Modulating Myogenesis: An Optimized In Vitro Assay to Pharmacologically Influence Primary Myoblast Differentiation
Cordell A. VanGenderen, Cordell A. VanGenderen, Jules A. Granet, Jules A. Granet, Romina L. Filippelli, Romina L. Filippelli, Yiyang Liu, Yiyang Liu, Natasha C. Chang, Natasha C. Chang
Abstract
The intentional pharmacological manipulation of myogenesis is an important technique for understanding the underlying mechanisms of muscle differentiation and disease etiology. Using the pharmacological agent metformin as an example molecule, we present a systematic approach to examine the impact of pharmacological agents on the myogenic program. This consists of optimizing the in vitro differentiation of primary myoblast cells followed by the generation of a dose-response curve for a respective pharmaceutical. To assess myogenic differentiation, we utilized three approaches (incorporating both transcriptional and protein techniques) to assess the effects of biologically active agents on the in vitro differentiation of primary myogenic progenitors. First, the immunofluorescent visualization of myosin heavy chain (MYHC), which is expressed in differentiated myofibers, is used to obtain the fusion index, a quantitative read-out of differentiation efficiency. Second, quantitative reverse transcription PCR (RT-qPCR) reveals the expression of myogenic factors (Pax7 , Myf5 , Myod , Myog , Myh2) at the transcript level. Third, western blotting is used to assess the protein expression levels of the myogenic markers (PAX7, MYF5, MYOD, MYOG, and MYHC). By monitoring the expression of these various myogenic factors during the differentiation process, the relative cellular state and differentiation status between samples can be determined. Combined, these approaches enable the successful assessment of the impact of pharmacological agents on myogenic differentiation. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Immunofluorescence assay for qualitative and quantitative assessment of pharmacological agents on in vitro myogenic differentiation
Support Protocol 1 : Evaluating myogenic gene expression by RT-qPCR
Support Protocol 2 : Evaluating myogenic protein expression by western blot
INTRODUCTION
The differentiation of muscle stem cells (also referred to as satellite cells) is an integral process in the development and regeneration of mature muscle fibers (Bentzinger, Wang, & Rudnicki, 2012). In healthy individuals, satellite cells are vital in ensuring proper regeneration of adult musculature as they act as a source of muscle-resident stem cells that can propagate and differentiate into mature muscle cells to aid in the recovery from muscle injury (Yin, Price, & Rudnicki, 2013). A critical aspect to investigating regenerative myogenesis is the intentional in vitro manipulation of the process. Commonly, this is performed through the addition of biologically active agents either prior to or during differentiation. To this end, a diverse array of agents have been reported to be capable of influencing myogenesis including glucocorticoids, pharmaceuticals, and endocannabinoids, which function through diverse mechanisms (Kim, Park, Kim, Park, & Whang, 2016; Montesano, Luzi, Senesi, & Terruzzi, 2013; Pavlidou et al., 2017; Zhao et al., 2010). Here, we utilize metformin (which has been previously described to impact myogenic differentiation through the alteration of cellular metabolism) to demonstrate methods of examining the effect of pharmacological agents on myogenesis (Pavlidou et al., 2017).
Satellite cells are a rare cell population within muscle tissue and in vivo analysis of their regenerative capacity can be time intensive and costly. Thus, complementary analysis of activated satellite cells (referred to as myoblasts) ex vivo may provide a feasible alternative perspective on myogenic processes (Hindi, McMillan, Afroze, Hindi, & Kumar, 2017). Data obtained from these in vitro studies may then be used to identify specific mechanisms related to myogenesis for further analyses in vivo. To track the progression of myoblast differentiation in vitro , a variety of techniques may be employed, including immunofluorescence (IF) microscopy, quantitative reverse transcription PCR (RT-qPCR), and western blotting to monitor cell fate changes from myoblasts to myotubes. Together, these approaches reveal the expression of myogenic genes (and, upon translation, their corresponding protein) that are differentially expressed during myogenesis. Knowledge of the temporal expression of these myogenic factors during differentiation allows us to infer information regarding cellular state within a differentiating cell population. The transcription factor paired box 7 (Pax7) is highly expressed in quiescent and activated satellite cells, thus serving as a marker of muscle stem cells (Seale et al., 2000). As satellite cells activate and commit to myogenesis, PAX7 activates the expression of myogenic factor 5 (Myf5), which acts as a marker of myogenic commitment (Kuang, Kuroda, Le Grand, & Rudnicki, 2007; McKinnell et al., 2008; Zammit, 2017). Activated satellite cells will induce expression of myoblast determination protein 1 (Myod) and enter the cell cycle, becoming myoblasts. Next, myoblasts differentiate into myocytes and downregulate Pax7 , while inducing transcriptional activation of myogenin (Myog ; Seale et al., 2000). Myocytes will then either fuse with each other or pre-existing myofibers to form terminally differentiated myotubes expressing myosin heavy chain (Myh2). Terminal differentiation into myotubes results in the downregulation of MYF5, MYOD, and MYOG (Zammit, 2017). The expression of these myogenic genes and proteins are quantifiable by RT-qPCR and western blot, thereby acting as additional measures to assess the identity of the cell population in addition to the commonly used cell morphology and fusion index approaches. Here, we outline methods to assess the impact of pharmacological modulation on the myogenic program by monitoring the expression of these myogenic factors. We also discuss the best practices for primary myoblast cell culturing prior to differentiation, characterization of myoblast differentiation efficacy, and the establishment of a pharmacological dose-response curve.
The protocols presented here are intended to aid in designing and subsequently performing assays that assess the pharmacological modulation of novel therapeutics on the in vitro myogenesis of primary myoblast cells. The Basic Protocol details how to perform an initial time-course differentiation assay to assess the basal differentiation efficacy of a given primary myoblast cell line. This information is then used to subsequently design pharmacological assays to establish a dose-response curve. To assess differentiation efficiency, IF microscopy is utilized to examine cell morphology and to determine the nuclear fusion index following differentiation. Support Protocol 1 and Support Protocol 2 provide additional methods for assessing myogenic differentiation by examining the relative gene and protein expression, respectively, of several myogenic markers that are expressed during different stages of the differentiation program. Taken together, this provides multiple complementary methods for evaluating the impact of pharmacological modulation on myogenic differentiation (Fig. 1).
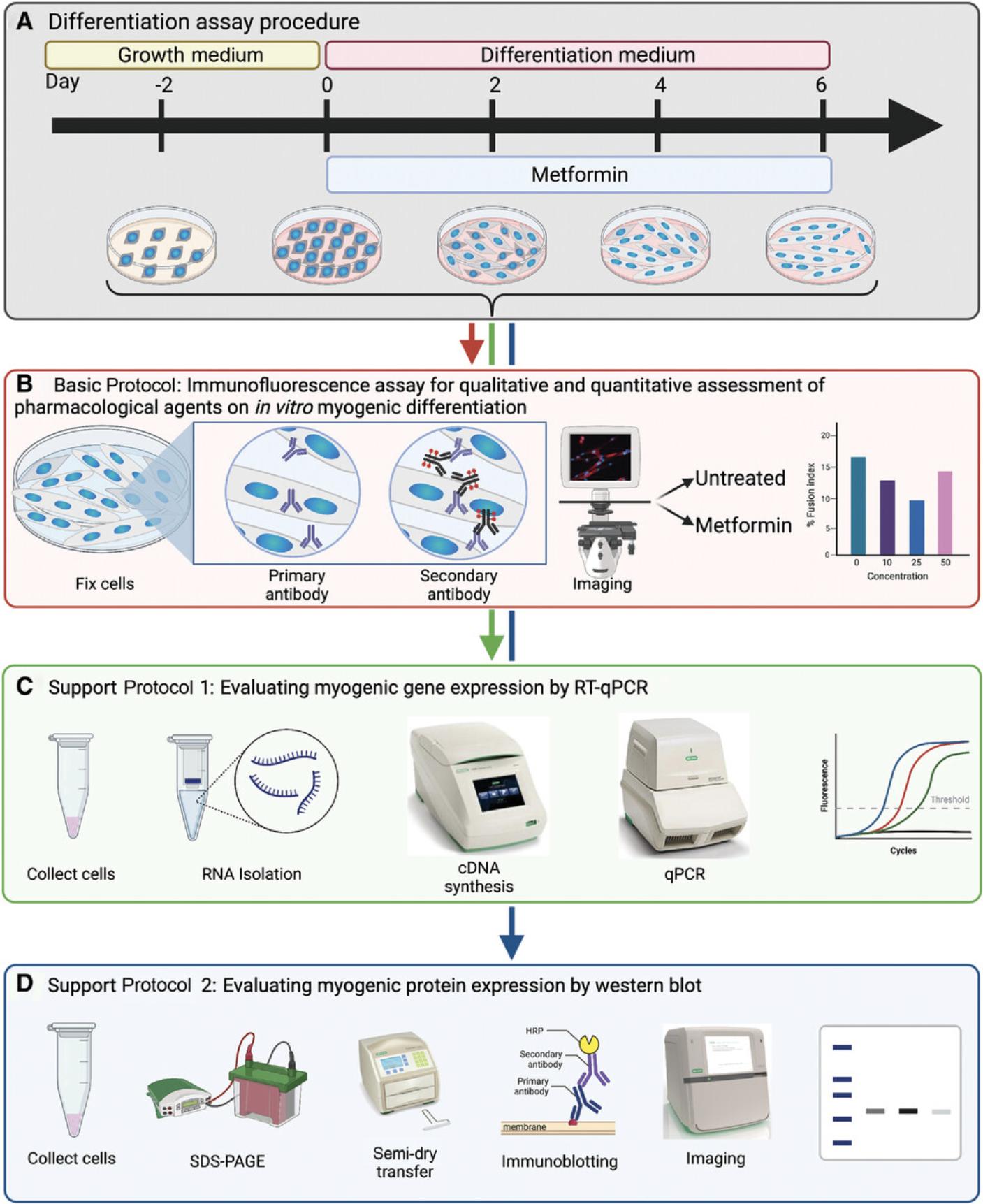
CAUTION : The primary myoblasts used in this protocol were isolated from mice. Other progenitor cell types or myoblasts isolated from other species may require different cell culturing and differentiation techniques as well as solutions.
Institutional animal care and use committee (IACUC) guidelines should be followed for all animal handling and primary cell isolations.
Basic Protocol: IMMUNOFLUORESCENCE ASSAY FOR QUALITATIVE AND QUANTITATIVE ASSESSMENT OF PHARMACOLOGICAL AGENTS ON IN VITRO MYOGENIC DIFFERENTIATION
The pharmacological manipulation of myogenic differentiation is an important tool for studying the mechanisms of differentiation. A variety of compounds have been reported to both positively and negatively influence differentiation kinetics in vitro (De Angelis et al., 1998; McRae et al., 2017; Montesano et al., 2013; Pavlidou et al., 2017; Zhao et al., 2010). One such compound is metformin which may inhibit or enhance differentiation depending on the presented dose (Pavlidou et al., 2017). Specifically, metformin acts to induce adenosine monophosphate (AMP) accumulation, which mimics an energy-deprived state, thereby inducing AMP-activated protein kinase (AMPK) activity and cellular autophagy (Viollet et al., 2012). This has both myogenic enhancing and impairing effects. As reported in Pavlidou et al. (2017) low concentrations of metformin have a slight myogenesis-promoting effect in the cell but are inhibitory at high concentrations. Here we demonstrate a methodology to assess the impacts of metformin across a range of concentrations on the differentiation of primary myoblasts isolated from the hind limb muscles of mice. Myogenic differentiation is assessed by immunostaining against myosin heavy chain (MYHC), a marker of terminal differentiation (Zammit, 2017), followed by microscopy to obtain phase contrast and fluorescence images of the samples. Subsequently, the fusion index, a measure of differentiation efficiency, is obtained by calculating the number of fused nuclei in developed myotubes divided by the total number of nuclei (Simionescu-Bankston et al., 2015). However, primary myoblast cell lines may exhibit distant differentiation capacities and kinetics, even if from the same genetic background. Therefore, prior to pharmacological treatment, an evaluation of the basal differentiation efficacy of the primary myoblast cell line is necessary to determine how to best implement a pharmacological dose-response curve. To achieve this, primary myoblasts are differentiated in the absence of the pharmacological agent over an extended time course in order to identify the optimal differentiation endpoint. Subsequently, using this established endpoint, a dose-response curve is performed to evaluate the impact of specific pharmacological agents on myogenic differentiation over a wide range of concentrations. The assessment of pharmacological modulation on the differentiation capacity in primary myoblast models provides insight into the therapeutic potential of these agents in enhancing muscle regeneration and treatment of myopathies.
Materials
-
Primary myoblasts (see Current Protocols article: Filippelli et al., 2021)
-
Collagen solution (see recipe)
-
Growth medium (GM; see recipe)
-
Differentiation medium (DM; see recipe)
-
Sterile 1× PBS (see recipe)
-
4% paraformaldehyde (PFA; see recipe)
-
Immunofluorescence (IF) blocking solution (see recipe)
-
IF washing solution (see recipe)
-
IF permeabilization solution (see recipe)
-
Mounting medium with DAPI (ProlongTM Gold Antifade Mountant with DAPI; Thermo Fisher Scientific, cat. no. P36935)
-
Anti-myosin heavy chain (MYHC) primary antibody (MilliporeSigma, cat. no. M4276)
-
Donkey anti-mouse IgG - Alexa Fluor 647 (secondary antibody; Invitrogen, cat. no. A31571)
-
35-mm tissue culture dishes (Falcon Scientific, cat. no. 08772A)
-
Cover slips (Thermo Fisher Scientific, cat. no. 12546)
-
Inverted microscope with fluorescent imaging capabilities (e.g., Thermo Fisher Scientific, EVOS M5000 microscope)
-
FIJI imaging software (Version 1.53s, Schindelin et al., 2012)
Primary myoblast culture and differentiation
NOTE : Steps 1-4 should be performed in a biosafety cabinet.
1.Isolate primary myoblasts as previously described (Current Protocols article: Filippelli et al., 2021).
2.Coat 35-mm tissue culture dishes with 500 µl collagen solution for 30 min, then remove collagen solution and let plates dry for at least 2 hr.
3.Culture myoblasts in GM on 35-mm dishes until they reach ∼95%-100% confluency. Subsequently, wash cells once with PBS. After washing, induce differentiation by replacing GM with DM for the desired differentiation period.
4.Collect samples at desired time points following differentiation. Aspirate DM and wash samples twice with PBS.
5.Fix samples by incubating each 35-mm dish with 750 µl of 4% PFA solution for 20 min at room temperature.
6.Gently aspirate PFA and wash twice for 5 min each time with PBS.
Immunofluorescence and phase contrast assay
7.Aspirate PBS and incubate cells with 750 µl IF permeabilization solution for 10 min at room temperature. Wash samples with 750 µl cold IF washing solution for 10 min and repeat twice for a total of three washes.
8.Block cells with 750 µl IF blocking solution for at least 1 hr.
9.Dilute anti-MYHC antibody 1:150 in IF blocking solution and incubate cells over night in 250 µl primary antibody solution at 4°C.
10.Aspirate primary antibody solution and wash three times for 10 min each in IF washing solution.
11.Dilute the secondary antibody 1:1,000 in IF blocking solution and incubate cells in 250 µl of secondary antibody solution for 1 hr at room temperature in the dark to prevent photobleaching.
12.Aspirate the secondary antibody solution and wash it three times for 10 min each time with IF washing solution, ensuring the samples are kept in the dark during the washes. Let dry 5 min at an angle, then remove any residual liquid from the corner of the dish.
13.Add 1 drop of mounting medium. Place a cover slip onto the drop of mounting medium using tweezers, removing any remaining air bubbles by gently tapping on the cover slip. Let sit overnight to dry at room temperature in the dark.
Image myotubes and determine nuclear fusion index
14.Using an inverted fluorescent microscope, acquire images of the DAPI, MYHC, and phase contrast channels with the 10× and 20× objective magnification.
15.To quantify the fusion index, we use FIJI software (Schindelin et al., 2012). Open DAPI, MYHC, and brightfield images into FIJI and merge the channels (Image → Color → Merge Channel) to produce a single image. Then, open the Channels Tool (Image → Color → Channels Tool), enabling the individual and combined viewing of each channel.
16.Open the cell counter tool (Plugins → Analyze → Cell counter) and click initialize to permit counting of the merged image. Count all nuclei (represented by DAPI staining) in the image within two categories: single nuclei that are unfused and nuclei within stained myotubes (represented by MYHC staining), which contain ≥2 nuclei (i.e., fused nuclei). The fusion index is then calculated using the following equation (Equation 1):
(1)
Performing a dose-response curve to assess pharmacological modulation of differentiation
17.Based on cell morphology and nuclear fusion index, select the desired differentiation time points as identified in steps 3-16 to assess the impact of a particular pharmacological agent on myogenesis. Repeat steps 3 and 4, with the purpose of seeding cells to perform a dose-response curve.
18.Repeat steps 5-16 to assess pharmacological modulation of myogenic differentiation at the different concentrations of the compound.
Support Protocol 1: EVALUATING MYOGENIC GENE EXPRESSION BY RT-qPCR
RT-qPCR is a simple method that can be used to evaluate changes in the transcriptional landscape. Myogenesis is controlled by dynamic and temporal expression of specific myogenic regulatory factors during distinct phases of the differentiation program. The dynamic expression of these myogenic proteins during differentiation are reflected at the level of transcription; in chronological order, Pax7 , Myf5 , MyoD , MyoG , and Myh2 are temporally expressed during myogenesis (Soleimani et al., 2012; Zammit, 2017). Thus, RT-qPCR can reveal changes in the levels of myogenic gene transcripts in samples during different stages of the differentiation process. To isolate RNA, Trizol allows for rapid cell lysis while protecting RNA from degradation (Meng & Feldman, 2010). Then, the Bio-Rad Aurum RNA Isolation kit provides a quick and clean method to isolate total RNA. This protocol combines both Trizol and the Bio-Rad Aurum Total RNA Mini Kit to obtain high quality RNA for analysis by RT-qPCR. Subsequently, complementary DNA (cDNA) is generated from RNA by reverse transcription. Lastly, the relative levels of transcripts across cDNA samples are assessed using qPCR. Methods outlined in this protocol are applicable for monitoring the expression of myogenic transcripts whose expression indicates different phases of differentiation and may be affected following treatment with a pharmacological agent. Here, we present gene expression analyses from primary myoblasts differentiated in the presence and absence of the pharmacological modulator metformin that have been collected at select time points (as described in the Basic Protocol).
Materials
-
Myoblast cells (see Basic Protocol)
-
Invitrogen Trizol Reagent (Thermo Fisher Scientific, cat. no. 15596-026)
-
Aurum Total RNA Mini Kit (Bio-Rad, cat. no. 732-6820)
-
Chloroform, molecular biology grade (Thermo Fisher Scientific, cat. no. C298500)
-
70% ethanol (EtOH; made with sterile or DEPC-treated water)
-
iScript™ Reverse Transcription Supermix for RT-qPCR (Bio-Rad, cat. no. 1708840)
-
RNase-free water (Wisent, cat. no. 809-115-CL)
-
Sterile 1× PBS (see recipe)
-
10 mM Tris, pH 7.5 (Bio Basic, cat. no. TB0165)
-
SsoFast™ EvaGreen® Supermix (Bio-Rad, cat. no. 1725200)
-
Cell scraper (Sarstedt, cat. no. 83.183)
-
15-ml conical centrifuge tubes (Corning, cat. no. 352097)
-
Sterilized pipet tips
-
Sterilized microcentrifuge tubes (FroggaBio, cat. no. LMCT1.7B10)
-
Benchtop centrifuge (to centrifuge 15-ml tubes, e.g., Eppendorf 5804)
-
Microcentrifuge (to centrifuge 1.7-ml microcentrifuge tubes, e.g., Eppendorf 5424R)
-
NanoDrop spectrophotometer (e.g., Thermo Fisher Scientific, Nanodrop OneC)
-
Thermocycler (e.g., Bio-Rad, T100™ Thermal Cycler 1861096EDU)
-
qPCR instrument (e.g., Bio-Rad, CFX Connect Real-Time PCR Detection System)
RNA isolation from differentiated cells
Bacteria are the primary source of RNases; therefore, prepare your RNAse free workspace by wiping down all equipment with 70% ethanol or RNAse wipes. Users should wear gloves and work with sterile technique.
1.Culture and differentiate myoblasts cells as described in steps 1-4 of Basic Protocol.
2.Following differentiation, harvest cells in DM with a cell scraper and transfer into a 15-ml conical centrifuge tube. Wash plate with PBS, collecting any remaining cells with the scraper, before pooling the PBS with the collected DM. Centrifuge in a benchtop centrifuge at 470 × g for 5 min.
3.Aspirate DM and wash cells by resuspending the cell pellet in 1 ml PBS. Transfer to a 1.7-ml microcentrifuge tube.
4.Centrifuge in a microcentrifuge at 220 × g for 5 min and aspirate PBS.
5.Add 500 µl Trizol to the cell pellet and break up cells by pipetting repeatedly. Vortex to ensure no clumps remain.
6.Let samples sit at room temperature for 5 min.
7.Add 100 µl chloroform (one-fifth the volume of Trizol) to each sample, then shake vigorously 30 to 60 s. Incubate samples at room temperature for 2 min.
8.Centrifuge at 11,000 × g or maximum speed for 30 min at 4°C. Collect the upper phase in a clean microcentrifuge tube.
9.Add 70% ethanol in equal volume to the collected upper phase to precipitate the RNA and mix well by inversion and/or vortex.
10.Pipet lysate onto an RNA binding column, which has been placed into a 2-ml capless wash tube (both tubes provided in the kit), then centrifuge column for 30 s at 8,000 × g.
11.Discard flow-through and replace column into the same wash tube. Add 700 µl of low stringency wash solution to the column, then centrifuge at 8,000 × g for 30 s. Discard flow-through and replace column into the same wash tube.
12.Add 80 µl DNase I mix to the membrane at the base of the column. Incubate at room temperature for 15 min.
13.Add 700 µl high stringency wash solution to the column, then centrifuge at 8,000 × g for 30 s.
14.Discard flow-through and replace column into the same wash tube. Add 700 µl low stringency wash solution to the column. Centrifuge at 8,000 × g for 1 min.
15.Discard flow-through and replace column into the same wash tube. Centrifuge at 8,000 × g for 2 min to remove residual wash solution.
16.Transfer the RNA binding column to a 1.5-ml capped microcentrifuge tube (provided in the kit). Pipet 50 µl RNase-free water onto the membrane at the bottom of the column. Incubate at room temperature 5 min then centrifuge at 8,000 × g for 2 min to elute the RNA.
cDNA synthesis
17.Determine the RNA concentration of each sample with a NanoDrop instrument, using RNase-free water as a blank. A 260/A 280 and A 260/A 230 ratios can also be obtained to assess purity.
18.Store RNA at −80°C.
19.For each sample, prepare a mix with 500 ng RNA, according to the manufacturer's protocol using the iScripttm Reverse Transcription Supermix and add water to bring the total volume to 20 µl, as per Table 1.
| Component | Volume per reaction (µl) |
|---|---|
| 5× iScript Reaction Mix | 4 |
| RNA (500 ng) | Variable |
| H2O | Up to 16 |
| Total volume | 20 |
20.Reverse transcribe RNA into cDNA using a thermocycler programmed to follow the protocol outlined in Table 2.Store cDNA at −20°C until ready to perform qPCR.
| Step | Temperature (°C) | Time (min) |
|---|---|---|
| Priming | 25 | 5 |
| Reverse transcription (RT) | 46 | 20 |
| RT inactivation | 95 | 1 |
| Optional step | 4 | Hold |
Quantitative PCR
21.Dilute cDNA 1:10 with RNase-free water.
22.Prepare a master mix for each set of primers as outlined in Table 3.
| Component | Volume per reaction (µl) |
|---|---|
| SsoFast EvaGreen Supermix | 5 |
| Forward Primer (10 µM) | 0.5 |
| Reverse Primer (10 µM) | 0.5 |
| cDNA template (1:10) | 2 |
| H2O | 2 |
| Total volume | 10 |
| Gene | Type | Sequence (5′-3′) | Tm (°C) | GC% | Length |
|---|---|---|---|---|---|
| Hprt | Forward | GGCCAGACTTTGTTGGATTTG | 54.7 | 47.6 | 21 |
| Reverse | CACAGGACTAGAACACCTGC | 55.2 | 55.0 | 20 | |
| Myf5 | Forward | TGACGGCATGCCTGAATGTA | 56.8 | 50 | 20 |
| Reverse | ATCTGCAGCACATGCATTTGATA | 55.4 | 39.1 | 23 | |
| Myh2 | Forward | GACCAGATCTTCCCCATGAA | 54.1 | 50 | 20 |
| Reverse | TAAGGGTTGACGGTGACACA | 56.1 | 50 | 20 | |
| Myod | Forward | TGATGGCATGATGGATTACAG | 52.6 | 42.9 | 21 |
| Reverse | GTAGTAGGCGGTGTCGTA | 53.3 | 55.6 | 18 | |
| Myog | Forward | CAACCCAGGAGATCATTTG | 50.8 | 47.4 | 19 |
| Reverse | CATATCCTCCACCGTGAT | 51.0 | 50 | 18 | |
| Pax7 | Forward | GACGACGAGGAAGGAGACAA | 56.3 | 55 | 20 |
| Reverse | ACATCTGAGCCCTCATCCAG | 56.5 | 55 | 20 | |
| Rps18 | Forward | AACGGTCTAGACAACAAGCTG | 54.9 | 47.6 | 21 |
| Reverse | AGTGGTCTTGGTGTGCTGAC | 57.5 | 55 | 20 |
23.Run the qPCR following the program in Table 5.
| Step | Temperature (°C) | Time (s) | Number of cycles |
|---|---|---|---|
| Enzyme activation | 95 | 30 | 1 |
| Denaturation | 95 | 5 | 40 |
| Annealing/extension | 59 | 5 | |
| Melt curve | 65-95 (in 0.5 increments) | 5 per increment | 1 |
Data analysis using the ΔΔCt method
Sample calculations for steps 24-29 can be found in the Supporting Information file.
24.Determine the average threshold cycle (Ct) value for each gene between technical replicates within a given sample.
25.Determine the average Ct (Av.Ct) of all samples for each gene (see Equation 2).
(2)
26.Calculate the ΔCt (Equation 3) by determining the difference in Ct values between the Av.Ct and the Ct of each sample.
(3)
27.Determine the relative quantity (RQ) of each gene for each sample (Equation 4).
(4)
28.Calculate the geometric mean (GeoMean) of the RQ values of all reference genes for each sample (Equation 5; Vandesompele et al., 2002).
(5)
29.To obtain the normalized expression for each gene within a given sample, calculate the ratio of the RQ value over the GeoMean RQ of the reference genes (Equation 6).
(6)
30.Data may be represented as a clustered bar chart.
Support Protocol 2: EVALUATING MYOGENIC PROTEIN EXPRESSION BY WESTERN BLOT
In this support protocol, we demonstrate the detection of myogenic protein expression using western blotting. In chronological order of expression during differentiation, these are: PAX7, MYF5, MYOD, MYOG, and MYHC (Seale et al., 2000; Zammit, 2017). Harvested cell populations are lysed to collect total protein extracts. Subsequently, lysates are resolved on an SDS-PAGE gel and transferred to a membrane, which undergoes immunoblotting to reveal protein expression. This analysis enables the tracking of protein expression changes that occur during differentiation, which serve as markers of distinct phases within the differentiation program. By comparing cells treated with pharmacological agents to control (vehicle treated) cells, the impact of pharmacological agents at discrete stages during the differentiation program can be determined. Here, we present the protein expression analyses of primary myoblasts differentiated in the presence and absence of metformin that have been collected at select time points (as described in the Basic Protocol).
Materials
-
Collagen solution (see recipe)
-
Primary myoblasts (see Current Protocols article: Filippelli et al., 2021)
-
Sterile PBS (see recipe)
-
Growth medium (GM; see recipe)
-
Differentiation medium (DM; see recipe)
-
Lysis buffer (see recipe)
-
Bradford reagent (Bio-Rad, cat. no. 5000006)
-
4× Laemmli buffer (Bio-Rad, cat. no. 1610747)
-
100× phosphatase inhibitor (Nacalai, cat. no. 07575-51)
-
100× protease inhibitor (Nacalai, cat. no. 25955-11)
-
12% acrylamide gel (see recipe)
-
Western blot protein ladder (Bio-Rad, cat. no. 1610374)
-
Polyvinylidene difluoride (PVDF; Bio-Rad, cat. no. 1620-77)
-
Semi-dry transfer buffer (see recipe)
-
Ponceau S solution (see recipe)
-
Polyoxyethylene-20 (Tween 20; Bio Basic, cat. no. TB0560)
-
Skim milk powder
-
Tris-buffered saline (TBS; see recipe)
-
BSA (Bio Basic, cat. no. ALB001)
-
Sterile deionized wate
-
Enhanced chemiluminescence (ECL) solution (Bio-Rad, cat. no. 170-5061)
-
Microcentrifuge tubes (FroggaBio, cat. no. LMCT1.7B10)
-
100-mm tissue culture dishes (Thermo Fisher Scientific, cat. no. 150466)
-
60-mm tissue culture dishes (Thermo Fisher Scientific, cat. no. 08772B)
-
Cell scraper (Sarstedt, cat. no. 83.183)
-
15-ml conical centrifuge tubes (Corning, cat. no. 352097)
-
Mini-PROTEAN Tetra Vertical Electrophoresis Cell (Bio-Rad, cat. no. 1658004)
-
Trans-Blot Turbo Machine (Bio-Rad, cat. no. 1704150)
-
Trans-Blot Turbo Midi-size Transfer Stacks (Bio-Rad, cat. no. 1704275)
-
Bio-Rad ChemiDoc Imaging System
Prepare total protein lysate from differentiated cells
1.Culture and differentiate cells as per steps 1-4 of Basic Protocol, with the following modifications: Seed primary myoblasts onto collagen-coated 60- or 100-mm dishes, depending on the desired quantity of protein.
| Cell culture dish | Growth area (cm2) | Number of cells (100% confluency) | Growth medium (ml) |
|---|---|---|---|
| 35 mm | 8 | 1 × 106 | 2.5 |
| 60 mm | 21 | 2.6 × 106 | 5 |
| 100 mm | 56 | 7 × 106 | 10 |
2.Following differentiation, harvest cells in DM with a cell scraper and transfer into a 15-ml conical centrifuge tube. Collect and wash cells as described in steps 2-4 of Support Protocol 1.
3.Lyse cells by resuspending the cell pellet in lysis buffer (typically 50 µl for a 100-mm plate and 30 µl for a 60-mm plate) containing 1× protease/phosphatase inhibitor cocktail and incubate on ice for 30 min.
4.Centrifuge in a microcentrifuge at 15,000 × g at 4°C for 20 min.
5.Perform a Bradford assay to assess protein concentrations and dilute lysates in sterile deionized water to obtain desired protein concentrations.
6.Add 4× Laemmli buffer to the lysates and denature samples at 95°C for 5 min.
Resolve proteins by SDS-PAGE
7.Polymerize SDS-PAGE gels, with the final acrylamide concentration depending on the molecular weight of target proteins.
8.Place gel into the running buffer tank, then fill it with 1× running buffer diluted from the 10× running buffer stock. Load sample lysate into the acrylamide gel alongside a protein ladder.
9.Run at 50 V until the dye front passes the stacking-resolving gel interface. Then increase voltage to 120 V until the dye front runs off.
10.In a Bio-Rad ChemiDoc imaging system, use the stain free imaging protocol to image the total protein load on the acrylamide gel.
11.Saturate the Turbo blot cassette and stacking papers with transfer buffer and assemble within the cassette. Assembly order from bottom to top is: stacking papers, PVDF membrane, acrylamide gel, stacking papers. Remove air bubbles by gently rolling over the top set of stacking papers with a roller.
12.Drain excess buffer and run the “Turbo” protocol, selecting the specific protocol most relevant for your transfer (mini or midi).
Immunoblotting
13.To confirm proper transfer of proteins to the PVDF membrane, remove membrane and stain with Ponceau S solution, then rinse thoroughly with distilled water.
14.De-stain with 0.05% Tween PBS (PBST) and block for 1 hr at room temperature in 10 ml of 5% skim milk PBST blocking solution.
15.Incubate the PVDF membrane in the primary antibody solution overnight at 4°C with agitation. The primary antibody solution consists of the primary antibody diluted to its respective optimal concentration (as identified in Table 7) in 5% skim milk powder (w/v) in PBST.
| Antigen | Vendor | Cat. no. | Source | Dilution |
|---|---|---|---|---|
| PAX7 | Developmental Studies Hybridoma Bank (DSHB) | N/A | Mouse | 1:500 |
| MYF5 | Santa Cruz | sc-518056 | Mouse | 1:500 |
| MYOD | Santa Cruz | sc-377460 | Mouse | 1:200 |
| MYOG | Santa Cruz | sc-12732 | Mouse | 1:200 |
| MYHC | MilliporeSigma | M4276 | Mouse | 1:1,000 |
| Tubulin | MilliporeSigma | T9026 | Mouse | 1:5,000 |
| AMPKα | Cell Signaling Technology | 2603S | Rabbit | 1:1,000 |
| p-AMPKα (T172) | Cell Signaling Technology | 2535S | Rabbit | 1:1,000 |
| Anti-mouse HRP | Bio-Rad | 1706516 | Goat | 1:10,000 |
| Anti-rabbit HRP | Bio-Rad | 1706515 | Goat | 1:10,000 |
16.Wash membrane three times for 5 min per wash using 10 ml PBST for each wash.
17.Incubate membrane in 10 ml secondary antibody solution (consisting of the appropriate secondary antibody depending on the species of the primary antibody, diluted to 1:10,000 in 5% skim milk powder in PBST) for 1 hr.
18.Wash membrane three times for 10 min per wash using 10 ml PBST for each wash.
19.Remove excess liquid from the membrane. Lay membrane on a Parafilm sheet. Mix ECL solution following the manufacturer's instructions and apply 1 ml to each membrane and incubate 5 min.
20.Remove excess liquid with a Kimwipe and image the membrane.
21.Image membrane using an appropriate detection system.
REAGENTS AND SOLUTIONS
Acrylamide gel, 12%
- Up to 6 ml of deionized H2O
- 0.375 M Tris·HCl, pH 8.8
- 12% acrylamide
- 0.1% SDS
- 0.5% 2,2,2-trichloroethanol (TCE)
- 0.1% ammonium persulfate (APS)
- 6 µl tetramethylethylenediamine (TEMED)
Store at 4°C for up to 1 week.
Collagen solution
- 998 ml of filter sterilized deionized H2O plus 2 ml glacial acetic acid using a 0.2-μm filter
- Add 1 bottle of 100 mg Collagen Type I, Rat Tail (Corning, cat. no. 354236)
- Mix well by inversion.
Store up to 6 months at 4°C.
The solution can be used up to five times.
Differentiation medium (DM)
- 95 ml Dulbecco's Modified Eagle Medium (DMEM; Gibco, cat. no. 11995-065)
- 95 ml Hams F-10 Nutrient Mix (Gibco, cat. no. 12390035)
- 10 ml Horse serum (Gibco, cat. no. 16050-122)
Store at 4°C for up to 1 month.
Growth medium (GM)
- 400 ml Ham's F-10 Nutrient Mix (F-10; Gibco, cat. no. 12390035)
- 100 ml FBS (Gibco, cat. no. 12483020; 20% final)
- 5 ml penicillin-streptomycin solution (Wisent, cat. no. 450-201-EL; 1% final)
- 25 µl of 50 ng/µl basic fibroblast growth factor (bFGF; Gibco, cat. no. PHG0261)
Store at 4°C for up to 1 month.
Solution should be prepared in a biological safety cabinet to maintain sterility. bFGF solution is prepared by dissolving 100 µg bFGF powder in 2 ml F-10. Solution is then filter sterilized with a 0.2-µm filter and dispensed into 25-µl aliquots for storage at − 20°C for ≤ 6 months or at − 80°C for longer.
IF blocking solution
- 5% donkey serum (Gibco, cat. no. 16050122)
- 2% BSA (BioShop, cat. no. ALB001)
- 0.1% Triton X-100 (Bio Basic, cat. no. TB0198)
- 0.05% Polyoxyethylene-20 (Tween 20; Bio Basic, cat. no. TB0560)
- in 1× PBS (keep in fridge)
Store at 4°C for up to 2 weeks.
IF permeabilization solution
- 0.1% Triton X-100 (Bio Basic, cat. no. TB0198)
- 0.1 M glycine (Bio Basic, cat. no. GB0235; CAS: 56-40-6)
- Dilute in 1× PBS.
Store at room temperature for up to 1 month.
IF washing solution
- 1% donkey serum (Gibco, cat. no. 16050122)
- Dilute in 1× PBS.
Store at 4°C for up to 2 weeks.
Lysis buffer
- 50 mM Tris·HCl, pH 7.5
- 150 mM NaCl
- 0.5% Triton X-100
- 0.5 mM EDTA
Store at 4°C for up to 1 year.
Paraformaldehyde solution, 4%
- One 10-ml vial of 8% (w/v) PFA aqueous solution (Electron Microscopy Sciences, cat. no. 175-8)
- 2 ml 10× PBS (Bio Basic, cat. no. PD8117)
- 8 ml Milli-Q H2O
Store at 4°C for 1 month.
PBS, sterile, 1×
- 100 ml 10× PBS (BioBasic, cat. no. PD8117)
- 900 ml Milli-Q H2O
- Sterilize by autoclaving
Store at room temperature indefinitely.
Ponceau S solution
- 0.2% (w/v) Ponceau S (MilliporeSigma, cat. no. P3504)
- 5% glacial acetic acid
- Store at room temperature indefinitely.
Semi-dry transfer buffer
- Typically prepare 500 ml at a time:
- 100 ml 5× Trans-blot Turbo Transfer Buffer (Bio-Rad, cat. no. 10026938)
- 300 ml deionized water and
- 100 ml 95% ethanol
Store at 4°C for 3 weeks.
Tris-buffered saline (TBS)
- 50 ml 20× TBS stock (Bio Basic, cat. no. UA0045)
- 950 ml deionized water
- Sterilize by autoclaving.
Store at room temperature indefinitely.
COMMENTARY
Background Information
Developmental myogenesis is the process of growing skeletal muscle tissue during embryonic development (Bentzinger et al., 2012). Once developed, the repair of adult muscle tissue (referred to as regenerative myogenesis) is dependent on a rare population of muscle stem cells referred to as satellite cells (Bentzinger et al., 2012). Located between the sarcolemma and basal lamina of skeletal muscle fibers, satellite cells become activated following an injury or when muscle fibers are otherwise damaged (such as through exercise). Subsequently, activated satellite cells rapidly proliferate and undergo differentiation, thereby repairing the damaged muscle tissue (Yin et al., 2013). In normal homeostatic muscle, satellite cells remain quiescent and express the transcription factor PAX7 which regulates the expression of genes involved in myogenic identity, proliferation, and inhibitors of differentiation (Seale et al., 2000; Soleimani et al., 2012). Following activation, satellite cells recruited to undergo differentiation express MYF5 and differentiate into myoblasts. MYF5 expression commits myogenic progenitors to upregulate MYOD expression, enter the cell cycle, and undergo rapid proliferation (Kuang et al., 2007; Yin et al., 2013). After proliferation, myoblasts will express MYOG, as they differentiate into myocytes (Yin et al., 2013). Finally, cells will terminally differentiate by either undergoing de novo formation of elongated MYHC expressing myotubes, or by donating their nuclei to pre-existing myofibers (Petrany & Millay, 2019; Stockdale, 1992; Yin et al., 2013).
The stages of regenerative myogenesis can be monitored in vitro using basic biochemical and molecular biology techniques, including IF staining, RT-qPCR, and western blotting. IF is the most common technique used as it allows for the visualization of cell morphology, particularly the terminally differentiated myotubes which are visualized by staining against MYHC (Stockdale, 1992). The immunofluorescent staining of myotubes ultimately enables the quantification of the ratio of nuclei within multi-nucleated myotubes to the total number of nuclei in a cell population (referred to as the fusion index) which serves as a quantitative indication of differentiation efficiency (Simionescu-Bankston et al., 2015). Alternatively, HEMA 3 or Jenner-Giesma staining can be used to assess in vitro myogenic differentiation through non-fluorescent myotube and nuclear staining (Current Protocols article: Levitt, Adler, & Simon, 2019). However, there are two limitations to morphological staining of myotubes for assessing myogenic differentiation. Notably, cell morphology is a qualitative measure of differentiation, while the fusion index is subject to human error when counting ambiguously fused nuclei. Thus, additional methods of examining cell differentiation status are useful in providing complementary approaches. To fulfill this role, RT-qPCR and western blotting can be utilized in combination with fusion index analysis to examine the cell state of the population by assessing the RNA and protein levels of the myogenic factors PAX7, MYF5, MYOD, MYOG, and MYHC (Zammit, 2017).
Although the myogenic factors listed above are the most commonly studied in myogenesis, the differential expression of myogenic factor 6 (MYF6), the myocyte enhancer factor 2 (MEF-2) family, and myostatin have been documented during differentiation and may be used as additional markers (Huang, Chen, & Chen, 2011; Jin, Shang, Peng, & Jiang, 2016; Langley et al., 2002; Naidu, Ludolph, To, Hinterberger, & Konieczny, 1995; Zammit, 2017). The pro-myogenesis protein MYF6 is a transcription factor expressed late during myogenesis to induce the terminal differentiation of myocytes and its expression remains in myotubes (Lazure et al., 2020; Zammit, 2017). Additionally, individual members of the MEF-2 transcription factor family (while alone are incapable of inducing myogenesis) augment the myogenic potential of cells by inducing the expression of additional myogenic factors (Jin et al., 2016; Naidu et al., 1995). Not all of these markers are pro-myogenic however, as myostatin is a negative regulator of skeletal muscle development that is expressed in the early stages of myogenesis and its accumulation may be indicative of impaired myogenesis (Huang et al., 2011; Langley et al., 2002).
Muscular pathologies are a diverse array of diseases that include the common age-related muscle loss known as sarcopenia to the severe and fatal Duchenne muscular dystrophy (Cruz-Jentoft & Sayer, 2019; Yiu & Kornberg, 2015). One potential therapeutic route of therapy is the direct induction of myogenesis to regenerate damaged and lost muscle tissue (Crist, 2017; Filippelli & Chang, 2021). The drug metformin, which is used as a treatment for individuals suffering from type II diabetes, may have potential for muscular disorders. In the literature, metformin has been described to act as both an activator and an inhibitor of muscle differentiation, depending on the concentration being used (Jiang & Liu, 2020; Pavlidou et al., 2017; Viollet et al., 2012). While its exact mechanism of action is not known, low doses of metformin in satellite cells mimic a calorie restricted state, thereby increasing the phosphorylation of AMPK, which results in enhanced expression of myogenic regulatory factors and enhanced myogenic differentiation (Jiang & Liu, 2020). Conversely, high concentrations (≥1,000 µM) of metformin inhibits myogenic differentiation through inhibiting MYOD and p21cip1 expression (Pavlidou et al., 2017).
Published studies assessing myogenic differentiation under different pharmacological contexts vary with respect to experimental parameters, including the composition of the differentiation medium (whose addition promotes the differentiation process of myoblasts in vitro) and the time point at which cells are collected for analysis. Inconsistencies between studies can lead to conflicting results, making it difficult to extrapolate results based on previous findings. One such example is dexamethasone, which has been reported to both induce and inhibit differentiation when used at similar concentrations (Kim et al., 2016; McRae et al., 2017). Notable differences between the two protocols are the durations of differentiation; Kim et al. (2016) differentiated C2C12 myoblast cells for 8 days, while McRae et al. (2017) differentiated for 3 days. Kim et al. (2016) also specified a lower level of potential confluence, stating their cells were between 80% and 90% confluent, while McRae et al. (2017) reported differentiating myoblasts only when above 90% confluence. Therefore, different differentiation methodologies may lead to incongruent results. By reporting a detailed methodology for establishing the pharmacological modulation of myogenic differentiation assays, we hope to provide a template for researchers to improve experimental reproducibility.
Critical Parameters
The most important parameter for an effective differentiation experiment is the quality of the primary myoblasts in culture. It is critical to keep myoblasts evenly dispersed and their confluency below 70% when passaging cells during expansion prior to experimentation. A high confluence (either across the plate or in local cell clusters) may induce premature differentiation which can be observed with an inverted cell culture microscope. Prematurely differentiated cells begin to take on a noticeably elongated cell morphology as compared to the round morphology of myoblasts. Even the presence of a few of these elongated cells is indicative that the cell population has begun to differentiate, and it is best to discard the cells and start with a fresh myoblast isolation or from frozen cell stocks. For new users not familiar with differentiation assays, it may be advantageous to seek the assistance of more seasoned users to aid in assessing confluency and the general health of the cells.
When passaging cells from 100-mm plates to 35-mm plates for differentiation, it is recommended that the plates are seeded at ∼70% to 80% confluence (see Table 6 for the conversions of cell number between plates). To prevent cell clustering when plating cells, we suggest swirling the plates around in a figure eight or plus sign pattern for 20 to 30 s to ensure an even distribution of cells. The number of times a cell line has been expanded in culture, referred to as the cell passage number, is also critical to the cells’ capacity to differentiate. Therefore, differentiation should consistently be performed using cells of low passage number. As well, differentiation should only be initiated in cells that have reached sufficient confluency (95% to 100% confluence). When in doubt, it is best to wait an additional day to ensure there are enough cells present on the dish to fuse into myotubes. Due to the number of factors that impact the consistency of myoblast differentiation, we recommend performing the control (vehicle) and pharmacologically treated differentiation assays concurrently with the same batch of cells. This will assist in minimizing the impact of cell culture conditions on the differentiation experiment.
For IF imaging, it is important to perform the antibody staining on all samples simultaneously, as antibody binding and signal intensity may vary over time. If all cells within a given experiment have not reached 95% to 100% confluency for differentiation, we recommend prioritizing differentiation on cells that have reached sufficient confluency. For downstream IF analysis, cells can be fixed and kept at 4°C in PBS until all cell lines can be stained simultaneously. We do not recommend leaving fixed cells for longer than 2 weeks in PBS as they may begin to detach from the plate, thus influencing fusion index results. During the counting of fused and unfused nuclei for fusion index analysis, it is important to be internally consistent when deciding what constitutes a nucleus fused within a myotube. Cells should be imaged promptly (ideally within a week of staining and mounting) and simultaneously to prevent the loss of fluorescence signal between samples imaged at different time points. Lastly, the same exposures and LED intensity settings should be used for each respective channel across the image acquisition process to ensure comparable images.
When performing RT-qPCR, the most critical step is the RNA extraction. During this step, avoiding RNase activity is critical to preserve the quality of the isolated RNA. Thus, it is important to clean the equipment and workspace with 70% ethanol. RNA will also be degraded during freeze/thaw cycles; therefore, it is best to perform the reverse transcription immediately following RNA isolation. Another important step is the design of the qPCR plate as all samples for a set of primers should be on the same plate. If this is not possible, an inter-plate calibrator (IPC) can be used to allow for comparison between plates (Taylor et al., 2019). The IPC is a qPCR reaction that will be identical between all plates, i.e., the same sample (which can be formed by pooling small amounts of various cDNA samples) with the same set of primers. The repetition of this specific reaction will allow for comparability between several plates. Another critical step is the loading of the plate; pipetting errors with small volumes will lead to a large variation in the results. After having distributed the master mix (that contains all reagents except for the cDNA) for each target in each well, we recommend pipetting up and down for each reaction when adding the cDNAs.
During western blot analysis, it may be difficult to find an optimal protein concentration and antibody dilution to use for different experimental parameters. Therefore, multiple attempts may be required to establish the optimal conditions for western blotting. A negative cell lysate control that is not of myogenic lineage (and therefore does not express myogenic markers) will assist in identifying the specific protein bands if antibodies detect non-specific proteins. Total protein controls (such as trichloroethanol) may also provide a more accurate indication of protein loading per lane, as cytoskeletal restructuring and metabolic perturbances may influence traditional single protein load controls (α-tubulin, β-actin, and GAPDH), as demonstrated in Figure 2D.
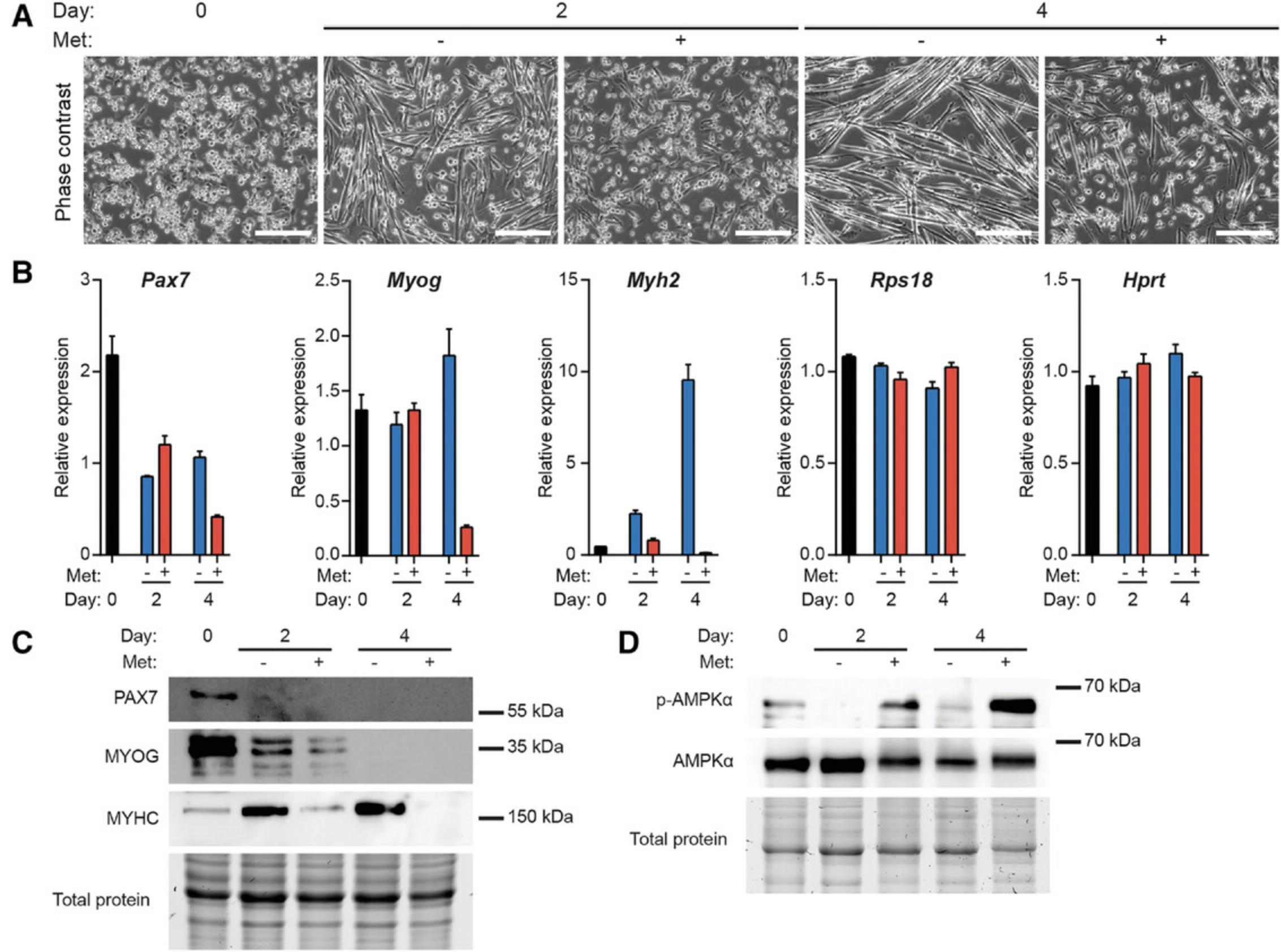
Finally, a functional readout of the pharmacological agent being examined is strongly advised to confirm that the drug is working through established molecular pathways. In this example study, we confirmed that treatment of differentiating myoblasts with metformin induced the activation of AMPK by assessing the phosphorylation status of AMPK at Thr 172 by western blot (Fig. 4D).
Troubleshooting
The in vitro differentiation of primary myoblasts can be technically challenging. Below are several common sources of poor differentiation.
- An insufficient cell confluence prior to initiating differentiation: In vitro myogenic differentiation is dependent on cell to cell contact between myoblasts and therefore efficient differentiation requires a confluent plate of cells. However, growing myoblasts to 100% confluence poses several challenges. We have observed that at ∼80% confluence, myoblasts begin detaching from the plate and therefore may be lost during medium changes while growing to full confluence. To prevent losing the detached cells during medium washes, allow the cells to re-attach on the plate prior to gently changing the medium. Myoblasts also exhibit slowed growth at high confluency. If the cells remain on the same plate for longer than 1 week, their behavior and differentiation are negatively impacted. Therefore, it is recommended that the plates are seeded for differentiation at relatively high confluence (∼70% to 80%). Ideally, differentiation and pharmacological treatment should be administered 3 to 4 days after seeding.
- Cells are of a high passage number: We have observed that primary myoblasts above passage number ten experience impaired differentiation. Poor differentiation may also be influenced by cell culturing techniques and the originating quantity and source (mouse strain) of the primary myoblasts. Therefore, we recommend performing differentiation experiments as soon as possible after isolation at the lowest passage number possible.
- Cell line specific behavior: We have observed that myoblasts isolated from mice of the same genetic strain exhibit variability in their capacity for differentiation. Therefore, when working with primary cell lines, inclusion of biological replicates (multiple cell lines) is recommended to ensure robustness of phenotypes observed.
- Inter-plate variability after seeding: After cells are seeded onto their experimental plates (e.g., from one 100-mm plate to three 35-mm plates for IF), the seeded plates may exhibit slight variations in cell confluency. This can be avoided by thoroughly resuspending cells to reduce cell clumping prior to seeding. Alternatively, use the smallest possible sized pipet to resuspend cells.
Other issues that are not related to differentiation are discussed in Table 8.
| Problem | Possible cause | Solution |
|---|---|---|
| Myoblast and myocyte clustering in IF images | Uneven cell distribution | When plating myoblasts for differentiation, swirl the plates in a figure eight or plus sign fashion to distribute cells for at least 30 s. Check plate under a light microscope after swirling to ensure cells are distributed evenly. |
| Low cell density after fixation | Cells lifting off plates after fixation | Do not aspirate solutions after fixation with a vacuum. Instead, use a micropipet (such as a P1000) to aspirate the solution to be removed. Subsequently, use a micropipet to add the next solution to be applied gently and aim towards the side of the plate. |
| Low IF signal intensity |
Photo bleaching and lack of signal during IF secondary antibody incubation Primary antibody concentration is too low |
Store immunostained plates in the dark at 4°C. Ideally image at the microscope 1 day post-mounting. Titrate primary antibody to determine the optimal concentration. |
| Low quantity of RNA |
Poor cell collection technique RNA degradation |
After the initial collection of cells with the scraper, wash the plate with PBS and collect this wash solution. Alternatively, grow the cells on 60-mm dishes which will provide sufficient RNA for extensive RT-qPCR analysis. Ensure that your workspace is RNase-free. You can also use filtered tips to prevent RNase contamination. |
| Low quality of RNA | Salt or protein contamination during RNA isolation |
When measuring the RNA concentration, the NanoDrop instrument will give two ratios: A260/A280: This ratio should be ∼2. If it is <1.7 the sample is most likely contaminated with protein. A treatment with proteinase K could improve that ratio. To avoid protein contamination, make sure to avoid the white interface when collecting the upper phase (Support Protocol 1; step 8). A260/A230: this ratio should be 2 or above. If the ratio is lower, you can re-precipitate RNA to remove contaminants. |
| Non-specific bands in protein lanes | Antibody non-specific binding | Depending on the lot, primary antibodies may exhibit non-specific bands. To identify the proper band, add a negative control lysate alongside myogenic samples. |
| Protein signal over/under exposure | Variability in antibody sensitivity |
Titrate the antibody concentrations at a given protein amount to identify the optimal antibody concentration. Titrate the protein amount against a given antibody concentration. Manually set the exposure time to obtain a series of images that correspond to a signal that is bright but not oversaturated. |
Understanding Results
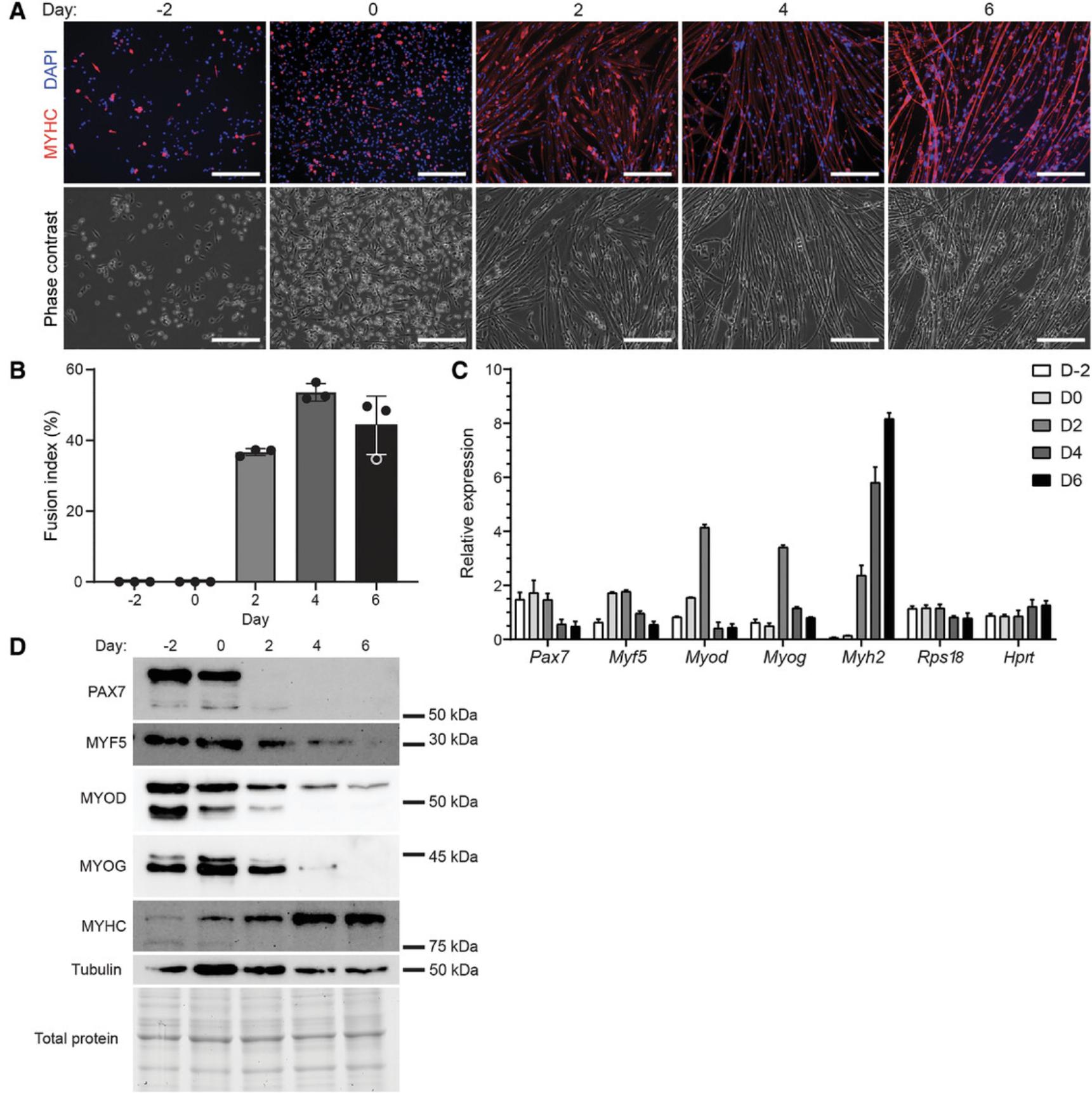
To examine the differentiation characteristics of a primary myoblast cell line, a 6-day differentiation assay was performed to assist in establishing the parameters for subsequent pharmacological modulation experiments with metformin. Every 2 days, we assessed changes to cell morphology (as determined by IF microscopy, Fig. 2A) along with alterations to the gene and protein expression of several myogenic factors using RT-qPCR (Fig. 2C) and western blotting (Fig. 2D). As shown in Figure 2A, prior to directed differentiation (at days −2 and 0) the cells exhibited primarily a round morphology and an absence of MYHC expression that is characteristic of undifferentiated myoblasts. Notably, the increase in cell density between days −2 and 0 induced differentiation, as many myoblasts adopted a spindle morphology at day 0. By day 2 of differentiation, myotube formation was observed, corresponding with the initiation of MYHC expression (represented in red, Fig. 2A), however numerous unfused myoblasts and myocytes remained. Between days 2 and 4, the myotubes continued to develop as evidenced by an increase in multi-nuclear fusions (from a fusion index of 36.5% at day 2 to 53.5% at day 4, Fig. 2B). Between days 4 and 6, a deterioration in myotube morphology was observed, which corresponded with a lower fusion index of 45.2% at day 6. Based on the cell morphology and calculated fusion index, we concluded that day 4 is the optimal time point for differentiation of this particular primary myoblast cell line.
The RT-qPCR and western blot analyses of the myogenic factors correlated with the observed changes in cell type and revealed the expression of these factors at the gene and protein level (Fig. 2C and D). At the transcript level, Pax7 expression was highest from day −2 to 2; Myf5 expression peaked from day 0 to 2; Myod and Myog expression were highest at day 2; while Myh2 expression steadily increased during differentiation (Fig. 2C). Expression of Rps18 and Hprt , which were used as reference genes, remained stable throughout the differentiation time course (Fig. 2C). Protein expression analyses correlated with mRNA transcript levels, with day −2 and 0 cells expressing the highest level of PAX7, MYF5, MYOD, and MYOG, which are indicative of the myoblast state (Fig. 2D). By day 2, PAX7 expression is absent, and MYF5, MYOD, and MYOG levels started to decrease, while MYHC expression increased. From day 2 to 4, these changes in protein abundance are indicative of cells that are no longer myoblasts and are transitioning from myocytes to myotubes. The robust expression of MYHC (and significant reduction of MYOD and MYOG) present at day 4 revealed that the cells at this stage are predominantly myotubes. Given the pronounced changes from day 2 to day 4 in the transcript and protein levels of the myogenic factors, and minimal differences between days 4 and 6, this further supports our conclusion drawn from the IF microscopy analyses that 4 days is the optimal differentiation duration for these cells.
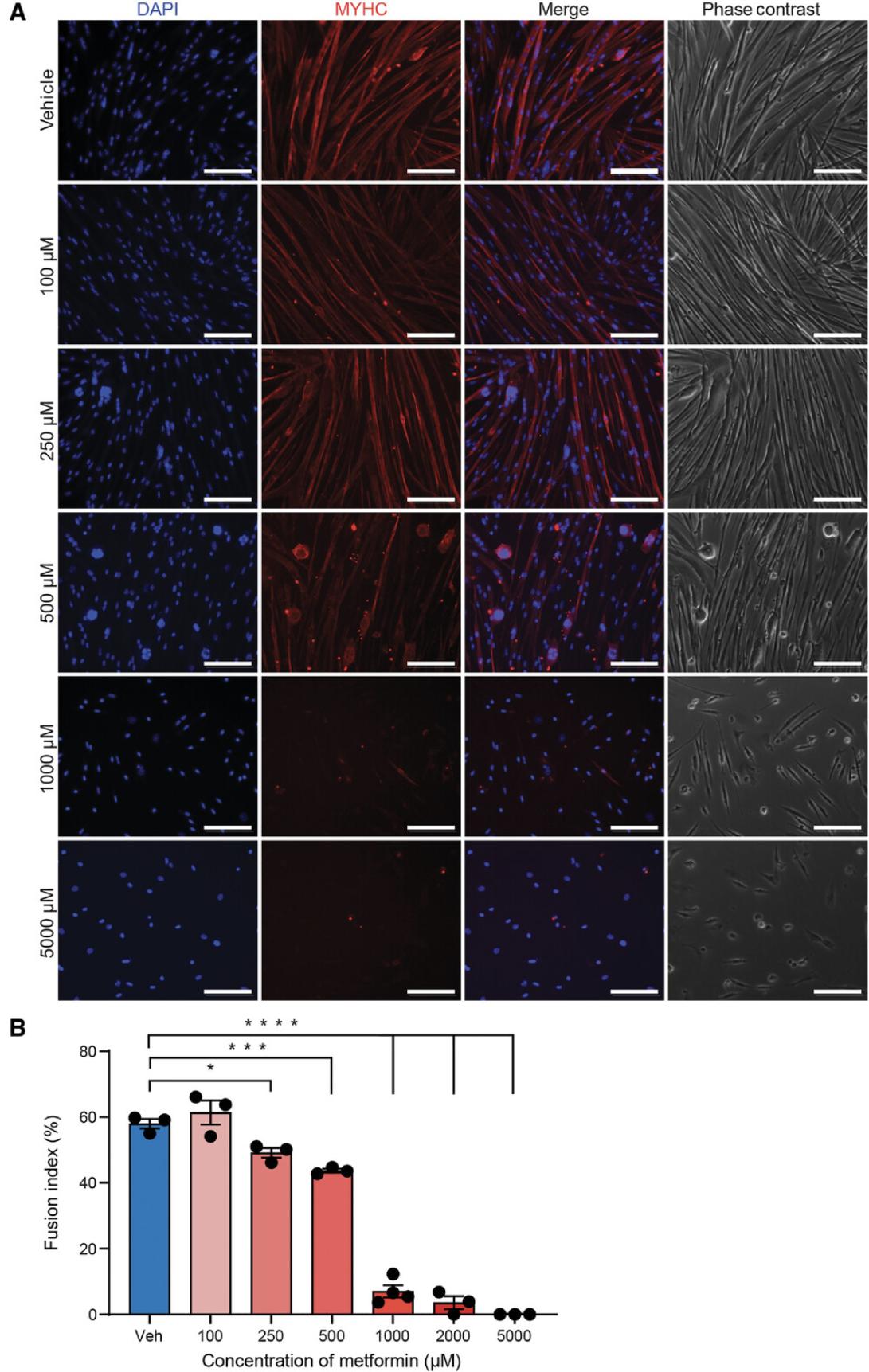
To establish an effective dose at which metformin inhibits differentiation, we treated cells concurrently during their differentiation for 4 days at the following concentrations: 100, 250, 500, 1,000, 2,000, and 5,000 µM. As shown by both cell morphology (Fig. 3A) and the fusion index (Fig. 3B), 100 µM of metformin did not significantly impact differentiation. Higher doses of metformin (250 µM and 500 µM) impeded differentiation but did not completely inhibit it, as the fusion indices were 49.1% and 43.9%, respectively, compared to 57.9% in vehicle-treated (DMSO) cells. Treatment with 1,000 µM of metformin significantly impaired differentiation with minimal cytotoxicity when compared to the higher doses of 2,000 and 5,000 µM. From these results, we determined that 1,000 µM is the optimal dose to investigate the negative impact of metformin on myogenic differentiation.
To further examine the effect of metformin on myogenesis we assessed cell morphology along with myogenic gene and protein expression following treatment with 1,000 µM metformin on differentiation at days 2 and 4. As revealed by phase contrast imaging, metformin severely inhibited the differentiation of myoblasts at days 2 and 4 (Fig. 4A). Gene expression analyses revealed that treatment with metformin inhibited the expression of Myog and Myh2 at day 4, which confirmed the impairment in terminal differentiation observed by microscopy (Fig. 4B). Similar results were observed at the protein level; MYOG was reduced following metformin treatment at day 2; and expression of MYHC was impaired at days 2 and 4 (Fig. 4C). Because MYHC expression corresponds with myotube development, its impaired expression in metformin treated cells indicated a defect in myotube formation. Thus, 1,000 µM metformin negatively impacts myogenesis; metformin treatment inhibits myotube formation and the expression of myogenic differentiation markers including MYOG and MYHC.
Time Considerations
Estimated times for procedures within each protocol are outlined in Table 9.
| Basic Protocol | |
| PFA fixation | 20 min |
| Washing off PFA | 10 min |
| Permeabilization | 10 min |
| Washing | 30 min |
| Blocking | 1-2 hr |
| Primary antibody incubation | Overnight |
| Secondary antibody incubation | 1 hr |
| Washing | 30 min |
| Mounting | Overnight |
| Microscopy imaging | 5 min per slide |
| Counting nuclei | 20-30 min per image |
| Support Protocol 1 | |
| Cell lysis | 1 hr |
| RNA isolation | Depends on the number of samples, typically ∼30 min |
| cDNA synthesis | 30 min |
| Preparing samples and master mixes | Depends on the number of samples and targets, typically ∼20 min |
| Loading plate | 45 min |
| Running plate | Depends on the qPCR, typically ∼1 hr |
| Support Protocol 2 | |
| Lysate preparation | Depends on the number of samples, typically ∼30 min |
| Gel electrophoresis | 2.5 hr |
| Semi-dry transfer | 15 min |
| Blocking | 1 hr |
| Primary antibody incubation | Overnight |
| Washing | 15 min |
| Secondary antibody incubation | 1 hr |
| Washing | 30 min |
| ECL treatment | 5 min |
| Imaging | Depends on the number of blots and signal intensity |
Acknowledgments
The authors thank Rebecca Robertson, Shulei Li, Anne-Marie Tremblay, and Alessio Cusmano for helpful discussions and critical reading of the manuscript. Research in the Chang laboratory is supported by research grants from the Stem Cell Network (ECR-C4R1-2), Defeat Duchenne Canada, Fonds de recherche du Québec - Santé (FRQS, 309857), and the Canadian Institutes of Health Research (469124). CAV is supported by a Faculty of Medicine and Health Sciences Internal Studentship from McGill University. RLF is supported by a Master's Training award from FRQS. NCC is supported by a Chercheurs-boursiers - Junior 1 award from FRQS. Figure 1 was created with BioRender.com.
Author Contributions
Cordell A. VanGenderen : Conceptualization, formal analysis, investigation, methodology, writing original draft, writing review and editing; Jules A. Granet : Conceptualization, formal analysis, investigation, methodology, writing original draft, writing review and editing; Romina L. Filippelli : Conceptualization, formal analysis, methodology, writing review and editing; Yiyang Liu : Investigation, writing review and editing; Natasha C. Chang : Conceptualization, funding acquisition, resources, supervision, writing original draft, writing review and editing
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
Data available on request from the authors.
Supporting Information
| Filename | Description |
|---|---|
| cpz1565-sup-0001-SuppInfo.pdf26.8 KB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Bentzinger, C. F., Wang, Y. X., & Rudnicki, M. A. (2012). Building muscle: Molecular regulation of myogenesis. Cold Spring Harbor Perspectives in Biology , 4(2), a008342. doi: 10.1101/cshperspect.a008342
- Crist, C. (2017). Emerging new tools to study and treat muscle pathologies: Genetics and molecular mechanisms underlying skeletal muscle development, regeneration, and disease. Journal of Pathology , 241(2), 264–272. doi: 10.1002/path.4830
- Cruz-Jentoft, A. J., & Sayer, A. A. (2019). Sarcopenia. The Lancet , 393(10191), 2636–2646. doi: 10.1016/S0140-6736(19)31138-9
- De Angelis, L., Borghi, S., Melchionna, R., Berghella, L., Baccarani-Contri, M., Parise, F., … Cossu, G. (1998). Inhibition of myogenesis by transforming growth factor β is density-dependent and related to the translocation of transcription factor MEF2 to the cytoplasm. Proceedings of the National Academy of Sciences of the United States of America , 95(21), 12358–12363. doi: 10.1073/pnas.95.21.12358
- Filippelli, R. L., & Chang, N. C. (2021). Empowering muscle stem cells for the treatment of Duchenne muscular dystrophy. Cells Tissues Organs , 211(6), 1–14. doi: 10.1159/000514305
- Filippelli, R. L., Omer, A., Li, S., van Oostende-Triplet, C., Gallouzi, I. E., & Chang, N. C. (2021). Automated quantification of subcellular particles in myogenic progenitors. Current Protocols , 1(12), e325. doi: 10.1002/cpz1.325
- Hindi, L., McMillan, J. D., Afroze, D., Hindi, S. M., & Kumar, A. (2017). Isolation, culturing, and differentiation of primary myoblasts from skeletal muscle of adult mice. Bio-Protocol , 7(9), e2248. doi: 10.21769/BIOPROTOC.2248
- Huang, Z., Chen, X., & Chen, D. (2011). Myostatin: A novel insight into its role in metabolism, signal pathways, and expression regulation. Cellular Signalling , 23(9), 1441–1446. doi: 10.1016/j.cellsig.2011.05.003
- Jiang, L. L., & Liu, L. (2020). Effect of metformin on stem cells: Molecular mechanism and clinical prospect. World Journal of Stem Cells , 12(12), 1455–1473. doi: 10.4252/wjsc.v12.i12.1455
- Jin, W., Shang, Y., Peng, J., & Jiang, S. (2016). Histone H3 methyltransferase suv39h1 prevents myogenic terminal differentiation by repressing MEF2 activity in muscle cells. International Journal of Molecular Sciences , 17(12), 1908. doi: 10.3390/ijms17121908
- Kim, J., Park, M. Y., Kim, H. K., Park, Y., & Whang, K. Y. (2016). Cortisone and dexamethasone inhibit myogenesis by modulating the AKT/mTOR signaling pathway in C2C12. Bioscience, Biotechnology and Biochemistry , 80(11), 2093–2099. doi: 10.1080/09168451.2016.1210502
- Kuang, S., Kuroda, K., Le Grand, F., & Rudnicki, M. A. (2007). Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell , 129(5), 999–1010. doi: 10.1016/j.cell.2007.03.044
- Langley, B., Thomas, M., Bishop, A., Sharma, M., Gilmour, S., & Kambadur, R. (2002). Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. Journal of Biological Chemistry , 277(51), 49831–49840. doi: 10.1074/jbc.M204291200
- Lazure, F., Blackburn, D. M., Corchado, A. H., Sahinyan, K., Karam, N., Sharanek, A., … Soleimani, V. D. (2020). Myf6/MRF4 is a myogenic niche regulator required for the maintenance of the muscle stem cell pool. EMBO Reports , 21(12), e49499. doi: 10.15252/embr.201949499
- Levitt, D. E., Adler, K. A., & Simon, L. (2019). HEMA 3 Staining: A simple alternative for the assessment of myoblast differentiation. Current Protocols in Stem Cell Biology , 51(1), e101. doi: 10.1002/cpsc.101
- McKinnell, I. W., Ishibashi, J., Le Grand, F., Punch, V. G. J., Addicks, G. C., Greenblatt, J. F., … Rudnicki, M. A. (2008). Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nature Cell Biology , 10(1), 77–84. doi: 10.1038/ncb1671
- McRae, N., Forgan, L., McNeill, B., Addinsall, A., McCulloch, D., Van Der Poel, C., & Stupka, N. (2017). Glucocorticoids improve myogenic differentiation in vitro by suppressing the synthesis of versican, a transitional matrix protein overexpressed in dystrophic skeletal muscles. International Journal of Molecular Sciences , 18(12), 2629. doi: 10.3390/ijms18122629
- Meng, L., & Feldman, L. (2010). A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnology Journal , 5(2), 183–186. doi: 10.1002/biot.200900211
- Montesano, A., Luzi, L., Senesi, P., & Terruzzi, I. (2013). Modulation of cell cycle progression by 5-azacytidine is associated with early myogenesis induction in murine myoblasts. International Journal of Biological Sciences , 9(4), 391–402. doi: 10.7150/ijbs.4729
- Naidu, P. S., Ludolph, D. C., To, R. Q., Hinterberger, T. J., & Konieczny, S. F. (1995). Myogenin and MEF2 function synergistically to activate the MRF4 promoter during myogenesis. Molecular and Cellular Biology , 15(5), 2707–2718. doi: 10.1128/mcb.15.5.2707
- Pavlidou, T., Rosina, M., Fuoco, C., Gerini, G., Gargioli, C., Castagnoli, L., & Cesareni, G. (2017). Regulation of myoblast differentiation by metabolic perturbations induced by metformin. PLoS ONE , 12(8), e0182475. doi: 10.1371/journal.pone.0182475
- Petrany, M. J., & Millay, D. P. (2019). Cell fusion: Merging membranes and making muscle. Trends in Cell Biology , 29(12), 964–973. doi: 10.1016/j.tcb.2019.09.002
- Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research , 29(9), E45. doi: 10.1093/nar/29.9.e45
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., … Cardona, A. (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods , 9(7), 676–682. doi: 10.1038/NMETH.2019
- Seale, P., Sabourin, L. A., Girgis-Gabardo, A., Mansouri, A., Gruss, P., & Rudnicki, M. A. (2000). Pax7 is required for the specification of myogenic satellite cells. Cell , 102(6), 777–786. doi: 10.1016/S0092-8674(00)00066-0
- Simionescu-Bankston, A., Pichavant, C., Canner, J. P., Apponi, L. H., Wang, Y., Steeds, C., … Pavlath, G. K. (2015). Creatine kinase B is necessary to limit myoblast fusion during myogenesis. American Journal of Physiology - Cell Physiology , 308(11), C919–C931. doi: 10.1152/ajpcell.00029.2015
- Soleimani, V. D., Punch, V. G., Kawabe, Y.- i., Jones, A. E., Palidwor, G. A., Porter, C. J., … Rudnicki, M. A. (2012). Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Developmental Cell , 22(6), 1208–1220. doi: 10.1016/j.devcel.2012.03.014
- Stockdale, F. E. (1992). Myogenic cell lineages. Developmental Biology , 154(2), 284–298. doi: 10.1016/0012-1606(92)90068-R
- Taylor, S. C., Nadeau, K., Abbasi, M., Lachance, C., Nguyen, M., & Fenrich, J. (2019). The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends in Biotechnology , 37(7), 761–774. doi: 10.1016/j.tibtech.2018.12.002
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., & Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology , 3(7), 1–12. doi: 10.1186/GB-2002-3-7-RESEARCH0034
- Viollet, B., Guigas, B., Sanz Garcia, N., Leclerc, J., Foretz, M., & Andreelli, F. (2012). Cellular and molecular mechanisms of metformin: An overview. Clinical Science , 122(6), 253–270. doi: 10.1042/CS20110386
- Yin, H., Price, F., & Rudnicki, M. A. (2013). Satellite cells and the muscle stem cell niche. Physiological Reviews , 93(1), 23–67. doi: 10.1152/physrev.00043.2011
- Yiu, E. M., & Kornberg, A. J. (2015). Duchenne muscular dystrophy. Journal of Paediatrics and Child Health , 51(8), 759–764. doi: 10.1111/jpc.12868
- Zammit, P. S. (2017). Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. In Seminars in cell and developmental biology (Vol. 72, pp. 19–32). New York: Academic Press. doi: 10.1016/j.semcdb.2017.11.011
- Zhao, D., Pond, A., Watkins, B., Gerrard, D., Wen, Y., Kuang, S., & Hannon, K. (2010). Peripheral endocannabinoids regulate skeletal muscle development and maintenance. European Journal of Translational Myology , 20(4), 167–180. doi: 10.4081/ejtm.2010.1814
Citing Literature
Number of times cited according to CrossRef: 1
- Jurandyr Pimentel Neto, Rodrigo Daniel Batista, Lara Caetano Rocha‐Braga, Marucia Chacur, Paula Oliveira Camargo, Adriano Polican Ciena, The telocytes relationship with satellite cells: Extracellular vesicles mediate the myotendinous junction remodeling, Microscopy Research and Technique, 10.1002/jemt.24549, 87 , 8, (1733-1741), (2024).

