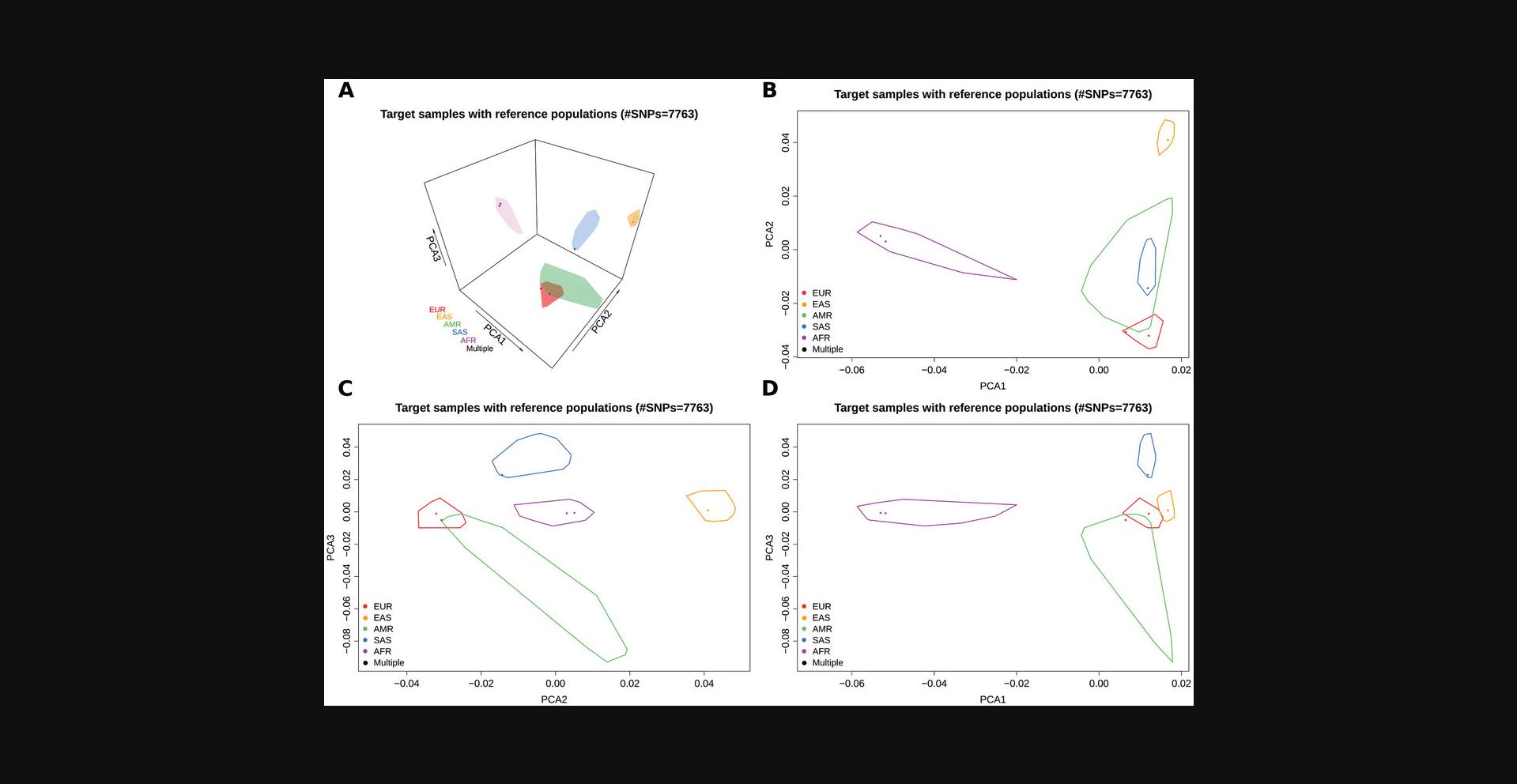
- Report.PCAcoord.txt
This file contains the principal component values for all target model's individuals. Each row represents the PCA space coordinates of an individual included in the target model.
- Report.txt
This file contains the results of the inferred ancestries. Table 3 shows an example of output reported in this file. The first column represents the individual id, the second column represents the inferred ancestry, and the third column represents the position of the individual with respect to the convex hulls. Individuals positioned inside a reference ancestry group are annotated with the corresponding reference ancestry and labeled as INSIDE. Individuals positioned outside all the reference ancestry groups are labeled as CLOSEST; in this case the fourth column reports the relative contribution of each reference ancestry group computed as the relative distance from the ancestry group centroid.
| Sample.id | Pop | Type | Contribution |
|---|---|---|---|
| Sample1 | AFR | INSIDE | |
| Sample2 | AFR | INSIDE | |
| Sample3 | EUR | INSIDE | |
| Sample4 | EUR | CLOSEST | EUR(86.96%)|AMR(13.04%) |
| Sample5 | SAS | INSIDE | |
| Sample6 | EAS | INSIDE |
- a The first column represents individuals ids, the second column contains the inferred ancestries, the third column contains the types of inferred ancestries, and the fourth column contains all reference model's populations contributions.
When any of these files is missing, this means that the analysis terminated prematurely (with the exception of Target.gds file that is not present when running Basic Protocol 1, b steps). In this case, the user can refer to the Troubleshooting section to deploy a possible cause of error. The main results of the ancestry analysis are saved in the Report.txt file. In our experience, when the reference model is suitable to represent the ancestries of the individuals in the target model, the user should expect to find in this file most of the inferred ancestries called as INSIDE. If something went wrong, the user may find the inferred ancestries mostly called as CLOSEST, with similar contributions across all reference ancestry groups. In this case, the design of the experiment and/or the parameters used in the ethseq.Analysis function are probably wrong (e.g., different genome assembly used).
When EthSEQ is run on a list sample1.bam, …, sampleN.bam of BAM files (Basic Protocols 1, c steps), the top-level output directory contains the same output files as described above, with two additional files and a subdirectory containing the ASEQ output files:
├──
├── sample1.genotype.vcf
├── sample1.GENOTYPE.ASEQ
├── sample1.heterozygous.vcf
├── …
├── sampleN.genotype.vcf
├── sampleN.GENOTYPE.ASEQ
└── sampleN.heterozygous.vcf
- ASEQ
ASEQ tool executable.
- ModelPosition.vcf
This file contains the list of SNP positions extracted from the reference model. This file is in VCF format and is used by ASEQ to call the genotypes at the specific positions listed in it.
- SampleX.genotype.vcf
These files in VCF format contain the genotype calls generated by ASEQ that are used by EthSEQ to perform the ancestry analysis. The number of these files corresponds to the number of input BAM files.
- SampleX.GENOTYPE.ASEQ
These files contain the pileup information for all SNP positions processed by ASEQ. For each SNP, the files report the read count for each of the 4 bases A, C, G and T, the allelic fraction (read count for the alternative bases divided by the total read count at that position), the total read count (e.g., coverage), the genomic coordinates (chromosome and position), and if available the unique identifier (dbSNP ID). This file is not used by EthSEQ. The number of these files correspond to the number of input BAM files.
- SampleX.heterozygous.vcf
These files contain the subset of SNPs that ASEQ called with heterozygous genotype. This file is not used by EthSEQ. The number of these files correspond to the number of input BAM files.
In detail, ASEQ performs all SNPs genotype calls for all input BAM files and generates all the output files into the ASEQGenotypes folder. If any of these files is missing or no genotypes are called, it means that an error occurred. In this case, the user can refer to the Troubleshooting section to deploy a possible cause of error. EthSEQ then uses the generated genotype calls to perform the ancestry analysis. The main results of the ancestry analysis are stored in the Report.txt file as described above. The user can re-run the ancestry analysis avoiding the re-generation of the genotypes by setting the parameter run.genotype to FALSE. In this case, the ASEQ step is not performed and EthSEQ performs the ancestry analysis using the files available in the expected ASEQGenotypes folder.
If EthSEQ is run to generate the reference model using the ethseq.RM function (Support Protocol 1), the out.dir directory contains the output files and all the intermediate files. For each input VCF genotype file, an intermediate GDS file is generated:
├── ReferenceModel.gds ├── RefSample1.vcf.gds └── RefSample2.vcf.gds
The ethseq.RM function creates in the output folder the final reference model named according to the name specified in model.name parameter. The file names of the created intermediate GDS files correspond to the names of the VCF input files. The resulting reference model is saved in the ReferenceModel.gds file. The user can perform an ancestry analysis using this file as reference model, setting the model.gds parameter of the ethseq.Analysis function with the path to this file as described in the Alternate Protocol.
If EthSEQ is run by performing the multi-step refinement analysis (Basic Protocol 2), the out.dir directory contains a different set of output files that depends on the refinement tree used. EthSEQ generates a set of files as explained above and renames the files reporting PCA coordinates (PCAcoord) and visual plots by adding the suffix ‘_Ref0 ’. Then, EthSEQ generates the same pair of files at each refinement step, increasing the number in the suffix by 1. Specifically, for each refinement step performed, a new set of PCAcoord and visual plot files are generated, and the global annotation file Report.txt is updated throughout the refinement steps. Of note, when multi-step refinement analysis is performed, no ancestry groups contributions are reported in the Report.txt file.
├── Aggregated.gds ├── Report.txt ├── Report_Ref0.PCAcoord ├── Report_Ref0.pdf ├── Report_Ref1.PCAcoord ├── Report_Ref1.pdf ├── … └── Target.pdf
- Target.gds and Aggregated.gds
As described above.
- Report.txt
This file contains the inferred ancestries. The multi-step refinement analysis updates this file at each refinement step. An example of file content is reported in Table 3, except for the fourth column with the contribution that in this mode is not generated.
- Report_Ref0.PCAcoord and Report_Ref0.pdf
These files contain the PCA coordinates and the PCA plots (Fig. 4A) result of the analysis that uses all the ancestry groups present in the reference model. The content of these two files is the same as the content of the files Report.PCAcoord and Report.pdf generated performing the analysis without the multi-step refinement. <img src="https://static.yanyin.tech/literature_test/cpz1663-fig-0004-m.jpg" alt="Example of output files performing a multi-step refinement analysis. The refinement tree used for this analysis has only one node defined as follows: "EUR|AFR|AMR". (A) Represents the Report_Ref0.pdf file showing the 2-dimensional representation of the first 2 principal components at step 0 of the multi-step procedure. (B) Represents the Report_Ref1.pdf file showing the 2-dimensional representation of the first 2 principal components at step 1 of the multi-step procedure." loading="lazy" title="Details are in the caption following the image"/>
- Report_RefX.PCAcoord and Report_RefX.pdf
These files contain the PCA coordinates and the PCA plots (Fig. 4B) result of the analysis performed at the X refinement step. EthSEQ generates pairs of these output files numbering them from 1 and increasing the numbering in the file name by 1 at each refinement step.
The user should find in the output folder a number of files proportional to the number of nodes in the refinement tree. For example, if the user runs Basic Protocol 2, the Report.PCAcoord and Report.pdf are replaced by three pairs of Report_RefX.PCAcoord and Report_RefX.pdf, where X is a number from 0 to 2. The main results of the ancestry analysis after all the refinement steps are available in the Report.txt file.
If ASEQ is run externally to generate genotype calls from a sample.bam BAM file (Support Protocol 2), the out directory specified when running the command will contain the following files:
├── sample.genotype.vcf ├── sample.GENOTYPE.ASEQ └── sample.heterozygous.vcf
- Sample.genotype.vcf
This file in VCF format contains the genotype calls generated for all input SNPs.
- Sample.GENOTYPE.ASEQ
As described above.
- Sample.heterozygous.vcf
As described above.
ASEQ performs genotype calls, and the result is saved in the sample.genotype.vcf file. The user can use this file as target model file input in VCF format to perform ancestry analysis.
COMMENTARY
Background Information
EthSEQ version 3 is an R package available at The Comprehensive Archive Network (https://cran.r-project.org) that includes significant improvements over the previous EthSEQ implementation. This is the result of its application to several genomics and clinical cohorts and to a variety of sequencing platforms, including different WES kits and different TS panels. In Carrot-Zhang et al. (2020), the EthSEQ approach was extended to exploit three PCA dimensions instead of two, improving its sensitivity and precision. In addition, due to its application across different scenarios (Beltran et al., 2015; Gandellini et al., 2019; Huang et al., 2017; Orlando et al., 2022; Sailer et al., 2019; Valentini et al., 2022), several computational and reporting improvements have been implemented. Specifically, EthSEQ version 3 requires 10 times less memory and provides a much larger collection of reference models across different human reference genome assemblies. Of note, although EthSEQ was designed for WES and TS data, its current ability to manage different types of input genotype data formats extends and generalizes its usability.
Critical Parameters
•composite.model.call.rate
EthSEQ PCA analysis uses this parameter considering the created aggregated GDS model. All SNPs with a call rate (calculated on the aggregated model) lower than this parameter are removed from the analysis. When this value is set to 1, EthSEQ performs the PCA analysis using only the SNPs with genotype calls available for all individuals. We suggest starting with this parameter set to 1.Sometimes setting the call rate to 1 is too restrictive, and a high number of SNPs are removed. As a consequence, the variance in the data could drastically drop. In those cases, the convex hulls tend to be bigger and closer to each other (Fig. 5A). Although in most cases the analysis is robust and able to define ancestries with good reliability also when a low number of SNPs is available, reducing the value of the parameter could improve the analysis precision. When this parameter is set to a number less than 1, the aggregated model includes SNPs with a call rate <1.When this parameter is too low, however, it is possible that several SNPs with missing genotype calls in the target samples (but not in the reference) are included; all samples in the pre-computed reference models have genotype calls for all SNPs. In these cases, the variance in target individuals might be too low with the PCA analysis tending to cluster target samples together in the center of the PCA space (Fig. 5B). Reasonable values of this parameter are usually >0.95.
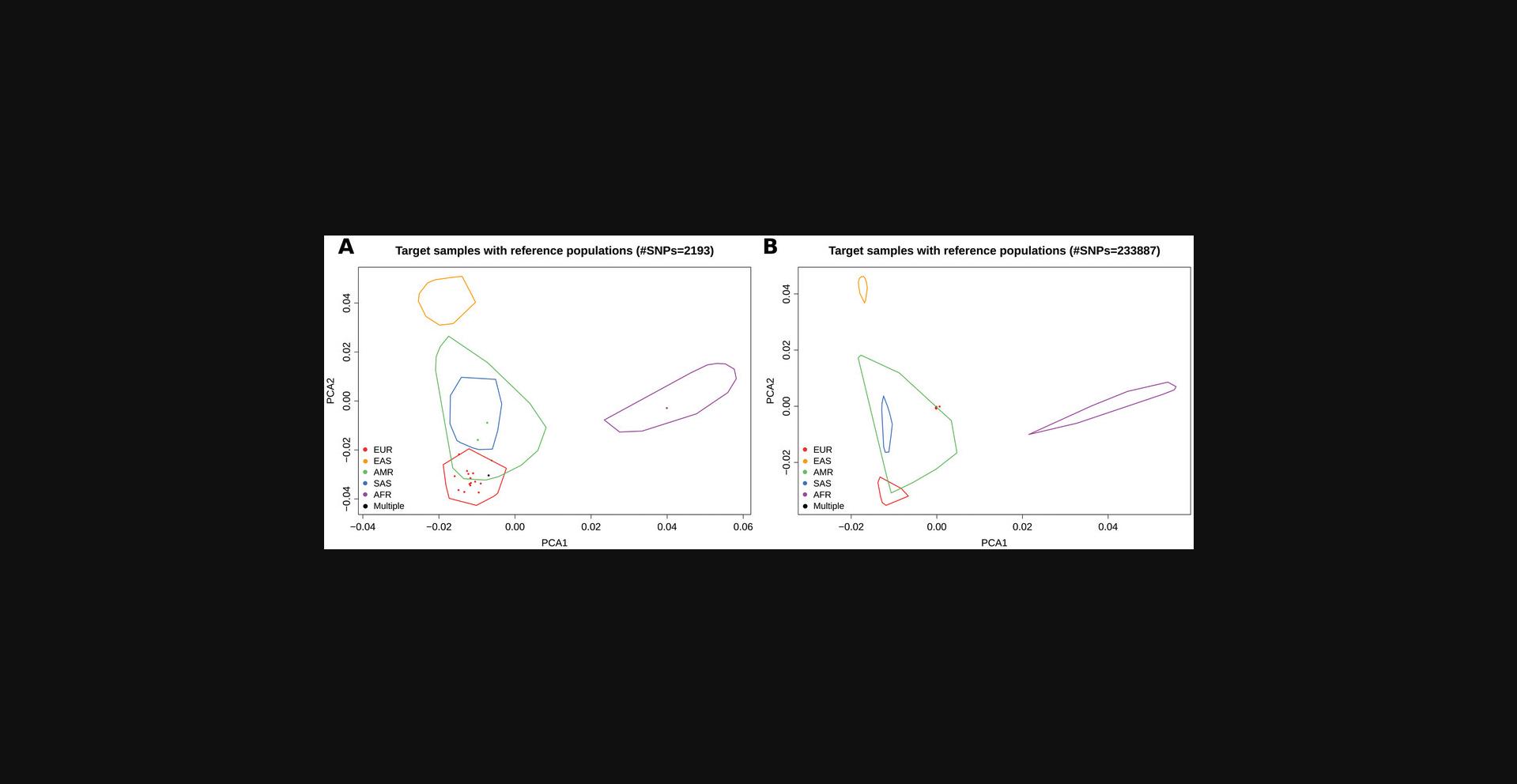
Troubleshooting
If the parameter verbose is set to ‘TRUE’, EthSEQ prints a detailed log. If an error occurs, carefully inspect this log and the step in which this occurs. See Table 4 for possible problems, causes and solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Analysis is prematurely terminated with no error printed during “Create Target model” step | The available RAM may be not enough | Increase the available RAM or the swap memory if working on Linux/MacOS system |
| All the target samples are in the middle of the PCA plots. | Several SNPs with missing calls are present in the aggregated model. This may due to the composite.model.call.rate parameter set with a too low a value. | Increase the value of the composite.model.call.rate parameter. |
| The convex sets are huge and overlap each other | A low number of SNPs is used to infer the ancestry | Decrease the value of the composite.model.call.rate parameter. Check how many SNPs are present in the input file and intersection with the reference. |
| All the SNPs are excluded during the aggregation step when performing Basic Protocol 1 | The assembly version and/or the chromosome encoding is different between reference and target models | Change the model.assembly and/or the bam.chr.encoding parameters according with input data |
| Analysis run exits with the message: Error occurs: Aggregated.gds has been created or opened | You are running EthSEQ in an interactive R session, and a previous analysis has failed with an error | Close the current R session and run the analysis again in a new R session |
Acknowledgments
The research leading to these results has received funding from Fondazione AIRC under MFAG 2017 - ID. 20621 project: P.I. Alessandro Romanel.
Open Access Funding provided by Università degli Studi di Trento within the CRUI-CARE Agreement.
Author Contributions
Davide Dalfovo : Conceptualization, data curation, formal analysis, methodology, software, validation, writing original draft, writing review and editing; Alessandro Romanel : Conceptualization, data curation, formal analysis, funding acquisition, methodology, software, supervision, validation, writing original draft, writing review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The EthSEQ R package is available at CRAN and at https://github.com/cibiobcg/EthSEQ.
Literature Cited
- Alexander, D. H., Novembre, J., & Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Research , 19, 1655–1664. doi: 10.1101/gr.094052.109
- Beltran, H., Eng, K., Mosquera, J. M., Sigaras, A., Romanel, A., Rennert, H., … Rubin, M. A. (2015). Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncology , 1, 466–474. doi: 10.1001/jamaoncol.2015.1313
- Carrot-Zhang, J., Chambwe, N., Damrauer, J. S., Knijnenburg, T. A., Robertson, A. G., Yau, C., … Beroukhim, R. (2020). Comprehensive analysis of genetic ancestry and its molecular correlates in cancer. Cancer Cell , 37, 639–654.e6. doi: 10.1016/j.ccell.2020.04.012
- Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., … Li, H. (2021). Twelve years of SAMtools and BCFtools. GigaScience , 10, giab008. doi: 10.1093/gigascience/giab008
- Frankish, A., Diekhans, M., Jungreis, I., Lagarde, J., Loveland, J. E., Mudge, J. M., … Flicek, P. (2021). GENCODE 2021. Nucleic Acids Research , 49, D916–D923. doi: 10.1093/nar/gkaa1087
- Gandellini, P., Casiraghi, N., Rancati, T., Benelli, M., Doldi, V., Romanel, A., … Zaffaroni, N. (2019). Core biopsies from prostate cancer patients in active surveillance protocols harbor PTEN and MYC alterations. European Urology Oncology , 2, 277–285. doi: 10.1016/j.euo.2018.08.010
- Garrison, E., & Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. Retrieved from https://arxiv.org/abs/1207.3907
- Huang, F. W., Mosquera, J. M., Garofalo, A., Oh, C., Baco, M., Amin-Mansour, A., … Garraway, L. A. (2017). Exome sequencing of African-American prostate cancer reveals loss-of-function ERF mutations. Cancer Discovery , 7, 973–983. doi: 10.1158/2159-8290.CD-16-0960
- Koboldt, D. C., Zhang, Q., Larson, D. E., Shen, D., McLellan, M. D., Lin, L., … Wilson, R. K. (2012). VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Research , 22, 568–576. doi: 10.1101/gr.129684.111
- Li, Y., Byun, J., Cai, G., Xiao, X., Han, Y., Cornelis, O., … Amos, C. I. (2016). FastPop: A rapid principal component derived method to infer intercontinental ancestry using genetic data. BMC Bioinformatics , 17, 122. doi: 10.1186/s12859-016-0965-1
- Orlando, F., Romanel, A., Trujillo, B., Sigouros, M., Wetterskog, D., Quaini, O., … Demichelis, F. (2022). Allele-informed copy number evaluation of plasma DNA samples from metastatic prostate cancer patients: The PCF_SELECT consortium assay. NAR Cancer , 4, zcac016. doi: 10.1093/narcan/zcac016
- Poplin, R., Ruano-Rubio, V., DePristo, M. A., Fennell, T. J., Carneiro, M. O., Van der Auwera, G. A., … Banks, E. (2017). Scaling accurate genetic variant discovery to tens of thousands of samples. Genomics , Retrieved from http://biorxiv.org/lookup/doi/10.1101/201178
- Privé, F., Luu, K., Blum, M. G. B., McGrath, J. J., & Vilhjálmsson, B. J. (2020). Efficient toolkit implementing best practices for principal component analysis of population genetic data. Bioinformatics , 36, 4449–4457. doi: 10.1093/bioinformatics/btaa520
- Raj, A., Stephens, M., & Pritchard, J. K. (2014). fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics , 197, 573–589. doi: 10.1534/genetics.114.164350
- Romanel, A., Lago, S., Prandi, D., Sboner, A., & Demichelis, F. (2015). ASEQ: Fast allele-specific studies from next-generation sequencing data. BMC Medical Genomics , 8, 9. doi: 10.1186/s12920-015-0084-2
- Romanel, A., Zhang, T., Elemento, O., & Demichelis, F. (2017). EthSEQ: Ethnicity annotation from whole exome sequencing data. Bioinformatics , 33, 2402–2404. doi: 10.1093/bioinformatics/btx165
- Sailer, V., Eng, K. W., Zhang, T., Bareja, R., Pisapia, D. J., Sigaras, A., … Beltran, H. (2019). Integrative molecular analysis of patients with advanced and metastatic cancer. JCO Precision Oncology , 3, PO.19.00047. doi: 10.1200/PO.19.00047
- Valentini, S., Gandolfi, F., Carolo, M., Dalfovo, D., Pozza, L., & Romanel, A. (2022). Polympact: Exploring functional relations among common human genetic variants. Nucleic Acids Research , 50, 1335–1350. doi: 10.1093/nar/gkac024
- Wang, C., Zhan, X., Liang, L., Abecasis, G. R., & Lin, X. (2015). Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. American Journal of Human Genetics , 96, 926–937. doi: 10.1016/j.ajhg.2015.04.018
- Yuan, J., Hu, Z., Mahal, B. A., Zhao, S. D., Kensler, K. H., Pi, J., … Zhang, L. (2018). Integrated analysis of genetic ancestry and genomic alterations across cancers. Cancer Cell , 34, 549–560.e9. doi: 10.1016/j.ccell.2018.08.019
- Zhang, F., Flickinger, M., Taliun, S. A. G., InPSYght Psychiatric Genetics Consortium, Abecasis, G. R., Scott, L. J., … Kang, H. (2020). Ancestry-agnostic estimation of DNA sample contamination from sequence reads. Genome Research , 30, 185–194. doi: 10.1101/gr.246934.118
- Zheng, X., Gogarten, S. M., Lawrence, M., Stilp, A., Conomos, M. P., Weir, B. S., … Levine, D. (2017). SeqArray-a storage-efficient high-performance data format for WGS variant calls. Bioinformatics , 33, 2251–2257. doi: 10.1093/bioinformatics/btx145
- Zheng, X., Levine, D., Shen, J., Gogarten, S. M., Laurie, C., & Weir, B. S. (2012). A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics , 28, 3326–3328. doi: 10.1093/bioinformatics/bts606

