MHC II–EGFP Knock-in Mouse Model
Jan Pačes, Jan Pačes, Valéria Grobárová, Valéria Grobárová, Zdeněk Zadražil, Zdeněk Zadražil, Karolina Knížková, Karolina Knížková, Nikola Malinská, Nikola Malinská, Liliana Tušková, Liliana Tušková, Marianne Boes, Marianne Boes, Jan Černý, Jan Černý
APCs
light-sheet fluorescence microscopy
lymphoid organs
MHC II–EGFP knock-in mouse
quantitative immunology
Abstract
The MHC II–EGFP knock-in mouse model enables us to visualize and track MHC-II-expressing cells in vivo by expressing enhanced green fluorescent protein (EGFP) fused to the MHC class II molecule under the MHC II beta chain promoter. Using this model, we can easily identify MHC-II-expressing cells, including dendritic cells, B cells, macrophages, and ILC3s, which play a key role as antigen-presenting cells (APCs) for CD4**+** T cells. In addition, we can also precisely identify and analyze APC-containing tissues and organs. Even after fixation, EGFP retains its fluorescence, so this model is suitable for immunofluorescence studies, facilitating an unbiased characterization of the histological context, especially with techniques such as light-sheet fluorescence microscopy. Furthermore, the MHC II–EGFP knock-in mouse model is valuable for studying the molecular mechanisms of MHC II gene regulation and expression by making it possible to correlate MHC II expression (MHC II–EGFP) with surface fraction through antibody detection, thereby shedding light on the intricate regulation of MHC II expression. Overall, this model is an essential asset for quantitative and systems immunological research, providing insights into immune cell dynamics and localization, with a tool for precise cell identification and with the ability to study MHC II gene regulation, thus furthering the understanding of immune responses and underlying mechanisms © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
This article was corrected on 14 April 2024. See the end of the full text for details.
Basic Protocol 1 : Characterization of antigen-specific MHC II loading compartment tubulation toward the immunological synapse
Basic Protocol 2 : Characterization of overall versus surface MHC II expression
Basic Protocol 3 : Identification and preparation of the lymphoid organs
Basic Protocol 4 : Quantification of APC content in lymphoid organs by fluorescence stereomicroscopy
Basic Protocol 5 : Quantification and measurement of intestinal lymphoid tissue by light-sheet fluorescence stereomicroscopy
Basic Protocol 6 : Visualization of corneal APCs
Basic Protocol 7 : Quantification of MHC II**+** cells in maternal milk by flow cytometry
Support Protocol 1 : Cell surface staining and flow cytometry analysis of spleen mononuclear cells
INTRODUCTION
Despite significant advances in immunological research and in the development of imaging methods, quantitative aspects of the immune system within complex, whole-organism contexts have been surprisingly overlooked thus far. This knowledge gap highlights the need for functional models that capture the general features of the immune system, especially for emerging systems immunology approaches aimed at characterizing complex immunological processes holistically. One of the key molecules driving adaptive responses is the MHC class II glycoprotein, which is expressed by antigen-presenting cells (APCs)—crucial regulators of helper and regulatory CD4**+** T cells—found in all tissues and in large numbers in primary and secondary lymphoid organs.
Developed two decades ago, the MHC II–EGFP knock-in mouse model holds significant promise in conjunction with the current trend toward quantitative analytical tools in immunology research (Boes et al., 2002; Pačes et al., 2022). These tools include comprehensive omics techniques and single-cell analysis and enable us to accurately and swiftly quantify events under dynamic and heterogeneous conditions. By introducing fluorescent trackers into mouse models and leveraging advanced microscopy techniques alongside powerful quantitative analysis tools, such as 3D sample imaging, we can overcome antibody-induced artifacts and collect data in situ , within specific histological contexts.
The MHC II–EGFP knock-in mouse model, which physiologically expresses enhanced green fluorescent protein (EGFP) fused to the MHC II beta chain, driven by the normal transcriptional regulatory elements, provides us with a valuable tool for identifying APCs in tissues while simultaneously avoiding potential artifacts associated with antibody-based approaches (Boes et al., 2002; Pačes et al., 2022). Using this model, we can also identify, quantify, and precisely excise not easily detectable APC-containing organs, such as lymph nodes or Peyer's patches, for subsequent histological and cytometric analysis. Furthermore, the fluorescence properties of EGFP are retained even after fixation with aldehyde fixatives, supporting a precise subcellular localization of MHC II class molecules, by differentiation of MHC-II-loading compartments from the plasma membrane, colocalization studies with markers of specific APC types, and examination of cellular interactions within tissues and organs.
The advantages of the MHC II–EGFP model also include the use of light-sheet fluorescence microscopy (LSFM) for visualizing APCs in situ without requiring sectioning in whole organisms or organs. This non-invasive imaging technique, particularly effective when applied to transparent organs such as the eye, facilitates a detailed characterization of APCs in the cornea and limbus. This technology, in combination with clearing methods, also enables 3D imaging of fixed naturally non-translucent organs (Pačes et al., 2022). In vitro studies using cells cultured from MHC II–EGFP mice have shown that they are similar to cells from normal, non-fluorescently-tagged mice in a physiological environment but differentiate APCs from bone marrow hematopoietic cells, thereby enabling us to study these cells using advanced microscopy techniques in vivo. In particular, videomicroscopy may unveil the cellular mechanisms involved in key immune events, such as antigen-dependent polarized exocytosis towards the immune synapse (Boes et al., 2002).
In addition to its applications in immunology research, the MHC II–EGFP knock-in mouse model opens up opportunities for studying milk composition, specifically the presence of professional APCs. With this model, researchers can gain insights into the functional morphology of immune organs, including the absolute quantification and characterization of individual variability. By advancing our understanding of the functional organization of not only the immune system but also other physiological processes, this model may contribute to rewriting textbooks and identifying new phenomena.
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals. Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol 1: CHARACTERIZATION OF ANTIGEN-SPECIFIC MHC II LOADING COMPARTMENT TUBULATION TOWARD THE IMMUNOLOGICAL SYNAPSE
The MHC II–EGFP knock-in mouse model provides a valuable platform for the in vivo characterization of antigen presentation phenomena, ranging from the molecular to whole-animal levels. Recent advancements in imaging techniques, combined with genetically engineered organismal models expressing fluorescent proteins, have revolutionized multiple disciplines in experimental biology. These developments have paved the way for rewriting textbooks and uncovering new phenomena, including the antigen-specific tubulation of the MHC II loading compartment in dendritic cells (DCs) toward the immunological synapse.
DCs are essential regulators of immune reactions and belong to the category of professional APCs. They have a unique ability to integrate danger and “stranger” molecular patterns. DC-mediated T cell activation is a pivotal event in the activation of immune reactivity. The MHC II–EGFP knock-in mouse serves as a suitable source of DCs, collected either directly as primary cells from relevant tissues or through differentiation from hematopoietic stem cells, such as those derived from the bone marrow. These cells can be loaded with a model antigen (e.g., ovalbumin) to examine their interaction with cognate T cells expressing the appropriate antigen-specific T cell receptor (OT II).
The high resolution of live-cell imaging microscopy provides us with the ability to not only perform long-term imaging of cell-to-cell interactions without causing significant photodamage, using routine methodologies such as spinning-disc technology, but also study immunological synapse formation and characterize fluorescently labeled MHC II loading compartment under various conditions. By acquiring multiple videomicroscopy datasets depicting diverse antigen-presentation scenarios (e.g., different types of material presented via MHC II and cognate T cell receptor specificity, the activation status of interacting cells, molecules interfering with the process), we can accurately quantify endosomal tubulation toward the immune synapse, including measuring tubule length and directionality.
Combining the MHC II–EGFP knock-in mouse model with advanced imaging techniques offers a powerful approach to investigating antigen presentation dynamics and uncovering intricate details of cellular interactions within the immune system. This knowledge fosters the development of immunological theory, with broader implications for understanding immune responses and designing targeted therapeutic strategies.
Materials
-
Mice: MHC II–EGFP knock-in mice (8 weeks old, males and females, bred in house)
-
D-PBS/2.5% FBS (see recipe)
-
RBC Lysis Buffer (Thermo Fisher Scientific, cat. no. J62150.AK)
-
DMEM culture medium (see recipe: phenol-red-free DMEM/10% FBS with L-glutamine and HEPES, supplemented with GM-CSF and IL-4)
-
Lipopolysaccharide (LPS; ultrapure from Escherichia coli strain 0111:B4 [LPS-EB ultrapure], InvivoGen, cat. no. tlrl-3pelps)
-
DMEM culture medium with OVA (see recipe)
-
8-week-old OT II mice (Charles River Laboratories)
-
Hoechst 33258 (Molecular Probes–Invitrogen, cat. no. H3569)
-
Phenol-red-free RPMI 1640 with L-glutamine (Life Technologies, cat. no. 11835030)
-
1.0 M HEPES (Thermo Fisher Scientific, cat. no. J16924.AE)
-
50-ml Falcon tubes (Eppendorf, cat. no. 0030122178)
-
25-G needle and 2-ml plastic syringe
-
Dissecting instruments (scissors, forceps)
-
1.5-ml microcentrifuge tubes
-
2.5-, 20-, 200-, and 1000-µl mechanical pipets
-
Cell strainer, 40 µm pore size (VWR, cat. no. 734-2760)
-
Centrifuge (Eppendorf, model no. 5810R, or similar)
-
6-well microplates (2000 µl/well medium capacity)
-
25-mm circular glass coverslips
-
CO2 incubator set to 5% CO2 and 37°C
-
Frost microscopy slides
-
Bürker counting chamber
-
Magnetic-activated cell sorter (MACS; Miltenyi Biotech)
-
Inverted Zeiss 200M microscope equipped with a 63× objective (1.4-NA PlanApo) and spinning-disc confocal head (PerkinElmer, Wellesley, MA)
-
Temperature-controlled open perfusion chamber (20/20 Technology, Wilmington, NC)
-
Slidebook (Intelligent Imaging Innovation, Denver, CO)
Preparation and culture of bone-marrow-cell-derived DCs (BMDCs)
1.Euthanize MHC II–EGFP knock-in mouse by cervical dislocation.
2.Working under sterile conditions, dissect the femurs and tibia using scissors and forceps, and cut the epiphyses out using scissors and opening the diaphysis.
3.Using a needle and syringe with 0.5 ml D-PBS/2.5% FBS, flush cells out from the femur and tibia diaphysis marrow cavity into a 40-μm nylon cell strainer placed in a 50-ml conical Falcon tube to remove tissue debris.
4.Remove erythrocytes by osmotic shock by adding 10 ml RBC Lysis Buffer per milliliter of cellular suspension and incubating for 10-15 min at room temperature. Centrifuge 5 min at 500 × g , room temperature, and resuspend the pellet in the appropriate volume of DMEM culture medium (phenol-red-free DMEM/10% FBS with 25 mM HEPES, 10 ng/ml GM-CSF, and 1 ng/ml IL-4).
5.Plate cells at a density of 1 × 106 cells (quantified first using a Bürker counting chamber) per well of a 6-well plate in 2 ml of the same DMEM culture medium.
6.Carefully insert a sterilized 25-mm circular coverslip into each well using forceps in the incubator (37°C, 5% CO2) and incubate for 5 days, changing the medium every second day (that is, on days 3 and 5).
7.On day 2, remove non-adherent cells by gently washing, and replace half of the medium.
8.If activated APCs are of interest, activate the cells by adding 10 μg of 5 μg/ml LPS to corresponding wells of the 6-well plate on day 4 for the last 20 h of culture.
Antigen presentation in real time
9.Culture dendritic cells to a concentration of 1 × 106 cell/well in a 6-well culture plate with one 25-mm circular glass coverslip in each well, as described in steps 1-7, but using 2 ml/well DMEM culture medium supplemented with 300 μM OVA.
10.Wash DCs three times with DMEM culture medium (without OVA), each time by gently removing ∼90% DMEM medium and washing using 2 ml medium.
11.Sacrifice one male or female 8-week-old OTII mouse (as ordered from Charles River Laboratories) by cervical dislocation.
12.Acquire the OVA-specific OT II T cells from the lymph nodes of the OT II mouse: dissect lymph nodes, e.g., subiliac lymph nodes, using scissors and forceps, prepare a cell suspension by crushing the lymph nodes between two frost microscopy slides and resuspending the cells into DMEM, and quantify them using a Bürker counting chamber.
13.Label OVA-specific T cells (0.5 × 106) isolated and pooled from different lymph nodes by adding Hoechst 33258 to 2 µg/ml and return to the CO2 incubator for 30 min. Next, wash the cells three times with phenol-red-free RPMI 1640, each time by gently removing ∼90% of the medium, adding 2 ml medium, resuspending, and centrifuging for 5 min at 300 × g.
14.Add the 1-ml aliquots of OT II T cell suspensions to each well of the 6-well plate from step 9 (containing DCs growing on 25-mm circular glass coverslips) and quickly centrifuge at 600 × g , room temperature, to bring the cells into contact with each other.
15.Transfer the 25-mm circular glass coverslips into the temperature-controlled open perfusion chamber.
16.Maintain cells in phenol-red-free DMEM medium with 25 mM HEPES and 10% FBS at 37°C in a temperature-controlled open perfusion chamber or use similar equipment compatible with your microscopic setup.
17.Perform videomicroscopy analysis immediately and after 2 and 6 hr. Imaging can be performed using an inverted Zeiss 200M microscope equipped with a × 63 objective (1.4-NA PlanApo) and a spinning-disc confocal head or any other suitable imaging system for long-term videomicroscopy.
18.Use Slidebook for image acquisition and data processing, or choose other software solutions.
19.Score of DC phenotypes on the acquired datasets for the ability to induce tubulation within 2 hr of addition of OTII T cells. At least 100 DCs should be analyzed independently, by two unbiased individuals, for the induction of tubulation (Bertho et al., 2003).
Basic Protocol 2: CHARACTERIZATION AND COMPARISON OF OVERALL AND SURFACE MHC II EXPRESSION
The MHC II–EGFP knock-in mouse model offers a unique tool for comparing the overall MHC II expression (based on EGFP fluorescence intensity) with the surface expression, which can reflect the differentiation or activation status of a particular cell population. The combination of the endogenous fluorescent tag with surface antibody-based labeling is compatible with in vivo conditions; the only alternative for overall MHC II expression quantification consists of cell fixation and permeabilization protocols. In this protocol we introduce a straightforward tool for identifying the overall versus surface MHC II expression in the mesenteric lymph nodes (MLNs). Our model enables a user to omit the steps of multiple staining with fluorophore-tagged antibodies, including the fixation and permeabilization steps, which are needed to identify intracellular markers for flow cytometry. This protocol enables further staining with other extracellular markers without compromising fluorescence performance, as may occur through fixation.
Materials
-
Mice: 8-week-old MHC II–EGFP knock-in (kindly provided by Marianne Boes, University Medical Center Utrecht, Utrecht, The Netherlands) and 8-week-old C57BL/6 female mice (Charles River Laboratories)
-
3% FBS RPMI (see recipe), ice cold
-
10 mg/ml collagenase D (see recipe)
-
0.5 M ethylenediaminetetraacetic acid calcium disodium salt hydrate (EDTA; Sigma-Aldrich, cat. no. 23411-34-9)
-
1000×-diluted Fc block solution (see recipe)
-
Cell staining buffer (see recipe)
-
APC/Fire™ 750 anti-mouse CD45 antibody (clone 30-F11, cat. no. 103154, Biolegend; see recipe)
-
APC anti-mouse CD19 antibody (clone 6D5, cat. no. 115512, Biolegend; see recipe)
-
PE-Cy™7 hamster anti-mouse CD11c (clone HL3; cat. no. 558079, BD Bioscience; see recipe)
-
MHC II single-stain control (APC/cyanine 7 [APC-Cy7] anti-mouse I-A/I-E antibody, clone M5/114.15.2, cat. no. 107628, Biolegend; see recipe)
-
Distilled H2O
-
1× PBS (see recipe)
-
200× Sytox Blue (see recipe)
-
Dissecting instruments (scissors, forceps)
-
Zeiss SteREO Lumar V12 stereomicroscope (objective: Zeiss ApoLumar S 1.2×, FWD 47 mm)
-
1.5-ml microcentrifuge tubes (Biologix, cat. no. 80-1500)
-
2.5-, 20-, 200-, and 1000-µl mechanical pipets
-
Thermoblock: Eppendorf ThermoMixer Comfort
-
Cell strainer, 40 µm pore size (VWR, cat. no. 734-2760)
-
Refrigerated centrifuge (Eppendorf, model no. 5810R) or similar
-
Vortex
-
96-well microplates (200 µl/well)
-
High Throughput Sampler (HTS; BD Biosciences)
-
LSR II flow cytometer (BD Biosciences)
-
FlowJo analysis software (https://www.flowjo.com/)
1.Euthanize the mice by cervical dislocation.
2.Under the fluorescence stereomicroscope, carefully remove the mesenteric lymph nodes (MLNs), and quickly transfer them into a small petri dish filled with 1× PBS.
3.On a petri dish, separate the individual nodules. Strip them of any residual adipose tissue (see Basic Protocol 3).
4.Transfer the individual MLNs into 1.5-ml microcentrifuge tubes containing 360 μl of 3% FBS RPMI, keeping them on ice.
5.Cut individual MLNs into small pieces with scissors.
6.Add 40 μl of 10 mg/ml collagenase solution and incubate the tubes at 37°C in the thermoblock (550 rpm) for 40 min.
7.Add 600 μl of 3% FBS RPMI to each sample.
8.Add 4 μl of 0.5 M EDTA to each sample, mix thoroughly, and then pipet the mixture through a cell strainer into a clean microcentrifuge tube.
9.Centrifuge the samples for 5 min at 300 × g , 4°C.
10.Discard the supernatant and resuspend the samples in 100 µl of 1000×-diluted Fc block solution to prevent nonspecific antibody binding. Incubate for 10 min.
11.Transfer the samples to a 96-well plate.
12.Centrifuge the samples again for 5 min at 300 × g , 4°C.
13.Prepare a mix of antibodies (CD45, MHC II, CD19, and CD11c) using 50 μl of solution per node. Also prepare samples for single-stain controls for CD45, MHC II, CD19, and CD11c, as well as unstained MHC II–EGFP control and wild-type (C57BL/6 mouse) control.
- For the CD45 single-stain control, add 0.3 μl APC/Fire™ 750 anti-mouse CD45 antibody to the 60 μl of cell staining buffer (to 1 μg/ml final).
- For the CD11c single-stain control, add 0.4 μl PE-Cy™7 hamster anti-mouse CD11c to the 60 μl of cell of staining buffer (to 1.33 μg/ml final).
- For the CD19 single-stain control, add 1.5 μl APC anti-mouse CD19 antibody to the 60 μl of cell staining buffer (to 5 μg/ml final).
- For the MHC II single-stain control, use pre-prepared 1000×-diluted APC-Cy7 anti-mouse I-A/I-E antibody as the staining buffer (to 0.2 μg/ml final); this avoids inaccuracy caused by adding too small amount of antibody.
14.Using 50 of antibody mix per node, stain the samples with the respective antibodies (single-stain controls + antibody mix) for 25 min, on ice, in the dark.
15.Wash the samples twice by centrifuging them for 5 min at 300 × g , 4°C, discarding the buffer, and resuspending them in 200 μl cell staining buffer.
16.Centrifuge samples again for 5 min at 300 × g , 4°C. Add 198 μl cell staining buffer to each sample.
17.Add 2 µl of 200× Sytox Blue to each sample.
18.Measure the samples using a BD High Throughput Sampler (HTS), using 160 μl of the 200 μl of sample at speed 1.5.
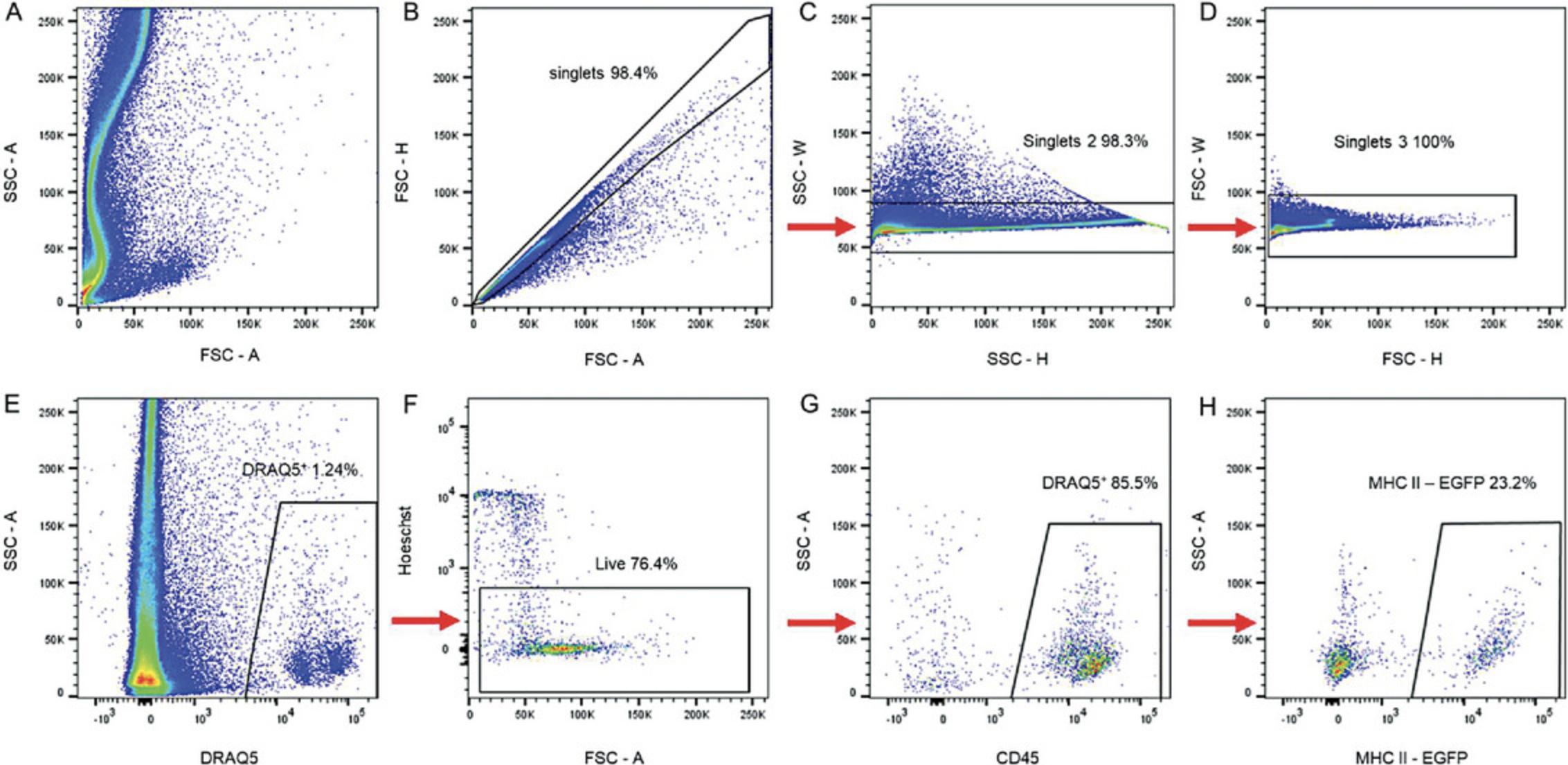
19.Analyze data using FlowJo™ v10.8.1 with the gating strategy shown in Figure 1.
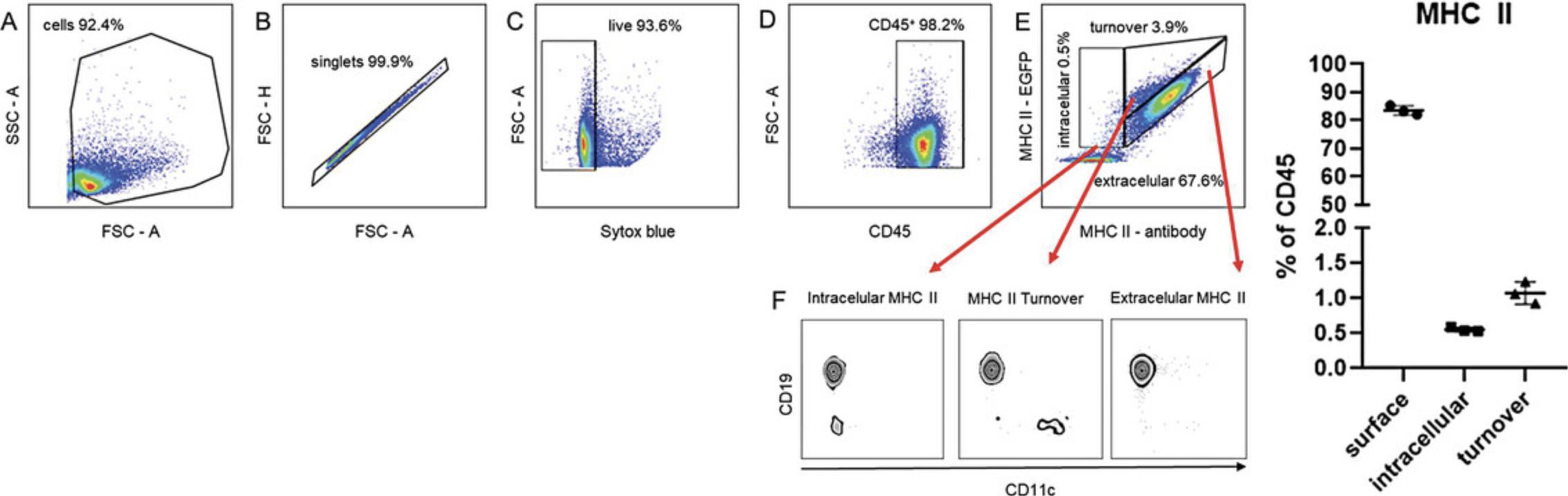
20.Gate cells according to the FSC and SSC parameters
21.Exclude singlets using FSC-H and FSC-A parameters.
22.Exclude death cells by gating Sytox-negative cells.
23.Gate CD45**+** cells.
24.Gate cells according to the EGFP/MHC II and anti-MHC II APC-Cy7 mean fluorescence intensity (MFI) ratio into (a) intracellular, i.e., no APC-Cy7 signal; (b) extracellular, i.e., linear ratio of EGFP and APC-Cy7 MFI = 1; and (c) turnover, i.e., GFP MFI/APC-Cy7 ratio > 1 (this population is clearly outside the extracellular population) (Fig. 1E and F).
Basic Protocol 3: IDENTIFICATION AND PREPARATION OF THE LYMPHOID ORGANS
The MHC II–EGFP knock-in mouse model provides us with a valuable tool for accurately identifying lymphoid organs, which typically contain MHC-II-expressing cells. These cells play a key role in local immune regulation via antigen presentation of exogenous or autophagy-derived antigens. However, lymphoid organs in mouse models are relatively small, often embedded within the surrounding tissue, and difficult to distinguish by the naked eye. Therefore, the precise identification and excision of these specific tissues are crucial for further processing, analysis, and data interpretation. Quantitative analysis requires extracting the entire lymphoid structure and identifying all relevant anatomical components. Simultaneously, flow cytometry and light-sheet fluorescence microscopy (LSFM) require completely removing adipose tissue residues. The MHC II–EGFP knock-in mouse model leverages the contrast between the fluorescence signal and background autofluorescence, making it possible to clearly identify all lymphoid tissues under a fluorescence stereomicroscope and to excise them precisely from surrounding tissue (Pačes et al., 2022).
Previous anatomic research has identified a total of 22 lymphatic nodes in the mouse body (Van den Broeck et al., 2006). By direct fluorescence labeling, we can efficiently and precisely identify all these lymph nodes. Additionally, this protocol provides detailed instructions for identifying and excising individual Peyer's patches (PPs) and for isolating the mesenteric lymph node (MLN) complex into separate nodes, which are responsible for draining specific segments of the intestine.
In this protocol, we introduce a tool for identifying lymphoid organs, making it possible to prepare the entire set for individual characterization using various techniques such as flow cytometry, histology, and whole-mount microscopy. This approach serves as a valuable method for comprehensive, quantitative immunological studies, particularly when simultaneously phenotyping the entire lymphoid system. With this protocol, researchers can accurately identify and analyze lymphoid organs, thereby gaining a deeper understanding of their structure, function, and immunological significance.
Materials
-
Mice: 8-week-old MHC II–EGFP knock-in female mice (kindly provided by Marianne Boes, University Medical Center Utrecht, Utrecht, The Netherlands)
-
1× PBS (see recipe)
-
Paraformaldehyde (Sigma-Aldrich, cat. no. 158127)
-
3% FBS RPMI (see recipe)
-
Dissecting plate
-
Ophthalmology scissors (MEDIN)
-
Pins
-
Straight-tip precision tweezers (P-lab)
-
Petri dishes
-
Zeiss SteREO Lumar.V12 stereomicroscope (objective: Zeiss ApoLumar S 1.2×, FWD 47 mm)
Identification of the lymphoid organs
1a. Euthanize the MHC II–EGFP knock-in mouse by cervical dislocation.
2a. Use scissors to make an incision along the skin from the tail to the neck, without puncturing any underlying structures.
3a. Open the skin and use pins to secure it onto the dissecting plate for stabilization.
4a. Position the mouse under the fluorescence stereomicroscope at 488-nm excitation and select a magnification between 9.6× and 20×, depending on the lymphoid organ under examination.
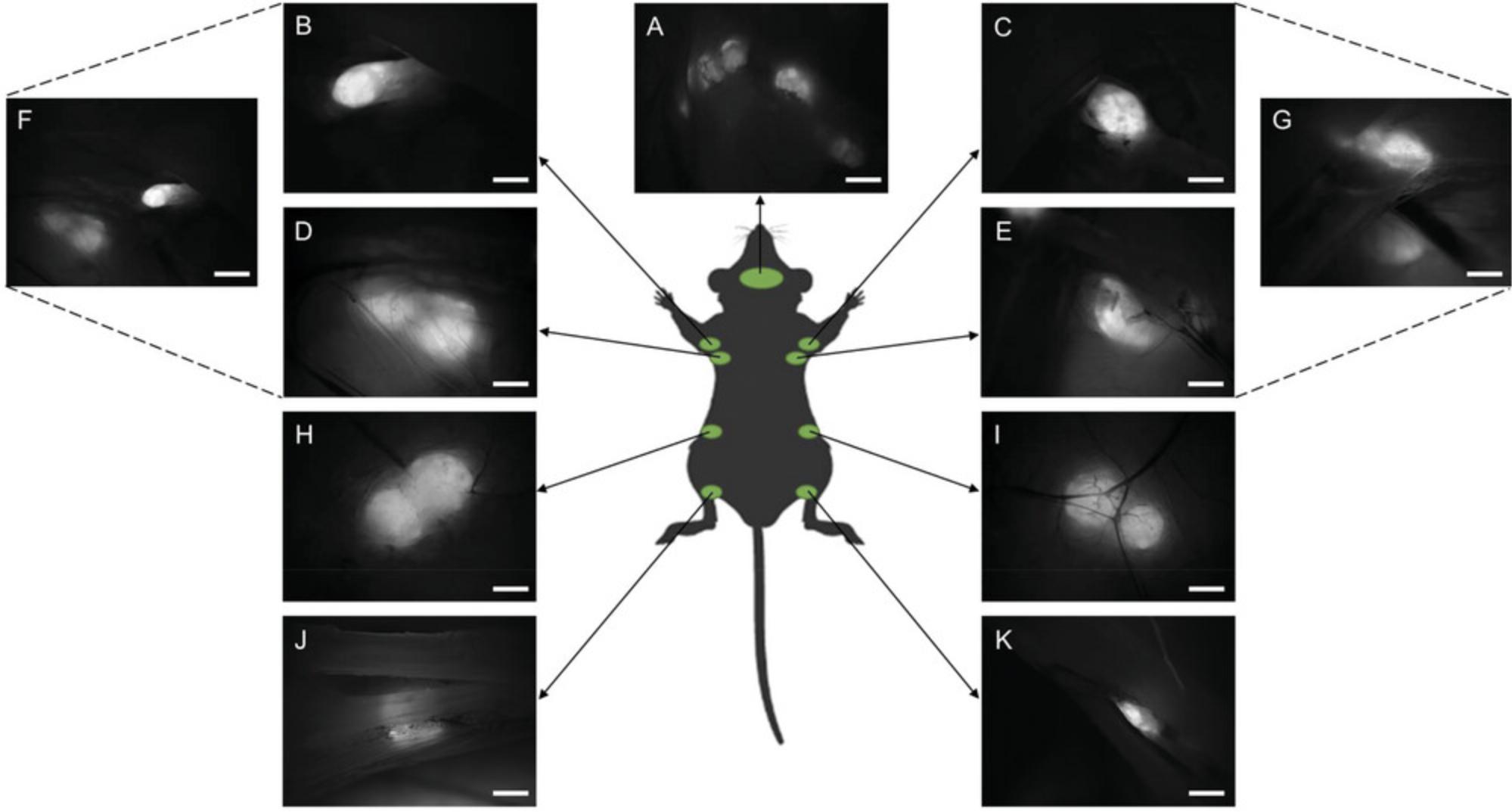
5a. Locate the subiliac lymph node, situated at the primary intersection of blood vessels on the exposed skin surface (see Fig. 2H and 2I).
6a. Identify the popliteal lymph node by rotating the leg 180° and observing this node in the popliteal region (see Fig. 3J and K).
7a. Identify the proper axillary and accessory axillary lymph nodes within the armpit area (see Fig. 2B-G).
8a. Shift focus to the neck region and individually locate the mandibular, accessory mandibular, and superficial parotid lymph nodes (see Figs. 2A and 3A and B).
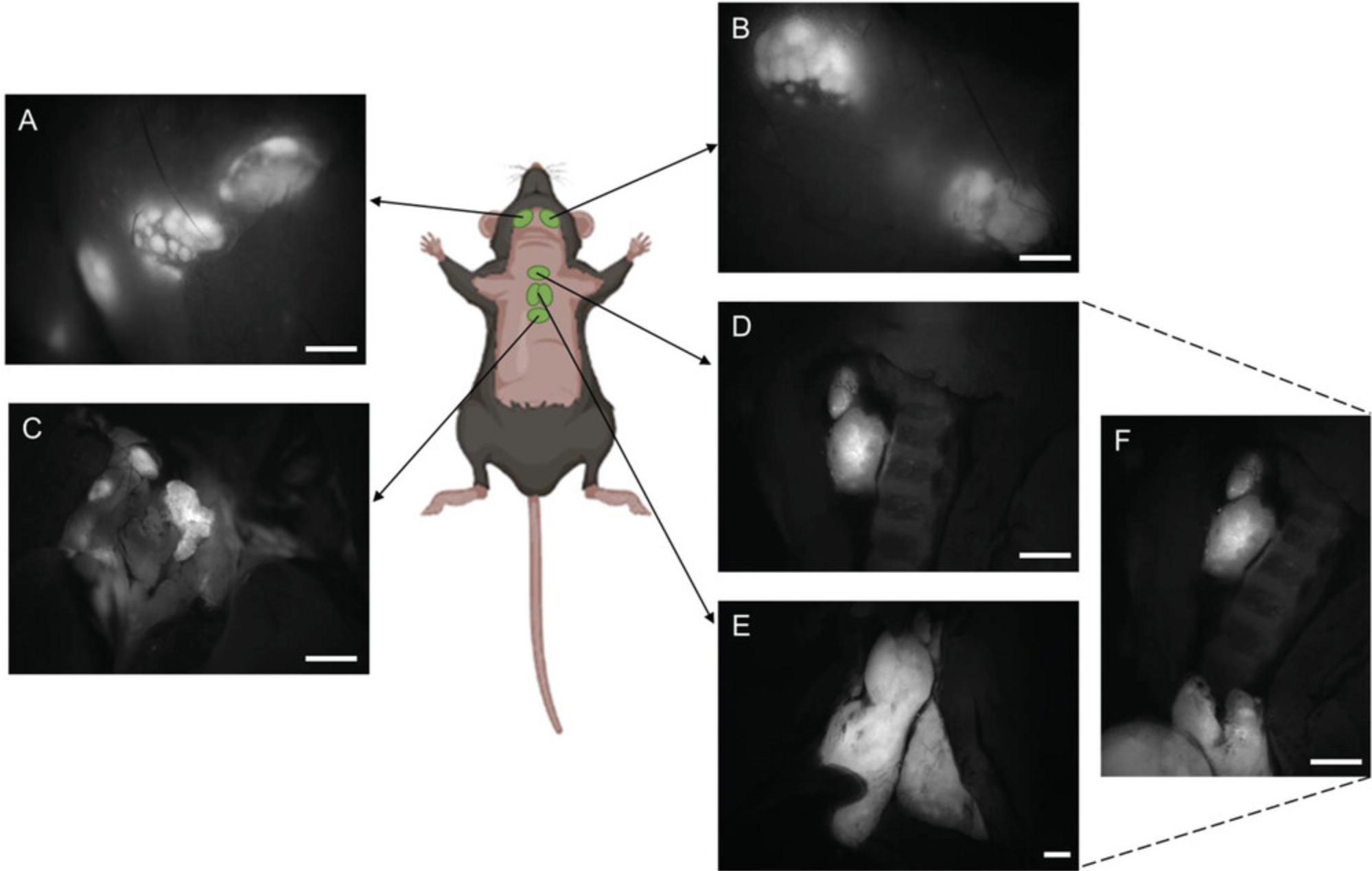
9a. Carefully cut the peritoneal cavity and identify the mesenteric lymph nodes (see Fig. 4).
10a. Remove the stomach and intestines from the peritoneal cavity.
11a. Identify the caudal mesenteric lymph nodes (see Fig. 4B) and the sciatic lymph nodes (see Fig. 4D).
12a. Identify the pancreaticoduodenal, gastric, and renal lymphoid complex (see Fig. 4A).
13a. Open the thoracic cavity and locate the thymus (see Fig. 3E and F).
14a. Identify the caudal deep cervical, cranial mediastinal, and tracheobronchial lymphoid complex (see Fig. 3D and F).
15a. Identify the caudal mediastinal lymphoid complex (see Fig. 3C).
Excision of individual mesenteric lymph nodes (MLN)
1b. Euthanize the mouse using cervical dislocation.
2b. Open the peritoneal cavity.
3b. Position the mouse under the fluorescence stereomicroscope at 488-nm excitation and set the magnification to 9.6×.
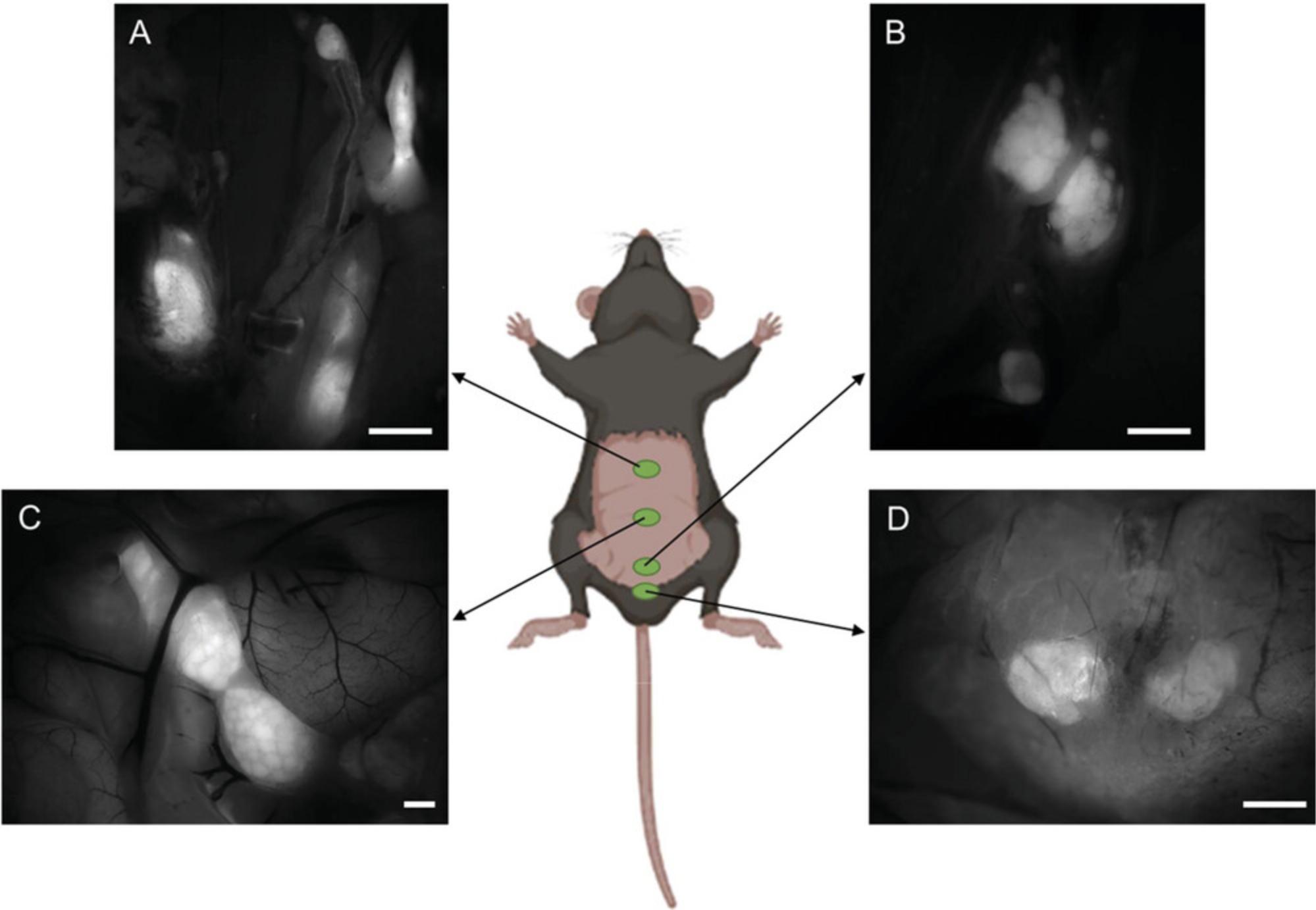
4b. Gently manipulate several intestinal folds from left to right until a distinct fluorescent mesenteric lymph node (MLN) complex becomes visible (see Fig. 5A).
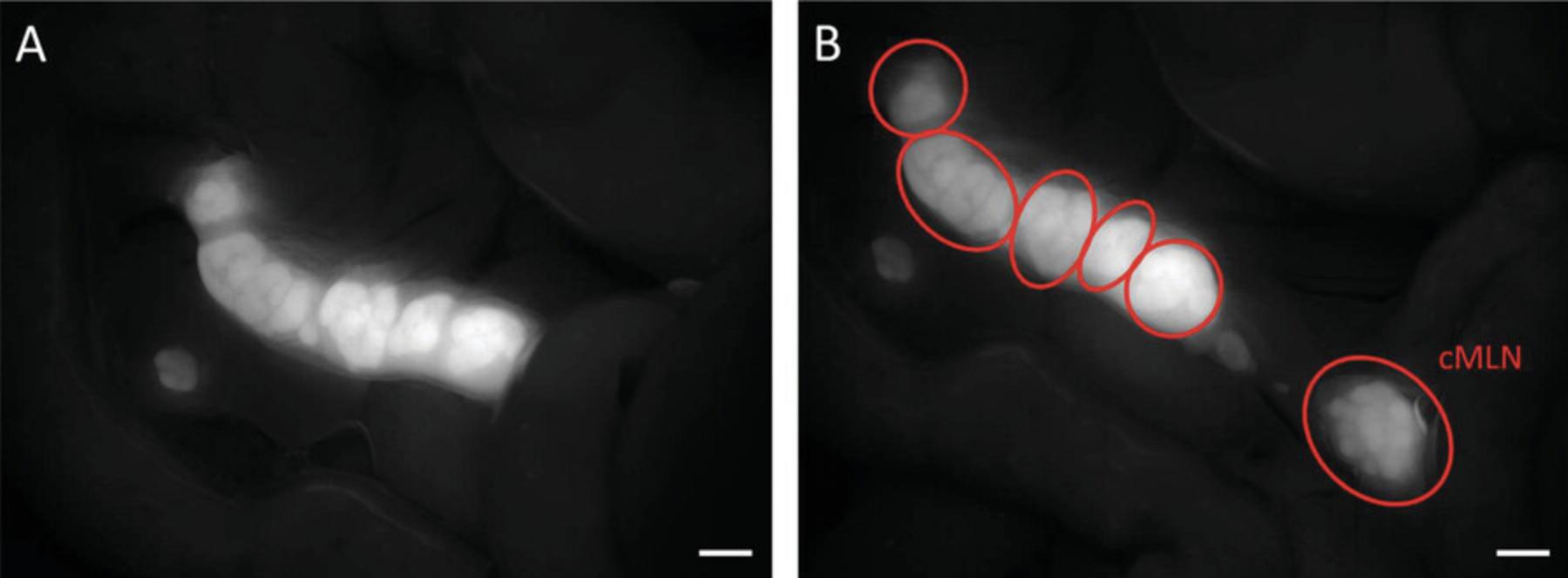
5b. Excise the entire MLN complex, making sure to remove the colon draining node as well; this node may not be closely attached to the rest of the complex and could be concealed beneath the intestinal fold on the right side.
6b. Transfer the excised complex to a petri dish containing 1× PBS. Carefully eliminate any remaining adipose tissue (non-fluorescent tissue surrounding the MLN) using sharp-tipped tweezers and scissors.
7b. Using the same tools, separate the individual nodes from the MLN complex (see Fig. 5B).
Excision of individual Peyer's patches and cecal patch
1c. Euthanize the mouse by cervical dislocation.
2c. Open the peritoneal cavity.
3c. Excise the entire gastrointestinal tract (GTI) between the stomach and rectum and transfer the GTI to a petri dish filled with PBS.
4c. Begin by slowly straightening the intestine from the beginning, removing the mesenterial and connective tissue, which hinder the unfolding of the folds (refer to Fig. 6A).
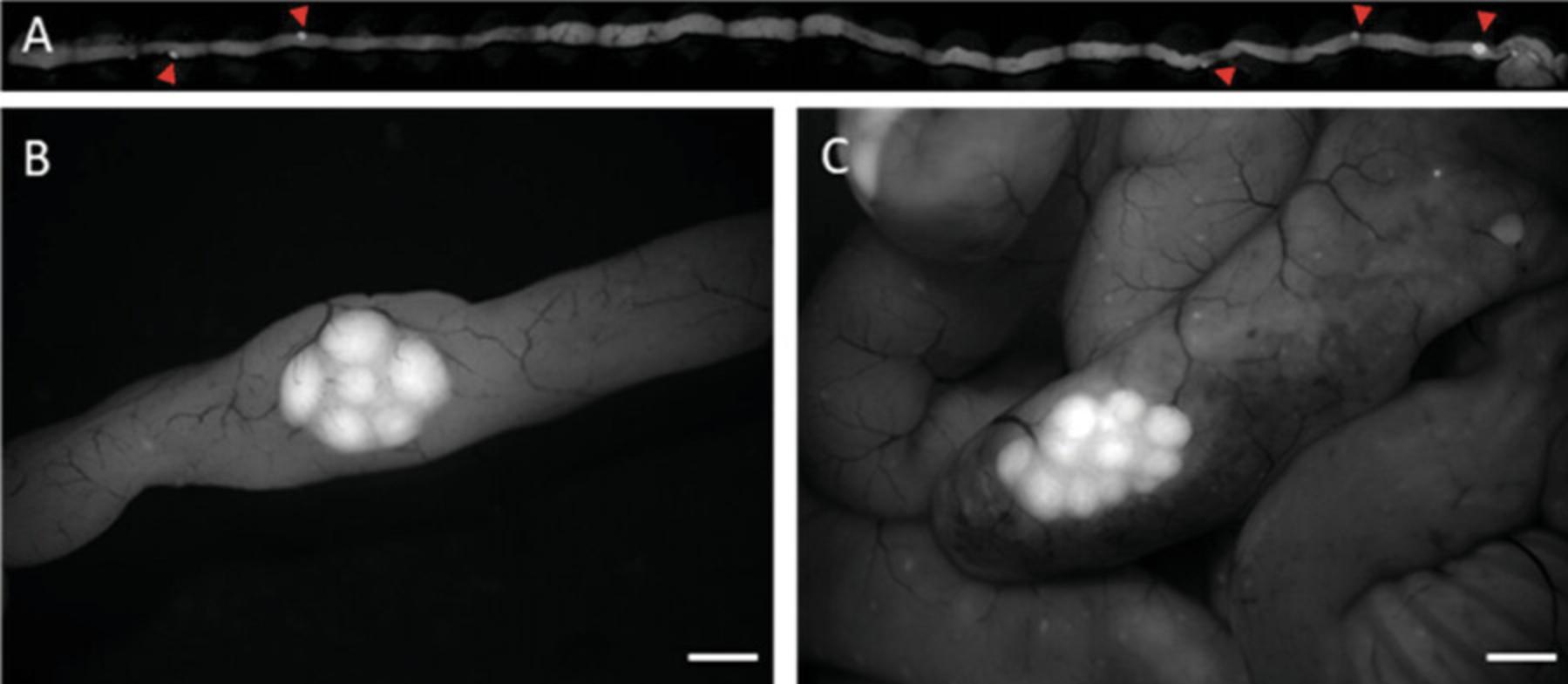
5c. Place the mouse under a fluorescence stereomicroscope at 488-nm excitation and set the magnification to 12×.
6c. Using sharp-tipped tweezers and scissors, precisely excise each Peyer's patch (PP) one by one, targeting the edge of the fluorescence signal (refer to Fig. 6B and C).
7c. Immediately wash the excised Peyer's patches in PBS to remove any remnants of intestinal content on the luminal side of the tissue.
8c. Depending on its planned further use, either fix the tissue in 3.8% paraformaldehyde for microscopy (it can potentially be stored at 4°C for up to 1 week, not more, due to the danger of enhance autofluorescence) or immediately transfer to 3% FBS RPMI for subsequent tissue dissociation and flow cytometry.
Basic Protocol 4: QUANTIFICATION AND MEASUREMENT OF INTESTINAL LYMPHOID TISSUE BY FLUORESCENCE STEREOMICROSCOPY
The MHC II–EGFP knock-in mouse model enables us to directly visualize antigen-presenting cells (APCs) and to accurately quantify individual lymphoid cell accumulations. This approach can thus be used not to study cell morphology but also to perform surface measurements without additional processing steps such as fixation, cutting, or staining. Avoiding these procedures mitigates a wide range of artifacts. However, the morphology of the immune tissue in the gastrointestinal tract (GIT) undergoes developmental changes, showing variations between individuals and mouse models (Gama et al., 2020). Additionally, the size and number of individual structures are affected by interactions with the microbiota (Kernbauer et al., 2014). These differences have only been sporadically studied even though the GIT immune system is quantitatively and functionally significant within the broader immune system. In this study, we present a methodological toolbox for quantifying the lymphoid tissue in the GIT, which reflects the hierarchical organization of the GIT, including the lamina propria, scattered intestinal lymphoid tissue, and Peyer's patches. This toolbox serves as a quick and straightforward experimental approach for phenotyping the entire lymphoid structures relevant to the mouse GIT and considers various factors such as development, sex, individual variability, infection, microbiota imbalance/modification, and genetics, including transgenic technologies. A notable advantage is the enhanced fluorescence observed in the lamina propria within individual villi, which enables a precise ex vivo quantification in situ without requiring standard histological methodologies prone to artifacts. By employing this methodological toolbox, researchers can gain deeper insights into the organization and tuning of the gastrointestinal immune system, which in turn has significant implications for understanding systemic immune responses.
Materials
-
Mice: 8-week-old MHC II–EGFP knock-in mice (kindly provided by Marianne Boes, University Medical Center Utrecht, Utrecht, the Netherlands)
-
1× PBS (see recipe)
-
Straight-tip precision tweezers (P-LAB)
-
Ophthalmology scissors (MEDIN)
-
Petri dishes
-
1000-µl micropipet
-
Zeiss SteREO Lumar.V12 stereomicroscope (objective: Zeiss ApoLumar S 1.2×, FWD 47 mm)
Extraction of small intestine
1.Euthanize the mouse by cervical dislocation.
2.Open the peritoneal cavity.
3.Excise the entire gastrointestinal tract (GIT) between the stomach and rectum and transfer the GIT to a petri dish filled with 1× PBS.
Quantification of intestinal villi
4a. Remove the small intestine and divide it into three approximately equally long segments for washing.
5a. Use an automatic pipet with a 1000-µl micropipet tip to repeatedly rinse each segment individually with PBS until most of the chyme and mucus is flushed out.
6a. Place the washed sample under a fluorescence stereomicroscope at 488-nm excitation and set the magnification to 16×.
7a. Once individual villi (GPF-tagged) are clearly visible (due to the high content of MHC II–EGFP+ cells), take pictures of the proximal jejunum and distal ileum (approximately corresponding to the area of the first and last Peyer's patches) under the stereomicroscope (refer to Fig. 7A and B).
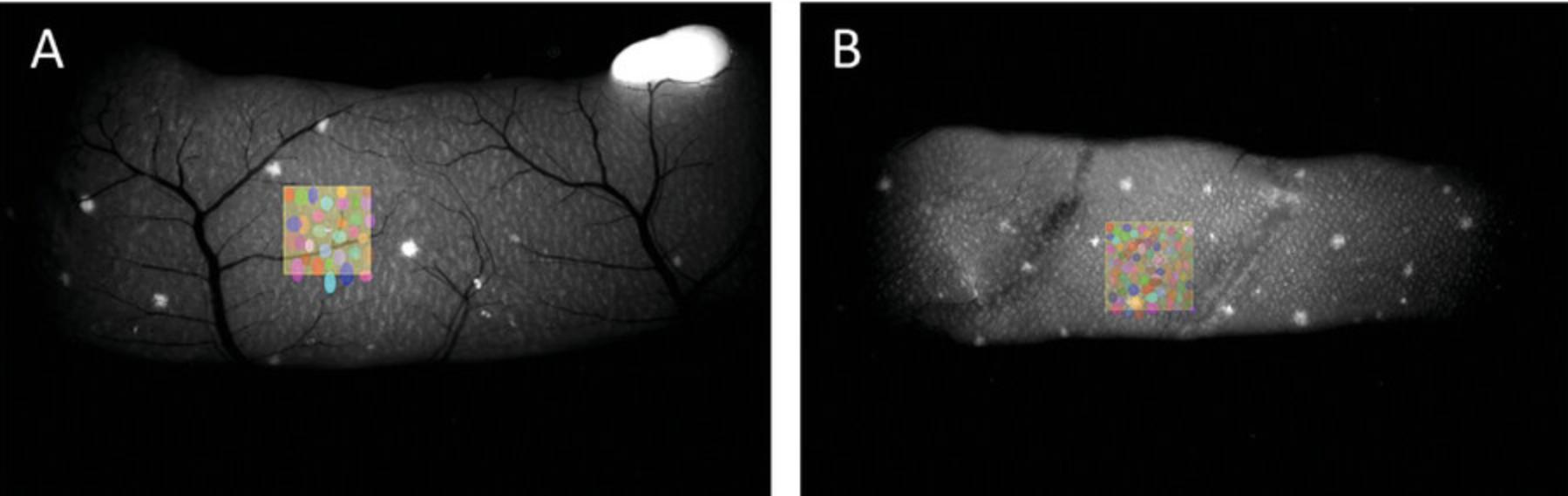
8a. Use the Arivis 4D software to quantify the intestinal villi.
9a. Adjust the image settings to ensure a clear distinction of individual villi.
10a. Randomly select three 1-mm² areas within each segment, i.e., three squares for the proximal jejunum (refer to Fig. 7A) and three squares for the distal ileum (refer to Fig. 7B).
11a. Manually mark the villi within each square using the object tool.
12a. Calculate the average number of villi per mm² for each segment. The average value for the entire intestine can be approximated as the average of these values.
Quantification of intestinal villi (alternate)
4b. Divide the intestine into six segments of equal lengths.
5b. Using an automatic pipet with a 1000-µl micropipet tip, repeatedly rinse each individual segment with PBS until flushing out most of the chyme and mucus.
6b. Place the sample under a fluorescence stereomicroscope at 488-nm excitation and set the magnification to 16×.
7b. Take pictures of all six segments when individual villi are clearly visible, under the stereomicroscope.
8b. Open the image files in Arivis 4D software and adjust the image settings to clearly distinguish individual villi.
9b. Randomly select three (or more) 1-mm² areas within each segment, resulting in a total of 18 squares.
10b. Manually mark the villi within each square using the object tool in the software.
11b. Calculate the average number of villi per mm² for each segment. The average value for the entire intestine can be approximated as the average of these values.
Quantification of solitary intestinal lymphoid tissue (SILT)
4c. Place the sample under a fluorescence stereomicroscope at 488-nm excitation and set the magnification to 12×.
5c. Take sequential pictures of the entire intestine, with at least 20% overlap between each image.
6c. Manually fuse the images using the Tile Sorter module in Arivis 4D software.
7c. Use the object tool in the software to accurately mark individual follicles (refer to Fig. 89A).
8c. Based on the surface area of individual follicles, their volume can also be estimated because the structure predominantly runs parallel to the surface (refer to Fig. 8A-C).
9c. Since the intestine does not have a specific lateral orientation, SITL data on one side of the intestine can be extrapolated to the other side.
Measurement of PPs and their subunits
4d. Place the sample under a fluorescence stereomicroscope at 488-nm excitation and set the magnification to 16×.
5d. Take sequential images of all Peyer's patches (PP) present.
6d. Using the measure tool in Arivis 4D software, manually measure the area of each PP and calculate the number of its subunits as shown in Figure 9.
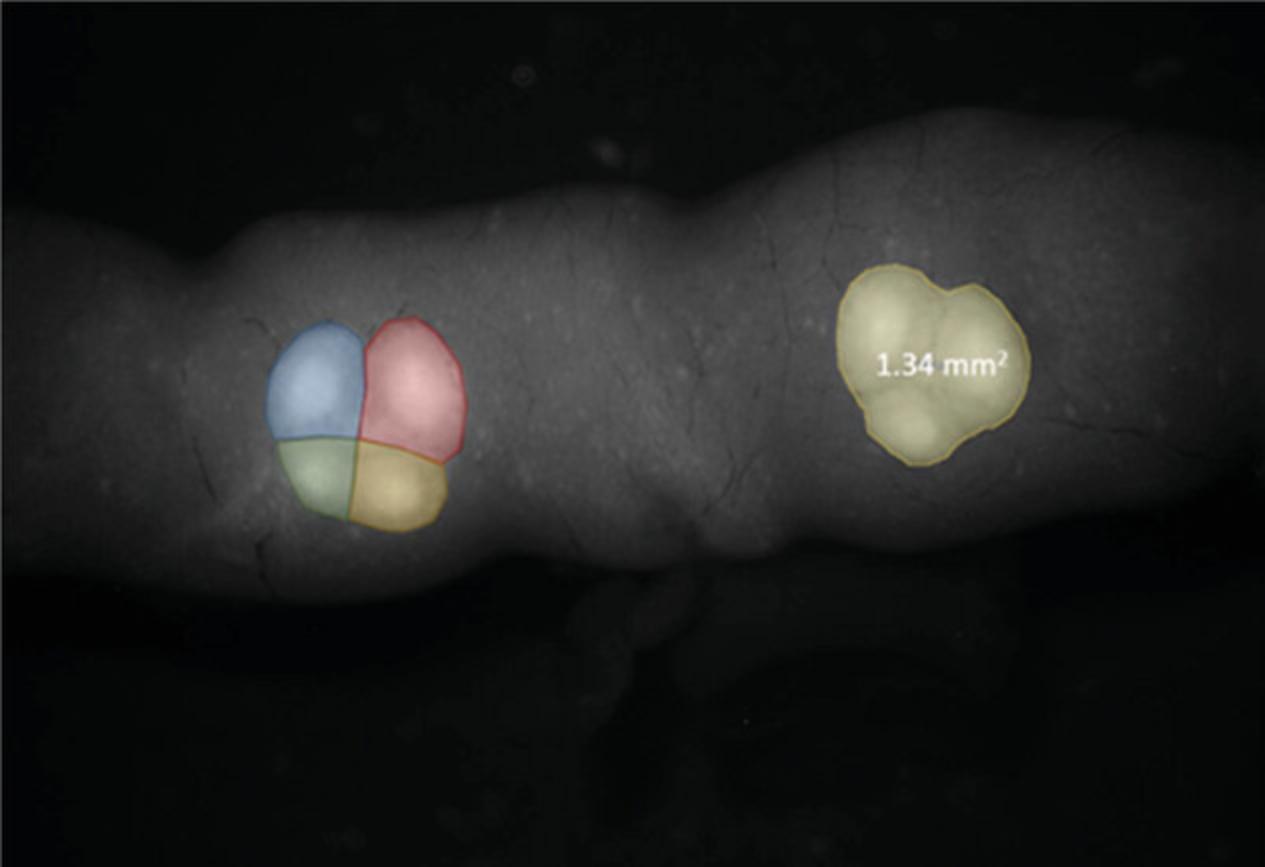
Measurement of intestinal surface
4e. Carefully cut away all ligaments to relax the intestinal folds and spirals precisely.
5e. Gently straighten the small intestine on the Petri dish filled with PBS, ensuring that the thickness of the layer approximately matches the thickness of the small intestine and avoiding stretching or deforming the small intestine in any way.
6e. Place the sample under a fluorescence stereomicroscope at 488-nm excitation and set the magnification to 12×.
7e. Capture sequential images of the entire intestine with at least 20% overlap between each image.
8e. Manually fuse the images using the Tile Sorter module in Arivis 4D software.
9e. Measure the length of the small intestine.
10e. Divide the small intestine into six segments of equal length and measure the thickness of the small intestine at the five points where the segments border each other. Calculate the average thickness of the small intestine from these five measurements.
Basic Protocol 5: QUANTIFICATION OF APC CONTENT IN LYMPHOID ORGANS BY LIGHT-SHEET FLUORESCENCE MICROSCOPY
Despite significant advancements in the field of quantitative immunology, the availability of quantitative data on physiological functions, particularly on immune cells, remains limited. For this reason, researchers often rely on relative data, analyzed and interpreted disregarding the broader context of the entire system. This lack of detailed quantitative studies is surprising given that even minor variations in cell counts or population proportions can have profound functional implications. Comprehensive quantitative research on specific immune cell populations must be thus conducted under both physiological and pathological conditions. For this purpose, quantitative systems immunological approaches are essential as they enable us to calculate exact and semi-exact values, which are imperative for accurate data interpretation. Light-sheet microscopy offers an unparalleled opportunity for studying whole cleared organs in situ , applying unique microscopic methods. These methods are particularly advantageous when using the MHC II–EGFP model, which exploits the bright and relatively photostable localization of endogenous MHC II expression through the EGFP tag. By leveraging such quantitative immunological approaches and innovative imaging techniques, researchers can gain unprecedented insights into the dynamics and functionality of the entire immune system, leading to a more comprehensive understanding of immune responses and their implications in various physiological and pathological contexts.
Materials
-
Mice: 8-week-old MHC II–EGFP knock-in mice (kindly provided by Marianne Boes, University Medical Center Utrecht, Utrecht, The Netherlands)
-
1× PBS (see recipe)
-
Paraformaldehyde (Sigma-Aldrich, cat. no. 158127)
-
CUBIC 1 clearing solution (see recipe)
-
CUBIC wash solution (see recipe)
-
DRAQ5 Fluorescent Probe Solution (5 mM; Thermo Fisher Scientific, cat. no. 62251)
-
Sodium azide (Sigma-Aldrich, cat. no. S8032)
-
CUBIC 2 clearing solution (see recipe)
-
Saccharose (Penta chemicals, cat. no. 24970-31000)
-
Straight-tip precision tweezers (P-LAB)
-
Ophthalmology scissors (MEDIN)
-
Petri dishes
-
2.5-, 20-, 200-, and 1000-µl micropipets
-
Zeiss SteREO Lumar.V12 stereomicroscope (objective: Zeiss ApoLumar S 1.2×, FWD 47 mm)
-
Analog Abbe refractometer AR4 (Krüss)
-
Zeiss Z.1 light-sheet microscope with a RI 1.45 clearing chamber (illumination objectives: Zeiss LSFM clearing 10×/0.2; detection objective: Zeiss Clr PN 20×/1.0K)
-
Krazy glue, Elmer's products
-
Huygens Professional software
-
Scientific Volume Imaging B.V. Arivis Vision4D software, version 3.6.2
Sample preparation for quantification of APCs in lymphoid organs
1.Euthanize the mouse by cervical dislocation.
2.Open the peritoneal cavity.
3.Excise the small intestine and transfer it to a petri dish.
4.Place the small intestine under a fluorescence stereomicroscope at 488-nm excitation and 16× magnification.
5.Dissect ∼6-mm-long sections from the proximal jejunum near the first Peyer's patch (PP) and the distal ileum surrounding the last PP.
6.Fix the sections in 3.8% paraformaldehyde at 4°C for 12 hr.
7.Wash the samples three times in 1× PBS.
8.Clear the samples using CUBIC 1 clearing solution for 7 days at 37°C, ensuring that the samples are protected from light to prevent bleaching.
9.Wash the samples three times in CUBIC wash solution at 37°C for at least 1 hr.
10.Stain the samples with DRAQ5™ Fluorescent Probe Solution, diluted 10,000× in 1 ml PBS with 0.01% sodium azide, for 7 days at 4°C. One again, protect the samples from light to prevent bleaching.
11.Wash the samples three times in PBS at 4°C for at least 1 hr.
12.Transfer the samples to CUBIC 2 clearing solution for 4 days at 37°C to match the refractive index. Ensure that the samples are shielded from light to prevent bleaching.
13.Measure the exact refractive index of the solution at the imaging temperature before imaging under the light-sheet fluorescence microscope. Set this precise refractive index value on the correction ring of the microscope objective.
14.Using glue, attach the cleared sample to a syringe and insert it into the sample holder.
Quantification of APCs in intestinal villi
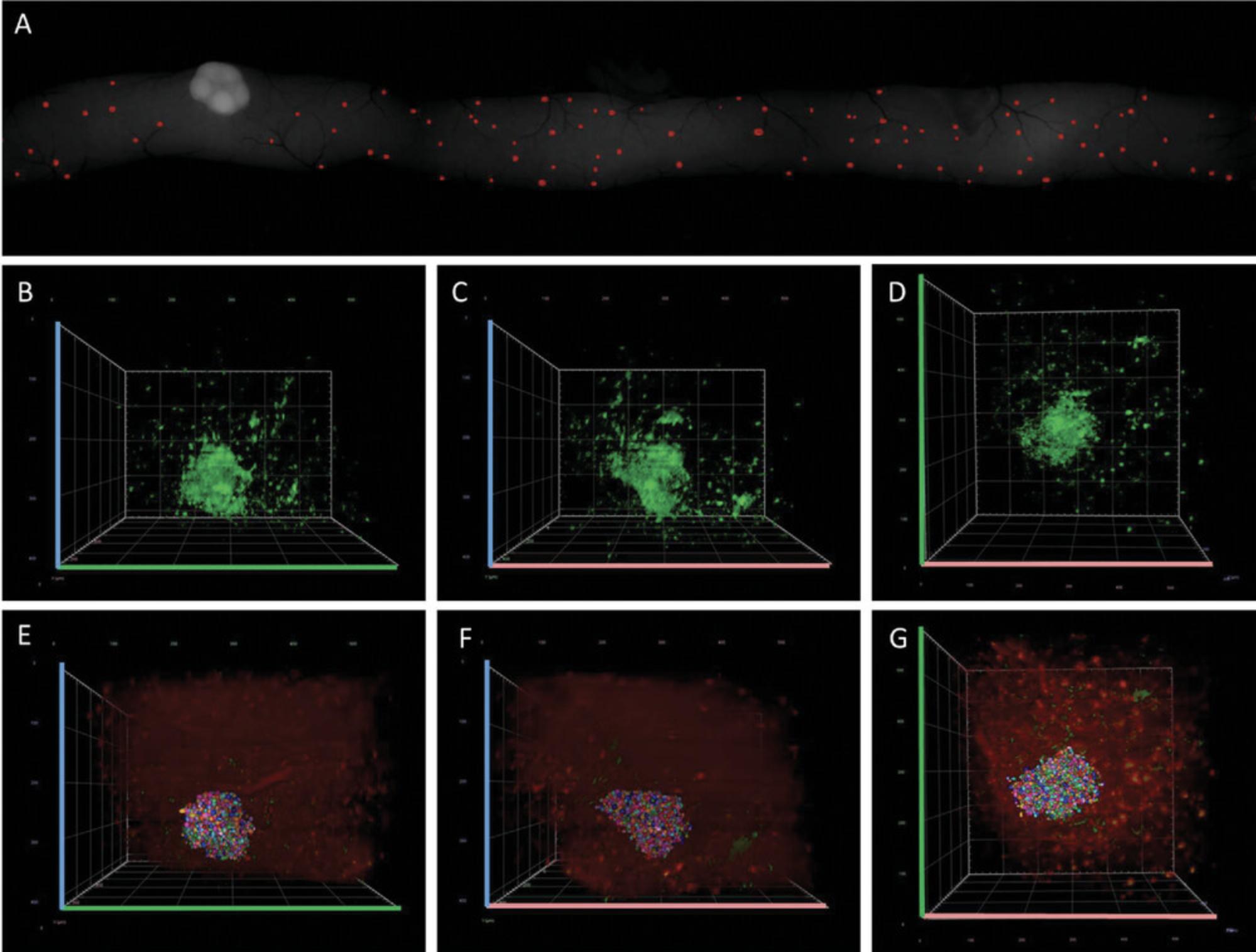
15a. Under a light-sheet microscope with a clearing setup of 1.45, acquire z -stacks of three randomly selected villi from a specific section. Perform scanning in three channels: one for DRAQ5, one for GFP, and one in the red end of the spectrum to detect autofluorescence.
16a. Open the image in Arivis Vision 4D software and adjust the contrast to clearly distinguish individual cells. Use the object tool to manually mark the nuclei of individual cells (Fig. 10A and B).
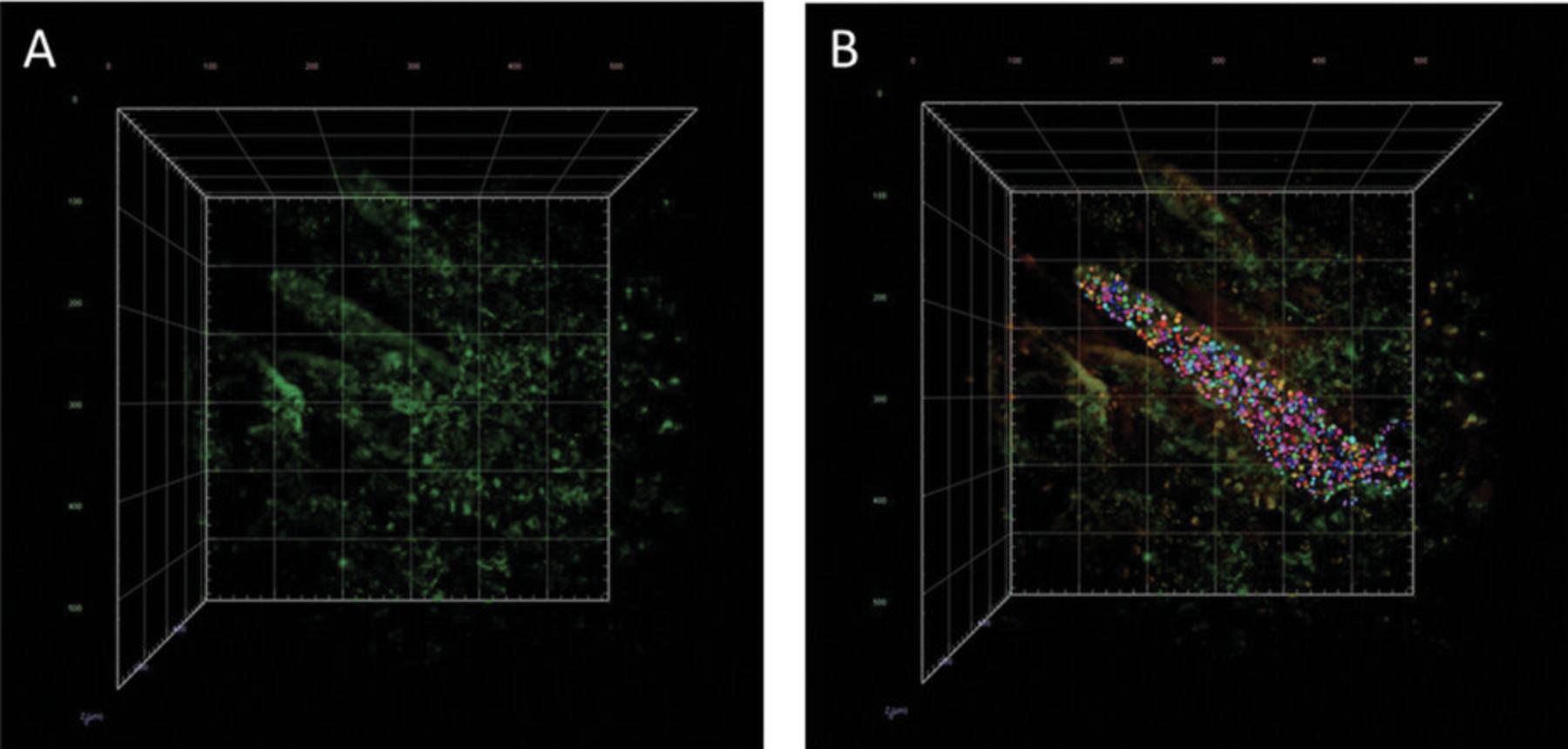
Quantification of APCs in SILT
15b. Using a light-sheet fluorescence microscope with a clearing setup of refractive index 1.45, acquire z -stacks of three randomly selected follicles from a specific section. Perform scanning in three channels: one for DRAQ5, one for GFP, and one in the red end of the spectrum to detect autofluorescence.
16b. Open the image in Arivis Vision 4D software and adjust the contrast to clearly distinguish individual cells. Use the object tool to manually mark the nuclei of individual cells (see Fig. 8). Please note that the settings required for this step will significantly differ from those used for quantification of villi.
Basic Protocol 6: VISUALIZATION OF CORNEAL APCs
The protocol described herein offers a valuable approach for promptly and effectively visualizing and quantifying immune cells in the mouse eye. The MHC II–EGFP knock-in mouse model provides a key advantage by enabling the visualization of immune cells within this naturally translucent organ in vivo without requiring staining procedures. The subsequent reconstruction and analysis of three-dimensional data further differentiates two types of antigen-presenting cells, namely macrophage- and Langerhans-like cells, based on their phenotype. This comprehensive protocol can be used to examine eyeballs immediately after dissection, capturing the in vivo status, or after fixation, expanding its applicability to postmortem analysis. By facilitating the characterization of immune cells in the eye through functional histology, this method helps us understand how the immune system is involved in ocular health and pathology.
Materials
-
Mice: 8-week-old MHC II–EGFP knock-in (kindly provided by Marianne Boes, University Medical Center Utrecht, Utrecht, the Netherlands)
-
1× PBS (see recipe), ice cold
-
Distilled water
-
Paraformaldehyde (Sigma-Aldrich, cat. no. 158127)
-
2-ml microcentrifuge tubes
-
Scissors
-
Tweezers
-
Petri dishes
-
Zeiss Z.1 light-sheet microscope with a water chamber (illumination objectives: Zeiss LSFM clearing 10×/0.2; detection objective: Zeiss Clr PN 20×/1.0K)
-
Huygens Professional software
-
Scientific Volume Imaging B.V. Arivis Vision4D software, version 3.6.2
1.Euthanize the mouse by cervical dislocation.
2.Remove the skin around the eyes.
3.Carefully extract the eyeball using tweezers and scissors, holding onto the optic nerve, and place it in 1× PBS, in a petri dish located on ice.
4.Rinse the removed eyeballs in PBS and distilled H2O to clean them off of residual tissue and hair.
5.Optional : If necessary, fix the eyeballs overnight in 2-ml microcentrifuge tubes with 3.8% paraformaldehyde in distilled water at 4°C.
6.Visualize the washed eyes immediately under a Zeiss Z.1 light-sheet fluorescence microscope with a water chamber. Scan from the limbus to the center of the cornea.
7.Optional : To facilitate the detection of the cells during analysis, deconvolve the data using Huygens Professional software.
8.Analyze the data in Arivis Vision4D software.
9.Manually mark and count macrophage- and Langerhans-like cells in the sample (Fig. 11A and B).
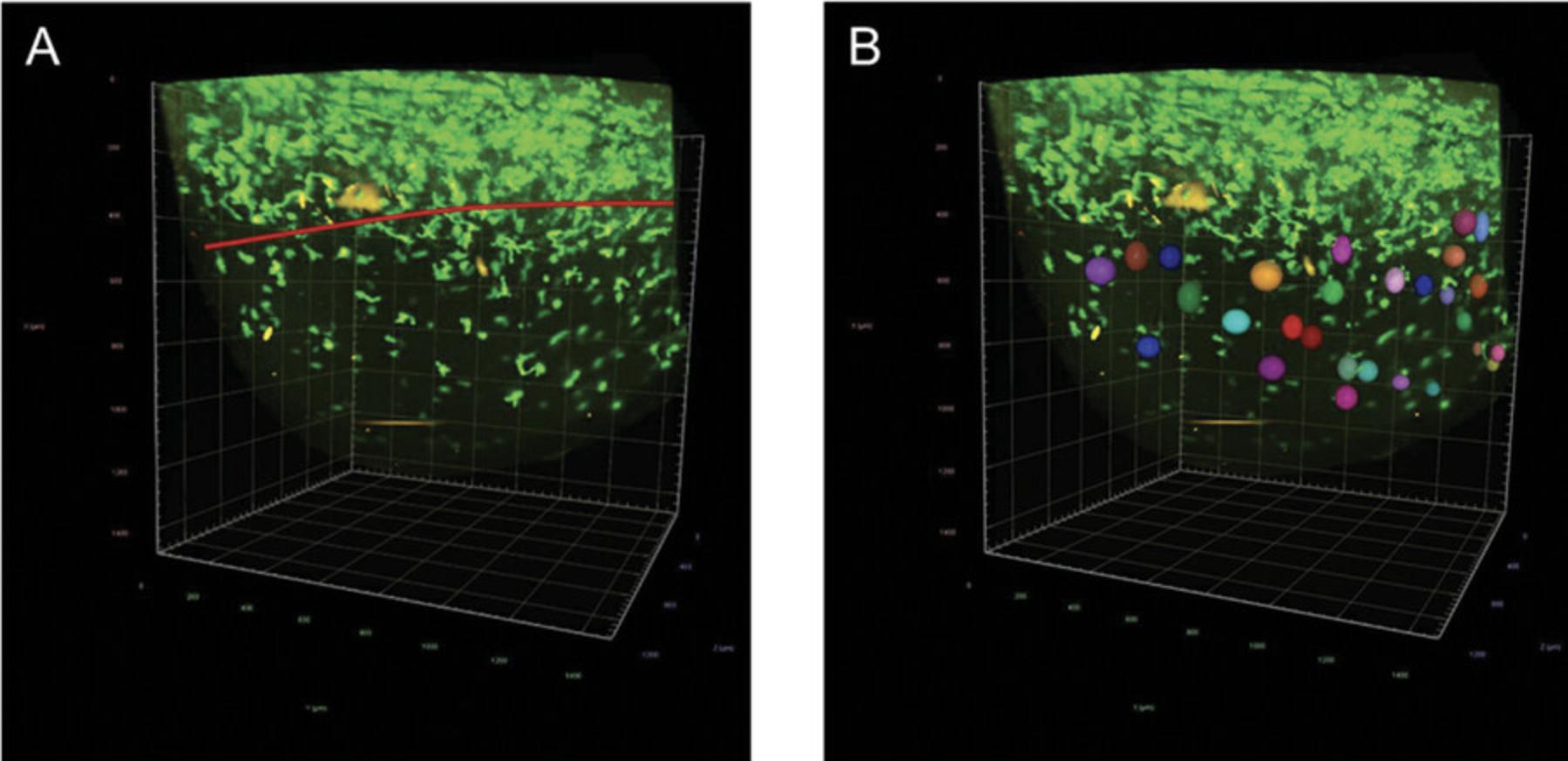
Basic Protocol 7: QUANTIFICATION OF MHC II+ CELLS IN MATERNAL MILK BY FLOW CYTOMETRY
Breastfeeding is widely recognized for its numerous health benefits to newborns, such as reducing the risk of respiratory infections and various diseases (Victora et al., 2016; Cristofalo et al., 2013). Apart from its nutritional value, maternal milk serves as a vital source of diverse bioactive factors, including immune cells (Witkowska-Zimny & Kaminska-El-Hassan, 2017; Laouar, 2020). Accurately and consistently characterizing the cellular composition of mouse milk requires using appropriate cell isolation techniques followed by cell staining and a precise analysis. However, mouse milk contains various components, such as fat globules, lipid droplets, and debris, that may prevent an accurate quantitative analysis of the cell content. With previous standard protocols, researchers have struggled to differentiate cellular from noncellular components. In this protocol, milk collection is performed under standard conditions without administering oxytocin, ensuring minimal impact on the physiological parameters of dams (Muranishi et al., 2016). Additionally, the following protocol presents a comprehensive strategy for accurate cell identification and discrimination of noncellular content based on a modified flow cytometric procedure described by Keller et al., 2019.Furthermore, this protocol facilitates the determination of not only relative but also absolute numbers of immune cells, enabling us to assess total leukocyte counts in mouse milk. Researchers can thus gain valuable insights into the cellular dynamics and immune components of maternal milk, thereby furthering our understanding of its significance for infant health and development.
Materials
-
Mice: 8-week-old MHC II–EGFP knock-in female mice (kindly provided by Marianne Boes, University Medical Center Utrecht, Utrecht, The Netherlands) and 8-week-old C57BL/6 female mice (Charles River Laboratories), all recently mated
-
Cell staining buffer (see recipe)
-
RPMI 1640 medium supplemented with L-glutamine and phenol red (Thermo Fisher Scientific, cat. no. 21875034), ice cold
-
10×- and 1000×-diluted Fc block solutions (see recipe)
-
APC/Fire™ 750 anti-mouse CD45 antibody (clone 30-F11, cat. no. 103154, Biolegend; see recipe)
-
10×-diluted DRAQ5 (see recipe)
-
TruStain FcX™ PLUS (anti-mouse CD16/32) antibody (Biolegend, cat. no. 165604)
-
1 mg/ml Hoechst solution (see recipe)
-
Metal boxes (for mouse pup isolation)
-
2.5-, 20-, 200-, and 1000-µl mechanical pipets
-
1.5-ml microcentrifuge tubes (Biologix, cat. no. 80-1500)
-
Refrigerated centrifuge (Eppendorf, model no. 5810R) or similar
-
Vortex
-
96-well microplates (500 µl/well)
-
LSR II flow cytometer (BD Biosciences)
-
FlowJo v10.8.0 analysis software (https://www.flowjo.com/)
Optional preparatory steps for milk collection from lactating mouse
The following optional steps should potentially increase milk yield, because the re-establishment of contact between mouse and pups could lead to the physiological release of oxytocin and higher milk production/release.
1.On the fourth day after parturition (1 day before the experiment), place sterile metal boxes in the mouse cages to allow the dams and their litters to become accustomed to them.
2.The next day (the fifth day after parturition), separate the dam from her litter by transferring the pups to the metal boxes located in the dam's cage.
3.After 3 hr of separation, open the isolation box to facilitate physical contact between the dams and their litter for 3-5 min.
4.Observe the dam's maternal behavior, including grooming and breastfeeding of the pups.
Milk collection from lactating mouse
5.Separate the dam from the litter for 3 hr and then euthanize the dam by cervical dislocation.
6.Gently massage the mammary glands to initiate milk collection. Collect drops of milk using a 200-µl pipet tip. When the amount of milk collected from a particular mammary gland decreases, move on to the next nipple.
7.Repeat step 6 several times to obtain milk from all 10 mammary glands, collecting the milk in a 1.5-ml microcentrifuge tube and keeping it on ice. Complete the milking process within 30-45 min.
8.Measure the volume of milk obtained with a 200-µl mechanical pipet, and store the collected milk on ice until ready to proceed with step 9a or 9b.
Cell surface staining for flow cytometry measurement
9a. Adjust the volume of the collected milk to 150 µl with cell staining buffer. Proceed to step 11.
9b. Add 1 ml ice-cold RPMI 1640 to each 1.5-ml microcentrifuge tube filled with the collected milk. Centrifuge the tube for 10 min at 400 × g , 4°C.
10a. Gently remove the supernatant by pipetting and resuspend the cell pellets by vortexing and/or pipetting.
11.Add 500 µl of 1000×-diluted Fc block solution to the cells to prevent nonspecific antibody binding. Incubate for 10 min.
12.Centrifuge the tube for 10 min at 400 × g , 4°C.
13.Carefully discard the supernatant, resuspend the cell pellet, and store it on ice for the next step.
14.Prepare an appropriate mix of 200×-diluted CD45 antibody with 1000×-diluted DRAQ5 in cell staining buffer (150 µl/sample). Store the mixture at 4°C in the dark until use.
15.Add 150 µl per sample of the prechilled CD45 antibody/DRAQ5 mix (from step 15) to the cells and incubate 25 min on ice in the dark.
16.Optional : Add 1.5 µl of 10×-diluted TruStain FcX™ PLUS (anti-mouse CD16/32) antibody and incubate it for 20 min on ice.
17.Optional : Add 0.75 µl CD45 antibody and 1.5 µl of 10×-diluted DRAQ5 directly to the cell suspension and incubate 25 min on ice in the dark.
18.Wash the cells by adding 1 ml of cell staining buffer.
19.Centrifuge the samples 10 min at 400 × g , 4°C.
20.Carefully remove the supernatant, resuspend the cells, and repeat the washing step by adding 1 ml of cell staining buffer.
21.Centrifuge the samples again for 10 min at 400 × g , 4°C.
22.Carefully remove the supernatant and resuspend the cells in 400 µl of cell staining buffer.
23.Add Hoechst 33258 to 1 µg/ml final (a 1000× dilution of the 1 mg/ml storage stock solution) to the appropriate samples to differentiate live from dead cells (as indicated in Table 1).
| Samples |
|---|
| 1. Negative/autofluorescence control – unstained cells (from C57BL/6 mouse milk) |
| 2. Mix (CD45 + DRAQ5) + viability staining (Hoechst 33258; from C57BL/6 mouse milk) |
| 3. Mix (CD45 + DRAQ5) + viability staining (Hoechst 33258; from MHC II–EGFP knock-in mouse milk) |
24.Transfer the samples to a 96-well microplate on ice (see Table 1 for specifications).
25.Store the samples on ice in the dark before proceeding to flow cytometry measurements.
Flow cytometry analysis
26.Analyze samples using flow cytometry analysis software, such as FlowJo v10.8.0.
27.After fluorochrome compensation, proceed to manual gating (Fig. 12).
28.For a proper gating strategy, use relevant controls (Fig. 13).
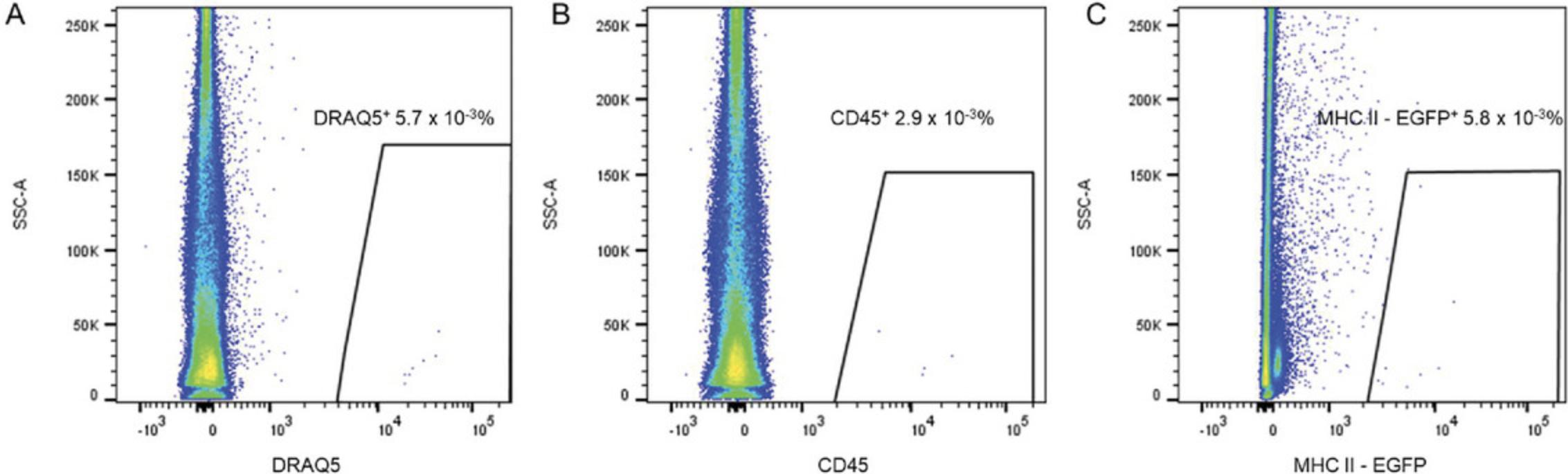
29.After doublet discrimination, plot SSC-A as a function of DRAQ5 to view the DRAQ5+ cells.
30.From the “DRAQ5+” gate, assess live cells by plotting Hoechst as a function of FSC-A.
31.Determine the population of CD45+ cells (plot showing SSC-A as a function of CD45).
32.From the “CD45+ cells” gate, plot the MHC II–EGFP+ cell population.
33.Count the absolute number of MHC II+ cells by recalculating the cell numbers by measuring the sample volume and relate the results to the volume of collected milk.
Support Protocol 1: CELL SURFACE STAINING AND FLOW CYTOMETRY ANALYSIS OF SPLEEN MONONUCLEAR CELLS
Compensation controls, also known as single-stain controls, play a key role in multicolor flow cytometry experiments. These controls are essential for accurately and precisely setting compensation values, particularly when measuring challenging samples such as milk, with high autofluorescence and nonspecific binding to oil droplets. The levels of compensation are commonly determined using splenocytes (spleen mononuclear cells, SMC) derived from the spleen for their complex cellular composition, encompassing various cell types at different activation or developmental stages. The cellular suspension derived from the spleen provides an excellent platform for optimizing the necessary compensation values. Moreover, the spleen is abundant in MHC II+ cells and, as such, ideal for achieving a precise compensation of spectral cross-talks with EGFP. By carefully setting the parameters for cell cytometry using the splenocyte suspension, researchers can ensure an accurate compensation and reliable interpretation of flow cytometry data.
Materials
-
Mice: 8-week-old MHC II–EGFP knock-in (kindly provided by Marianne Boes, University Medical Center Utrecht, Utrecht, The Netherlands) and 8-week-old C57BL/6 female mice (Charles River Laboratories)
-
RPMI 1640 medium supplemented with L-glutamine and phenol red (Thermo Fisher Scientific, cat. no. 21875034), ice cold
-
RPMI 1640/10% FBS (see recipe)
-
Cell staining buffer (see recipe)
-
500×-diluted Fc block solution (see recipe)
-
Single-stain controls (CD45, DRAQ5) for compensation (see recipes)
-
APC/Fire™ 750 anti-mouse CD45 antibody (clone 30-F11, cat. no. 103154, Biolegend)
-
DRAQ5™ Fluorescent Probe Solution (5 mM; Thermo Fisher Scientific, cat. no. 62251)
-
Dissecting instruments (scissors, forceps)
-
1.5-ml microcentrifuge tubes (Biologix, cat. no. 80-1500)
-
2.5-, 20-, 200-, and 1000-µl mechanical pipets
-
50-ml conical centrifuge tubes (Jet Biofil, CFT-011-500)
-
Cell strainer, 40-µm pore size (VWR, cat. no. 734-2760)
-
Plunger from Inject Luer Solo, 2 ml (B. Braun, cat. no. 4606027V) single-use syringe
-
Refrigerated centrifuge (Eppendorf, model no. 5810R) or similar
-
Vortex
-
Thermoblock: Thermomixer Comfort (Eppendorf)
-
96-well microplates (500 µl/well)
-
LSR II flow cytometer (BD Biosciences)
-
FlowJo v10.8.0 analysis software (https://www.flowjo.com/)
Spleen mononuclear cells isolation
1.After collecting milk (Basic Protocol 7), remove the mouse spleen from C57BL/6 and MHC II–EGFP knock-in mice using scissors and forceps. Transfer the spleen to 1.5-ml microcentrifuge tubes containing 1 ml RPMI/10% FBS and keep it on ice until the next step.
2.Prepare 50-ml conical centrifuge tubes and label them accordingly. Place a 40-µl cell strainer on the top of each tube.
3.Transfer the spleen along with 1 ml RPMI/10% FBS onto the cell strainer.
4.Mince the spleen using the flat end of a plunger from a sterile 2-ml syringe.
5.Wash the cell strainer with 10 ml RPMI/10% FBS and then discard it.
6.Prepare new 50-ml conical tubes with new 40-µl cell strainers.
7.Transfer the cell suspension to a new tube and discard the strainer.
8.Fill the cell suspension up to 20 ml with RPMI/10% FBS and resuspend the cells.
9.Repeat steps 1-8 for all samples and keep them on ice for flow cytometry staining.
Cell surface staining
10.Prepare single-stain controls for CD45 and DRAQ5, mix CD45 antibody (200× dilution) with DRAQ5 (1000× dilution) in cell staining buffer, and store these solutions at 4°C in the dark (see recipes in Reagents and Solutions).
11.Transfer 200 µl of the cell suspension isolated from mouse spleens to 1.5-ml microcentrifuge tubes. Repeat this step for all samples, as indicated in Table 2.Keep the cells on ice at all times.
| Samples |
|---|
| 1. Negative/autofluorescence control – unstained cells (SMC from C57BL/6 mouse) |
| 2. Positive control – viability staining (Hoechst 33258+; SMC from C57BL/6 mouse) |
| 3. Positive control – CD45+ (SMC from C57BL/6 mouse) |
| 4. Positive control – DRAQ5+ (SMC from C57BL/6 mouse) |
| 5. Positive control – EGFP+ (SMC from MHC II–EGFP knock-in mouse) |
| 6. Mix (CD45 + DRAQ5) + viability staining (Hoechst 33258; SMC from MHC II–EGFP knock-in mouse) |
12.Add 200 µl of 500×-diluted Fc block solution to each sample and incubate them for 10 min on ice.
13.Centrifuge the samples at 400 × g for 10 min at 4°C, along with the milk samples from Basic Protocol 7, step 11.
14.Discard the supernatant and resuspend the cell pellet by vortexing.
15.Incubate the cells with the mixture of antibodies for 25 min on ice in the dark, together with the milk samples from Basic Protocol 7, step 18.
16.Add 100 µl cell staining buffer to the samples that are not being stained.
17.Optional : During the staining procedure, divide the positive control sample for viability staining (refer to Table 2) in half. Incubate one half (50 µl) in a new 1.5-ml microcentrifuge tube at 70°C for 10 min in a thermoblock. After incubation, add the suspension of dead cells to the original sample.
18.Wash the cells by adding 500 µl cell staining buffer.
19.Centrifuge the samples for 10 min at 400 × g , 4°C, along with the milk samples from Basic Protocol 7, step 19.
20.Aspirate the supernatant, resuspend the cells, and repeat the washing step by adding 500 µl cell staining buffer.
21.Centrifuge the samples again for 10 min at 400 × g , 4°C, along with the milk samples from Basic Protocol 7, step 22.
22.Aspirate the supernatant and resuspend the cells in 200 µl cell staining buffer.
23.Add 1 µg/ml Hoechst 33258 (a 1000× dilution of the 1 mg/ml storage stock solution) to the appropriate samples (as indicated in Table 2).
24.Transfer the samples to the 96-well microplate used for the milk samples in Basic Protocol 7, step 24.Keep the samples on ice.
25.Store the samples on ice in the dark for subsequent flow cytometry measurements.
REAGENTS AND SOLUTIONS
CD11c single-stain control
- 60 μl cell staining buffer (see recipe)
- 0.4 μl PE-Cy™7 Hamster Anti-Mouse CD11c (clone HL3, cat. no. 558079, BD Bioscience)
- Use immediately; do not store
CD19 single-stain control
- 60 μl cell staining buffer (see recipe)
- 1.5 μl APC anti-mouse CD19 antibody (clone 6D5, cat. no. 115512, Biolegend)
- Use immediately; do not store
CD45 single-stain control
- Mix:
- 147.75 μl cell staining buffer (see recipe)
- 0.75 μl APC/Fire™ 750 anti-mouse CD45 antibody (clone 30-F11, cat. no. 103154, Biolegend)
- 1.5 μl of 10×-diluted DRAQ5
- Prepare before starting an experiment.
- Do not store.
Cell staining buffer (D-PBS supplemented with 0.02% gelatin and 0.05% sodium azide)
- 1 L 1× PBS (see recipe)
- 400 μl gelatin from cold water fish skin, 40%-50% in H2O (Sigma-Aldrich, cat. no. G7765-250ML)
- 5 ml 10% sodium azide (Sigma-Aldrich, cat. no. S8032)
- Store up to 6 months at 4°C
Collagenase D, 10 mg/ml
- 100 mg collagenase D (cat. no. COLLD-RO, Roche)
- 10 ml HBSS without Ca+ or Mg+
- Store in 200-µl aliquots up to 1 year at –20°C
CUBIC 1 clearing solution
Dissolve 125 g urea (Penta Chemicals, cat. no. 21420-31000) in 175 ml dH2O and then add 124 ml of N,N,N′,N′ -tetrakis(2-hydroxypropyl)ethylenediamine (4NTEA; Sigma-Aldrich, cat. no. 122262) and 70 ml Triton X-100 (Sigma-Aldrich, cat no. T8787). Store up to 6 months at room temperature. Discard if the solution smells of ammonia.
CUBIC 2 clearing solution
First, dissolve 62.5 g urea in 65 ml of dH2O; then add and dissolve 125 g sucrose; and, finally, add 22.3 ml triethanolamine (TEA; Sigma-Aldrich, cat. no. 90279) and 190 μl Triton X-100 (Sigma-Aldrich, cat no. T8787). Store up to 2 weeks at room temperature. Discard if the solution shows crystals.
CUBIC wash solution
Dissolve 2.5 g bovine serum albumin (BSA; Sigma-Aldrich, cat. no. A7906)in 500 ml dH2O. Add 500 μl of 10% sodium azide (Sigma-Aldrich, cat. no. S8032) and 500 μl Triton X-100 (Sigma-Aldrich, cat no. T8787). Store up to 6 months at 4°C. Discard if precipitation occurs.
DMEM culture medium
- 12.775 ml DMEM medium supplemented with L-glutamine (Life Technologies, cat. no. 21063029)
- 375 μl 1.0 M HEPES buffer (Thermo Fisher Scientific, cat. no. J16924.AE)
- 1.5 ml fetal bovine serum (FBS; Gibco™, Thermo Fisher Scientific, cat. no. 10270106)
- 150 μl 1.0 mg/ml murine GM-CSF (see recipe)
- 150 μl 1.0 mg/ml murine IL-4 (see recipe)
- Store up to 1 month at 4°C
DMEM culture medium + OVA
To 15 ml DMEM culture medium (see recipe), add 4 μg ovalbumin (Sigma-Aldrich, cat. no. S7951, or Roche Molecular Biochemicals) or the equivalent amount of OVA in the form of crude egg white (extracted from fresh eggs; crude egg white contains 54% OVA).
Do not store.
DMEM/10% FBS
- 450 ml DMEM medium supplemented with L-glutamine (Life Technologies, Grand Island, NY)
- 50 ml fetal bovine serum (FBS; Gibco™, Thermo Fisher Scientific, cat. no. 10270106)
- Do not store.
DRAQ5, 10× diluted
- 9 μl cell staining buffer (see recipe)
- 1 μl DRAQ5™ Fluorescent Probe Solution (5 mM; Thermo Fisher Scientific, cat. no. 62251)
- Prepare fresh before starting an experiment.
- Do not store.
DRAQ5 single-stain control, 1000× diluted
- 150 μl cell staining buffer (see recipe)
- 0.3 μl DRAQ5™ Fluorescent Probe Solution (5 mM; Thermo Fisher Scientific, cat. no. 62251)
- Do not store.
3% FBS RPMI
- 9.7 ml RPMI 1640 medium supplemented with L-glutamine and phenol red (Thermo Fisher Scientific, cat. no. 21875034)
- 0.3 ml fetal bovine serum (FBS; Gibco™, Thermo Fisher Scientific, cat. no. 10270106)
- Do not store.
Fc block solutions
- 10×-diluted :
- 9 μl cell staining buffer (see recipe)
- 1 μl TruStain FcX™ PLUS (anti-mouse CD16/32) antibody (Biolegend, cat. no. 165604)
- 500×-diluted :
- 2 ml cell staining buffer (see recipe)
- 4 μl TruStain FcX™ PLUS (anti-mouse CD16/32) antibody (Biolegend, cat. no. 165604)
- 1000×-diluted :
- 1 ml cell staining buffer
- 1 μl TruStain FcX™ PLUS (anti-mouse CD16/32) antibody (Biolegend, cat. no. 165604)
- Prepare fresh before starting an experiment.
GM-CSF, murine, 1.0 mg/ml
Reconstitute GM-CSF (PeproTech, Rocky Hill, NJ) at 1.0 mg/ml concentration in sterile distilled water immediately before the experiment. Only short-term storage (1 week) at 4°C is recommended; for extended storage, the protein solution should be stored with a carrier protein (e.g., 0.1% BSA) in working aliquots at –20°C to –80°C.
Hoechst 33258, 1 mg/ml
- 1 ml distilled H2O
- 1 mg Hoechst 33258 (Thermo Fisher Scientific, cat. no. H21491)
IL-4, murine, 1.0 mg/ml
Reconstitute IL4 (Roche Molecular Biochemicals, Somerville, NJ) in sterile distilled water at 1.0 mg/ml concentration immediately before the experiment. Only short-term (1 week) storage at 4°C is recommended. For extended storage, the protein solution should be stored with a carrier protein (e.g., 0.1% BSA) in working aliquots at –20°C to –80°C.
MHC II single-stain control
- 60 μl cell staining buffer (see recipe)
- 1000× diluted APC/cyanine 7 anti-mouse I-A/I-E antibody (clone M5/114.15.2, cat. no. 107628, Biolegend)
Ovalbumin, 10 mg/ml
Add 1 ml room-temperature water to 10 mg ovalbumin (Sigma-Aldrich). Do not store.
Paraformaldehyde, 3.8%
- 1.9 g paraformaldehyde
- Distilled water to 500 ml
- Store up to 6 months at 4°C
PBS, 1×
- 900 ml distilled H2O
- 100 ml 10× D-PBS (see recipe)
- Store up to 6 months at 4°C
PBS, 10×
- 1 L distilled H2O
- 96 g Dulbecco's Phosphate Buffered Saline (D-PBS; Sigma-Aldrich, D5652), pH 7.2-7.8
PBS/2.5% FBS solution
- 975 μl 1× PBS (see recipe)
- 25 μl fetal bovine serum (FBS; Gibco™, Thermo Fisher Scientific, cat. no. 10270106)
- Use immediately; do not store
RPMI 1640/10% FBS
- 45 ml phenol-red-free RPMI 1640 medium supplemented with L-glutamine (Thermo Fisher Scientific, cat. no. 21875034)
- 5 ml fetal bovine serum (FBS; Gibco™, Thermo Fisher Scientific, cat. no. 10270106)
- Store up to 1 week at 4°C
Sodium azide, 10%
- 10 ml distilled H2O
- 1 g sodium azide (Sigma-Aldrich, cat. no. S8032)
- Store at room temperature; discard if precipitates occur
Sytox Blue, 200×
- 0.5 μl SYTOX™ Blue Dead Cell Stain (cat. no. S34857, Invitrogen)
- 99.5 μl 1× PBS (see recipe)
- Prepare fresh before starting the experiment
COMMENTARY
Background Information
Dendritic cells (DCs) are crucial regulators of immune reactions and belong to the category of professional antigen-presenting cells (APCs) typically expressing MHC II (which can be directly visualized in vivo using flow cytometry or fluorescence microscopy/stereomicroscopy when the MHC II–EGFP knock in mouse is utilized). DCs have the unique ability to integrate danger and “stranger” molecular patterns. DC-mediated T cell activation plays a pivotal role in immune reactivity. The MHC II–EGFP knock-in mouse serves as a valuable source of DCs, which are obtained either directly as primary cells from relevant tissues or through differentiation from hematopoietic stem cells, specifically those derived from the bone marrow. These DCs can be loaded with a model antigen (e.g., ovalbumin) to examine their interaction with cognate T cells expressing the appropriate antigen-specific T cell receptor (OT II).
High-resolution live-cell imaging microscopes, combined with the ability to perform long-term imaging without causing significant photodamage (using techniques such as spinning-disc technology), enable us to study of immunological synapse formation and to characterize the fluorescently labeled MHC II loading compartment under various conditions. By acquiring multiple videomicroscopy datasets depicting diverse antigen-presenting scenarios, such as different types of presented material and the specificity of the cognate T cell receptor, we can accurately quantify endosomal tubulation towards the immune synapse, including measuring the length and directionality of the tubules.
Typically, flow cytometry involves staining surface markers, combined in some experimental setups by cell fixation, permeabilization, and intracellular marker staining. However, MHC II–EGFP knock-in mouse offers an alternative approach. By utilizing the EGFP tag on MHC II (Boes et al., 2002), we can directly visualize intracellular MHC II without fixation, permeabilization, or additional intracellular staining steps. Comparing the staining of surface MHC II and EGFP fluorescence, we are able to compare intracellular and extracellular MHC II expression levels, which could exceptionally reflect differentiation/activation status of the particular cell population.
The MHC II–EGFP knock-in mouse model enables direct visualization and identification the various lymphoid tissues thanks to its specific fluorescence signal. As such, this mouse model is an ideal tool for accurately dissecting lymphoid organs, e.g., individual Peyer's patches (PPs), or precisely separating individual mesenteric lymph nodes (MLN). This separation is crucial for interpreting immune phenomena relevant to the gastrointestinal tract (GIT) along its intestinal gradient. Each mesenteric lymph node drains specific segments of the gastrointestinal tract, resulting in distinct immunocharacteristics of the respective subunit of the complex (Houston et al., 2015). Therefore, the ability to precisely dissect and analyze MLNs using MHC II–EGFP knock-in mouse is of the utmost importance for gaining a comprehensive understanding of GIT-related immune processes.
Transgenic fluorescent mouse models could help to revise handed down morphological/histological facts. Traditionally, for example, the total counts of villi and individual lymphoid formations have primarily been estimated by extrapolating numbers from sectioned samples. However, direct in situ visualization provides a more comprehensive perspective and enables a detailed examination of specific locations, all without potential artifacts introduced by tissue processing. This approach has the advantage of both providing us with an overall view and with the ability to conduct precise investigations at specific sites.
Recently, revolutionary imaging techniques have been developed that open up new possibilities for imaging complex histological contexts. Particularly suitable is light-sheet fluorescence microscopy (LSFM), which enables support imaging of large 3D samples by positioning the detection plane perpendicular to the illumination plane and illuminating the sample from two sides. However, when imaging tissue samples, lipids, pigments, and other light-scattering molecules hinder deeper light penetration. Therefore, these molecules must be removed for clearer imaging. Various clearing methods are currently available, employing different principles, such as hydrogel polymerization, organic solvents, or diffusion (Susaki et al., 2014). In Clear, Unobstructed Brain/Body Imaging Cocktails and Computational Analysis (CUBIC), hyperhydration and slight tissue expansion are followed by the removal of lipids and light- scattering using strong detergents (Susaki et al., 2015). Subsequently, the refractive index of the tissue is matched with a second solution that easily penetrates the tissue, has a refractive index compatible with the proteins, and is suitable for the microscope, objective, and chamber used in this protocol.
The only naturally fully transparent vertebrate morphological structure is eye (the structures involving visual path, including cornea), which could be directly visualized using LSFM. The diverse immune cells present in the cornea play a key role in its defense against pathogens (Foulsham et al., 2018). However, there is limited information available regarding the detailed characterization of these cells. In previous literature, murine corneal cells were visualized similarly to ours, in the direction from the corneal center to the periphery, using EGFP but under the control of CD11c promoter. CD11c**+** LC-like cells present in the central part of the cornea do not express MHC II proteins, which may lead to different results when quantifying the cells (Knickelbein et al., 2009). With our fluorescent MHC II EGFP model, we can differentiate two types of cells, namely LC-like cells and MΦ-like cells, based on their distinct cell shapes. In line with previous research, we demonstrate that cell density is highest in the peripheral region of the cornea and gradually decreases towards the center (Knickelbein et al., 2009).
The immature neonatal adaptive immune system lacks antigen education and relies on maternal assistance for infant immunity. Maternal immune cells are transferred through the milk, which plays a key role in the transgenerational transfer of maternal immunity (Darby et al., 2019). To understand these processes accurately, immune cell populations must be precisely characterized by gathering quantitative data. Collecting sufficient amounts of mouse milk for subsequent analysis is far more challenging than collecting human milk. To overcome this difficulty, oxytocin injection is typically used to stimulate milk production (Depeters & Hovey, 2009; Gómez-Gallego et al., 2014; Willingham et al., 2014). However, our protocol describes a method for collecting a sufficient volume of milk without using oxytocin and, hence, without affecting the results.
Detecting cells in the milk poses challenges given its composition–autofluorescent lipid droplets, cell debris, apoptotic and dead cells, which all have a considerable impact on data evaluation. Moreover, mouse milk has a higher (20-30%) fat content (Görs et al., 2009) than human milk (3%-5%; Jenness, 1979), which further complicates the analysis. Traditional centrifugation alone is insufficient for separating lipid droplets from cells. Flow cytometry analysis, plotting the SSC-A by FSC-A graph, cannot accurately identify cells based on size and granularity among other particles. Various approaches have been attempted, such as Nile red staining of lipid droplets (Keller et al., 2019) and fluorochrome-labeled CD45 staining (Hassiotou et al., 2013; Keller et al., 2019). However, these methods have not provided a correct and reliable cell identification.
Thus far, standard protocols have failed to differentiate between noncellular (e.g., oil droplets) and cellular milk content, leading to irreproducible results due to the aforementioned discrepancies. To address these challenges, Keller et al. introduced a novel and reliable flow-cytometry-based approach to quantify cells in human milk, utilizing DRAQ5 and Sytox Blue DNA staining for proper cell identification (Keller et al., 2019).
In this protocol, we optimized the approach established by Keller et al. and performed cell content analysis of mouse milk based on centrifugation and a staining protocol using DRAQ5 and Hoechst 33258 for DNA staining. Additionally, we introduced the MHC II–EGFP mouse model into this protocol as an antibody-label-free system to prevent incorrect cell identification caused by antibody binding to noncellular content. Overall, we have introduced an easy-to-use, non-invasive method for collecting mouse milk and established a reliable and reproducible flow cytometry analysis for quantifying the absolute number of cells in mouse milk.
Critical Parameters
DCs are extremely sensitive to the contamination, and therefore the purity of the cultivation medium for in vitro experiments is essential. The cultivation medium must be free of pathogen-associated molecular patterns (PAMPS), such as LPS, to allow reproducible assessment of the differentiation status without excessive activation of developing dendritic cells. The visualization techniques should be performed without measurable photodamage to avoid inducing cellular pathology on both sides of the immunological synapse—APC and T cell. For quantification purposes, all experiments must be performed with blinding to avoid bias during the interpretation. Alternatively, unbiased data-mining algorithms may be applied by a bioinformatician to prevent biased results.
Achieving optimal antibody staining for both flow cytometry and microscopy necessitates the use of reagents kept at ice-cold temperatures and performance of all procedures at 4°C, while minimizing exposure to light. It is advisable to include sodium azide in the staining buffer to aid in preventing the modulation and internalization of surface markers, as well as the capping of antigens and antibodies. These processes can potentially diminish the intensity of fluorescence. Before commencing the experiment, it is essential to titrate all antibodies to determine the optimal dilutions.
When designing a flow cytometry panel comprising multiple colors, it is recommended that antibodies labeled with bright fluorochromes (fluorophore brightness index of 4 or 5) be used for the detection of low-abundance markers. Conversely, dim fluorochromes are suggested for highly expressed antigens. The selection of fluorochromes must be compatible with the flow cytometry instrument, taking into account factors such as the instrument's configuration, including laser and filters. It is advisable to employ fluorochromes with no fluorescence spillover, thereby eliminating the need for single-stain controls.
In experiments requiring time-sensitive procedures, such as the characterization of live cells by flow cytometry, tissue dissection must be shortened as much as possible, which requires using a stereomicroscope equipped with precise 3D vision and a lens featuring a long working distance.
For accurate analysis of the lymphoid organs, a precise excision must be combined with a thorough washing of the samples. This step is particularly crucial for PP as the cellular lifespan after euthanasia is limited and could vary in different cell populations. Epithelial cells are the first to die, followed by myeloid cells, whereas lymphocytes, specifically T lymphocytes, have the longest survival time. Consequently, T lymphocytes contribute the most to the final cell population, significantly affecting the measurements and introducing complexity to the interpretation of results.
Acquiring a high-quality image is crucial for clearly differentiating the positive signal from background autofluorescence. For example, when quantifying villi, the intestine must be thoroughly washed to remove any food residues and large mucus accumulations. Scanning the entire intestine requires immersing it in PBS while maintaining a relaxed state to avoid deformation, such as stretching. To facilitate the subsequent fusion of specific views, ensure that there is at least 20% overlap between them.
During the staining process using DRAQ5 to label nuclei for LSFM, the dye gradually binds from the surface to the center of the sample. The decrease in fluorescence intensity is further amplified by the absorption of excitation and emission light within the tissue. Therefore, it is crucial to ensure uniform staining throughout the sample, which requires determining the optimal dye concentration suitable for the specific tissue type and sample volume. However, this assessment is challenging due to adipose tissue, which binds a significant amount of dye, resulting in insufficient staining of the target tissue.
The clearing solutions should be prepared in advance because the individual components take several hours to dissolve completely. For the modified CUBIC 2 solution, following the specific order of steps is essential to prevent rapid cooling of the solution upon dissolution of urea, which can cause the concentrated sucrose solution to solidify. When matching the refractive index, the same batch of CUBIC 2 solution must be used for sample preparation and sensing. Using a solution that is older than ∼2 weeks should be avoided to prevent crystallization during scanning. Before imaging, the solution must be checked for any small crystals or bubbles.
When attaching the samples to the syringe inside LSFM equipment, caution should be exercised to prevent the glue from coming into contact with the sample. Before imaging, the exact refractive index must be measured at the imaging temperature and the correction ring adjusted on the microscope objective accordingly. Maintaining a consistent temperature throughout the imaging process is preferable. Applying 3D deconvolution may help to enhance the resolution, but potential artifacts may be associated with this method. In particular, if the image quality varies significantly within the sample, 3D deconvolution may complicate the quantification.
To accurately characterize the native eyeball without fixation using LSFM, the fluorescence signal must be visualized as soon as possible to prevent cell death and EGFP degradation. When quantifying only the corneal antigen-presenting cells (APCs), the border with the limbus must be clearly defined, as illustrated in Figure 11. Care should be taken to avoid optical distortion when imaging the eyeball too deeply, reaching the lens with its unique optical conditions, such as refractive index. To prevent this, keep in mind that the mouse cornea has a thickness of ∼100 μm.
Several factors can affect the volume of mouse milk collected, including the number of pups, the number of days after birth, and general stress conditions. The reliability of subsequent analysis may be affected by the small volumes. The pellet fraction should be visible after the centrifugation step.
Leukocytes should be centrifuged at 300 × g. However, to properly separate cream and skim milk from the pellet, centrifugation at a higher relative centrifugal force (RCF) is necessary. Based on our previous results, we found that adding 12% cream compounds to a suspension of spleen mononuclear cells, which partially mimics the average fat content of mouse milk (Görs et al., 2009), protects cells from cell death at higher centrifugation speeds (600 g ; data not shown). Therefore, all centrifugation steps are performed at 400 × g as a compromise between obtaining the highest number of cells and maintaining their viability. After the centrifugation, the supernatant should be carefully aspirated to avoid disturbing the pellet fraction and to obtain a sufficient amount of cells for flow cytometry analysis.
The emission spectrum of DRAQ5 should be properly checked for the emission overlay. For a positive control of cell viability, optional step 17 in Support Protocol 1 could be used. To accurately measure the milk volume, samples should be measured with a BD High Throughput Sampler (HTS) for a precise cell quantification and recalculated to determine the sample volume. But the whole staining procedure should be performed in microcentrifugation tubes to allow better handling of the sample during supernatant aspiration steps to minimize cell loss. The samples should also be prepared for measurement in a higher volume of cell staining buffer (400 μl in our case) to reduce the concentration of non-cellular milk components, which can impact the analysis.
Troubleshooting
It takes ∼1 week to differentiate DCs from mouse bone marrow stem cells. If the DCs at day 5-6 after in vitro cultivation are highly positive for surface MHC II (easily detectable using flow cytometry or microscopy detecting the EGFP fluorescence), the cultivation medium is most likely contaminated with LPS, and the induction of the immunological synapse and characterization of the corresponding tubulation is not reproducible. Thus, identify an LPS-free batch of medium for further experimentation. If cells change shape during videomicroscopy (typically, round up and potentially detach from the surface), modify the imaging conditions (minimize the intensity of the excitatory light and exposure time) to gather the most physiologically relevant data.
In instances where individual nodes are smaller in size (when preparing the lymphoid-organ-derived leukocytes), the visibility of the corresponding pellet may be limited. It is crucial to ensure proper separation of adipose tissue residues to prevent cell loss during the washing process. Therefore, it is advisable to follow the procedure outlined in Basic Protocol 3. The volume of the staining mix is adjusted considering the potential significant variation in cell numbers among individual nodes, as using a lower volume could adversely affect the staining of larger samples. To maintain the accuracy of the gating strategy (Basic Protocol 2), it is recommended that additional markers be incorporated to identify distinct subpopulations of dendritic cells (DCs). These markers should effectively demonstrate that the populations obtained through the gating strategy, which relies on both intracellular and extracellular MHC II, genuinely represent different subpopulations of DCs. This approach should also be applied to other cell types where the biologically relevant translocation of MHC II from intracellular stores (MHC II loading compartment) occurs. To confirm correct gating, it is also advisable to back-gate given populations for other markers used in the staining process.
Considerable variation in lymphoid organs among mice makes it challenging to identify all lymph nodes in every mouse. Moreover, the number and size of lymph nodes within the complexes differ among individual mice.
Thoroughly removing residual adipose tissue is crucial for an accurate flow cytometry and 3D microscopy analysis. Fat droplets can significantly complicate working with single-cell suspensions and reduce sample transparency, even with clearing methods. Additionally, fat droplets can bind hydrophobic dyes and absorb dead cells or fragments. However, adipose tissue can be easily distinguished from immune tissue due to its minimal autofluorescence, and its residues can be readily removed as well.
The last mesenteric lymph node (MLN), which drains the colon, is often separated from the rest of the complex and may not be immediately visible. In such cases, a thorough search of the mesentery is necessary. Typically, the node is located to the right of the remaining complex and may be concealed by intestinal folds.
For flow cytometry experiments, the processing time can significantly impact cell counts and the proportion of individual populations. Therefore, when harvesting both MLN and PPs from the same animal, the protocols should be divided between two researchers. One researcher can focus on splitting individual nodes, whereas the other can extract individual PP.
To effectively identify individual villi using fluorescence stereomicroscopy, the intestine must be thoroughly washed to minimize the interference of high autofluorescence caused by food remains. The autofluorescence of the mucus surrounding the villi can also pose challenges. If washing out the mucus is not feasible, an alternative approach consists of carefully mechanically cutting the intestine and removing the mucus from the luminal side. Automatic quantification is not viable due to variations in autofluorescence intensity and overall intestinal transparency and to the considerable variation in the positive signal of the villi at different locations within the intestine.
The distribution of scattered lymphoid tissue, including cryptopatches and isolated lymphoid follicles, throughout the small intestine, along with the presence of villi in areas with highly diverse fluorescence intensities, hinders simple, automatic SLT segmentation. However, recent advancements in machine learning have allowed us to initiate training for segmentation using Ilastik software albeit not yet at the level of manual quantitation. Manual alignment is needed to avoid artifacts such as follicle or SLT duplication that may occur with inaccurate automatic alignment. Therefore, any automatic fusion and alignment results must be carefully verified.
Analyzing individual PPs is comparatively easier because they are larger and show intense fluorescence. The risk of missing any of them is negligible. To minimize the chance of overlooking PPs, we can wrap the intestine around forceps to visualize all sides simultaneously. By holding the end of the intestine with a second pair of forceps and pulling the entire intestine around the first forceps, we can inspect the entire surface of the intestine without missing any PPs. Even when performing automatic segmentation of PPs, accurately segmenting the entire area of all PPs without artifacts can be challenging. Differentiating individual follicles poses an even greater challenge.
Performing 3D deconvolution is the best approach to enhance resolution, but potential artifacts may arise from this method. More specifically, if there is significant variability in image quality across the sample, 3D deconvolution may even complicate the quantification process. Switching between 2D and 3D views may help to more easily distinguish individual cells from each other.
In cases where scanning using LSFM extends beyond 24 hr, CUBIC 2 may gradually evaporate, degrading its optical properties. To prevent evaporation, the solution should be periodically changed and replenished with fresh CUBIC 2 using a filling syringe.
Because of the heterogeneous structure of the sample and the potential formation of bubbles, it is necessary to rotate the sample for LSFM to identify the optimal direction of illumination and detection. Adjusting the right and left illuminations and subsequently fusing both views may enhance the contrast and help to optimize the image.
The tissues must be generally handled with care to avoid perforating or exerting excessive pressure especially when extracting the eyes. Furthermore, specimens must be thoroughly washed to remove any blood and eliminate excess tissue and hairs that may cause artifacts during LSFM scanning. Once collected, the eyes should be imaged promptly. Although fixation is an option, it can increase background autofluorescence and make cell identification more challenging. Preserving the optic nerve can make it easier to handle the sample. When attaching the sample to the light-sheet microscope holder, only a small amount of glue should be used, and caution should be exercised to prevent the glue from spreading over the eye, as this hinders imaging.
The volume of mouse milk varies individually depending on the factors and circumstances mentioned above. We collected milk on the 5th day after parturition, based on our experience. An adequate volume of milk must be collected to ensure a suitable number of cells for subsequent analysis. If low volumes of milk are collected, the number of cells will be very low, making the analysis challenging. Therefore, carefully aspirate the supernatant (skim milk layer) to avoid disturbing the cell pellet and to maintain a sufficient number of cells.
During the staining procedure, the samples must be protected from direct light to prevent any impact on fluorescence intensity. When samples for compensation have lower expression of a marker than the examined samples, compensation beads must be used to ensure accurate compensation.
Understanding Results
Once the physiological immune synapse is formed, dendritic cells tend to form tubular fluorescent protrusions (due to MHC II positivity) toward the T cell. This phenomenon is clearly distinguishable from other forms of vesicular trafficking.
The ratio between surface/overall/intracellular MHC II expression reflects the activation/differentiation status of the characterized cell population. The flow cytometry results should clearly distinguish the intracellular and surface MHC II expression and possible relative quantification expressed as mean fluorescence intensity. Using this strategy, the increase in the ratio between the mean fluorescence intensity (MFI) of EGFP and APC-Cy7-conjugated anti-MHC II antibody should be linear and stable in cells primarily with MHC II extracellularly. Cells with a higher MFI of EGFP than MHC II also display intracellular MHC II (we term them “turnover,” as some of the MHC II molecules are only being retained intracellularly), which cannot be stained with the anti-MHC-II antibody. A population of cells negative for MHC II and positive only for EGFP shows that there is no surface MHC II expression (Fig. 14)
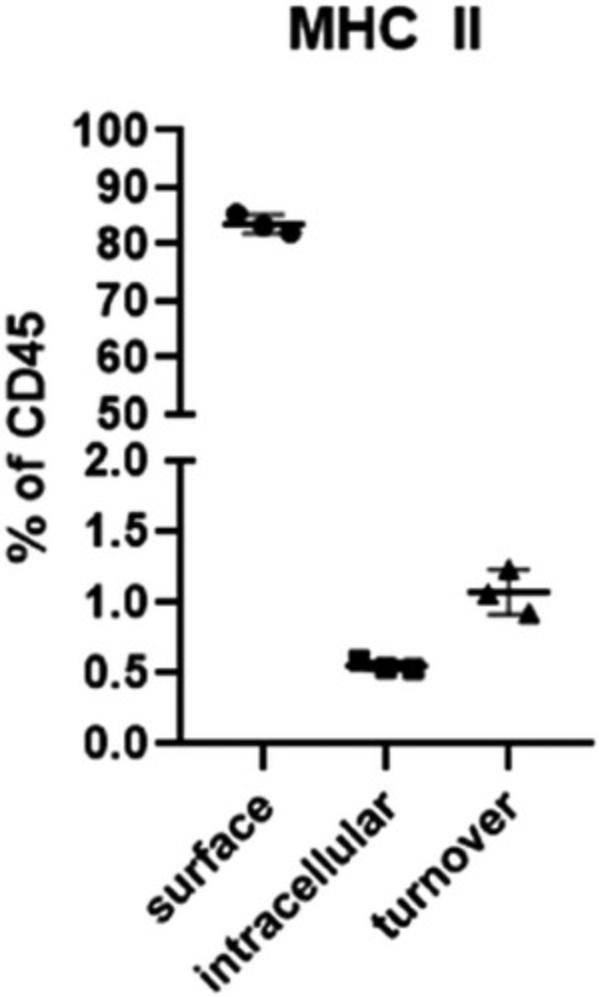
By identifying lymph nodes in multiple mice, we were able to observe significant variability among individuals. When examining lymph nodes on the skin, we discovered variability in size, but not in number. On the neck, however, lymph nodes varied both in size and number. Furthermore, we noted an apparent asymmetry between the left and right sides of the neck. Mesenteric and caudal mesenteric lymph nodes were easily found in every mouse, whereas sciatic lymph nodes were identified only in a single mouse. The most substantial variability among individual mice in terms of lymph node numbers and significant left-right asymmetry were found in lymphoid complexes in the peritoneal and the thoracic cavity surrounding pancreas and trachea. In contrast to Van den Broeck et al. (2006), we were unable to identify all lymph nodes in the peritoneal and thoracic cavities.
When an appropriate illumination wavelength that optimally excites enhanced green fluorescent protein (EGFP) is used, the fluorescence signal emitted by lymphoid organs will be significantly higher than the background fluorescence of other tissues (green autofluorescence). To accurately identify individual lymph nodes or lymph node complexes, we recommend referring to corresponding figures and schemes provided as a suitable guide.
Using the fluorescence stereomicroscopy, the average number of both MLNs and PPs in the female MHC II–EGFP mouse is ∼7 (Pačes et al., 2022)
Individual villi can be easily identified by the intensified fluorescence signal at their center, which corresponds to the presence of antigen-presenting cells (APCs) in the lamina propria of their lumen. The scattered lymphoid tissue is also distinguishable as bright perivascular dots found across the length of the small intestine (∼500 units per gastrointestinal tract; Pačes et al., 2022). PPs, on the other hand, are readily recognizable for their significant size and strong fluorescence signal, typically ranging from 6 to 9 per small intestine.
Using LSFM, there are on average 422 APCs per villus in the proximal jejunum and 200 APCs per villus in the distal ileum (Pačes et al., 2022). An average of 672 cells were counted in each SILT follicle (Pačes et al., 2022). The data also enable us to analyze the shape and measure the dimensions of the individual structures, as shown in Figures 8B-G and 10A and B.
When utilizing LSFM, an appropriate illumination wavelength that optimally excites EGFP, the fluorescence signal of the corneal APCs becomes clearly visible. We quantified the immune cell populations in the cornea through manual analysis of light-sheet microscope data. The cell counts of the samples were converted into counts per square millimeter (mm2). Our experiments consistently demonstrated a significantly higher number of MΦ-like cells (35 ± 7) per mm2 than LC-like cells (12 ± 4) per mm2 (Pačes et al., 2022).
In our experiments, the litters typically consisted of 6-9 pups (Fig. 15), although 10 pups in a litter is not uncommon. For milk collection, we selected the 5th day after parturition as the beginning of the collection period for obtaining sufficient volume of the mature milk. The milking procedure yielded ∼50-140 µl of milk (Fig. 16B). Muranishi et al. (2016) conducted milk collection on the tenth and eleventh day after parturition and reported the best results, with an average of 130.5 ± 25.1 µl (n = 20).
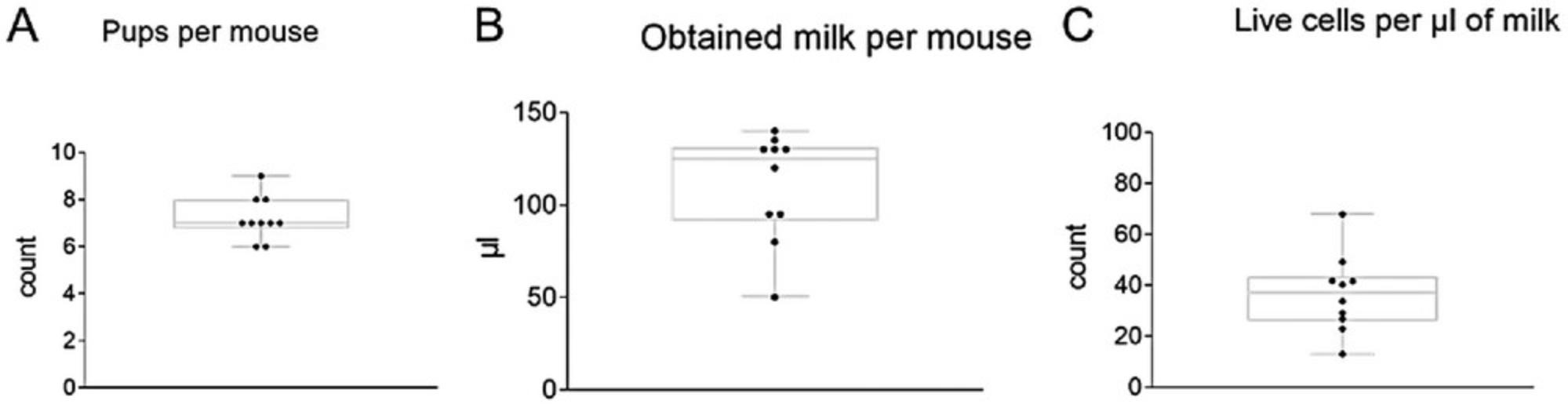
Through our quantitative flow cytometry analysis, we were able to detect a range of 12.94-67.88 live cells per 1 µl of mouse milk (average: 36.62 ± 15.32/µl; Fig. 15). We found that, on average, 85.35 ± 4.94% of live cells were CD45**+** cells, and their absolute numbers ranged from 11.89 to 58.12/µl (average: 31.37 ± 13.58 CD45**+** /µl) in the milk. Using this approach, we were able to identify ∼25.33 ± 3.39% of MHC II**+** cells (out of CD45**+** cells), with an absolute cell number ranging from 2.62 to 13.45 MHC II**+** cells/µl (average: 7.99 ± 3.65 MHC II**+** cells/µl) in mouse milk (Fig. 16A-D).
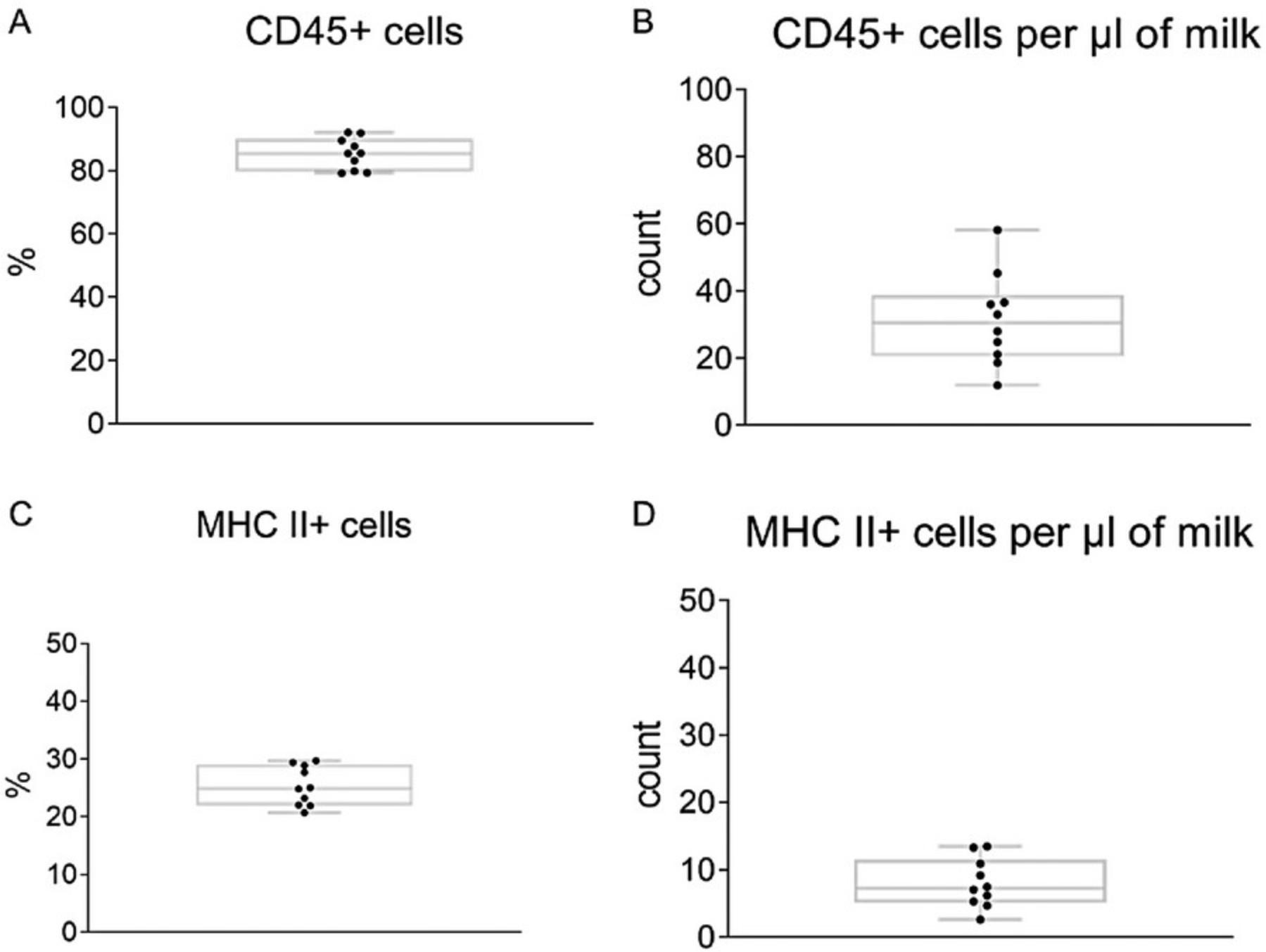
Time Considerations
For the characterization of the antigen-specific tubulation of the intracellular MHC II positive compartment towards immune synapse, 1 h is needed for the preparation of the bone marrow cell suspension, including plating into 6-well plates, and 5-6 days for the differentiation of the dendritic cells in vitro. At least one full day of live-cell microscopy using the heated cultivation chamber (optimally also controlling the atmospheric CO2 concentration to 5%) is needed. The times should be multiplied when applying more conditions (different antigenic peptides in combination with various TCR specificities.
The process of identifying all lymphoid organs typically takes 30-60 min per mouse. However, with practice, this identification may be shortened to ∼30 min.
It takes 25 min to dissect the MLN complex, divide it into individual nodes, and precisely remove all remaining fatty tissue. Individual Peyer's patches can be dissected within 15 min after cervical dislocation.
It takes 25 min to dissect the MLN complex, divide it into individual nodes, and precisely remove all remaining fatty tissue. Digestion, with washing and filtration, takes ∼70 min. Antibody staining including washing steps and subsequent viability staining takes ∼40 min. The whole process of sample preparation for flow cytometry takes 2.5 hr. However, time for preparing dye solutions must also be taken into consideration.
For the quantification of intestinal villi, dissecting one mouse takes ∼30 min and washing the sample ∼20 min. Imaging requires ∼30 min, and manual segmentation takes ∼1 hr.
For the quantification of scattered intestinal lymphoid tissue (SILT), dissecting one mouse takes ∼30 min. Imaging requires ∼2 hr and manual alignment takes ∼1.5 hr. Manual segmentation takes ∼2 hr.
For the measurement of PPs and their subunits, dissecting one mouse takes ∼30 min. Imaging requires ∼30 min and manual segmentation takes ∼1 hr.
For the measurement of the intestinal surface, dissecting one mouse takes ∼30 min. Imaging requires ∼2 min; manual alignment, 1.5 hr; and manual measurement, 30 min or more.
Preparing the samples for LSFM, dissection is followed by a 1-day fixation and subsequent 7-day clearing. Washing can easily be performed in 1 day, and the sample will then be stained by nuclear staining for 1 week. The refractive index must be matched for at least 4 days. Imaging takes ∼10 hr per mouse. Due to the time required for instrument setting and the final scanning adjustment, more samples should be imaged in 2-3 consecutive days. Image processing and 3D deconvolution requires 3 hr per 3D stack, which means ∼18 hr per mouse. Manual segmentation takes ∼2 hr per 3D stack, or 12 hr per mouse.
The duration of LSFM imaging may vary with the number of samples and the length of the microscope scan. The sampling process, including cleaning and rinsing, typically takes ∼30 min. The scanning time is affected by factors such as settings, area size, number of color channels, and z -stack thickness. On average, imaging one eye takes ∼2 hr. The time needed for manual analysis depends on the sample size and on the operator experience. Counting the cells in one sample usually takes ∼30 min.
Characterization of the cellular composition of the mother's milk can take ∼7 hr to complete, but the actual duration may vary with the number of mice/samples. With two MHC II–EGFP knock-in mouse dams and 9-10 samples for flow cytometry, the time breakdown is as follows:
The separation of dams from their pups will take ∼3 hr. Milk collection requires ∼1 hr.
Sample preparation and antibody staining take ∼2 hr. Flow cytometry takes ∼1 hr.
These procedures may become shorter as the operator gains experience, so the overall time may decrease accordingly.
Acknowledgments
The authors acknowledge the support by the Grant Agency of Charles University in Prague, No. 1250120.
Author Contributions
Jan Pačes : Conceptualization; data curation; formal analysis; investigation; methodology; project administration; software; validation; visualization; writing—original draft; writing—review and editing. Valéria Grobárová : Conceptualization; data curation; formal analysis; investigation; methodology; project administration; software; validation; writing—original draft. Zdeněk Zadražil : Data curation; formal analysis; writing—original draft. Karolina Knížková : Conceptualization; data curation; formal analysis; investigation; methodology; software; validation; visualization; writing—original draft. Nikola Malinská : Methodology; visualization; writing—original draft. Liliana Tušková : Data curation; formal analysis; investigation; methodology. Marianne Boes : Conceptualization; investigation; methodology; project administration; supervision; validation; writing—original draft. Jan Černý : Conceptualization; funding acquisition; project administration; resources; writing—original draft.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Litrature Cited
- Awade, A. C. (1996). On hen egg fractionation: Applications of liquid chromatography to the isolation and the purification of hen egg white and egg yolk proteins. Zeitschrift Fur Lebensmittel-Untersuchung Und -Forschung , 202(1), 1–14. https://doi.org/10.1007/BF01229676
- Bertho, N., Cerny, J., Kim, Y.-M., Fiebiger, E., Ploegh, H., & Boes, M. (2003). Requirements for T cell-polarized tubulation of class II+ compartments in dendritic cells. Journal of Immunology , 171(11), 5689–5696. https://doi.org/10.4049/jimmunol.171.11.5689
- Boes, M., Cerny, J., Massol, R., Op Den Brouw, M., Kirchhausen, T., Chen, J., & Ploegh, H. L. (2002). T-cell engagement of dendritic cells rapidly rearranges MHC class II transport. Nature , 418(6901), 983–988. https://doi.org/10.1038/nature01004
- Cristofalo, E. A., Schanler, R. J., Blanco, C. L., Sullivan, S., Trawoeger, R., Kiechl-Kohlendorfer, U., Dudell, G., Rechtman, D. J., Lee, M. L., Lucas, A., & Abrams, S. (2013). Randomized trial of exclusive human milk versus preterm formula diets in extremely premature infants. Journal of Pediatrics 163 , 6, P1592–P1595.e1. https://doi.org/10.1016/j.jpeds.2013.07.011
- Darby, M. G., Chetty, A., Mrjden, D., Rolot, M., Smith, K., MacKowiak, C., Sedda, D., Nyangahu, D., Jaspan, H., Toellner, K. M., Waisman, A., Quesniaux, V., Ryffel, B., Cunningham, A. F., Dewals, B. G., Brombacher, F., & Horsnell, W. G. C. (2019). Pre- conception maternal helminth infection transfers via nursing long-lasting cellular immunity against helminths to offspring. Science Advances , 5(5), 3058–3087. https://doi.org/10.1126/sciadv.aav3058
- Depeters, E. J., & Hovey, R. C. (2009). Methods for collecting milk from mice. Journal of Mammary Gland Biology and Neoplasia , 14(4), 397–400. https://doi.org/10.1007/s10911-009-9158-0
- Foulsham, W., Coco, G., Amouzegar, A., Chauhan, S. K., & Dana, R. (2018). When clarity is crucial: Regulating ocular surface immunity. Trends in Immunology , 39(4), 288–301. https://doi.org/10.1016/j.it.2017.11.007
- Gama, L. A., Rocha Machado, M. P., Beckmann, A. P. S., Miranda, J. R., de, A., Corá, L. A., & Américo, M. F. (2020). Gastrointestinal motility and morphology in mice: Strain-dependent differences. Neurogastroenterology & Motility, 32(6), e13824. https://doi.org/10.1111/nmo.13824
- Gómez-Gallego, C., Ilo, Jaakkola, U., Salminen, S., Periago, M. J., Ros, G., & Frias, R. (2014). A method to collect high volumes of milk from mice (Mus musculus). Anales de Veterinaria de Murcia , 29, 55–61.
- Görs, S., Kucia, M., Langhammer, M., Junghans, P., & Metges, C. C. (2009). Technical note: Milk composition in mice—methodological aspects and effects of mouse strain and lactation day. Journal of Dairy Science , 92(2), 632–637. https://doi.org/10.3168/jds.2008-1563
- Hassiotou, F., Geddes, D. T., & Hartmann, P. E. (2013). Cells in human milk: State of the science. Journal of Human Lactation , 29(2), 171–182. https://doi.org/10.1177/0890334413477242
- Houston, S. A., Cerovic, V., Thomson, C., Brewer, J., Mowat, A. M., & Milling, S. (2015). The lymph nodes draining the small intestine and colon are anatomically separate and immunologically distinct. Mucosal Immunology , 9(2), 468–478. https://doi.org/10.1038/mi.2015.77
- Jenness, R. (1979). The composition of human milk. Seminars in Perinatology , 3, 225–239.
- Keller, T., Wengenroth, L., Smorra, D., Probst, K., Kurian, L., Kribs, A., & Brachvogel, B. (2019). Novel DRAQ5TM/SYTOX® blue based flow cytometric strategy to identify and characterize stem cells in human breast milk. Cytometry Part B - Clinical Cytometry , 96(6), 480–489. https://doi.org/10.1002/cyto.b.21748
- Kernbauer, E., Ding, Y., & Cadwell, K. (2014). An enteric virus can replace the beneficial function of commensal bacteria. Nature , 516(7529), 94–98. https://doi.org/10.1038/nature13960
- Knickelbein, J. E., Watkins, S. C., Mcmenamin, P. G., & Hendricks, R. L. (2009). Stratification of antigen-presenting cells within the normal cornea. Ophthalmology and Eye Diseases , 1, 45–54. https://doi.org/10.4137/oed.s2813
- Laouar, A. (2020). Maternal leukocytes and infant immune programming during breastfeeding. Trends in Immunology , 41(3), 225–239. https://doi.org/10.1016/j.it.2020.01.005
- Muranishi, Y., Parry, L., Averous, J., Terrisse, A., Maurin, A. C., Chaveroux, C., Mesclon, F., Carraro, V., Bruhat, A., Fafournoux, P., & Jousse, C. (2016). Method for collecting mouse milk without exogenous oxytocin stimulation. BioTechniques , 60(1), 47–49. https://doi.org/10.2144/000114373
- Pačes, J., Knížková, K., Tušková, L., Grobárová, V., Zadražil, Z., Boes, M., & Černý, J. (2022). MHC II–EGFP knock-in mouse model is a suitable tool for systems and quantitative immunology. Immunology Letters , 251–252, 75–85. https://doi.org/10.1016/j.imlet.2022.10.007
- Susaki, E. A., Tainaka, K., Perrin, D., Kishino, F., Tawara, T., Watanabe, T. M., Yokoyama, C., Onoe, H., Eguchi, M., Yamaguchi, S., Abe, T., Kiyonari, H., Shimizu, Y., Miyawaki, A., Yokota, H., & Ueda, H. R. (2014). Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell , 157(3), 726–739. https://doi.org/10.1016/j.cell.2014.03.042
- Susaki, E. A., Tainaka, K., Perrin, D., Yukinaga, H., Kuno, A., & Ueda, H. R. (2015). Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nature Protocols , 10(11), 1709–1727. https://doi.org/10.1038/nprot.2015.085
- van den Broeck, W., Derore, A., & Simoens, P. (2006). Anatomy and nomenclature of murine lymph nodes: Descriptive study and nomenclatory standardization in BALB/cAnNCrl mice. Journal of Immunological Methods , 312(1–2), 12–19. https://doi.org/10.1016/j.jim.2006.01.022
- Victora, C. G., Bahl, R., Barros, A. J. D., França, V. A., Horton, S., Krasevec, J., Murch, S., Sankar, M. J., Walker, N., & Rollins, N. C. (2016). Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet , 387, 475. https://doi.org/10.1016/S0140-6736(15)01024-7
- Willingham, K., McNulty, E., Anderson, K., Hayes-Klug, J., Nalls, A., & Mathiason, C. (2014). Milk collection methods for mice and Reeves’ Muntjac deer. Journal of Visualized Experiments: JoVE , 89, 51007. https://doi.org/10.3791/51007
- Witkowska-Zimny, M., & Kaminska-El-Hassan, E. (2017). Cells of human breast milk. In Cellular and Molecular Biology Letters , 22, 11. https://doi.org/10.1186/s11658-017-0042-4
CORRECTIONS
In this publication, the following correction has been made:
A missing Acknowledgments section has been added.
The current version online now includes this correction and may be considered the authoritative version of record.

