Mapping Maize Mutants Using Bulked-Segregant Analysis and Next-Generation Sequencing
Norman B. Best, Norman B. Best, Paula McSteen, Paula McSteen
Abstract
Forward genetics is used to identify the genetic basis for a phenotype. The approach involves identifying a mutant organism exhibiting a phenotype of interest and then mapping the causative locus or gene. Bulked-segregant analysis (BSA) is a quick and effective approach to map mutants using pools of mutants and wild-type plants from a segregating population to identify linkage of the mutant phenotype, and this approach has been successfully used in plants. Traditional linkage mapping approaches are outdated and time intensive, and can be very difficult. With the highly evolved development and reduction in cost of high-throughput sequencing, this new approach combined with BSA has become extremely effective in multiple plant species, including Zea mays (maize). While the approach is incredibly powerful, careful experimental design, bioinformatic mapping techniques, and interpretation of results are important to obtain the desired results in an effective and timely manner. Poor design of a mapping population, limitations in bioinformatic experience, and inadequate understanding of sequence data are limitations of these approaches for the researcher. Here, we describe a straightforward protocol for mapping mutations responsible for a phenotype of interest in maize, using high-throughput sequencing and BSA. Specifically, we discuss relevant aspects of developing a mutant mapping population. This is followed by a detailed protocol for DNA preparation and analysis of short-read sequences to map and identify candidate causative mutations responsible for the mutant phenotype of interest. We provide command-line and perl scripts to complete the bioinformatic analysis of the mutant sequence data. This protocol lays out the design of the BSA, bioinformatic approaches, and interpreting the sequencing data. These methods are very adaptable to any forward genetics experiment and provide a step-by-step approach to identifying the genetic basis of a maize mutant phenotype. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Bulked-segregant analysis and high-throughput sequencing to map maize mutants
INTRODUCTION
Forward genetic approaches are effective methods to identify the genetic locus responsible for a mutant phenotype. This protocol describes a bulked-segregant analysis (BSA) approach coupled with next-generation sequencing to map and identify the mutation that causes the mutant phenotype. Identifying the causative locus for a mutant phenotype is difficult, especially in Zea mays due to genome size, repetitive sequences, and long life cycle. This classical approach requires a large number of polymerase chain reactions (PCR) or restriction digest reactions on a genome-wide scale, which takes a large amount of time. Traditional linkage mapping techniques are time intensive, require large mapping populations (a population that is effective for linkage mapping of genetic markers), and may need development of novel markers (Gallavotti & Whipple, 2015). The development of high-throughput whole-genome re-sequencing has allowed for novel approaches to map mutant genes in a very rapid time frame (Austin et al., 2011; James et al., 2013; Schneeberger & Weigel, 2011). The cost reductions in whole-genome re-sequencing make this approach cost-effective compared to classical linkage-mapping experiments. Further, the development of highly efficient bioinformatic tools (Bolger, Lohse, & Usadel, 2014; Danecek et al., 2021; Li & Durbin, 2009; Li et al., 2009; Quinlan & Hall, 2010; Wall, 1994) allows for easy alignment of short-read sequences and identification of polymorphisms for mapping mutant genes. Effectively utilizing these bioinformatic software applications to align sequence data and mine the data to map a causative mutation can be daunting to a novice researcher, so this protocol lays out how to do this efficiently and effectively.
BSA involves screening for genomic differences between a pooled DNA sample of segregating mutants, usually in an F2 population, and the wild-type sequence (Michelmore, Paran, & Kesseli, 1991). In this article, we describe a BSA and high-throughput sequencing method to map and identify causative mutations (Fig. 1) in Zea mays (maize) (Best et al., 2021; Yao et al., 2019). Generation of maize mutants can be done by chemical mutagenesis or by utilizing transposon-activated populations. An effective chemical mutagen, ethyl methanesulfonate (EMS), interacts with guanine in DNA to create an O6-ethylguanine (Amano & Smith, 1965; Settles, 2020). During DNA replication, the abnormal base pair is paired with thymine instead of cytosine. This results in a transition mutation where the original G:C pair has been replaced with an A:T pair. Structural mutations and insertion and deletion (indels) mutations, as well as natural variants, can also be mapped using this protocol. Identification of these causative mutations, rather than causative SNP mutations, is more difficult with this approach. Similar to a classical mapping approach, creation of a segregating F2 population (or further back-crossed line) is necessary. To map a mutant, the mapping population is formed from a cross between a plant exhibiting a mutant phenotype of interest and a maize inbred from a separate heterotic group and self-pollinated, so that the mapping population has different segregating sequences and is vigorous. Genetic mapping or linkage mapping is used to identify the order of genes on a chromosome, and is determined by recombination frequency. Physical mapping identifies a map region in physical base pairs where variants of a certain haplotype associate with a mutant phenotype in a segregating population. This allows the researcher to identify a map region as the genomic location of the mutation responsible for the mutant phenotype. Briefly, the approach is as follows, and is described in the Basic Protocol. Mutants exhibiting the phenotype of interest in the F2 generation are identified, and tissue is collected and pooled. DNA is then extracted and sequenced using a short-read high-throughput sequencer. Sequenced reads are then aligned to the maize reference genome in which the mutant was originally identified. Alternatively, the inbred line that was used for out-crossing can also be used as the reference genome. Single-nucleotide polymorphisms (SNPs) are then identified in coding regions using publicly available bioinformatic software, and the allele frequency is calculated in a 100-SNP sliding window to physically map the causative mutation. Only the SNPs in the coding regions are used as intergenic SNPs, and SNPs in repetitive sequences such as transposable elements have a higher false positive rate (Ribeiro et al., 2015). EMS is used for mutagenesis, and homozygous SNPs are identified and filtered for G:C to A:T nucleotide transitions (Amano & Smith, 1965; Settles, 2020). SNPs within the identified map window are characterized for effects on coding-sequence changes to identify a candidate causative mutation. This protocol provides the scripts to complete the analysis and explains how to interpret the results. Lastly, development of additional alleles through reverse genetic approaches, such as CRISPR/Cas9 , Uniform Mu (Settles et al., 2007), or Bonn Mu (Marcon et al., 2020), will help confirm the causative mutation and characterization of the mutant phenotype. Further characterization of the mutant with biochemical, genetic, and/or molecular biology experiments that can be designed based upon the biological function of the candidate gene will further confirm the causative mutation. This protocol is much quicker and more effective than traditional linkage mapping approaches for mapping the causative locus responsible for a mutant phenotype, especially in maize.
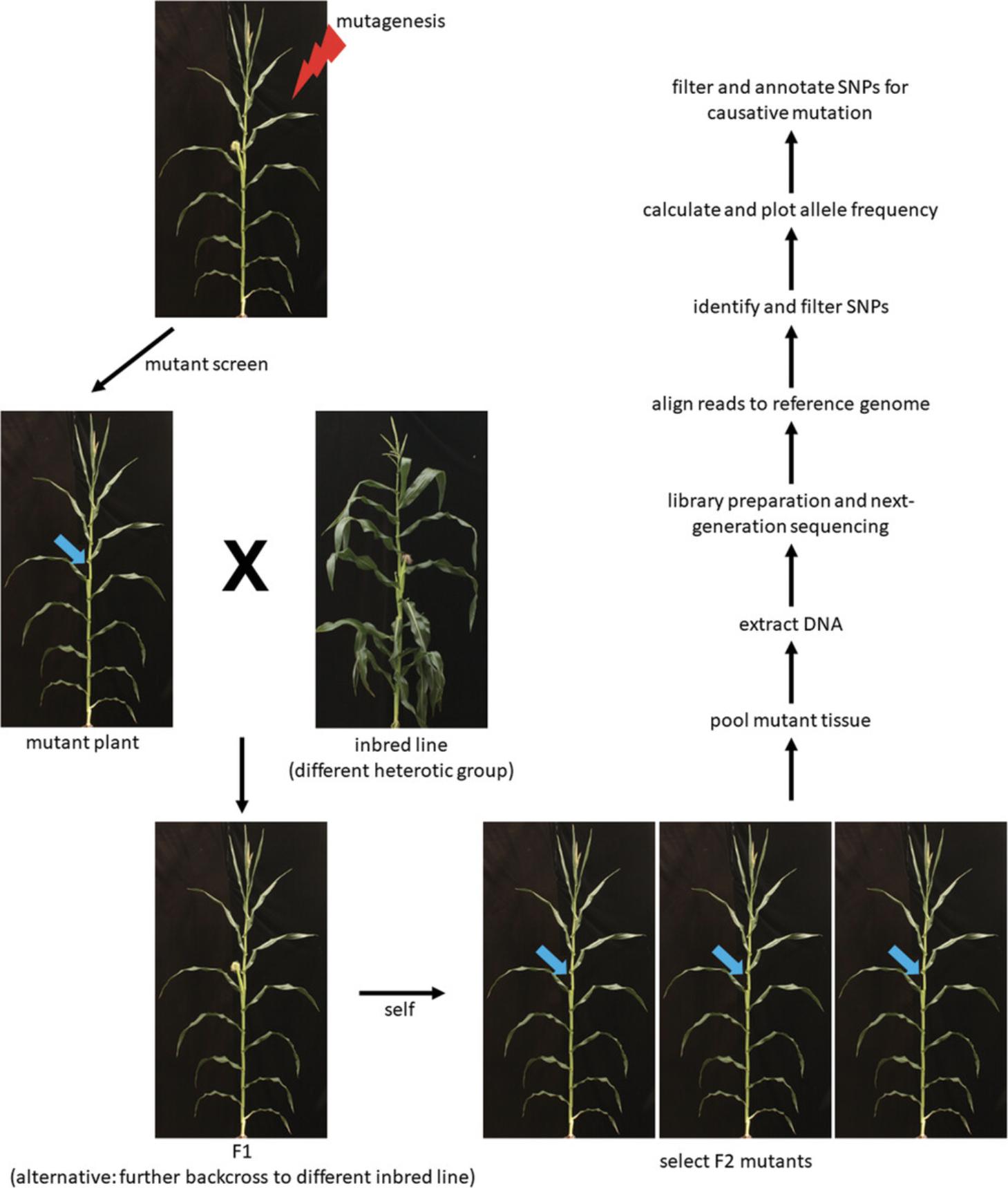
STRATEGIC PLANNING
Before beginning, identification of an easily recognizable mutant phenotype is necessary. Knowing the original genetic background on which the mutant phenotype was identified is beneficial, as well as knowing the nature of the mutation (i.e., EMS mutagenesis, Robertson's Mutator , etc.), but neither is necessary for mapping the causative mutation. Knowing the nature of the mutation (semi-dominant, dominant, or recessive) is necessary to ensure that the mapping population is correctly designed (i.e., creating F3 families for a semi-dominant mutant to identify bona fide homozygote plants compared to only needing F2 families for a recessive mutant), and downstream computational hypotheses are accurately tested. A mutant phenotype can become suppressed in a particular genetic background, and knowing which lines do this is important (Coe, 1994). Preliminary test crosses, and subsequent self-pollination of the F1 generation to create segregating F2 plants, with different inbred lines from various heterotic groups (at least three), will allow you to identify the genetic nature of the mutation and detect possible interference by the genetic background of the mutant phenotype. The F2 segregating plants will be the mapping population from which plants with the mutant phenotype will be selected for BSA. Obtaining a segregating mutant population that contains homozygous mutants is necessary to have effective mapping data, as well as for narrowing down candidate gene lists. Not following these important criteria will result in poor results in which no peaks, or multiple peaks, will be identified; therefore, the causative locus responsible for the mutant phenotype will not be identified by a single peak. Designing mapping populations for dominant versus recessive mutants will require alternative approaches.
Recessive mutant approach
To verify that the mutation is recessive, you will observe that the mutant phenotype will not be visible in the F1 generation but will then reappear in the F2 generation. After identification and verification of a recessive mutant phenotype, outcross the mutant to a different genetic background that does not suppress the mutant phenotype. Segregation ratios should be calculated in the F2 generation to ensure that this is the case. The alternative genetic background should primarily be from another heterotic group, such as synthetic stiff stalk crossed with non–stiff stalk or tropical crossed with a synthetic stiff stalk. Until other maize inbred genomes have been better annotated, using B73 as one of the lines is beneficial to help with alignment and SNP calling of sequenced reads. Creation of an F2 population is effective for mapping the causative mutation. The higher the number of individuals the better, to allow for higher amounts of recombination near the locus of interest. Accurate phenotyping is necessary to ensure no contamination of the mutant pool by non-mutant samples. Contamination by one non-mutant will ultimately affect downstream hypotheses when analyzing sequence data. Having fewer individuals is better than having one non-mutant in your pool. The number of mutants in an F2 pool should be no less than 10, but as the number increases, the map window will decrease due to increased chances for recombination.
Further back-crossing to the alternative contrasting inbred line for additional generations can be beneficial to decrease background noise (i.e., increase the homozygosity of the genetic background in regions not linked to the causative mutation). However, multiple generations of back-crossing will only narrow the mapping window slightly. Due to the long life cycle of maize, additional generations are not beneficial due to the time it takes to introgress the mutant into another background for sequence mapping. One could use Fast-Flowering Mini-Maize to speed up the introgression process, but only if you know the original mutant background and this has a reference genome (McCaw, Wallace, Albert, Buckler, & Birchler, 2016).
Dominant/semi-dominant approach
To map dominant or semi-dominant mutants, obtaining homozygous plants is necessary to effectively map by sequencing. Generation of mapping populations should be done using the same precautions as listed in the recessive mutant approach for heterotic groups and background effects on phenotype. For semi-dominant mutants where the phenotype can be distinguished between heterozygous and homozygous mutants, the F2 generation can be used. Selection of only homozygous mutants is necessary for downstream bioinformatic analysis, so careful selection of mutants is key. With a semi-dominant mutant, testing the segregation ratio in the F3 generation may be highly beneficial to ensure that non-mutants are not selected for the mutant pool.
For fully dominant mutants, this will necessarily require going to the F3 generation and selecting F3 families that only contain the mutant with no wild-type plants. Increasing the number of separate F3 families, derived from separate F2 plants, from which to collect mutant tissue will increase the rate of recombination and narrow the mapping window. Mutant tissue should be collected from at least 10 separate homozygous F3 families.
Basic Protocol: BULKED-SEGREGANT ANALYSIS AND HIGH-THROUGHPUT SEQUENCING TO MAP MAIZE MUTANTS
This protocol will allow the researcher to map and identify the region responsible for a maize mutant phenotype using BSA and high-throughput sequencing of genomic DNA. For this, a segregating mutant population must first be developed (Strategic Planning). Then, mutant tissue is pooled, and is then DNA-extracted and sequenced using a short-read sequencer. Collecting young leaf tissue that is snap frozen in liquid nitrogen is the best way to obtain the mutant tissue. We then describe how to use publicly available bioinformatic tools to align the short-read sequences to a reference genome to obtain files containing SNP positions in the mutant sequences. These files are then filtered to identify the map position of the causative mutation using a 100-SNP sliding window. If EMS mutagenesis was used to create the mutant phenotype, further downstream analysis of SNPs for G:C to A:T transitions will help identify a set of candidate genes within the map window. As an example for the protocol, we show the mapping and identification of the causative mutation responsible for the ba1-GN135 mutant allele that has a barren stalk phenotype (i.e., no ear branches on the stalk) and a weak reduction in tassel branching. This mutant was identified in A619 and backcrossed to B73 for four generations.
Materials
-
Maize inbred line that was used to create reference genome (obtained from https://www.ars-grin.gov)
-
Frozen pooled maize mutant tissue from F2 or F3 population (see Strategic Planning)
-
Liquid nitrogen
-
Urea extraction buffer (see recipe)
-
25:24:1 phenol/chloroform/isoamyl alcohol, pH 8.0 (Sigma-Aldrich, 77617)
-
Isopropanol
-
Ammonium acetate (see recipe)
-
70% (v/v) ethanol in water
-
TE buffer, pH 8.0 (see recipe)
-
Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Q32850)
-
Tru-Seq DNA PCR-Free Kit (Illumina, 20015962)
-
Leaf punch
-
50-ml conical tubes
-
Mortar and pestle
-
2-ml microcentrifuge tubes
-
Vortex
-
Benchtop centrifuge
-
Nanodrop spectrophotometer (Thermo Fisher Scientific, 13-400-519)
-
Qubit fluorometer (Thermo Fisher Scientific, Q33238)
-
Reference genome file in FASTA format (https://plants.ensembl.org/Zea_mays/Info/Index)
-
GFF3 file for reference genome (https://plants.ensembl.org/Zea_mays/Info/Index)
-
Perl scripts (https://doi.org/10.5281/zenodo.7041941)
-
Linux server
-
Microsoft Excel
Extract genomic DNA
A DNA prep with urea extraction buffer is described below. Other approaches using commercial kits or home-made solutions may also be used.
1.Collect a single leaf punch from each F2 mutant into a 50-ml conical tube and snap-freeze in liquid nitrogen.
2.Homogenize all mutant leaf tissue into a fine powder using a mortar and pestle with liquid nitrogen.
3.Add ∼100 mg of homogenized plant tissue to a 2-ml microcentrifuge tube along with 0.6 ml of urea extraction buffer. Vortex well for 30 s.
4.Add 0.5 ml of 25:24:1 phenol/chloroform/isoamyl alcohol to the tube and vortex for 15 min.
5.Centrifuge the sample to a benchtop centrifuge for 30 min at 18,000 × g , room temperature.
6.Carefully transfer the supernatant (upper layer) to a new 2-ml microcentrifuge tube. Be sure not to include any of the lower phase, as this can lower DNA quality.
7.Slowly add 0.6 ml of isopropanol and 60 µl of ammonium acetate. Mix gently by slowing inverting the tube back and forth.
8.Spool DNA with clean pipette tip and add to new 2-ml centrifuge tube with 1 ml of 70% (v/v) ethanol.
9.Centrifuge tube for 10 min at 18,000 × g , room temperature, to pellet DNA.
10.Let pellet air dry to remove remaining ethanol, but do not completely dry pellet.
11.Resuspend in 50 µl of TE buffer.
12.Check quality (i.e., A 260/A 280 and A 260/A 230 ratio) of DNA on a Nanodrop spectrophotometer and quantity of DNA on Qubit fluorometer using Qubit dsDNA BR Assay Kit with manufacturer's recommended protocol.
13.Create sequencing library using Tru-Seq DNA PCR-free kit following the manufacturer's recommended protocol. Then, sequence library on short-read sequencer as 150-bp paired-end reads.
Align and investigate sequenced reads
14.Download and install the most recent version of the required software packages and genome files shown in Table 1 to your Linux server environment.
| Program | Purpose | Source |
|---|---|---|
| Trimmomatic (current version 0.39) | Adapter trimming | http://www.usadellab.org/cms/index.php?page=trimmomatic (Bolger et al., 2014) |
| Java (current version 8.3) | Run Trimmomatic software | https://www.java.com/en/download/manual.jsp |
| BWA (current version 0.7.17) | Alignment | http://bio-bwa.sourceforge.net/bwa.shtml (Li & Durbin, 2009) |
| SAMtools (current version 1.10) | File conversion and SNP calling | http://www.htslib.org/doc/1.10/samtools.html (Li et al., 2009) |
| BEDtools (current version 2.30) | Genome coverage | https://bedtools.readthedocs.io/en/latest/content/tools/genomecov.html (Quinlan & Hall, 2010) |
| Bcftools (current version 1.15) | SNP filtering | https://samtools.github.io/bcftools (Danecek et al., 2021) |
| Perl (current version 5.36.0) | Run scripts to create allele frequency files | https://www.perl.org/ (Wall, 1994) |
| SnpEff (current version 5.1) | Filter SNP file for effects on protein coding changes | http://pcingola.github.io/SnpEff/ (Cingolani et al., 2012) |
15.Create a bwa alignment index of the most current reference genome of your selected inbred line using bwa with the following command:
- bwa index -a bwtsw <reference_genome.fa>
This will create files in the same directory with the same name as the reference file with .amb, .ann, .bwt, .pac, and .sa extensions.
16.Create a samtools index of the most current reference genome of your selected inbred line using samtools with the following command:
- samtools faidx <reference_genome.fa>
This will create a file in the same directory with the same name as the reference file but with a .fai extension.
17.Trim adapters off reads and filter for paired reads only using the Trimmomatic software package:
- java -jar trimmomatic-0.39.jar PE -<phred_quality_score> <forward_read.fastq> <reverse_read.fastq> <forward_read_pairtrim.fastq> <forward_read_unpairtrim.fastq> <reverse_read_pairtrim.fastq> <reverse_read_unpairtrim.fastq> ILLUMINACLIP :<adapter_sequences.fa>:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36
Select the correct quality score: phred+33 or phred+64.This depends on the Illumina pipeline used. All Illumina pipelines 1.8 and later are phred+33, whereas earlier Illumina pipelines use a variation of phred+64. Select the correct adapter sequences from library preparation. Trimmomatic has a number of adapter sequences available. If the Tru-Seq DNA PCR-free kit was used with paired-end sequencing, then use the TruSeq3-PE.fa.
18.Align sequenced reads to the most current reference genome. Multiple threads can be used to increase alignment time. Using the following commands to output a SAM file:
- bwa aln -t
-n 0.04 <reference_genome.fa> <forward_read_pairtrim.fastq> > - bwa aln -t
-n 0.04 <reference_genome.fa> <reverse_read_pairtrim.fastq> > - bwa sampe -P <reference_genome.fa> <forward_read.sai> <reverse_read.sai> <forward_read.fastq> <reverse_read.fastq> >
Alignment of the sequenced reads by BWA will output a Sequence Alignment/Map format (SAM) file. This is a tab-delimited file that has a header section and an alignment section.
19.Convert SAM file to BAM file:
- samtools view -bS <aligned_reads.sam> > <aligned_reads.bam>
The SAM file is then converted into a BAM file, a binary version of the SAM file. Both files should be large in size depending on sequencing depth, with the SAM file being larger than the BAM file.
20.Sort the BAM file:
- samtools sort -T <align_reads.sorted> -o <aligned_reads.sorted.bam> <aligned_reads.bam>
The -T command will create temporary files that will then be merged into a single BAM file to minimize computing memory requirements.
21.Calculate genome coverage for aligned reads using the following command:
- samtools view -u -q 20 <aligned_reads.sorted.bam> | genomeCoverageBed -ibam stdin -g <reference_genome.fa.fai > <coverage.genome.q20.cov>
- for gcov in <coverage.genome.q20.cov>
- do
- echo $gcov
- grep "^genome" $gcov > z.txt
- awk 'BEGIN{total=0} {total += (
3)} END{total=total/$4;print "\nAverage - Coverage: "total}' < z.txt >> $gcov
- done
- rm -f z.txt
The calculated genome coverage will be on the last line of the coverage.genome.q20.cov file written as: Average Coverage: ##.####. The genome coverage can be used to make predictions about false positives at the end of the analysis, as the total read counts at a given location should be relatively close to the genome coverage value.
22.Create BED file from the GFF3 file that contains all CDS locations in the genome using the perl file reference_genome_CDS.pl.
23.Create pileup file for all CDS positions using the samtools function mpileup with a read quality score greater than 20 and position score greater than 20.This will create an output .bcf file:
- samtools mpileup -l $BED -Q20 -q20 -t AD -t ADF -t ADR -t DP -t SP -uf <reference_genome.fa> <aligned_reads.sorted.bam> | bcftools view > <CDS_pileup.bcf>
The read quality score filters are used to eliminate sequencing and alignment errors.
24.Convert .bcf file to .vcf file using bcftools:
- bcftools view <CDS_pileup.bcf> > <CDS_pileup.vcf>
25.Filter .vcf variant file so that only the positions of SNPs are included using bcftools:
- bcftools view -c -V indels -v <CDS_pileup.vcf> > <CDS_pileup_variant_noindels.vcf>
26.Create separate .tsv files for each chromosome using the perl file allelefrequency.pl.
27.To calculate the allele frequency in a 100-SNP sliding window, open each chromosome .tsv file in Microsoft Excel. Average the values in cells C1:C100 and then C2:C101, etc., in column 4 to calculate the average allele frequency of 100 SNPs using the “AVERAGE” command in Microsoft Excel. The sliding window is created by moving one SNP at a time across the chromosome.
28.Plot column 2 (SNP positions) on the x -axis and column 4 (100-SNP sliding window) on the y -axis to map the location of the causative mutation.
29.To identify candidate mutations within the identified map window, re-run the samtools function mpileup without the CDS .bed file with the following command:
- samtools mpileup -Q20 -q20 -t AD -t ADF -t ADR -t DP -t SP -uf <reference_genome.fa> <aligned_reads.sorted.bam> | bcftools view > <ALL_pileup.bcf>
This step will identify all variants in the sequenced sample compared to the reference genome. The putative causative mutation for the mutant phenotype will be in this file.
30.Convert .bcf file to .vcf file using bcftools:
- bcftools view <ALL_pileup.bcf> > <ALL_pileup.vcf>
31.Filter .vcf variant file so that only the position of single-nucleotide polymorphisms (SNPs) and insertions and deletions (indels) are included using bcftools:
- bcftools view -c -v <ALL_pileup.vcf> > <ALL_pileup_variants.vcf>
32.Filter the .vcf variant file for SNP/indel strand bias (having reads on both strands) using the snpEff tool SnpSift.jar:
- cat <ALL_pileup_variants.vcf> | java -jar SnpSift.jar filter "(DP4[2] >0)& (DP4[3]>0)" > <ALL_pileup_variants_strandbias.vcf>
This is an important step to filter out false positives, as a subset of sequencing errors will occur on only one strand.
33.Filter the .vcf variant strand bias file for only homozygous non-reference mutations using the snpEff tool SnpSift.jar:
- cat <ALL_pileup_variants_strandbias.vcf> | java -jar SnpSift.jar filter "(DP4[0]=0) & (DP4[1]=0)" > <ALL_pileup_variants_strandbias_homozygous.vcf>
34.Annotate the final .vcf file for predicted functional effects on gene coding sequence using the snpEff.jar program as part of the SnpEff software:
- java -jar snpEff.jar eff -ud 0 -c snpEff.config <Zea_mays> <ALL_pileup_variants_strandbias_homozygous.vcf> > <ALL_pileup_variants_strandbias_homozygous_snpEff.vcf>
Be sure to check that the current version of the maize genome database is loaded in snpEff.
Look for HIGH effect mutations within the previously identified map window by filtering column 8 in Excel to only contain “HIGH” text.
If the mutagen ethyl methanesulfonate (EMS) was used to create the mutant, then the list can be further filtered within Microsoft Excel to only have G to A or C to T transitions.
35.Investigate each candidate variant for effect on coding sequence. Interpret the candidate variants in terms of the biological question, e.g., that the candidate gene should function in a pathway responsible for the mutant phenotype.
- cat <ALL_pileup_variant_noindels_strandbias.vcf> | java -jar SnpSift.jar filter "(DP4[0] <2)& (DP4[1]<2)" > <ALL_pileup_variant_noindels_strandbias.vcf>
This is most likely caused by contamination from including a wild-type plant in the mutant pool, operator error by mixing up samples, or sequencing errors by the sequencing machine.
REAGENTS AND SOLUTIONS
Ammonium acetate (pH 5.2), 4.4 M
- 105 ml of PCR-quality water
- 50.5 ml of glacial acetic acid
- 45 ml of 14.8 N ammonium hydroxide
- Store at room temperature for up to 12 months
CAUTION: All reagents should be added slowly in a fume hood as the glacial acetic acid and ammonium hydroxide are caustic and can burn the skin or lungs if exposed.
From Leach, McSteen, & Braun (2016).
TE buffer (pH 8.0), 10 mM
- 1 ml of 1 M Tris·HCl (pH 8.0)
- 0.2 ml of 0.5 M EDTA (pH 8.0)
- Make up to 100 ml with deionized water.
- Store at room temperature for up to 12 months
Urea extraction buffer
- 420 g urea
- 87.5 ml of 4 M sodium chloride
- 50 ml 1 M Tris⋅HCl (pH 8.0)
- 40 ml of 0.5 M ethylenediaminetetraacetic acid (EDTA) (pH 8.0)
- 10 g N-lauroylsarcosine (VWR; 76204-306)
- Fill to 1 L with deionized water and put on stir plate for 2 hr.
- Store at room temperature for up to 6 months.
CAUTION: A mask should be worn when adding n-lauroylsarcosine to prevent inhalation of powder.
From Chen & Dellaporta (1994); Leach et al. (2016).
COMMENTARY
Background Information
Maize is extremely pliable to forward genetic screens using methods such as chemical mutagenesis, radiation, transposon tagging, or even natural variation (Candela & Hake, 2008). Identifying the causative mutation for a maize mutant phenotype is a difficult task due to genome size, repetitive sequences, and time constraints (Bortiri, Jackson, & Hake, 2006). Classical positional mapping approaches require a large number of recombinants to narrow down a gene window by identifying polymorphisms between two parent lines (Gallavotti & Whipple, 2015). This approach requires independent DNA extractions from each segregant and a large amount of genotyping on a genome-wide scale, which is highly time intensive and difficult. Segregants are primarily genotyped using traditional methods such as single sequence repeat (SSR) markers, cleaved amplified polymorphic sequences (CAPS), or derived CAPS (dCAPS). The inception of next-generation sequencing has allowed for novel approaches to map-based cloning that are quick and efficient, which have replaced the classical positional mapping approach (Austin et al., 2011; James et al., 2013; Schneeberger & Weigel, 2011).
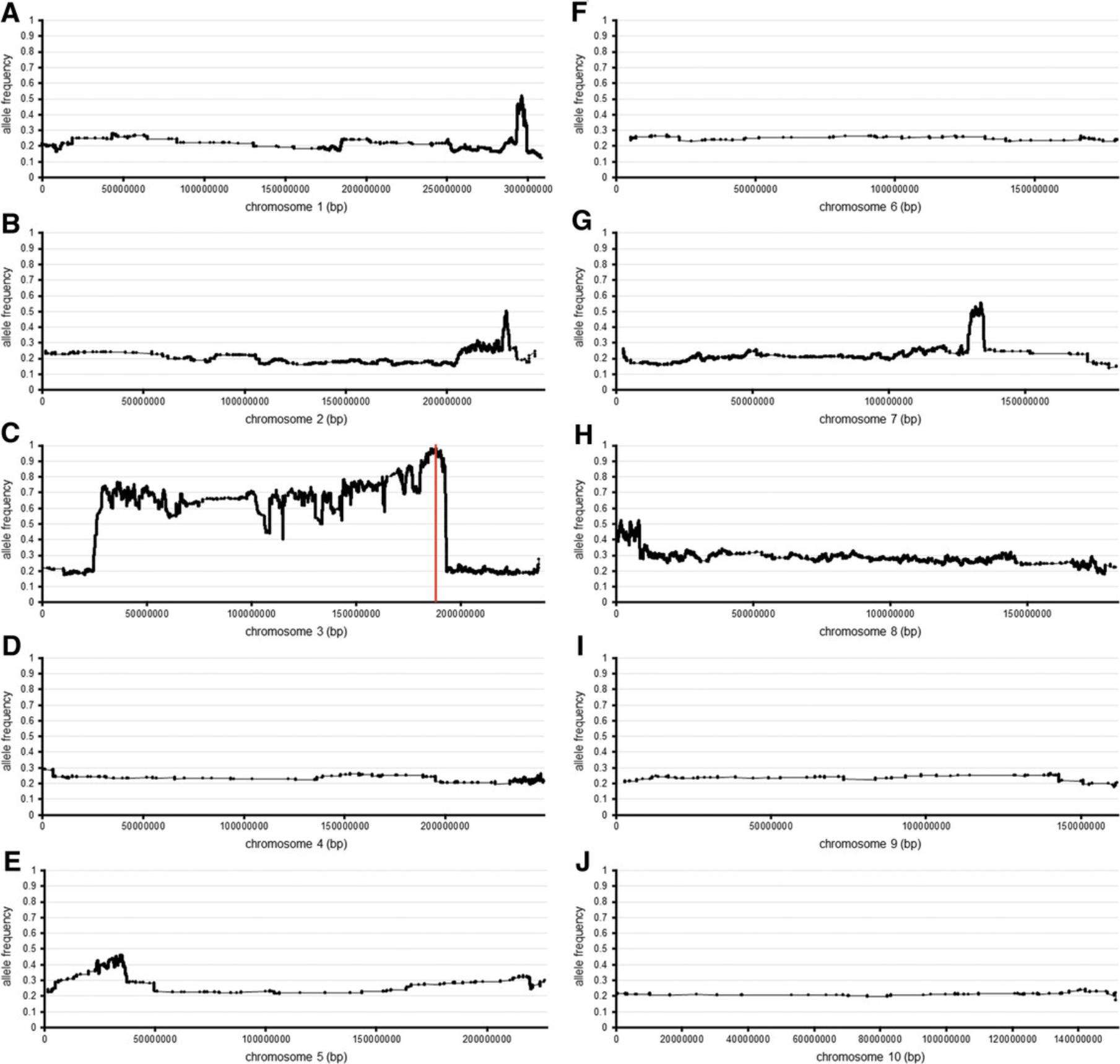
Techniques in maize like the one presented in this article have been published previously; however, they have used transcriptomic approaches (Best et al., 2021; Li et al., 2013; Liu, Yeh, Tang, Nettleton, & Schnable, 2012; Tang et al., 2014) or alternative mapping protocols (Klein et al., 2018; Nestler et al., 2014). Transcriptomic mapping approaches are effective in that the researcher gains an extra level of knowledge by conducting differential gene expression analysis if the wild-type siblings are also sequenced; however, they make the experiment more costly. This approach is an advantage over the protocol proposed here if the mutation is in a non-coding regulatory element, as it is likely that the expression of the transcript will be altered in the mutant sample compared to the wild type. Careful timing in collection of plant tissue, however, can be a pitfall of this approach. If the causative gene is not expressed at the time of tissue collection, then there will be no sequence data to identify the mutation. The bioinformatic analysis presented in this protocol can still be used to analysis RNA sequence data, as SNPs linked to the causative gene will still be identified. The added level of risk in transcriptomic mapping experiments makes genomic approaches superior. Alternative BSA and next-generation genomic sequencing approaches to mapping of SNPs have also been developed. Only mapping homozygous SNPs and binning the number of SNPs into chromosomal segments is an effective method to map maize mutant genes (Klein et al., 2018), and will result in similar results. The sliding window approach is advantageous over the bin approach, as it will better portray recombination breakpoints (Fig. 2).
There are alternative sequence alignment and variant calling tools available for use. ZOOM (Lin, Zhang, Zhang, Ma, & Li, 2008), and similar software applications that operate by hashing the sequences (transforming the sequence into a smaller value) and then scanning the reference, have a flexible memory footprint and require long run times. SOAPv1 (Li, Li, Kristiansen, & Wang, 2008), and similar software applications that operate by hashing the reference, have a large memory requirement. The BWA software was selected, as it is typically faster and utilizes a string-matching approach that improves alignment of sequences with indels and reduces misalignments resulting from sequencing errors. Bowtie2 operates similarly to BWA, but BWA has a better alignment rate and gene coverage when aligning DNA sequences (Musich, Cadle-Davidson, & Osier, 2021). There are also alternative variant calling software applications publicly available. SAMtools is used in this protocol because of its ease of inclusion in a pipeline, ability to easily convert file types, and exceptional variant calling (Kumar, Banks, & Cloutier, 2012; Li et al., 2009).
Classical BSA approaches compared DNA from a mutant pool to DNA from wild-type siblings in the same population. This protocol only investigates the mutant pool and compares it to a reference genome sequence. The rationale for only investigating the mutant pool is that the reference genome sequence acts as the wild-type sample. Collecting and sequencing a wild-type pool would not add benefit for the researcher besides further confirming the map location, and would increase the cost of the experiment. One advantage of sequencing a wild-type pool would be if multiple peaks are identified in the mutant pool sequence data. Multiple peaks could occur if the reference genome has genomic regions that are different from the inbred line used in the experiment (i.e., B73 inbred lines used to create reference genome are not exactly the same as the B73 inbred line used to make the mapping population). The wild-type sample would then be compared to the mutant sample, and if they share similar peaks, these could be eliminated as the location of the causative mutation and are instead due to differences between the reference genome and inbred line used in the protocol.
Additional applications of this method could be to map single-gene differences between two different inbred lines. Recently, the 25 Nested Association Mapping (NAM) founders were sequenced, and reference genomes have been created for each (Hufford et al., 2021). These NAM founders contain a large amount of diversity, and any of these sequenced lines can be used as a reference. Structural variants that are responsible for a particular phenotype could be mapped using this protocol, but would likely result in a large map window. Translocations will be very difficult to identify using this protocol, since short reads are sequenced and aligned to a reference genome. It is likely that reads spanning the translocation breaking points will not be aligned by the bioinformatic software. Natural genetic modifiers of a mutant phenotype have been effective in characterizing gene function and identifying differences between diverse maize lines (Anderson et al., 2019; Lopes & Larkins, 1995). The bioinformatic techniques presented in this protocol can also be used to map these modifiers. However, it is best if the genetic location of the original mutation that is being modified has been identified as a peak at the modifier location; the original mutation will likely result and the researcher could then discern the multiple peaks.
Critical Parameters
Development of superior mapping populations and selection of mutants to pool
Creating a robust F2 population (see Strategic Planning) is critical to obtain the expected results. Knowing the original genetic background in which the mutant was identified is highly beneficial to the design of the mapping population. Selecting the alternate inbred line from another heterotic group to create the initial F1 cross will increase the number of SNPs between the two lines and result in better mapping data for the F2 segregants. Identifying an inbred line that does not affect the mutant phenotype will ensure that no other loci are being selected in the segregant sequence analysis. Therefore, crossing with multiple inbred lines and testing for modification of the phenotype is beneficial. Backcrossing additional generations will lower the background noise of non-linked SNPs, but time limitations should be considered. As shown in Figure 2C, there is still significant linkage of the original background towards the centromere after four generations of backcrossing, thus only narrowing the map window minimally. Increasing the number of individuals in the segregant pool is more beneficial to narrow the map window (Best et al., 2021). Selection of bona fide mutants for BSA is critical for hypothesis testing of the bioinformatic data. Thus, one should be very stringent when selecting mutants to pool. One contaminant in the mutant tissue pool will negatively affect the mapping data and downstream SNP analysis.
As shown in Figure 2, the average allele frequency for the 100-SNP sliding window plots is around 0.2 to 0.3, except for chromosome 3 (Fig. 2C). The average allele frequency of non-linked genomic regions is around 0.2 to 0.3, and not 0.0, because there is still a low frequency of SNPs in the sequence data from differences between the inbred line and reference genome, sequencing errors, and incorrect alignment of sequences to the reference genome, as well as only SNP position allele frequencies being calculated and plotted. The low allele frequency (0.2 to 0.3) is observed due to the GN135 mutant being identified in A619 and then introgressed into B73, and B73 was used as the reference genome. If the mapping population were designed so that the mutant being mapped was identified in the same background as used for the reference genome, then these plots would be inverted. The allele frequency would be around 1.0 for any region where the mutant locus was not linked, and the researcher would see a dip (towards 0.0) where the causative mutation was located.
Quality of DNA and library preparation, and sequencing depth
Preparation of good-quality DNA (i.e., A 260/A 280 ratio is around 1.8 to 2.0) is always necessary. DNA preps can be difficult, and many times contamination and/or operator error cannot be avoided. Preparing new extraction buffers before beginning the protocol can help alleviate potential problems. Careful mixing and pipetting of solvents can minimize opportunities for contamination by phenol. The mutant phenotype should also be considered when collecting tissue. Proper identification of mutant segregants is necessary, but if the mutant phenotype results in poor development of tissue to collect (i.e., disease susceptibility, chlorosis, etc.), this can result in poor DNA preps. Collection of young, vibrant plant tissue will yield higher-quantity and -quality DNA. Furthermore, collecting plant tissue before pollen shed will help decrease the chances for contamination, as pollen could be present on the leaves.
Do not skip any steps in the manufacturer-recommended library preparation. Sequencing of paired-end reads is better than single-end reads. Paired-end reads will increase the alignment rate, especially in repetitive regions of the genome. Current short-read sequencing technology can effectively sequence up to 400-bp reads. Longer reads can increase the alignment rate, but also have a higher error rate resulting in problems with SNP calling and the introduction of false positives. Sequencing 50 Gb will result in 15× coverage, and is sufficient to map and identify causative mutations. Sequencing at higher depths will increase the quality of the mapping, but can result in more sequencing errors. Sequencing at higher depths may require allowing for non-homozygous SNPs at step 33 to alleviate the presence of sequencing errors.
Strength of bioinformatic scripts
Multiple maize reference genomes have been developed and well annotated (Ge et al., 2022; Haberer et al., 2020; Hirsch et al., 2016; Hufford et al., 2021; Jiao et al., 2017; Springer et al., 2018; Sun et al., 2018; Yang et al., 2019), and more are being developed every day. The genetic material used to create the reference genome may be different from that utilized in your experiment. This may affect the mapping approach if large sections of the genome are dissimilar to the reference, and can result in multiple peaks that are not linked to the causative mutation. Individual natural mutations will not affect the mapping portion of the experiment, but could poorly affect the downstream causative mutation filtering. Starting with inbred material that is very similar to the material used to create the reference genome will be highly beneficial. If there are multiple peaks due to differences in the reference and inbred line used, then a wild-type sibling pool can be sequenced to identify similarities between the two samples to eliminate false positives. Stocks of most of the lines used to develop the reference genomes are available at the Germplasm Resources Information Network (GRIN; https://www.ars-grin.gov).
As newer versions of bioinformatic tools are developed, some commands may be changed or become outdated. Reviewing manuals for each bioinformatic package, especially in updated versions, will ensure that proper commands are used. Annotation of SNPs by snpEff requires the correct genome database to correspond to the current version of the maize genome build. Checking the pre-loaded databases can save time if your reference genome is available. Refer to the snpEff manual on how to build a genome database if your reference genome is not offered.
Confirmation of causative locus
Once a candidate gene is identified, confirmation of the causative mutation is necessary. Co-segregation of the mutation with the mutant phenotype should be tested on additional individuals, rather than pools, using Sanger sequencing (Sanger, Nicklen, & Coulson, 1977), CAPS (Konieczny & Ausubel, 1993), or dCAPS (Neff, Neff, Chory, & Pepper, 1998) genotyping protocols. Development of these genotyping protocols is also beneficial to confirm the next-generation sequencing results. Ultimately, a second allele of the mutation will have to be identified either through additional forward genetic mutant screens for the same mutant phenotype or utilizing reverse genetic approaches, as previously described. Allelism tests should be performed between the multiple alleles to confirm that they fail to complement each other.
Troubleshooting
See Table 2 for a list of common problems with the protocols, their causes, and potential solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Poor DNA yield | Buffers are bad, tissue is bad, or tissue was poorly ground | Make new buffers and finely grind the tissue samples |
| Poor-quality DNA | Phenol, salt, or protein contamination | Carefully pipette off supernatant to not include phenol (step 6). Add an extra 70% ethanol rinse (repeat step 8). |
| No mapping peaks are identified | Non-mutant samples were collected or there are not enough genetic variants between the two lines in the mapping population. |
Re-plant mapping population and carefully select only mutants Use alternative mapping population with more genetically distinct parents (i.e., evolutionary distant heterotic groups) |
| Multiple mapping peaks are identified | A genetic modifier of mutant phenotype is segregating in the mapping population, or the inbred line has genomic regions that are different from the reference sequence |
Investigate mutant phenotype in other genetic backgrounds to identify linkage between phenotype and peak locations Sequence wild-type pool to identify and eliminate similar peaks in both samples |
| Did not find candidate gene | Contamination or sequencing error. Causative mutation is in non-coding region. | Allow non-homozygous mutations in filtered list. Check for indel mutations. Investigate promoter regions for mutations using PlantPAN 3.0. Do RNA-Seq on mutant and wild-type pools to identify differences in transcript abundance in mapped region. |
Understanding Results
Users will obtain allele frequency plots, which will be plotted from 0 to 1 on the y -axis, and will include chromosome position on the x -axis (Fig. 2). An allele frequency near 1 indicates linkage of the causative mutation. An allele frequency near 0 would indicate that the region is similar to the reference genome. As seen in Figure 2C, there is a plateau on chromosome 3, with the highest peak being around 190 Mb. The presence of the plateau is due to recombination events occurring at a lower frequency when closer to the centromere and being more prevalent towards the end of the chromosome. This phenomenon will commonly be observed when plotting the 100-SNP sliding window data for the chromosome on which the causative mutation is located (i.e., linkage towards the centromere from the location of the causative mutation).
If a large mapping region is identified, additional F2 mutants can be selected and sequenced as a pool, and the same protocol can be used to increase the chances for recombination and narrow the mapping window. Creating F3 families to sequence is also feasible, but the number of F3 families (individual F2 plants self-pollinated) should be as large as possible to increase the opportunity for independent recombination events. As seen in Figure 2, the allele frequency never reaches zero. This is due to only SNP positions being calculated and, thus, the program is only plotting false positives, sequencing errors, and differences in sequence between the stocks used to create the mapping population and the stocks used to make the reference genome. If the reference genome used to align the sequences was the same as the line in which the mutant was identified, then this allele frequency would be inverted, and linkage of the causative mutation would be where the allele frequency was closer to zero.
To identify candidate causative mutations for the mutant phenotype, the SNP positions are re-filtered to identify homozygous SNPs and annotated for effect on gene sequence using the SnpEff software. The output VCF file can be opened in Excel in a tab-delimited fashion. There is a header section and variant call section. In the variant call section, there are 10 columns. Column 1 is chromosome number, column 2 is genomic position, column 3 is ID, column 4 is the reference allele, column 5 is the non-reference allele, column 6 is the quality score for the variant, column 7 is the filter applied to the variant, column 8 contains information about the variant, and column 9 defines the format of the output in column 10. If EMS mutagenesis were used, the reference and non-reference columns can be filtered in Microsoft Excel for only G to A or C to T transitions to be included. Column 8 can then be text-filtered to only contain the word “HIGH.” These are high-effect mutations on gene sequence, such as a stop gained or stop lost mutation. The researcher should focus on the genomic region identified and the gene id written in column 8. The gene annotations at MaizeGDB.org can be referenced to narrow down candidate genes based upon biological relevance to the mutant phenotype. If a good candidate gene (i.e., “HIGH” effect mutation) is not identified, then the list can be text-filtered to contain “MODERATE,” which primarily includes missense mutations. Finally, if no good candidate genes are identified, then step 33 can be changed to allow for a few reference reads. The same filtering steps can be taken on this file as previously stated. Filtering the GN135 file to allow for a few reference reads and filtering for EMS SNPs (i.e., G to A or C to T) identified three “HIGH” effect mutations in the map window, of five total, all of which were on chromosome 3. One stop-gained mutation in gene “Zm00001eb148990” encodes barren stalk1 (ba1) (Gallavotti et al., 2004). The GN135 mutant used in the example data has a barren stalk phenotype with effects on tassel branching, and an allelism test confirmed this as the causative gene. This mutant has now been designated as ba1-GN135.
Again, if no good candidate genes are identified, then the causative mutation could be in non-coding regions. Identifying mutations in promotor regions is quite difficult, especially in maize. Careful investigation of promotor regions of candidate genes for mutation of transcription factor binding sites in the mapped window can be conducted using PlantPAN 3.0 (Chow et al., 2019). This is more likely for semi-dominant or dominant mutations than recessive mutations.
Time Considerations
The overall procedure should take 2 years, or can take longer. The most time-consuming step is the development of the mutant populations for tissue collection. The initial outcross of the mutant takes a full maize-growing season. Typically, only two growing seasons of maize can be accomplished in a single year. Therefore, to obtain F2 segregating mutant plants, this would take 1.5 years. Creating the first F1 cross in the summer will allow for selfing of the F1 in the winter (via greenhouse or winter nursery), and then the F2 plants can be planted and tissue collected under large field conditions in the summer. Additional time is required for additional back-cross generations. Preparation of DNA and libraries takes ∼1 week. Sequencing takes ∼1 month depending on queuing. Lastly, the bioinformatic analysis takes ∼2 weeks. Additional time may be required to generate and test additional mutant alleles.
Acknowledgments
This work was supported by funds from a USDA-NIFA fellowship to N.B.B (#2019-67012-29655). Work was also supported by funds from the National Science Foundation (NSF) Plant Genome Research Program (PGRP) to P.M. (IOS-1546873). This research used resources provided by the SCINet project of the USDA Agricultural Research Service, ARS project number 0500-00093-001-00-D. The authors would like to thank B. Dilkes and C. Addo-Quaye for support through the learning process. The authors would also like to thank the Maize Inflorescence Architecture Project and Gerry Neuffer for creating the ba *GN135 mutant. This manuscript is dedicated to Gerry Neuffer for all the amazing maize mutants he created over the years.
Author Contributions
Norman Best : Conceptualization, Data curation, Formal analysis, Investigation, Writing original draft; Paula McSteen : Resources, Writing review and editing.
Conflict of Interest
The authors declare no conflict of interest. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The U.S. Department of Agriculture is an equal opportunity provider and employer.
Open Research
Data Availability Statement
All scripts used in this protocol are published on Github at https://doi.org/10.5281/zenodo.7041941. Sample data files are available at https://doi.org/10.6084/m9.figshare.c.6180031.v1. Raw sequence reads used to develop sample data files are available at the NCBI short read archive under BioProject number PRJNA848842. Mutant stocks of ba1-GN135 that were used for the example data are available from the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu/) as stock 6516B.
Literature Cited
- Amano, E., & Smith, H. H. (1965). Mutations induced by ethyl methanesulfonate in maize. Mutation Research , 2(4), 344–351. doi: 10.1016/0027-5107(65)90070-9
- Anderson, A., St Aubin, B., Abraham-Juárez, M. J., Leiboff, S., Shen, Z., Briggs, S., … Hake, S. (2019). The second site modifier, sympathy for the ligule, encodes a homolog of arabidopsis ENHANCED DISEASE RESISTANCE4 and rescues the Liguleless narrow maize mutant. The Plant Cell , 31(8), 1829–1844. doi: 10.1105/tpc.18.00840
- Austin, R. S., Vidaurre, D., Stamatiou, G., Breit, R., Provart, N. J., Bonetta, D., … Guttman, D. S. (2011). Next-generation mapping of Arabidopsis genes. Plant Journal , 67(4), 715–725. doi: 10.1111/j.1365-313X.2011.04619.x
- Best, N. B., Addo-Quaye, C., Kim, B.-S., Weil, C. F., Schulz, B., Johal, G., & Dilkes, B. P. (2021). Mutation of the nuclear pore complex component, aladin1, disrupts asymmetric cell division in Zea mays (maize). G3 Genes, Genomes, Genetics , 11(7), jkab106. doi: 10.1093/g3journal/jkab106
- Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics , 30(15), 2114–2120. doi: 10.1093/bioinformatics/btu170
- Bortiri, E., Jackson, D., & Hake, S. (2006). Advances in maize genomics: The emergence of positional cloning. Current Opinion in Plant Biology , 9(2), 164–171. doi: 10.1016/j.pbi.2006.01.006
- Candela, H., & Hake, S. (2008). The art and design of genetic screens: Maize. Nature Reviews Genetics , 9(3), 192–203. doi: 10.1038/nrg2291
- Chen, J., & Dellaporta, S. L. (1994). Urea-based plant DNA miniprep. In M. Freeling & V. Walbot (Eds.), The maize handbook (pp. 526–527). New York, NY: Springer-Verlag.
- Chow, C. N., Lee, T. Y., Hung, Y. C., Li, G. Z., Tseng, K. C., Liu, Y. H., … Chang, W. C. (2019). PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Research , 47(D1), D1155–D1163. doi: 10.1093/nar/gky1081
- Cingolani, P., Platts, A., le Wang, L., Coon, M., Nguyen, T., Wang, L., … Ruden, D. M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) , 6(2), 80–92. doi: 10.4161/fly.19695
- Coe, E. H. (1994). Genetic experiments and mapping. In M. Freeling & V. Walbot (Eds.), The maize handbook (pp. 189–197). New York, NY: Springer New York.
- Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., … Li, H. (2021). Twelve years of SAMtools and BCFtools. GigaScience , 10(2), giab008. doi: 10.1093/gigascience/giab008
- Gallavotti, A., & Whipple, C. J. (2015). Positional cloning in maize (Zea mays subsp. mays, Poaceae). Applied Plant Sci , 3(1), 1400092. doi: 10.3732/apps.1400092
- Gallavotti, A., Zhao, Q., Kyozuka, J., Meeley, R. B., Ritter, M. K., Doebley, J. F., … Schmidt, R. J. (2004). The role of barren stalk1 in the architecture of maize. Nature , 432(7017), 630–635. doi: 10.1038/nature03148
- Ge, F., Qu, J., Liu, P., Pan, L., Zou, C., Yuan, G., … Shen, Y. (2022). Genome assembly of the maize inbred line A188 provides a new reference genome for functional genomics. The Crop Journal , 10(1), 47–55. doi: 10.1016/j.cj.2021.08.002
- Haberer, G., Kamal, N., Bauer, E., Gundlach, H., Fischer, I., Seidel, M. A., … Mayer, K. F. X. (2020). European maize genomes highlight intraspecies variation in repeat and gene content. Nature Genetics , 52(9), 950–957. doi: 10.1038/s41588-020-0671-9
- Hirsch, C. N., Hirsch, C. D., Brohammer, A. B., Bowman, M. J., Soifer, I., Barad, O., … Mikel, M. A. (2016). Draft assembly of elite inbred line PH2O7 provides insights into genomic and transcriptome diversity in maize. Plant Cell , 28(11), 2700–2714. doi: 10.1105/tpc.16.00353
- Hufford, M. B., Seetharam, A. S., Woodhouse, M. R., Chougule, K. M., Ou, S. J., Liu, J. N., … Dawe, R. K. (2021). De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science , 373(6555), 655–+. doi: 10.1126/science.abg5289
- James, G. V., Patel, V., Nordstrom, K. J., Klasen, J. R., Salome, P. A., Weigel, D., & Schneeberger, K. (2013). User guide for mapping-by-sequencing in arabidopsis. Genome Biology , 14(6), R61. doi: 10.1186/gb-2013-14-6-r61
- Jiao, Y., Peluso, P., Shi, J., Liang, T., Stitzer, M. C., Wang, B., … Ware, D. (2017). Improved maize reference genome with single-molecule technologies. Nature , 546(7659), 524–527. doi: 10.1038/nature22971
- Klein, H., Xiao, Y., Conklin, P. A., Govindarajulu, R., Kelly, J. A., Scanlon, M. J., … Bartlett, M. (2018). Bulked-segregant analysis coupled to whole genome sequencing (BSA-Seq) for rapid gene cloning in maize. G3 (Bethesda) , 8(11), 3583–3592. doi: 10.1534/g3.118.200499
- Konieczny, A., & Ausubel, F. M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. The Plant Journal , 4(2), 403–410. doi: 10.1046/j.1365-313X.1993.04020403.x
- Kumar, S., Banks, T. W., & Cloutier, S. (2012). SNP discovery through next-generation sequencing and its applications. International Journal of Plant Genomics , 2012, 831460. doi: 10.1155/2012/831460
- Leach, K. A., McSteen, P. C., & Braun, D. M. (2016). Genomic DNA isolation from maize (Zea mays) leaves using a simple, high-throughput protocol. Current Protocols in Plant Biology , 1(1), 15–27. doi: 10.1002/cppb.20000
- Li, H., & Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics , 25(14), 1754–1760. doi: 10.1093/bioinformatics/btp324
- Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., … 1000 Genome Project Data Processing Subgroup. (2009). The sequence alignment/map format and SAMtools. Bioinformatics , 25(16), 2078–2079. doi: 10.1093/bioinformatics/btp352
- Li, L., Li, D., Liu, S., Ma, X., Dietrich, C. R., Hu, H. C., … Schnable, P. S. (2013). The maize glossy13 gene, cloned via BSR-Seq and Seq-walking encodes a putative ABC transporter required for the normal accumulation of epicuticular waxes. PLoS One , 8(12), e82333. doi: 10.1371/journal.pone.0082333
- Li, R., Li, Y., Kristiansen, K., & Wang, J. (2008). SOAP: Short oligonucleotide alignment program. Bioinformatics , 24(5), 713–714. doi: 10.1093/bioinformatics/btn025
- Lin, H., Zhang, Z., Zhang, M. Q., Ma, B., & Li, M. (2008). ZOOM! Zillions of oligos mapped. Bioinformatics , 24(21), 2431–2437. doi: 10.1093/bioinformatics/btn416
- Liu, S., Yeh, C. T., Tang, H. M., Nettleton, D., & Schnable, P. S. (2012). Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One , 7(5), e36406. doi: 10.1371/journal.pone.0036406
- Lopes, M. A., & Larkins, B. A. (1995). Genetic analysis of opaque2 modifier gene activity in maize endosperm. Theoretical and Applied Genetics , 91(2), 274–281. doi: 10.1007/BF00220889
- Marcon, C., Altrogge, L., Win, Y. N., Stocker, T., Gardiner, J. M., Portwood, J. L. 2nd., … Hochholdinger, F. (2020). BonnMu: A sequence-indexed resource of transposon-induced maize mutations for functional genomics studies. Plant Physiology , 184(2), 620–631. doi: 10.1104/pp.20.00478
- McCaw, M. E., Wallace, J. G., Albert, P. S., Buckler, E. S., & Birchler, J. A. (2016). Fast-flowering mini-maize: Seed to seed in 60 days. Genetics , 204(1), 35–42. doi: 10.1534/genetics.116.191726
- Michelmore, R. W., Paran, I., & Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis—a rapid method to detect markers in specific genomic regions by using segregating populations. Proceedings of the National Academy of Sciences of the United States of America , 88(21), 9828–9832. doi: 10.1073/pnas.88.21.9828
- Musich, R., Cadle-Davidson, L., & Osier, M. V. (2021). Comparison of short-read sequence aligners indicates strengths and weaknesses for biologists to consider. Frontiers in Plant Science , 12, 657240. doi: 10.3389/fpls.2021.657240
- Neff, M. M., Neff, J. D., Chory, J., & Pepper, A. E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. The Plant Journal , 14(3), 387–392. doi: 10.1046/j.1365-313/.1998.00124.x
- Nestler, J., Liu, S., Wen, T. J., Paschold, A., Marcon, C., Tang, H. M., … Hochholdinger, F. (2014). Roothairless5, which functions in maize (Zea mays L.) root hair initiation and elongation encodes a monocot-specific NADPH oxidase. The Plant Journal , 79(5), 729–740. doi: 10.1111/tpj.12578
- Quinlan, A. R., & Hall, I. M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics , 26(6), 841–842. doi: 10.1093/bioinformatics/btq033
- Ribeiro, A., Golicz, A., Hackett, C. A., Milne, I., Stephen, G., Marshall, D., … Bayer, M. (2015). An investigation of causes of false positive single nucleotide polymorphisms using simulated reads from a small eukaryote genome. BMC Bioinformatics , 16, 382. doi: 10.1186/s12859-015-0801-z
- Sanger, F., Nicklen, S., & Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America , 74(12), 5463–5467. doi: 10.1073/pnas.74.12.5463
- Schneeberger, K., & Weigel, D. (2011). Fast-forward genetics enabled by new sequencing technologies. Trends in Plant Science , 16(5), 282–288. doi: 10.1016/j.tplants.2011.02.006
- Settles, A. M. (2020). EMS mutagenesis of maize pollen. Methods in Molecular Biology , 2122, 25–33. doi: 10.1007/978-1-0716-0342-0_3
- Settles, A. M., Holding, D. R., Tan, B. C., Latshaw, S. P., Liu, J., Suzuki, M., … McCarty, D. R. (2007). Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics , 8, 116. doi: 10.1186/1471-2164-8-116
- Simbolo, M., Gottardi, M., Corbo, V., Fassan, M., Mafficini, A., Malpeli, G., … Scarpa, A. (2013). DNA qualification workflow for next generation sequencing of histopathological samples. PLoS One , 8(6), e62692. doi: 10.1371/journal.pone.0062692
- Springer, N. M., Anderson, S. N., Andorf, C. M., Ahern, K. R., Bai, F., Barad, O., … Brutnell, T. P. (2018). The maize W22 genome provides a foundation for functional genomics and transposon biology. Nature Genetics , 50(9), 1282–1288. doi: 10.1038/s41588-018-0158-0
- Sun, S., Zhou, Y., Chen, J., Shi, J., Zhao, H., Zhao, H., … Lai, J. (2018). Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nature Genetics , 50(9), 1289–1295. doi: 10.1038/s41588-018-0182-0
- Tang, H. M., Liu, S., Hill-Skinner, S., Wu, W., Reed, D., Yeh, C. T., … Schnable, P. S. (2014). The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. The Plant Journal , 77(3), 380–392. doi: 10.1111/tpj.12394
- Wall, L. (1994). The Perl programming language. Prentice Hall Software Series.
- Yang, N., Liu, J., Gao, Q., Gui, S., Chen, L., Yang, L., … Yan, J. (2019). Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nature Genetics , 51(6), 1052–1059. doi: 10.1038/s41588-019-0427-6
- Yao, H., Skirpan, A., Wardell, B., Matthes, M. S., Best, N. B., McCubbin, T., … McSteen, P. (2019). The barren stalk2 gene is required for axillary meristem development in maize. Molecular Plant , 12(3), 374–389. doi: 10.1016/j.molp.2018.12.024
Citing Literature
Number of times cited according to CrossRef: 1
- Christian Damian Lorenzo, David Blasco‐Escámez, Arthur Beauchet, Pieter Wytynck, Matilde Sanches, Jose Rodrigo Garcia del Campo, Dirk Inzé, Hilde Nelissen, Maize mutant screens: from classical methods to new CRISPR‐based approaches, New Phytologist, 10.1111/nph.20084, 244 , 2, (384-393), (2024).

