Histone Purification Combined with High-Resolution Mass Spectrometry to Examine Histone Post-Translational Modifications and Histone Variants in Caenorhabditis elegans
Christian G. Riedel, Christian G. Riedel, Lluís Millan-Ariño, Lluís Millan-Ariño, Zuo-Fei Yuan, Zuo-Fei Yuan, Marlies E. Oomen, Marlies E. Oomen, Simone Brandenburg, Simone Brandenburg, Alexey Chernobrovkin, Alexey Chernobrovkin, Jérôme Salignon, Jérôme Salignon, Lioba Körner, Lioba Körner, Roman A. Zubarev, Roman A. Zubarev, Benjamin A. Garcia, Benjamin A. Garcia
aging
Caenorhabditis elegans
epigenetics
histone post-translational modifications
histone variants
mass spectrometry
Abstract
Histones are the major proteinaceous component of chromatin in eukaryotic cells and an important part of the epigenome, affecting most DNA-related events, including transcription, DNA replication, and chromosome segregation. The properties of histones are greatly influenced by their post-translational modifications (PTMs), over 200 of which are known today. Given this large number, researchers need sophisticated methods to study histone PTMs comprehensively. In particular, mass spectrometry (MS)−based approaches have gained popularity, allowing for the quantification of dozens of histone PTMs at once. Using these approaches, even the study of co-occurring PTMs and the discovery of novel PTMs become feasible. The success of MS-based approaches relies substantially on obtaining pure and well-preserved histones for analysis, which can be difficult depending on the source material. Caenorhabditis elegans has been a popular model organism to study the epigenome, but isolation of pure histones from these animals has been challenging. Here, we address this issue, presenting a method for efficient isolation of pure histone proteins from C. elegans at good yield. Further, we describe an MS pipeline optimized for accurate relative quantification of histone PTMs from C. elegans. We alkylate and tryptically digest the histones, analyze them by bottom-up MS, and then evaluate the resulting data by a C. elegans −adapted version of the software EpiProfile 2.0. Finally, we show the utility of this pipeline by determining differences in histone PTMs between C. elegans strains that age at different rates and thereby achieve very different lifespans. © 2020 The Authors.
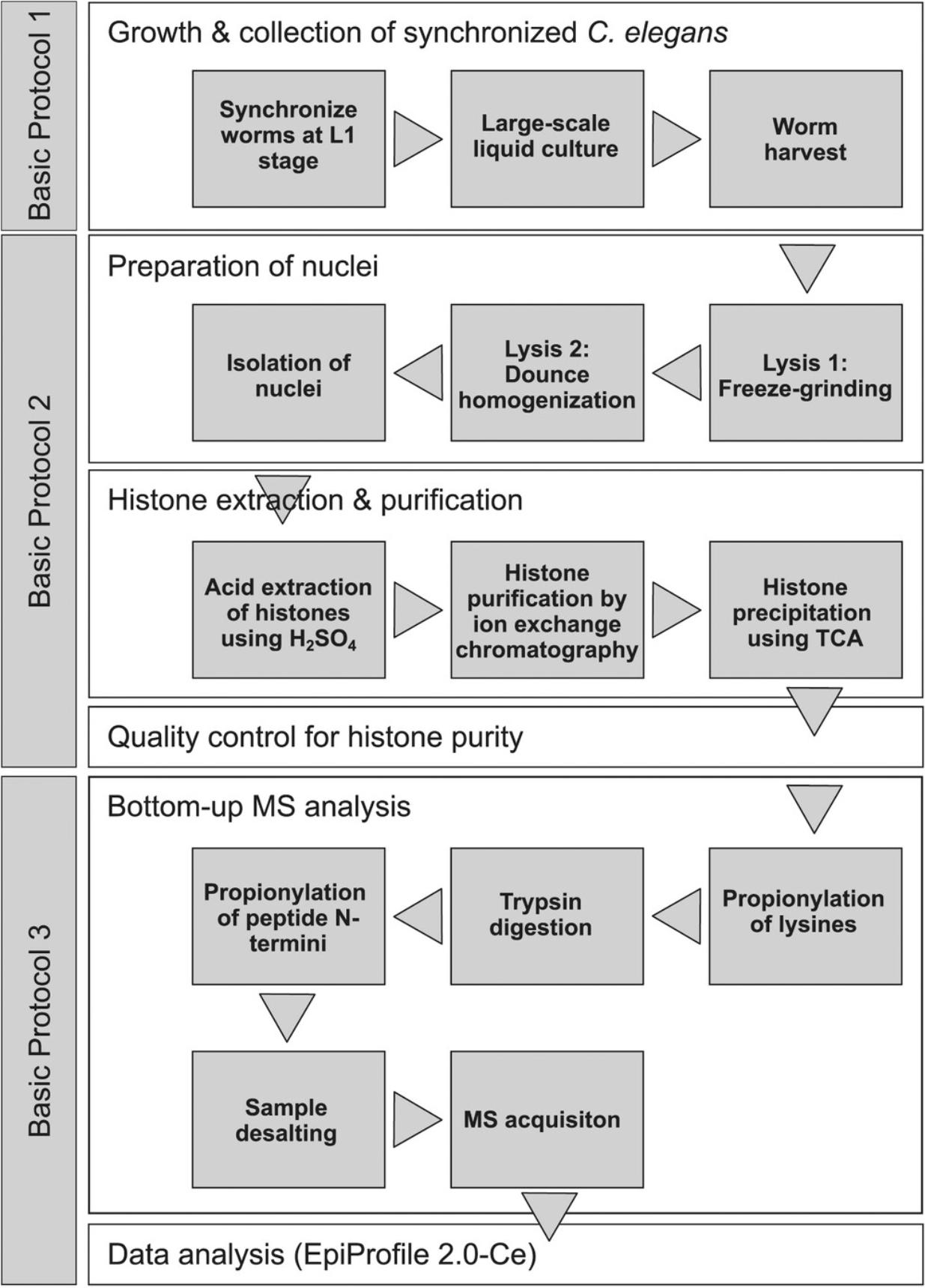
Basic Protocol 1 : Large-scale growth and harvesting of synchronized C. elegans
Basic Protocol 2 : Nuclear preparation, histone extraction, and histone purification
Basic Protocol 3 : Bottom-up mass spectrometry analysis of histone PTMs and histone variants
INTRODUCTION
This article provides a detailed set of protocols for the efficient extraction and purification of histone proteins from whole animals of the model organism Caenorhabditis elegans , achieving unprecedented purity at sufficient yield for comprehensive analysis of histone post-translational modifications (PTMs) and histone variants by bottom-up mass spectrometry. First, we describe how to grow this popular nematode model organism to a large scale by culturing it in liquid medium (Basic Protocol 1). Then, we present a method to isolate and lyse nuclei from the harvested animals, and to eventually purify nuclear histones by cation-exchange chromatography (Basic Protocol 2). This method achieves sufficient purity so that the obtained histones can be used directly for mass spectrometric analysis, avoiding any additional purification steps (e.g., by SDS-PAGE or HPLC) that would otherwise lower the yield. Quantification of histone PTMs and histone variants is then achieved by bottom-up mass spectrometry and subsequent analyses using a C. elegans −adapted version of EpiProfile 2.0, a recently developed software to analyze histone PTMs from MS measurements (Yuan et al., 2015, 2018) (Basic Protocol 3). Hereafter, we will refer to our C. elegans −adapted version of this software as EpiProfile 2.0-Ce. Figure 1 depicts a schematic workflow of the procedure in this article, and Tables 1 and 2 summarize all the histone PTMs and their combinations, as well as the histone variants that EpiProfile 2.0-Ce quantifies. To demonstrate the utility of our method, we determine the histone PTM composition of three C. elegans strains that each age at a different rate due to different levels of signaling through the insulin/IGF signaling (IIS) pathway and its downstream transcription factor DAF-16/FOXO. It has recently been reported that aging is associated with specific changes in histone PTM abundance (Booth & Brunet, 2016; Zhou, Sen, Lin, & Riedel, 2018). Using our method, we identify several expected but also some new histone PTMs whose abundance differs between these three strains.
| Histone | Peptide START | Peptide END | Peptide sequence | Peptide modifications analyzed |
|---|---|---|---|---|
| H3 | 3 | 8 | TKQTAR | unmod; K4me1; K4me2; K4me3; K4ac |
| H3 | 9 | 17 | KSTGGKAPR | unmod; K9me1; K9me2; K9me3; K9ac; K14ac; K9me1K14ac; K9me2K14ac; K9me3K14ac; K9acK14ac |
| H3 | 9 | 17 | KSTGGKAPR | S10ph; K9me1S10ph; K9me2S10ph; K9me3S10ph; K9acS10ph; S10phK14ac; K9me1S10phK14ac; K9me2S10phK14ac; K9me3S10phK14ac; K9acS10phK14ac |
| H3 | 18 | 26 | KQLATKAAR | unmod; K23me1; K18me1; K18me1K23me1; K18ac; K23ac; K18acK23ac; K23me2; K23me3; K18acK23me1; K18acK23me2; K18acK23me3 |
| H3.3V1 | 18 | 26 | KALATKAAR | unmod; K23me1; K18me1; K18me1K23me1; K18ac; K23ac; K18acK23ac |
| H3 | 27 | 40 | KSAPASGGVKKPHR | unmod; K27me1; K27me2; K27me3; K27ac; K36me1; K27me1K36me1; K27me2K36me1; K27me3K36me1; K27acK36me1; K36me2; K27me1K36me2; K27me2K36me2; K27me3K36me2; K27acK36me2; K36me3; K27me1K36me3; K27me2K36me3; K27me3K36me3; K27acK36me3; K36ac; K27me1K36ac; K27me2K36ac; K27me3K36ac; K27acK36ac |
| H3.3V1 | 27 | 40 | KSAIVTGSVKKVHR | unmod; K36me1; K27me1; K27me2; K36me2; K27me3; K36me3; K27me2K36me1; K27me1K36me2; K27me1K36me1; K27me3K36me1; K27me1K36me3; K27me2K36me2; K27me3K36me2; K27ac |
| H3.3V2 | 27 | 40 | KSAPTTGGVKKPHR | unmod; K27me1; K27me2; K27me3; K27ac; K36me1; K27me1K36me1; K27me2K36me1; K27me3K36me1; K27acK36me1; K36me2; K27me1K36me2; K27me2K36me2; K27me3K36me2; K27acK36me2; K36me3; K27me1K36me3; K27me2K36me3; K27me3K36me3; K27acK36me3; K36ac; K27me1K36ac; K27me2K36ac; K27me3K36ac; K27acK36ac |
| H3 | 54 | 63 | YQKSTELLIR | unmod; K56ac |
| H3 | 64 | 69 | KLPFQR | unmod; K64ac |
| H3 | 73 | 83 | EIAQDFKTDLR | unmod; K79me1; K79me2; K79me3; K79ac |
| H3 | 117 | 128 | VTIMPKDIQLAR | unmod; K122ac |
| H4 | 4 | 17 | GKGGKGLGKGGAKR | unmod; K5ac; K8ac; K12ac; K16ac; K5acK8ac; K5acK12ac; K5acK16ac; K8acK12ac; K8acK16ac; K12acK16ac; K5acK8acK12ac; K5acK8acK16ac; K5acK12acK16ac; K8acK12acK16ac; K5acK8acK12acK16ac |
| H4 | 20 | 23 | KVLR | unmod; K20me1; K20me2; K20me3; K20ac |
| H4 | 24 | 35 | DNIQGITKPAIR | unmod; K31ac |
| H4 | 40 | 45 | RGGVKR | unmod; K44me1 |
| H4 | 68 | 78 | DAVTYCEHAKR | unmod; T71ph |
| H4 | 79 | 92 | KTVTAMDVVYALKR | unmod; K79ac |
| H1 | 90 | 107 | GVTSKALVQAAGSGANGR | unmod; K94me1; K94ac |
| H2A | 4 | 18 | GKGGKAKTGGKAKSR | unmod; K5ac; K8ac; K10ac; K16ac; K5acK8ac; K5acK10ac; K5acK16ac; K8acK10ac; K8acK16ac; K10acK16ac; K8acK10acK16ac; K5acK10acK16ac; K5acK8acK16ac; K5acK8acK10ac; K5acK8acK10acK16ac |
| H2AV | 1 | 21 | AGGKGKAGKDSGKSKSKVVSR | unmod; K4ac; K9ac; K13ac; K17ac; K4acK9ac; K4acK13ac; K4acK17ac; K9acK13ac; K9acK17ac; K13acK17ac; K9acK13acK17ac; K4acK13acK17ac; K4acK9acK17ac; K4acK9acK13ac; K4acK9acK13acK17ac |
| H2A | 37 | 43 | KGNYAQR | unmod; K37ac |
| H2A | 73 | 78 | DNKKTR | unmod; K75ac |
| H2B | 31 | 40 | KESYSVYIYR | unmod; K31me1; K31me2; K31me3; K31ac |
| H2B | 77 | 83 | LAHYNKR | unmod; K82ac |
| Histone | Peptide sequence | Coding gene | Histone variant | Description |
|---|---|---|---|---|
| H3 | YRPGTVALR | several | H3 | Consensus sequence for canonical histone H3 |
| VTIMPKDIQLAR | several | H3 | Consensus sequence for canonical histone H3 | |
| IRGER | several | H3 | Consensus sequence for canonical histone H3 | |
| FRPGTVALR | his-69, his-74 | H3.3V | Ortholog of human H3.3B | |
| VTIMPKDMQLAR | his-69, his-74, his-72 | H3.3V | Ortholog of human H3.3B | |
| H2A | AGLQFPVGR | several | H2A | Consensus sequence for canonical histone H2A |
| HLQLAVR | several | H2A | Consensus sequence for canonical histone H2A | |
| FLKQR | htz-1 | H2AV | Ortholog of human H2A.Z | |
| HLHLAIR | htz-1 | H2AV | Ortholog of human H2A.Z | |
| H1 | QALKR | his-24 | H1.0 | Ortholog of human H1.0 |
- a Histone peptides analyzed by EpiProfile 2.0-Ce can be further customized according to user's need to analyze other histone variants than those presented here as an example.
Basic Protocol 1: LARGE-SCALE GROWTH AND HARVESTING OF SYNCHRONIZED C. ELEGANS
Our protocol has been optimized to obtain histones of high purity and sufficient yield for mass spectrometry analyses while preserving the histones’ post-translational modifications (PTMs). Since the histone content in relation to body weight is rather low in C. elegans and since some loss occurs throughout the protocol, a substantial amount of starting material is needed. Thus, large quantities of animals need to be grown, which is most easily achieved using liquid culture.
The actual required amount of animals will depend on the purpose of the experiment. Normally, 0.5−1 million animals are sufficient to study most of the histone PTMs in adult wild-type animals. However, if one would like to detect particularly rare PTMs or analyze a C. elegans mutant that bears lower histone content, a higher starting amount is required. Here, it is important to know that in adult C. elegans , the tissue contributing the most to histones is the germ line. Thus, any mutant that lacks or has an impaired germline may need to be grown on an increased scale to ultimately yield the same histone amount as wild-type animals.
Materials
-
Desired worm strains, grown on 100-mm-diameter Nematode Growth Medium (NGM) plates (see recipe), e.g.:
- Wild-type C. elegans strain (N2)
- Slow-aging C. elegans strain with a hypomorphic insulin/IGF receptor allele (daf-2(e1370ts)III) (CB1370)
- Fast-aging C. elegans strain with a null allele for DAF-16/FOXO (daf-16(mu86lf)I) (CF1038)
-
150-mm NGM plates (see recipe)
-
50× OP50-1: Stationary OP50-1 E.coli culture (obtained from the CGC), 50-fold concentrated relative to an overnight LB culture, and resuspended in S-Basal (see recipe)
-
M9 buffer (see recipe)
-
Hypochlorite solution (see recipe)
-
1 M MgSO4 stock solution
-
S-Complete medium (see recipe)
-
5-fluoro-2′-deoxyuridine (FUDR; Sigma-Aldrich, F0503)
-
Sterile Milli-Q water or equivalent
-
Nuclear Isolation Buffer (NIB; see recipe)
-
200 mM AEBSF (4-(2-aminoethyl) bezenesulfonyl fluoride hydrochloride; Sigma-Aldrich, A8456)
-
2.5 µM microcystin (Sigma-Aldrich, M4194)
-
5 M sodium butyrate (Sigma-Aldrich, B5887)
-
0.9 M dithiothreitol (DTT; ThermoFisher Scientific, R0861)
-
100× cOmpleteTM EDTA-free Protease Inhibitor Cocktail (Roche, 11873580001)
-
0.1 M spermine (Sigma-Aldrich, S3256)
-
0.1 M spermidine (Sigma-Aldrich, 05292)
-
Liquid nitrogen
-
15-ml conical centrifuge tubes
-
Bench-top centrifuge for 15-, 50-, and 500-ml tubes (e.g., ThermoFisher Scientific MegaFuge 40R)
-
Vortex (e.g., Vortex Genie 2)
-
Stereomicroscope (e.g., Olympus SZX7)
-
Rotary mixer
-
2-L Erlenmeyer flasks
-
Spectrophotometer (e.g., ThermoFisher Scientific NanoDrop 2000c) and disposable 1-ml cuvettes
-
Shaking incubator (e.g., New Brunswick Innova 44R)
-
500-ml conical centrifuge tubes (Corning, 431123)
-
40-µm cell strainers (if needed, e.g., BD Falcon, 352340)
-
50-ml conical centrifuge tubes
Egg preparation to synchronize C. elegans animals at the L1 larval stage
1.For each sample/condition, start with five 100-mm-diameter NGM plates that are crowded with unsynchronized animals. Transfer these animals to ten 150-mm-diameter NGM plates each seeded with 1 ml of 50× OP50-1 E. coli. Incubate plates at the desired temperature until the plates are crowded with gravid adults. Typically, the growth temperature for wild-type animals is 20°C. However, depending on the strain used (e.g., when it carries a temperature-sensitive allele), other temperatures in the range from 15°C to 25°C may be preferred.
2.Wash worms off the plates using 12 ml of M9 buffer.
3.Collect worms in a 15-ml tube and centrifuge 1 min at 1500 × g to pellet the worms. Discard the supernatant.
4.Repeat this harvesting/washing step until all worms have been removed from the plate and the supernatant from step 3 is clear (approximately three washes).
5.After the final wash, resuspend worms in 7.5 ml sterile M9 buffer.
6.Add 2.5 ml of fresh hypochlorite solution.
7.Vortex tube for 30 s.
8.Examine under a stereomicroscope to determine if most worm corpses have disappeared and many released eggs can be seen.
9.Repeat steps 7 and 8 until all eggs have been released and barely any corpses remain. Overall, this bleaching (steps 7 to 9) will take approximately 6 min.
10.Centrifuge the tube 1 min at 1500 × g to pellet the released eggs.
11.Carefully take off the supernatant and discard it.
12.Wash eggs at least five times, each time with 10 ml M9 buffer, centrifuging 1 min at 1500 × g between washes.
13.Spin down eggs 1 min at 1500 × g , and resuspend eggs in 10 ml M9 buffer supplemented with 1 mM MgSO4.
14.Rotate the tube containing the eggs for 1 day at room temperature or 2 days at 15°C.
Large-scale growth of C. elegans in liquid culture
15.Check that there are no unhatched eggs left, and determine the concentration of L1s (first stage larvae) by placing three spots of 5 or 10 µl of the C. elegans suspension on an NGM plate and counting the L1 larvae under a stereomicroscope.
16.For each sample/condition, prepare a 2-L Erlenmeyer flask containing 1 L of S-Complete Medium.
17.Seed the 1 L of S-Complete Medium with ∼80 ml of 50× OP50-1 bacteria.
18.Check the OD at 600 nm (OD600), e.g., using a spectrophotometer. The OD should be between 2 and 4.For accurate results, use disposable cuvettes, use S-Complete Medium as blank, and dilute your culture 1:10.
19.Inoculate the culture with synchronized L1s from the hatched egg preparation. The L1 concentration should not exceed 1000 animals/ml. (e.g., for 1 L of culture, inoculate with 0.5−1 million L1s).
20.Incubate worm culture in a shaking incubator at the desired temperature and 140 rpm. Typically, the growth temperature for wild-type animals is 20°C. However, depending on the strain used (e.g., when it carries a temperature-sensitive allele), other temperatures in the range from 15°C to 25°C may be preferred.
21.Check the OD600 of the culture daily and keep it between 2 and 4 A. Add 50× OP50-1 bacteria when needed.
22.Keep worms in culture until the desired developmental or adult stage. Up to that point, replace S-Complete Medium every 2-3 days with fresh medium and new bacteria. This is done by transferring the culture into 500-ml conical centrifuge tubes, allowing the worms to settle by gravity for ∼10 min, removing the supernatant, and replacing it with new S-Complete Medium and 50× OP50-1 E. coli as in steps 16-18.
Harvest worms at a defined time point
From this point on, use cold buffers and cold centrifuges, and work on ice or in a cold room.
23.Transfer the culture into 500-ml conical centrifuge tubes.
24.Allow the worms to settle for 10 min on ice, then centrifuge 3 min at 1500 × g , 4°C. Discard the supernatant.
25.Transfer the worms to 50-ml centrifuge tubes.
26.Wash the worms with M9 buffer and collect them by spinning them down 2 min at 1500 × g , 4°C. Repeat this wash step until the supernatant is cleared of bacteria (approximately three washes).
27.Transfer the worms to 15-ml tubes.
28.Wash the worms once with 10 ml Milli-Q water, then centrifuge 2 min at 1500 × g , 4°C.
29.Take off supernatant to just above the worm pellet.
30.Mix one volume of worm pellet with two volumes of ice-cold NIB+ buffer.
To make NIB+ buffer, freshly supplement Nuclear Isolation Buffer (NIB) with protease inhibitors and stabilizing agents:
- 500 µM AEBSF
- 5 nM microcystin
- 10 mM sodium butyrate
- 1 mM DTT
- 1× cOmplete protease inhibitor cocktail
- 0.15 mM spermine
- 0.15 mM spermidine.
31.Write down the total volume of the worm suspension (worm pellet plus NIB+ buffer; used later in Basic Protocol 2) and then snap-freeze the worms by slowly dripping the suspension into a bucket filled with liquid nitrogen (to make beads).
Basic Protocol 2: NUCLEAR PREPARATION, HISTONE EXTRACTION, AND HISTONE PURIFICATION
The following acid extraction protocol is optimized to universally isolate all histone proteins and their variants. However, for the more efficient extraction of certain histone variants with unique biochemical and biophysical properties, other methods might be considered. For instance, extraction by diluted perchloric acid is a powerful tool to specifically extract more unstructured histone H1 variants (Zougman & Wiśniewski, 2006).
After acid extraction, we achieve histone purification and enrichment by subjecting the extracted material to ion-exchange chromatography. Histone proteins are rich in basic amino acid residues, and are thus positively charged. In vivo, this promotes their interaction with the negatively charged phosphate backbone of DNA. In vitro, we can make use of this property. Due to their positive charge, histones have a high affinity for cation-exchange resins, which allows for their efficient separation from other contaminants (see Rodriguez-Collazo, Leuba, & Zlatanova, 2009).
Materials
-
Frozen worm beads in NIB+ buffer (Basic Protocol 1, step 31)
-
Nuclear Isolation Buffer (NIB; see recipe)
-
200 mM AEBSF (4-(2-aminoethyl) bezenesulfonyl fluoride hydrochloride; Sigma-Aldrich, A8456)
-
2.5 µM microcystin (Sigma-Aldrich, M4194)
-
5 M sodium butyrate (Sigma-Aldrich, B5887)
-
0.9 M dithiothreitol (DTT; ThermoFisher Scientific, R0861)
-
100× cOmpleteTM EDTA-free Protease Inhibitor Cocktail (Roche, 11873580001)
-
0.1 M spermine (Sigma-Aldrich, S3256)
-
0.1 M spermidine (Sigma-Aldrich, 05292)
-
2× Buffer H (see recipe)
-
5 M NaCl
-
B-Hypo-Lys Buffer (see recipe)
-
0.4 N H2SO4 (see recipe)
-
SP Sepharose High Performance resin (GE Healthcare, 17-1087-01)
-
Equilibration buffer (see recipe)
-
1 M Tris·Cl, pH 8.0 (Current Protocols, 1998)
-
0.5 M EDTA, pH 8.0 (Current Protocols, 1998)
-
Histone wash buffer (see recipe)
-
Histone elution buffer (see recipe)
-
Trichloroacetic acid (TCA)
-
Acetone
-
50 mM NaHCO3
-
PierceTM BCA Protein Assay (ThermoFisher Scientific, 23225; also see Current Protocols article: Gallagher, 2012)
-
1 M Tris base
-
Milli-Q water or equivalent
-
2× Laemmli sample buffer (see Current Protocols article: Gallagher, 2012)
-
4%-20% pre-cast Tris-Glycine gels (e.g., ThermoFisher Scientific, EC6026BOX)
-
Tris-Glycine SDS (TGS) running buffer (e.g., BioRad, 1610732)
-
Coomassie solution (see recipe)
-
Destaining solution (see recipe)
-
Cryogenic grinder: e.g., CryoMill (Retsch)
-
Stereomicroscope (e.g., Olympus SZX7)
-
15-ml Dounce homogenizer set (Sigma-Aldrich, D9938)
-
7-ml Dounce homogenizer set (Sigma-Aldrich, D9063)
-
15-ml centrifuge tubes
-
Benchtop refrigerated centrifuge for 15- and 50-ml tubes (e.g., ThermoFisher Scientific MegaFuge 40R)
-
Benchtop refrigerated microcentrifuge (e.g., Eppendorf 5424R)
-
Large refrigerated centrifuge with rotor for round-bottom tubes (e.g., Beckman Avanti with JA-15.50 rotor)
-
13-ml round-bottom centrifuge tubes with lid (Sarstedt, 55.518 and 65.816)
-
Rotary mixer
-
1.5-ml microcentrifuge tubes
-
Poly-Prep chromatography columns (Bio-Rad, 7311550)
-
pH test paper
-
50-ml centrifuge tubes
-
Additional reagents and equipment for protein assay (see Current Protocols article: Olson & Markwell, 2007) and gel electrophoresis (see Current Protocols article: Gallagher, 2012)
Freeze-grinding and nuclear preparation
1.To fragment the frozen worms, freeze-grind the worm beads from Basic Protocol 1 using a cryogenic grinder. Using this device, grind until the worms are entirely destroyed and no worm fragments remain visible upon checking a small amount of the ground material under a stereomicroscope after thawing.
2.Add four volumes of NIB+ buffer relative to the total volume of worm suspension in Basic Protocol 1, step 31.
To make NIB+ buffer, freshly supplement Nuclear Isolation Buffer (NIB) with protease inhibitors and stabilizing agents:
- 500 µM AEBSF
- 5 nM microcystin
- 10 mM sodium butyrate
- 1 mM DTT
- 1× cOmplete protease inhibitor cocktail
- 0.15 mM spermine
- 0.15 mM spermidine.
Thaw on ice.
3.Transfer the sample to a pre-chilled Dounce homogenizer. Use either a 7-ml or 15-ml Dounce homogenizer depending on the sample volume.
4.Continue the lysis of the animals and their cells using 50 strokes in the Dounce homogenizer on ice (10 strokes with pestle A and 40 strokes with pestle B).
5.Transfer the sample to a pre-chilled 15-ml tube. Split into several tubes if needed.
6.Take 50 µl of the sample and keep it at −80°C for later analysis.
7.Spin the sample for 2 min at 200 × g , 4°C, to get rid of any debris.
8.Transfer the supernatant to a pre-chilled 13-ml round-bottom tube. Split into several tubes if needed. Keep pellets at −80°C for later optional analysis.
9.Collect the nuclei by centrifuging the 13-ml round-bottom tube 10 min at 10,000 × g , 4°C.
10.Remove the supernatant containing the cytoplasmic fraction and store it at −80°C for later optional analysis.
11.Disrupt the pellet containing the intact nuclei by resuspending in 1.67 ml of Buffer H+ per 1 ml of total volume of worm suspension that was snap-frozen (see last step of Basic Protocol 1).
To prepare Buffer H+, freshly supplement Buffer H with:
- 400 mM NaCl
- 500 µM AEBSF
- 5 nM microcystin
- 10 mM sodium butyrate
- 2 mM DTT
- 1× cOmplete protease inhibitor cocktail.
12.Transfer homogenate to a pre-chilled Dounce homogenizer and break the nuclei (10 strokes with pestle A, and 40 strokes with pestle B). Use either a 7-ml or 15-ml Dounce homogenizer depending on the sample volume.
13.Transfer the homogenate to a 13-ml round-bottom tube and rotate it for 30 min at 4°C on a rotary mixer.
14.Spin the sample for 10 min at 10,000 × g , 4°C, to pellet the chromatin.
15.Collect the supernatant and store it at −80°C for later optional analysis.
Acid extraction of histones
16.Wash the chromatin pellet three times in 1 to 3 ml B-Hypo-Lys Buffer freshly supplemented with inhibitors:
- 500 µM AEBSF
- 5 nM microcystin
- 10 mM sodium butyrate
- 1 mM DTT
- 1× cOmplete™ protease inhibitor cocktail.
Mix well until the pellet is dissolved. For easier pellet resuspension, resuspend first with 100 µl and then add the remaining buffer.
17.Spin 10 min at 10,000 × g , 4°C.
18.Collect the supernatant and store it at −80°C for later optional analysis.
19.Re-spin for 1 min and take off any remaining supernatant after the last wash.
20.Resuspend the pellet in 1 ml 0.4 N H2SO4 and transfer the sample to a 1.5-ml microcentrifuge tube.
21.Rotate on a rotary mixer overnight at 4°C.
Histone purification by cation-exchange chromatography
Column preparation
For your convenience, you can prepare the resin and column already on the day before the histone purification. The prepared column should be stored at 4°C with the top and bottom of the column properly closed to avoid drying of the resin.
22.Prepare a Poly-Prep chromatography column by placing it in a stand.
23.Using the end of a Pasteur pipette, push the filter disc to the bottom of the column.
24.Close the bottom of the column using a cap.
25.Add 1 ml Milli-Q water and mark the top water level on the outside of the column. Open the bottom of the column and allow the water to drain.
26.Close the bottom of the column and add 1 ml of SP Sepharose High Performance resin (gently resuspend the resin before pipetting). Keep adding SP Sepharose until the settled resin reaches the 1-ml mark made on the column.
27.Place a tube or container underneath the column to collect flow-through.
28.Add 3 ml of Milli-Q water, open the bottom of the column, let it drain by gravity flow, then close the bottom of the column. Discard the flow-through.
29.Repeat step 28 two more times for a total of three washes.
30.Add 3 ml of equilibration buffer, open the bottom of the column, let it drain by gravity flow, then close the bottom of the column. Discard the flow-through.
31.Repeat step 30 two more times for a total of three washes.
32.Close also the top of the column and store it at 4°C until the histone sample is added.
Histone purification
Perform all subsequent steps on ice or at 4°C to avoid enzymatic activities that may alter histone PTMs.
33.After the overnight incubation, spin down the acid extract (from step 21) for 5 min at 14,000 × g , 4°C.
34.Collect the supernatant in a 15-ml centrifuge tube. Keep the pellet at −80°C for later optional analysis.
35.Also take 50 µl of the supernatant and store at −80°C for later optional analysis.
36.Neutralize the acid extract by adding an equal volume of 1 M Tris·Cl, pH 8.0.
37.Measure the pH by pipetting 5-10 µl of neutralized extract onto a pH test paper. At this stage, the pH should be between 7 and 8.If the pH remains lower, add some 1 M Tris base to raise the pH.
38.Supplement the extract to a final concentration of:
- 200 mM NaCl
- 2 mM EDTA, pH 8.0
- 1 mM DTT
- 500 µM AEBSF
- 5 nM microcystin
- 10 mM sodium butyrate
- 1× cOmplete™ protease inhibitor cocktail.
39.Take 50 µl of the extract and store it at −80°C for later optional analysis.
40.Take the caps off the equilibrated SP-resin column and place it in a 50-ml tube, fixing the column in place with tape.
41.Spin 3 min at 100 × g , 4°C, to completely remove the equilibration buffer.
42.Close the column with the bottom cap and add 500 µl of the extract.
43.Let the SP-resin settle on ice for 10 min.
44.Take the bottom cap off again and add the remaining extract.
45.Allow the extract to pass through the column by gravity flow.
46.Keep the flow-through for later optional analysis.
47.Wash the SP-resin with at least 10 ml histone wash buffer by gravity flow.
48.Keep flow-through for later optional analysis.
49.Add 1 ml histone elution buffer to the column and collect the eluate. Repeat this step two more times and collect each eluate into a different tube (eluate 1, eluate 2, and eluate 3).
Histone precipitation
50.Spin down eluates 15 min at 20,000 × g , 4°C.
51.Transfer supernatants to fresh tubes. Make sure not to touch any pellets; it is better to leave a bit of the supernatant behind rather than disturb the pellet.
52.Store pellets at −80°C for later optional analysis.
53.Spin down again 10 min at 20,000 × g , 4°C.
54.Transfer supernatants to fresh tubes. Make sure not to touch any pellets; it is better to leave a bit of the supernatant behind rather than disturb the pellet.
55.Store pellets at −80°C for later optional analysis.
56.Add 1/3 volume of 100% trichloroacetic acid drop by drop to the supernatants, then invert tube 10 times to precipitate the histones.
57.Incubate on ice for 30 min at 4°C.
58.Spin down for 20 min at 20,000 × g , 4°C.
59.Remove supernatant and add 500 µl of ice-cold acetone (kept at −20°C).
60.Invert three times.
61.Spin down 10 min at 20,000 × g. 4°C. Discard the supernatant.
62.Repeat this wash step.
63.Take off all supernatant and let the pellet dry at room temperature for 20 min.
64.Resuspend the histone pellet in 50 µl of 50 mM NaHCO3.
65.Using a P10 pipette tip, spot some sample onto pH paper. If the pH is below 7-8, immediately add some 1 M Tris base to increase the pH accordingly.
Quality control of the histone extraction by SDS-PAGE and Coomassie staining
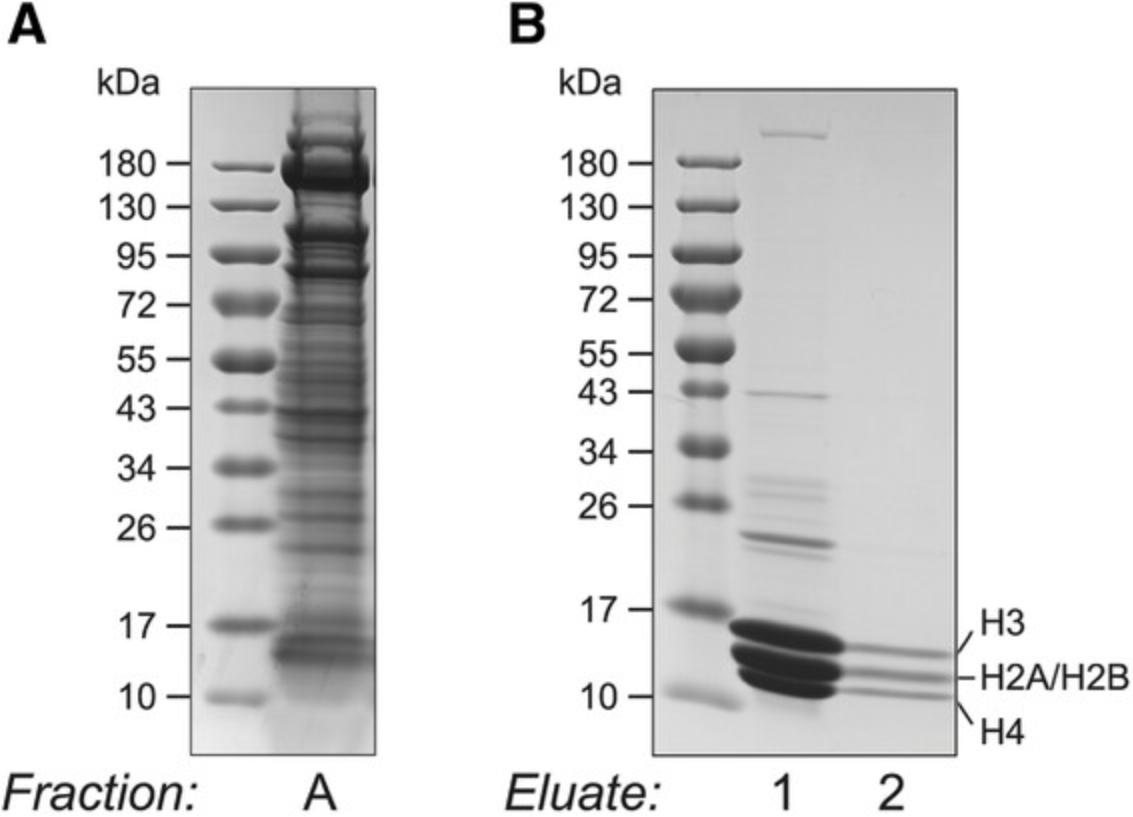
To assess the purity of the extracted histones before proceeding to MS analysis, samples are subjected to SDS-PAGE and Coomassie staining. Core histones are low-molecular-weight proteins, migrating between ∼11 and 15 kDa. High-percentage (15%) or gradient (4%-20%) Tris-glycine gels are recommended to efficiently resolve them. Starting material, purified histones, and any optional samples collected throughout the purification can be run together on the same gel. See Figure 2B for an example of how the final histone sample purity compares with the original starting material (Fig. 2A).
66.Estimate histone concentration using a BCA or Bradford assay (see Current Protocols article: Olson & Markwell, 2007).
67.Mix approximately 1 µg of the histone extract with 2× Laemmli buffer.
68.Denature the proteins for 5 min at 95°C.
69.Run the sample on a pre-cast 15% or 4%-20% pre-cast Tris-glycine gel using Tris-glycine SDS (TGS) running buffer (also see Current Protocols article: Gallagher, 2012).
70.Stain the gel by slowly shaking in Coomassie solution for 1 hr at room temperature.
71.Destain the gel by slowly shaking in destaining solution. Replace the destaining solution every 30 min until the histone bands have become visible and the gel background is fully destained.
Basic Protocol 3: BOTTOM-UP MASS SPECTROMETRY ANALYSIS OF HISTONE PTMS AND HISTONE VARIANTS
In this protocol, the histone samples from Basic Protocol 2 are prepared and analyzed by MS. We use a bottom-up MS approach, where we first cleave the histones by trypsin into small peptides (5-15 amino acids in length), separate them by nano-scale reversed-phase chromatography (nanoLC), and then quantify them by tandem MS (MS/MS) (Karch, Sidoli, & Garcia, 2016; Sidoli, Bhanu, Karch, Wang, & Garcia, 2016a). However, our approach differs from standard protocols by the addition of two alkylation steps. First, we know that trypsin cuts after lysine and arginine residues and that histones are very basic, containing many of these amino acids. As a consequence, trypsin would normally cleave histones into peptides that are too small for optimal MS/MS analysis (Garcia et al., 2007). To address this, we use propionic acid to alkylate the ζ-amino groups of unmodified or monomethylated lysine residues, which prevents tryptic digestion at those sites, resulting in peptides that are longer and much more suitable. Second, once tryptic digestion is complete, we again use propionic acid, this time to alkylate each peptide's newly generated N-terminus. This aids the separation of peptides during nanoLC and further improves MS/MS results. To learn more, also about alternative MS approaches to study histone PTMs and variants, please consult Karch et al. (2016).
Once the MS/MS data have been collected, their analysis is challenging, most importantly because histones carry so many PTMs that can occur alone or in combination with others in a given peptide. This results in a large number of possible peptides that need to be distinguished from others. Another challenge is the existence of isobaric peptides that have the same m/z and similar elution times. Thus, we have developed a software application called EpiProfile (Yuan et al., 2015, 2018) to overcome these issues and provide a straightforward solution for the analysis of histone MS/MS data. Here, we use a C. elegans −adapted version of this software, called EpiProfile 2.0-Ce.
Materials
-
Purified histone samples in 50 mM NaHCO3 or Milli-Q water (Basic Protocol 2, step 64)
-
100 mM ammonium bicarbonate (NH4HCO3), pH 8.0
-
Glacial acetic acid
-
Propionic anhydride
-
Isopropanol
-
Ammonium hydroxide (NH4OH)
-
Trypsin (Promega)
-
Wash buffer: 0.1% acetic acid in water
-
Methanol
-
Elution buffer: 75% (v/v) acetonitrile/5% acetic acid/20% water
-
nanoLC buffer A: 0.1% (v/v) formic acid in water
-
nanoLC buffer B: 0.1% (v/v) formic acid in 75% acetonitrile/25% water
-
Vacuum centrifuge (e.g., Eppendorf Vacufuge)
-
pH paper
-
Vortex (e.g., Vortex Genie 2)
-
Thermal block (e.g., Eppendorf Thermomix)
-
1.5- or 2-ml microcentrifuge tubes
-
Home-made C18 columns or stage-tips
-
nano C18 column (length: 25 cm, silica tip (outer diameter: 360 µm, inner diameter: 75 µm; New Objective)
-
Orbitrap Fusion Mass Spectrometer (ThermoFisher Scientific)
Propionylation of lysine residues
1.Use ∼20 µg of purified histones from Basic Protocol 2, dry samples down in a vacuum centrifuge and resuspend them in 25 µl of 50 mM NH4HCO3, pH 8.0. Adjust sample concentration with concentrated NH4HCO3 if samples were in Milli-Q water, or dry samples down in a vacuum centrifuge and resuspend with NH4HCO3 if samples were in NaHCO3.
2.Use a P10 pipette and spot some sample onto pH paper, to ensure that the pH is around 8.0. Otherwise adjust it by adding glacial acetic acid or powdered NH4HCO3.
3.Make fresh propionylation reagent by combining propionic anhydride and isopropanol in a 1:3 (v/v) ratio.
4.Add 10 µl of propionylation reagent to the sample and vortex briefly.
5.Check the pH with pH paper. It now should be between 4 and 6.Immediately add 3-7 µl NH4OH to increase the pH to ∼8.
6.Incubate samples 15 min at 37°C.
7.Dry down to less than 5 µl in a vacuum centrifuge. This might take several minutes.
8.Repeat steps 1-7 once again with fresh reagents to ensure that >95% of the reactions are complete.
9.Resuspend samples in 100 µl of 100 mM NH4HCO3, pH 8.0.
Tryptic digestion of the propionylated histones
10.Add trypsin in a 1:20 ratio, i.e., 1 µg trypsin per 20 µg histones.
11.Incubate 6 hr to overnight at 37°C.
12.Quench trypsin by freezing at −80°C or by adding 5 µl glacial acetic acid to lower the pH to ∼4.
13.Dry sample down to 5 µl in a vacuum centrifuge.
Propionylation of the N-termini of the tryptic peptides
14.Add 15 µl 100 mM NH4HCO3, pH 8.0, for a final volume of 20 µl and confirm that the pH has remained around 8.
15.Repeat the derivatization described in steps 1-7, using fresh reagents.
16.Dry sample down to less than 5 µl in a vacuum centrifuge. This might take several minutes.
17.Add 200 µl of wash buffer to the sample. Check that the pH is below 4.0 with pH paper. If needed, adjust the pH by adding glacial acetic acid.
Sample desalting with C18 Stage-Tip
This step will remove salt from the digested sample, which otherwise may interfere with the nanoLC-MS/MS analysis.
18.Construct home-made C18 columns or stage-tips as described in Sidoli et al. (2016a). Briefly, punch a C18 material disk with a P1000 pipette tip previously cut to make the hole larger, push the minidisk out of the P1000 tip with the help of a fused silica capillary, and place it firmly to the bottom of a P100/200 pipette tip. If desalting more than 25 µg of protein, use two C18 minidisks in the same P100/P200 tip.
19.Place the column or stage-tips in a 1.5-ml or 2-ml microcentrifuge tube. Use a centrifuge adapter to hold the column or tip in place.
20.Activate the resin by adding 50 µl methanol to the column/tip and spinning 30-60 s at 500 × g.
21.Repeat step 20.
22.Equilibrate the column by adding 200 µl wash buffer and spinning 30-60 s at 500 × g.
23.Discard flow-through in the collection tube and repeat step 22.
24.Apply sample from step 17 to the column/stage-tip.
25.Centrifuge 2-5 min at 200 × g , until all the sample has passed through the column.
26.Wash the column with 50 µl wash buffer and centrifuge 30-60 s at 500 × g.
27.Repeat step 26.
28.Place the column into a new 1.5-ml microcentrifuge tube and elute the sample with 75 µl elution buffer by spinning 2-5 min at 200 × g.
29.Repeat step 28.
30.Dry down the desalted sample to less than 5 µl in a vacuum centrifuge.
MS acquisition
In this protocol, bottom-up MS data are acquired through a combination of data-dependent acquisition (DDA) with targeted MS2 scans to distinguish coeluting isobaric peptides (see Table 3). To eliminate the need for targeting coeluting isobaric peptides, data-independent acquisition (DIA) methods can be a good alternative to DDA, although they are not recommended for identifying unknown peptides. For details on how to perform DIA consult Karch et al. (2016).
| Mass list of targeted isobaric peptides | ||||
|---|---|---|---|---|
| Targeted isobaric peptide | m/z | Z | t start (min) | t stop (min) |
| H3K36me3/K27me2K36me1 | 543.9860 | 3 | 20 | 40 |
| H3.3K36me3/K27me2K36me1 | 558.6674 | 3 | 20 | 40 |
| H3K9ac/K14ac | 528.2988 | 2 | 20 | 40 |
| H3S10phK14ac/S10phK9ac | 568.2790 | 2 | 25 | 40 |
| H3K18me1/K23me1 | 584.8561 | 2 | 30 | 50 |
| H2A 4_18 3ac | 862.9863 | 2 | 30 | 50 |
| H2A 4_18 2ac | 869.9941 | 2 | 30 | 50 |
| H4 4_17 3ac | 754.9308 | 2 | 30 | 50 |
| H2A 4_18 1ac | 877.0019 | 2 | 30 | 50 |
| H3K27me1/K36me1 | 829.4728 | 2 | 30 | 50 |
| H3.3L2K36me3/K27me2K36me1 | 574.0200 | 3 | 30 | 60 |
| H4 4_17 2ac | 761.9386 | 2 | 33 | 50 |
| H3.3K27me1/K36me1 | 851.4859 | 2 | 35 | 50 |
| H4 4_17 1ac | 768.9465 | 2 | 35 | 55 |
| H2AV 1_21 3ac | 794.7711 | 3 | 35 | 60 |
| H2AV 1_21 2ac | 799.4430 | 3 | 40 | 60 |
| H3K18ac/K23ac | 570.8404 | 2 | 40 | 60 |
| H2AV 1_21 1ac | 804.1148 | 3 | 40 | 60 |
| H3.3L2K27me1/K36me1 | 874.5251 | 2 | 50 | 70 |
- a
All the isobaric peptides targeted for MS2 are listed, including their m/z, their charge state (z), and the retention time coordinates (start and end) during which elution of the respective peptide is expected. Retention coordinates were determined empirically in previous test runs using comparable equipment.
31.Prepare nanoLC Buffers A (0.1% formic acid in water) and B (0.1% formic acid in a solution of 75% acetonitrile/25% water).
32.Program a 70-min HPLC gradient: starting at 4% B, then linearly increasing B to 35% over 60 min, followed by a linear increase to 95% B over 10 min. The flow rate should be 300 nl/min.
33.We use a nano C18 column, 25 cm long with a silica tip. The eluate is electro-sprayed into the mass spectrometer directly via the column's tip.
34.Program the MS acquisition method for DDA combined with targeted scans. Targeted peptides take priority. See Table 3 for a detailed mass list of the targeted isobaric peptides.
35.Resuspend the sample to ∼1 µg/µl in Buffer A.
36.Load 1-3 µg of sample onto the nanoLC column.
37.Run the nanoLC-MS/MS method as programmed in steps 31 and 32.
Data analysis using EpiProfile 2.0-Ce
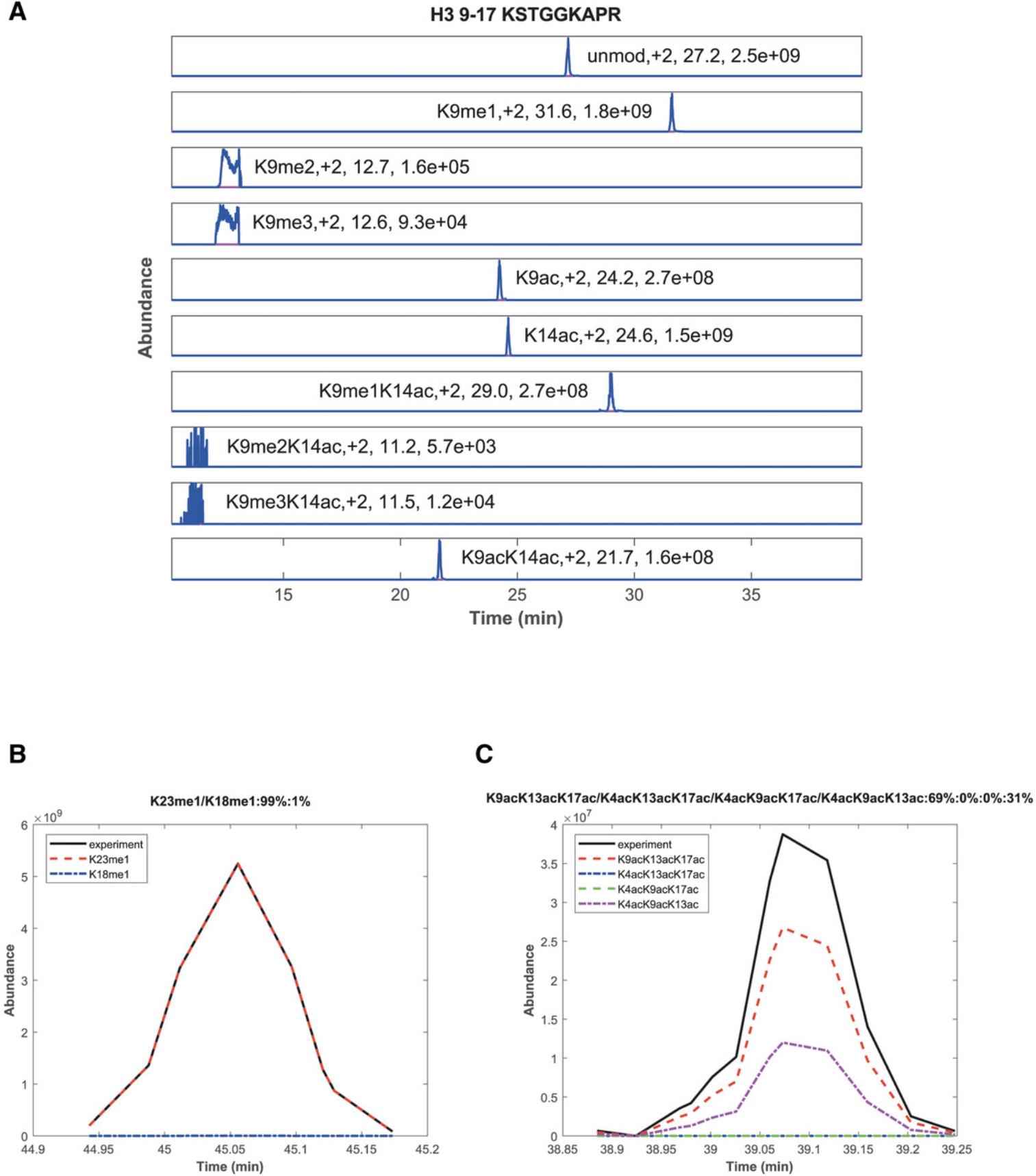
Although quantification of histone peptides can be achieved manually, e.g., by using the Xcalibur QualBrowser (Thermo) or by using the freeware Skyline (MacLean et al., 2010), we recommend the use of EpiProfile. This software has been specifically developed for the quantification of histone PTMs and variants from MS/MS data, including both DDA and DIA approaches (Yuan et al., 2015, 2018). EpiProfile is a freely available Matlab-based automated tool that reads raw data files and outputs tables of quantified histone peptides (see Table 4 for an example), layouts (MS1 elution profiles, Fig. 3A), and annotated MS/MS spectra used for identification. EpiProfile is very easy to use and can quantify coeluting isobaric peptides (Fig. 3B and 3C). Since its original release, EpiProfile has been adapted for the study of histones in different organisms including humans, mice, C. elegans , and yeast. We recommend consulting previous publications if you would like to obtain a deeper understanding of EpiProfile (Yuan et al., 2015, 2018).
| Quantification of endogenous histone peptides | |||
|---|---|---|---|
| Wild-type strain (N2) | |||
| Peptide | RT (min) | Area | Ratio |
| KSTGGKAPR(H3_9_17) | |||
| Unmod | 27.17 | 1.02E + 11 | 0.446667 |
| K9me1 | 31.61 | 6.59E + 10 | 0.288417 |
| K9me2 | 12.67 | 3.23E + 07 | 0.000141 |
| K9me3 | 12.61 | 2.04E + 07 | 0.000089 |
| K9ac | 24.25 | 8.89E + 09 | 0.038923 |
| K14ac | 24.61 | 3.62E + 10 | 0.158326 |
| K9me1K14ac | 29.02 | 1.48E + 10 | 0.064777 |
| K9me2K14ac | 11.23 | 3.42E + 04 | 0.000000 |
| K9me3K14ac | 11.46 | 2.15E + 05 | 0.000001 |
| K9acK14ac | 23.00 | 4.09E + 09 | 0.001792 |
- a
This is information taken from the EpiProfile Output File histone_ratios.xlsx. The retention time, area under the extracted ion chromatogram (XIC), and relative abundance (ratio) for each (un)modified form of the H3 9-17 peptide is shown. These data were obtained from histones of wild-type worms.
REAGENTS AND SOLUTIONS
B-Hypo-Lys
- 10 mM HEPES, pH 7.5
- 1 mM KCl
- 1.5 mM MgCl2
B-Hypo-Lys stock can be stored at 4°C. Just before its use, freshly add additives and inhibitors.
Buffer H, 2×
- 50 mM HEPES, pH 7.5
- 0.2 mM EDTA, pH 8.0
- 30% (v/v) glycerol
- 20 mM MgCl2
- 4% (v/v) NP-40 (Merck, 492016)
Buffer H (2×) stock can be stored at 4°C. Just before its use, further dilute it to 1× and freshly add additives and inhibitors to obtain Buffer H+.
Coomassie solution
- Prepare in Milli-Q water:
- 10% (v/v) acetic acid
- 25% (v/v) 2-propanol
- 0.25% (w/v) Coomassie Brilliant Blue R250
- Store up to several months at room temperature
Destaining solution
- Prepare in Milli-Q water:
- 10% (v/v) acetic acid
- 10% (v/v) 2-propanol
- Store up to several months at room temperature
Equilibration buffer
- 50 mM Tris·Cl, pH 8.0 (Current Protocols, 1998)
- 2 mM EDTA, pH 8.0
- Store up to several months at 4°C
H2SO4 , 0.4 N
- Bring 2.75 ml of concentrated H2SO4 to 250 ml with water. Solution can be stored at 4°C.
Histone elution buffer
- 50 mM Tris·Cl, pH 8.0 (Current Protocols, 1998)
- 2 mM EDTA, pH 8.0
- 2 M NaCl
- Store up to several months at 4°C
Histone wash buffer
- 50 mM Tris·Cl, pH 8.0 (Current Protocols, 1998)
- 2 mM EDTA, pH 8.0
- 0.5 M NaCl
- Store up to several months at 4°C
Hypochlorite solution
- 1 volume 10 M NaOH
- 4 volumes 10%-15% sodium hypochlorite
- 5 volumes sterile Milli-Q water
- Prepare it fresh before use
M9 buffer (2 L)
- 6 g KH2PO4
- 12 g Na2HPO4
- 10 g NaCl
- 2 L sterile Milli-Q water or equivalent
- Sterilize by autoclaving
- Store up to several months at room temperature
Nematode growth medium (NGM) plates (high peptone, with streptomycin and Nystatin, 1 L)
- 3 g NaCl
- 17 g agar
- 7.5 g Bacto peptone
- 975 ml sterile Milli-Q water or equivalent
- Autoclave
- After cooling, add the following:
- 1 ml 5 mg/ml cholesterol in ethanol
- 1 ml 1 M CaCl2
- 1 ml 1 M MgSO4
- 25 ml 1 M KPO4, buffer pH 6.0 (108.3 g KH2PO4, 35.6 g K2HPO4, H2O to 1 L)
- 1.25 ml 4400 U/ml Nystatin suspension
- 2 ml 100 mg/ml streptomycin
- Pour into appropriately sized Petri dishes while maintaining sterility
- Store up to 1 month at 4°C
Nuclear isolation buffer (NIB) and NIB+buffer
- Prepare NIB stock :
- 25 mM HEPES, pH 7.5
- 0.5 M sucrose
- 25 mM KCI
- 10 mM MgCl2
- 0.1 mM EDTA, pH 8.0
- Store NIB stock with the above ingredients up to several months at 4°C
S-Basal
- 5.85 g NaCl
- 1 g K2HPO4
- 6 g KH2PO4
- 1 ml 5 mg/ml cholesterol in ethanol
- Milli-Q water (or equivalent) to 1 L
- Sterilize by autoclaving
- Store up to several months at room temperature
S-Complete medium
- 1 L S Basal (see recipe)
- 10 ml 1 M potassium citrate, pH 6
- 10 ml trace metals solution (1.86 g disodium EDTA; 0.69 g FeSO4•7 H2O; 0.2 g MnCl2•4 H2O; 0.29 g ZnSO4 •7 H2O; 0.025 g CuSO4•5 H2O; add Milli-Q water (or equivalent) to 1 L; sterilize by autoclaving and store in the dark)
- 3 ml 1 M CaCl2
- 3 ml 1 M MgSO4
- 2 ml 100 mg/ml streptomycin
- 10 ml 4400 U/ml Nystatin suspension
- Mix components while maintaining sterility (do not autoclave)
- Prepare it fresh before use
COMMENTARY
Background Information
DNA is packaged as chromatin inside the eukaryotic nucleus. The simplest structural unit of chromatin is the nucleosome, composed of ∼147 bp of DNA that wrap around a histone octamer, comprising two copies each of the four core histones H2A, H2B, H3, and H4 (Kornberg, 1974; Luger, Mäder, Richmond, Sargent, & Richmond, 1997). A fifth type of histone, the linker histone H1, sits outside the nucleosome and helps to further stabilize and condense chromatin (Fyodorov, Zhou, Skoultchi, & Bai, 2018; Thoma & Koller, 1977). Chromatin structure and its level of compaction have a direct impact on most DNA-associated events, including transcriptional regulation, DNA repair, DNA replication, and cell division (Hauer & Gasser, 2017; Hübner, Eckersley-Maslin, & Spector, 2013; MacAlpine & Almouzni, 2013).
Similar to other proteins in the cell, histones can be post-translationally modified by different enzymes that deposit or remove chemical groups at individual amino acid residues. Most of these PTMs occur in histones’ exposed N- and C-terminal tails, but some also exist in their globular domains (Kouzarides, 2007). Methylation, acetylation, and phosphorylation are the most common and best-studied histone PTMs. Nevertheless, there is increasing evidence for other modifications (i.e., ADP-ribosylation, crotonylation, SUMOylation, or citrullination) that are now studied as well (Zhao & Garcia, 2015). Canonical histones are expressed during S phase of the cell cycle, transcribed from multiple gene copies that are organized in clusters within the genome. During evolution, many histone variants emerged from these canonical histones, all of which have altered biochemical and biophysical properties due to minor sequence differences, and thereby have acquired specific functions needed in specific contexts (Millán-Ariño, Izquierdo-Bouldstridge, & Jordan, 2016; Zink & Hake, 2016). In contrast to canonical histones, these variants are typically encoded only from a single gene locus and expressed throughout the cell cycle. Dynamics of both histone PTMs and histone variants are associated with different physiological processes such as cellular differentiation or the aging process (Atlasi & Stunnenberg, 2017; Booth & Brunet, 2016), and their imbalance or dysregulation is tightly linked with many pathologies (Feinberg, Koldobskiy, & Göndör, 2016). This also makes them a potential target to treat such pathologies (Morel, Jeffery, Aspeslagh, Almouzni, & Postel-Vinay, 2019).
The conventional methods of studying histone PTMs via western blotting or Luminex approaches are limited by the availability of specific antibodies recognizing all these modified residues. Although a broad palette of antibodies is currently available from different vendors, good antibodies are not available for all PTMs, PTM combinations, and variants. Moreover, differences in histone amino acid sequence between species can further limit the availability of good antibodies for less commonly studied organisms. Mass spectrometry methods have recently helped to overcome these issues. They make it possible to study and quantify dozens of histone PTMs in a single experiment, to quantify combinatorial occurrences of PTMs within the same peptides (Karch, DeNizio, Black, & Garcia, 2013), and to obtain this information in any organism. New modifications can even be discovered (Britton, Gonzales-Cope, Zee, & Garcia, 2011). By now, several studies have described mass spectrometry methods to quantify histone PTMs and variants (Huang, Lin, Garcia, & Zhao, 2015). However, what is crucial for the success of all these approaches is the quality of the histone material to be analyzed. While from some species, e.g., human cells, histones can be obtained rather easily by mere acid extraction, in other species like C. elegans this can be very challenging. Particular effort needs to be exercised in order to obtain high-purity nuclear histone preparations at sufficient yield while preserving their PTMs.
Critical Parameters and Troubleshooting
Synchrony and reproducibility of C. elegans cultures
The methodology described in this article has been developed to study differences in histone PTMs or variant abundance between different C. elegans samples. Since the epigenome, and thus these marks, can drastically change depending on environmental influences, including food availability (Berger & Sassone-Corsi, 2016), or on the developmental stage of the organism (Sidoli, Vandamme, Salcini, & Jensen, 2016b), it is important to avoid any variability in the culturing conditions and to make sure that all samples are perfectly synchronized to the same developmental stage or age. Thus, the protocols provided here for C. elegans liquid culture should be strictly followed, and culturing conditions (i.e., temperature and food availability) should be carefully monitored and kept as constant as possible between replicates and throughout the experiment. In particular, we recommend using a temperature tracker, which is placed inside the shaking incubator and checked daily. Furthermore, the abundance of OP50-1 E. coli in the medium is checked daily by OD600 measurement, to remain between 2 and 4 absorbance units. For this, 10 ml of culture are transferred to a 15-ml tube, the C. elegans are settled by gravity for 10 min, and then an aliquot of the supernatant is subjected to the OD measurement at 600 nm. Finally, the developmental stage of the animals and thus synchrony between samples and replicates should be checked daily during the first days of the experiment. Should any sample show delays in progressing through development, this delay should be quantified and the sample harvested at an accordingly later time.
Input material
The above set of protocols grows 0.5-1 million C. elegans and eventually recovers 20-30 µg of pure histones. However, sometimes more animals should be used. This is particularly true for certain strains, e.g., strains that have a retarded germline, for the analysis of animals from earlier developmental stages, or when you have an interest in quantifying particularly rare histone PTMs.
Achieving optimal yield
Besides increasing the amount of C. elegans used, there are a few additional points to consider in order to maximize your histone yield and preserve the histones’ integrity and PTMs. First, it is important to maintain an optimal ratio of biological material and extraction/washing buffers throughout the extraction procedure. Closely follow the ratios indicated in the protocol and scale the buffer amounts depending on the amount of starting material that you use (worm pellet after harvesting). Second, for optimal histone extraction, animals need to be efficiently lysed. In C. elegans , this requires breakage of the cuticle, which we achieve by grinding in a cryogenic mill. Grinding results should be checked under a microscope. Upon optimal grinding, all animals should be broken. Ideally, no worm structures should be visible anymore. Further homogenization and lysis is achieved by a Dounce homogenizer. Make sure to use a Dounce homogenizer appropriate for the sample volume and with the correct pestle type.
Histone preservation
Proteins and their PTMs are more stable at lower temperatures, when proteases or enzymes that alter their PTMs tend to be less active. Thus, the histone extraction and purification should always be performed on ice or inside a cold room. For similar reasons, we recommend storing all collected samples at −80°C rather than 4°C or −20°C. It is furthermore important to use protease inhibitors and stabilizing agents during the extraction procedure. The choice of stabilizing agents may be adjusted, depending on the histone modifications that you are interested in. In the protocols presented here, microcystin is used to preserve phosphorylations and sodium butyrate is used to preserve acetylations.
Ion-exchange chromatography
One of the main benefits of the protocols described above is the purity of the resulting histone samples. This is achieved by incorporation of an ion-exchange chromatography step, which removes most non-histone contaminants. As the purity is so high, no further purification (e.g., gel-purification or HPLC) is required before trypsin digestion and nanoLC-MS/MS. However, errors in executing the ion-exchange chromatography may lead to impurities that interfere with nanoLC-MS/MS. We therefore recommend always evaluating the purity of your final histone sample by analytical SDS-PAGE and Coomassie staining. If substantial contamination from non-histone proteins can be seen, further purification by ion-exchange chromatography or gel purification should be considered; for future purifications, the ion-exchange chromatography should be conducted more carefully. Most importantly, make sure to properly neutralize the acid-extracted samples to a pH of 7-8 before loading them onto the column.
Histone alkylation and digestion
For a good analysis by MS/MS, histones should be optimally propionylated and the tryptic digestion should go to completion. To achieve this, multiple measures should be taken. In the purification protocol's final precipitation step, make sure to properly wash the histone pellet with acetone as described. Any traces of acid that remain with the sample would critically interfere with derivatization and digestion. We also suggest performing the propionylation quickly, not treating too many samples at the same time. And, before initiating the trypsin digestion, carefully confirm that the pH is ∼8.0. Issues with efficient alkylation and digestion would otherwise lead to low detection of the desired peptides.
Statistical Analysis
For the creation of the heat map in Figure 4, the following linear model was used: log2(mark) ∼ date + condition. This allowed us to account for batch effects and to obtain log2 fold-change values. To account for the high number of tests performed, we corrected our p -values for multiple testing using the False Discovery Rate method. Corrected p -values were computed using the q -value function from the R package q -value. Contrasts were manually set in order to compare different conditions. Clusters for the heatmap were computed by agglomerative hierarchical clustering using the hclust function. All analyses were performed in R version 3.6.3 (R Core Team, 2013) using the packages tidyverse (Wickham, 2016), magrittr (Milton Bache & Wickham, 2014), and Biobase (Huber et al., 2015).
![Details are in the caption following the image Bottom-up mass spectrometry uncovers differences in the relative abundance of histone PTMs between wild-type (N2), long-lived [daf-2(e1370ts)], and short-lived [daf-16(mu86lf)] C. elegans. (A) A heatmap of the changes in H3 and H4 PTM abundances for the comparisons of daf-2(e1370ts) versus wild-type and wild-type versus daf-16(mu86lf) animals. Significance of up- or downregulated PTMs is indicated by asterisks (* = 0.1 < p > 0.05; ** = 0.05 < p > 0.01; *** = 0.01 < p > 0.001; **** = 0.001 < p > 0.0001; ***** = 0.0001 < p > 0; p-values were corrected for multiple testing by the False Discovery Rate method). (B) Combinatorial histone PTM landscape of the H3 18-23 peptide in the indicated C. elegans strains. Relative ratios of the unmodified and modified forms of the peptide are shown, as calculated by EpiProfile 2.0-Ce. Error bars represent the standard error across three MS runs.](https://static.yanyin.tech/literature_test/cpps114-fig-0004-m.jpg)
Understanding Results
The quantification of histone post-translational modifications in C. elegans can be a powerful tool to study how the epigenome contributes to specific aspects of this organism's physiology. To provide an example, we used it to determine, how histone PTMs differ between animals that age either quickly, normally, or slowly.
The nutrient-sensing insulin/IGF-like signaling (IIS) pathway is one of the best characterized signaling pathways that regulates the rate at which we age (Kimura, Tissenbaum, Liu, & Ruvkun, 1997; Tatar et al., 2001). When there are plenty of nutrients and IIS is high, aging occurs rather quickly, but when IIS is low, aging is slowed down. Mostly, the latter situation is caused by a transcription factor called DAF-16/FOXO. This transcription factor is negatively regulated by IIS and drives the expression of many aging-preventive genes (Murphy et al., 2003; Zhou et al., 2018).
For this study, we chose wild-type C. elegans , which age at a normal rate, a hypomorph of the insulin/IGF-like receptor daf-2 (_daf-_2(e1370ts)), which ages slowly and eventually reaches a very long lifespan, and a null mutant of daf-16 (daf-16(mu86lf)), which ages quickly and reaches only a very short lifespan. We brought these animals to scale, synchronized them, added 50 µM 5-fluoro-2′-deoxyuridine (FUDR) starting at the L4 stage to prevent progeny production, and eventually grew them until day 2 of adulthood. Animals were then harvested and lysed, and histones extracted, purified, and analyzed by nanoLC-MS/MS—all according to our protocols above. We then used EpiProfile 2.0-Ce to analyze the MS/MS data and quantify the histone PTMs and variants. EpiProfile uses previous knowledge about relative elution times for the identification and quantification of histone peptides [for details consult (Yuan et al., 2015, 2018)]. For a given modification on a peptide backbone, EpiProfile calculates its relative abundance or ratio by dividing its area under the MS1 chromatogram by the sum of areas of all (un)modified forms of this same peptide backbone. This information is found in the output file histone_ratios.xlsx (see Table 4 for an example). It allows for the study of all possible modified forms of each peptide, and thus also the study of combinatorial histone PTMs and PTM switches within a given peptide. Besides that, EpiProfile also calculates the relative abundance of individual histone PTMs by summing the relative abundance of all peptides that contain a given modification. These data are found in the output file histone_ratios_single_PTMs.xlsx.
Relative quantification of PTMs on histones H3 and H4 (found in the histone_ratios_single_PTMs.xlsx file) revealed substantial differences in the abundance of certain histone marks in slow-, normally, and fast-aging animals. The heat map in Figure 4A illustrates these differences on a log2-fold scale. Certain marks are positively associated with animals that age more slowly (e.g., H3K18ac or H4K20ac), while others are rather negatively associated (e.g., H3K23me2/3 or H4K20me1). Our data indicate dynamic changes in histone residues H3K18, H3K23, and H4K20, from methylated to acetylated states in animals that age slowly. When looking at the relative abundance of all forms of the H3 18-26 peptide (found in the histone_ratios.xlsx file), we observed that the increasing acetylation at H3K18 coincides with acetylation of H3K23 of the same peptide (Fig. 4B). We conclude that H4K20ac and likely double-acetylation of H3 at K18 and K23 are marks associated with slow aging and longevity.
Time Considerations
The time to grow strains on a large scale before harvesting (Basic Protocol 1) is variable and depends on diverse parameters, e.g., the temperature, the strains used, the desired worm stage to harvest, etc. Whatever the case, the following should give you some guidance. The initial growth of C. elegans until they fill multiple 15-cm plates may take approximately 2 weeks. Egg preparation from gravid adults takes around 2 hr. Hatching of the embryos out of their eggshell and their arrest as L1 larvae is reached after 18 hr at room temperature or 2 days at 15°C. The growth in liquid culture will strongly depend on the stage of animals that you would like to harvest, and can take anywhere from 1 to many days. The harvesting of the liquid culture and freezing of the animals will take about 3 hr.
For Basic Protocol 2, the grinding procedure may take 1 to 2 hr. The resulting worm powder can be stored at −80°C or directly used for nuclear preparation and histone acid extraction. Those steps take 1 full day before leaving samples rotating overnight at 4°C with H2SO4. Another full day is required for the histone purification by cation-exchange chromatography and for the histone precipitation step. Quality control of histone samples by SDS-PAGE and Coomassie staining takes around 4 to 6 hr, including the time it takes to run the protein gels and the time for staining and destaining of the gel.
Finally, sample preparation for nanoLC-MS/MS and the actual nanoLC-MS/MS run (Basic Protocol 3) take 2-3 days. Data analysis by EpiProfile 2.0-Ce is highly automated and takes only a few minutes to run. However, additional analyses for data representation might take longer.
Acknowledgments
We thank the Proteomics Biomedicum core facility at Karolinska Institute for MS measurements. We thank Stefan Kubicek for experimental advice. We thank the Caenorhabditis Genetic Center (CGC) for C. elegans strains. We thank all Riedel lab members for helpful discussions. Open access funding was provided by Karolinska Institute. B.A.G. was supported by the NIH grants AI118891, CA196539, and GM110104. C.G.R. was supported by the Swedish Research Council (VR) grants 2015-03740, 2017-06088, and 2019-04868, the COST grant BM1408 (GENiE), and an ICMC project grant.
Author Contributions
Lluís Millan-Ariño : Data curation; formal analysis; investigation; methodology; writing-original draft; writing-review & editing. Zuo-Fei Yuan : Data curation; formal analysis; methodology; software; validation; writing-review & editing. Marlies E. Oomen : Formal analysis; investigation; methodology. Simone Brandenburg : Investigation; methodology. Alexey Chernobrovkin : Data curation; formal analysis; investigation; methodology. Jérôme Salignon : Data curation; formal analysis; investigation. Lioba Körner : Investigation; methodology. Roman A. Zubarev : Resources; supervision. Benjamin A. Garcia : Resources; software; supervision. Christian G. Riedel : Conceptualization; funding acquisition; resources; supervision; writing-original draft; writing-review & editing.
Literature Cited
- Atlasi, Y., & Stunnenberg, H. G. (2017). The interplay of epigenetic marks during stem cell differentiation and development. Nature Reviews Genetics , 18(11), 643–658. doi: 10.1038/nrg.2017.57.
- Berger, S. L., & Sassone-Corsi, P. (2016). Metabolic signaling to chromatin. Cold Spring Harbor Perspectives in Biology , 8, a019463. doi: 10.1101/cshperspect.a019463.
- Booth, L. N., & Brunet, A. (2016). The aging epigenome. Molecular Cell , 62, 728–744. doi: 10.1016/j.molcel.2016.05.013.
- Britton, L. M. P., Gonzales-Cope, M., Zee, B. M., & Garcia, B. A. (2011). Breaking the histone code with quantitative mass spectrometry. Expert Review of Proteomics , 8, 631–643. doi: 10.1586/epr.11.47.
- Current Protocols (1998). Commonly used reagents. Current Protocols in Protein Science , 12, A.2E.1–A.2E.5. doi: 10.1002/0471140864.psa02es12.
- Feinberg, A. P., Koldobskiy, M. A., & Göndör, A. (2016). Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nature Reviews Genetics , 17, 284–299. doi: 10.1038/nrg.2016.13.
- Fyodorov, D. V., Zhou, B. R., Skoultchi, A. I., & Bai, Y. (2018). Emerging roles of linker histones in regulating chromatin structure and function. Nature Reviews Molecular Cell Biology , 19, 192–206. doi: 10.1038/nrm.2017.94.
- Gallagher, S. (2012). One-dimensional SDS gel electrophoresis of proteins. Current Protocols in Protein Science , 68, 10.1.1–10.1.44. doi: 10.1002/0471140864.ps1001s68.
- Garcia, B. A., Mollah, S., Ueberheide, B. M., Busby, S. A., Muratore, T. L., Shabanowitz, J., & Hunt, D. F. (2007). Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nature Protocols , 2, 933–938. doi: 10.1038/nprot.2007.106.
- Hauer, M. H., & Gasser, S. M. (2017). Chromatin and nucleosome dynamics in DNA damage and repair. Genes and Development , 31, 2204–2221. doi: 10.1101/gad.307702.117.
- Huang, H., Lin, S., Garcia, B. A., & Zhao, Y. (2015). Quantitative proteomic analysis of histone modifications. Chemical Reviews , 115, 2376–2418. doi: 10.1021/cr500491u.
- Huber, W., Carey, V. J., Gentleman, R., Anders, S., Carlson, M., Carvalho, B. S., … Morgan, M. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods , 12, 115–121. doi: 10.1038/nmeth.3252.
- Hübner, M. R., Eckersley-Maslin, M. A., & Spector, D. L. (2013). Chromatin organization and transcriptional regulation. Current Opinion in Genetics and Development , 23, 89–95. doi: 10.1016/j.gde.2012.11.006.
- Karch, K. R., DeNizio, J. E., Black, B. E., & Garcia, B. A. (2013). Identification and interrogation of combinatorial histone modifications. Frontiers in Genetics , 4, 264. doi: 10.3389/fgene.2013.00264.
- Karch, K. R., Sidoli, S., & Garcia, B. A. (2016). Identification and quantification of histone PTMS using high-resolution mass spectrometry. Methods in Enzymology , 574, 3–29. doi: 10.1016/bs.mie.2015.12.007.
- Kimura, K. D., Tissenbaum, H. A., Liu, Y., & Ruvkun, G. (1997). Daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science , 277, 942–946. doi: 10.1126/science.277.5328.942.
- Kornberg, R. D. (1974). Chromatin structure: A repeating unit of histones and DNA. Science , 184(4139), 868–871. doi: 10.1126/science.184.4139.868.
- Kouzarides, T. (2007). Chromatin modifications and their function. Cell , 128, 693–705. doi: 10.1016/j.cell.2007.02.005.
- Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F., & Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature , 389, 251–260. doi: 10.1038/38444.
- MacAlpine, D. M., & Almouzni, G. (2013). Chromatin and DNA replication. Cold Spring Harbor Perspectives in Biology , 5, a010207–a010207. doi: 10.1101/cshperspect.a010207.
- MacLean, B., Tomazela, D. M., Shulman, N., Chambers, M., Finney, G. L., Frewen, B., … MacCoss, M. J. (2010). Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics , 26, 966–968. doi: 10.1093/bioinformatics/btq054.
- Millán-Ariño, L., Izquierdo-Bouldstridge, A., & Jordan, A. (2016). Specificities and genomic distribution of somatic mammalian histone H1 subtypes. Biochimica et Biophysica Acta−Gene Regulatory Mechanisms , 1859, 510–519. doi: 10.1016/j.bbagrm.2015.10.013.
- Milton Bache, S., & Wickham, H. (2014). Magrittr: A forward-pipe operator for R. Available at https://cran.r-project.org/web/packages/magrittr/index.html.
- Morel, D., Jeffery, D., Aspeslagh, S., Almouzni, G., & Postel-Vinay, S. (2019). Combining epigenetic drugs with other therapies for solid tumours—past lessons and future promise. Nature Reviews Clinical Oncology , 17(2), 91–107.
- Murphy, C. T., McCarroll, S. A., Bargmann, C. I., Fraser, A., Kamath, R. S., Ahringer, J., … Kenyon, C. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature , 424, 277–284. doi: 10.1038/nature01789.
- Olson, J. S. C., & Markwell, J. (2007). Assays for determination of protein concentration. Current Protocols in Protein Science , 48, 3.4.1–3.4.29. doi: 10.1002/0471140864.ps0304s48.
- R Core Team (2013). R: A language and environment for statistical computing. Retrieved from http://www.r-project.org/.
- Rodriguez-Collazo, P., Leuba, S. H., & Zlatanova, J. (2009). Robust methods for purification of histones from cultured mammalian cells with the preservation of their native modifications. Nucleic Acids Research , 37, e81. doi: 10.1093/nar/gkp273.
- Sidoli, S., Bhanu, N. V., Karch, K. R., Wang, X., & Garcia, B. A. (2016a). Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: From histone extraction to data analysis. Journal of Visualized Experiments , 54112. doi: 10.3791/54112.
- Sidoli, S., Vandamme, J., Salcini, A. E., & Jensen, O. N. (2016b). Dynamic changes of histone H3 marks during Caenorhabditis elegans lifecycle revealed by middle-down proteomics. Proteomics , 16, 459–464. doi: 10.1002/pmic.201500285.
- Tatar, M., Kopelman, A., Epstein, D., Tu, M. P., Yin, C. M., & Garofalo, R. S. (2001). A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science , 292, 107–110. doi: 10.1126/science.1057987.
- Thoma, F., & Koller, T. (1977). Influence of histone H1 on chromatin structure. Cell , 12, 101–107. doi: 10.1016/0092-8674(77)90188-X.
- Wickham, H. (2016). tidyverse: Easily Install and Load “Tidyverse” Packages. Available at https://tidyverse.tidyverse.org/.
- Yuan, Z. F., Lin, S., Molden, R. C., Cao, X. J., Bhanu, N. V., Wang, X., … Garcia, B. A. (2015). Epiprofile quantifies histone peptides with modifications by extracting retention time and intensity in high-resolution mass spectra. Molecular and Cellular Proteomics , 14, 1696–1707. doi: 10.1074/mcp.M114.046011.
- Yuan, Z. F., Sidoli, S., Marchione, D. M., Simithy, J., Janssen, K. A., Szurgot, M. R., & Garcia, B. A. (2018). EpiProfile 2.0: A computational platform for processing epi-proteomics mass spectrometry data. Journal of Proteome Research , 17, 2533–2541. doi: 10.1021/acs.jproteome.8b00133.
- Zhao, Y., & Garcia, B. A. (2015). Comprehensive catalog of currently documented histone modifications. Cold Spring Harbor Perspectives in Biology , 7, a025064. doi: 10.1101/cshperspect.a025064.
- Zhou, X., Sen, I., Lin, X.-X., & Riedel, C. G. (2018). Regulation of age-related decline by transcription factors and their crosstalk with the epigenome. Current Genomics , 19, 464–482. doi: 10.2174/1389202919666180503125850.
- Zink, L. M., & Hake, S. B. (2016). Histone variants: Nuclear function and disease. Current Opinion in Genetics and Development , 37, 82–89. doi: 10.1016/j.gde.2015.12.002.
- Zougman, A., & Wiśniewski, J. R. (2006). Beyond linker histones and high mobility group proteins: Global profiling of perchloric acid soluble proteins. Journal of Proteome Research , 5, 925–934. doi: 10.1021/pr050415p.
Citing Literature
Number of times cited according to CrossRef: 4
- Kazuya Okami, Shintaro Fumoto, Mana Yamashita, Moe Nakashima, Hirotaka Miyamoto, Shigeru Kawakami, Koyo Nishida, One-Step Formation Method of Plasmid DNA-Loaded, Extracellular Vesicles-Mimicking Lipid Nanoparticles Based on Nucleic Acids Dilution-Induced Assembly, Cells, 10.3390/cells13141183, 13 , 14, (1183), (2024).
- Weilai Wang, Tong Liu, Xiujuan Wang, Fengming Chen, Xuesong Feng, Feng Zhang, Fully Integrated Recognition and Enrichment Electrospray Ionization Source for High-Sensitivity Mass Spectrometry Determination of Bioamine, Analytical Chemistry, 10.1021/acs.analchem.4c02607, (2024).
- Valerie J. Robert, Matthieu Caron, Loic Gely, Annie Adrait, Victoria Pakulska, Yohann Couté, Manon Chevalier, Christian G. Riedel, Cecile Bedet, Francesca Palladino, SIN-3 acts in distinct complexes to regulate the germline transcriptional program in Caenorhabditis elegans , Development, 10.1242/dev.201755, 150 , 21, (2023).
- Jiawei Liu, Fengming Chen, Yang song, Yiping Chen, Feng Zhang, Construction of a Highly Selective Enrichment, Ionization, and Detection Platform Based on a Broad-Spectrum Antibody, Analytical Chemistry, 10.1021/acs.analchem.3c01098, 95 , 29, (10984-10991), (2023).

