Exploring Chemical Information in PubChem
Sunghwan Kim, Sunghwan Kim
cheminformatics
chemical structure search
drug discovery
molecular similarity
PubChem
public database
Abstract
PubChem (https://pubchem.ncbi.nlm.nih.gov) is a public chemical database that serves scientific communities as well as the general public. This database collects chemical information from hundreds of data sources and organizes them into multiple data collections, including Substance, Compound, BioAssay, Protein, Gene, Pathway, and Patent. These collections are interlinked with each other, allowing users to discover related records in the various collections (e.g., drugs targeting a protein or genes modulated by a chemical). PubChem can be searched by keyword (e.g., a chemical, protein, or gene name) as well as by chemical structure. The input structure can be provided using popular line notations or drawn with the PubChem Sketcher. PubChem supports various types of structure searches, including identity search, 2-D and 3-D similarity searches, and substructure and superstructure searches. Results from multiple searches can be combined using Boolean operators (i.e., AND, OR, and NOT) to formulate complex queries. PubChem allows the user to quickly retrieve a list of records annotated with a particular classification or ontological term. This paper provides step-by-step instructions on how to explore PubChem data with examples of commonly requested tasks. © 2021. This article is a U.S. Government work and is in the public domain in the USA. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Finding genes and proteins that interact with a given compound
Basic Protocol 2 : Finding drug-like compounds similar to a query compound through a two-dimensional (2-D) similarity search
Basic Protocol 3 : Finding compounds similar to a query compound through a three-dimensional (3-D) similarity search
Support Protocol : Computing similarity scores between compounds
Basic Protocol 4 : Getting the bioactivity data for the hit compounds from substructure search
Basic Protocol 5 : Finding drugs that target a particular gene
Basic Protocol 6 : Getting bioactivity data of all chemicals tested against a protein.
Basic Protocol 7 : Finding compounds annotated with classifications or ontological terms
Basic Protocol 8 : Finding stereoisomers and isotopomers of a compound through identity search
INTRODUCTION
PubChem (https://pubchem.ncbi.nlm.nih.gov; Kim, 2016; Kim et al., 2019; Kim et al., 2021; Kim et al., 2016) is a public chemical database created by the National Library of Medicine (NLM), an institute within the U.S. National Institutes of Health (NIH). With millions of unique users every month, PubChem is a very popular chemistry information resource for biomedical research communities in many areas, including cheminformatics, chemical biology, medicinal chemistry, and drug discovery. Importantly, PubChem also serves as a source of big data in chemistry, used in many machine learning and data science projects for virtual screening, computational toxicology, drug repurposing, etc.
PubChem's information content, collected from hundreds of data sources, is organized into multiple data collections, including Substance, Compound, BioAssay, Gene, Protein, Pathway, and Patent (Kim et al., 2021). Substance archives the chemical data submitted by individual data sources and Compound stores the unique chemical structures extracted from Substance through chemical structure standardization (Hähnke, Kim, & Bolton, 2018; Kim et al., 2016). BioAssay contains biological assay descriptions and test results deposited by assay data providers. The record identifiers (IDs) used in Substance, Compound, and BioAssay are called Substance ID (SID), Compound ID (CID), and Assay ID (AID), respectively. The other data collections (i.e., Gene, Protein, Pathway, and Patent) provide alternative views of PubChem data, related to a specific gene, protein, pathway, and patent document, respectively. Each record in the data collections has a dedicated web page (called a Summary page), which presents information available in PubChem for that record. This page also presents relevant annotations collected by PubChem from authoritative data sources.
PubChem's search interface, available on the PubChem homepage (https://pubchem.ncbi.nlm.nih.gov), allows users to simultaneously search the data collections using a text query. A chemical structure query can be used to perform various types of chemical structure searches, including identity, two-dimensional (2-D) and three-dimensional (3-D) similarity, and substructure and superstructure searches. In addition, PubChem provides various tools and services that help users to exploit PubChem data, which are described in detail in previous papers (Kim et al., 2019; Kim et al., 2021; Kim et al., 2016).
This article provides step-by-step instructions on how to perform common tasks in PubChem. In Basic Protocol 1, losartan (an antihypertensive drug) is used as an example to explain how to search PubChem by chemical name and find genes and proteins that interact with that chemical. Basic Protocols 2 and 3 focus on 2-D and 3-D similarity searches, respectively, which are described in detail in Background Information. Basic Protocol 2 shows how to find compounds structurally similar to losartan based on 2-D similarity and how to filter them based on molecular properties to identify drug-like compounds. Basic Protocol 3 demonstrates how to find compounds similar to losartan in terms of 3-D similarity. In the Support Protocol, similarity scores between compounds are computed using the PubChem Score Matrix Service. In Basic Protocol 4, a substructure search is performed to identify compounds that share a common scaffold with losartan, and their bioactivity data is downloaded. Basic Protocol 5 shows how to search drugs that target a particular gene, and Basic Protocol 6 explains how to retrieve the bioactivity data for compounds tested against a given protein. In Basic Protocol 7, the PubChem Classification Browser is used to find compounds annotated with a classification or ontological term (e.g., antihypertensive agents). Finally, Basic Protocol 8 details how to perform an identity search to find stereoisomers and isotopomers of a given compound, using valsartan as an example.
Basic Protocol 1: FINDING GENES AND PROTEINS THAT INTERACT WITH A GIVEN COMPOUND
The most common use of PubChem is to search for a specific piece of information on a chemical. This is typically done by performing a text search with a chemical name as a query, going to the Summary page of the best hit compound returned from the search, and locating the desired information on that page. This process is shown in Basic Protocol 1, which demonstrates how to find proteins and genes known to interact with losartan (CID 3961), a widely used antihypertensive drug.
The chemical-protein and chemical-gene interaction data in PubChem originate from multiple sources, such as DrugBank (Wishart et al., 2018), Comparative Toxicogenomics Database (CTD; Davis et al., 2021), Drug-Gene Interaction Database (DGIdb; Freshour et al., 2021), IUPHAR/BPS Guide to PHARMACOLOGY (Armstrong et al., 2020), ChEMBL (Mendez et al., 2019), and RCSB Protein Data Bank (PDB; Burley et al., 2019). The biological test results for a chemical can also be a good source for its interactions with macromolecules. While the interaction data from DrugBank is retrieved in Basic Protocol 1 as an example, the chemical-macromolecule associations from one data source are not necessarily the same as those from other sources. Therefore, it is recommended to access the data from all relevant sources and review the variances in the related records.
Materials
- An up-to-date Web browser, such as Google Chrome, Microsoft Edge, Safari, or Firefox, is required for this protocol (and all other protocols in this article)
1.Go to the PubChem homepage (https://pubchem.ncbi.nlm.nih.gov).
2.Type losartan in the search box and click on the search (magnifying glass) button (‘1’ in Fig. 1).
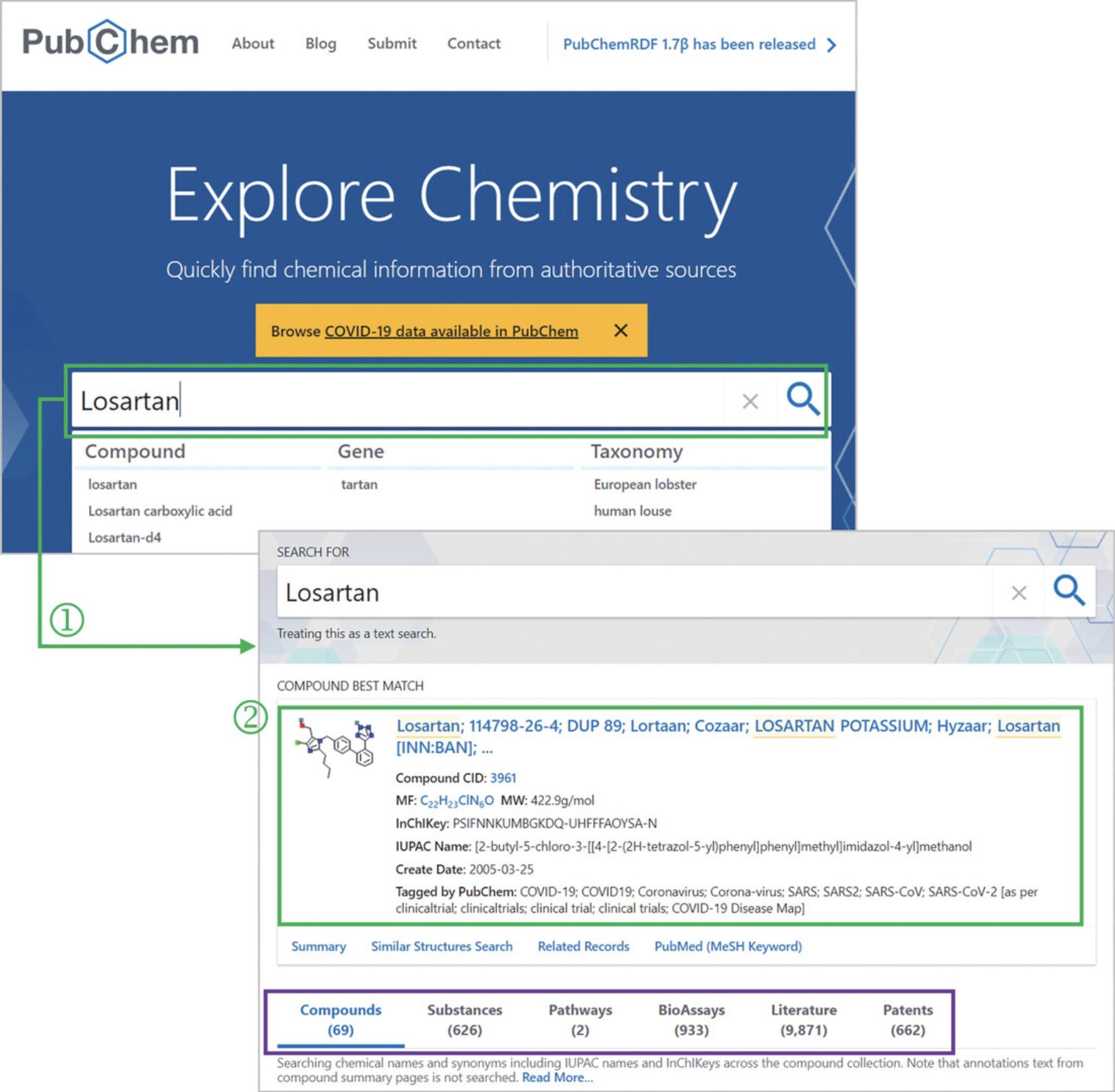
3.Click the best match shown at the top of the search results (‘2’ in Fig. 1) to go to the Summary page for the selected compound.
4.Go to the “DrugBank Interactions” subsection under the “Biomolecular Interactions and Pathways” section (‘2’ in Fig. 2), using the Table of Contents on the right column (‘1’ in Fig. 2).
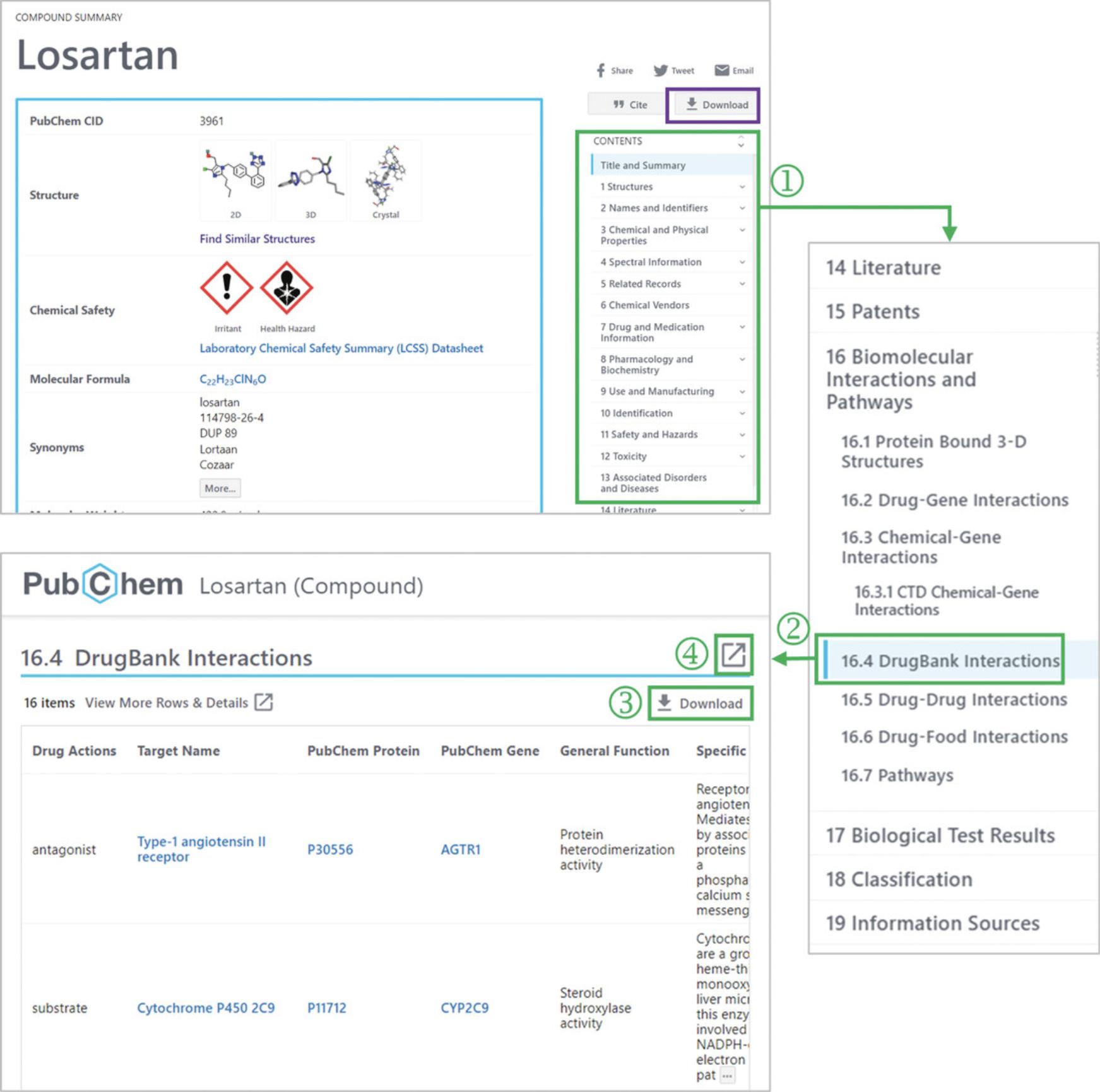
5.Click the “Download” button to download the list of the macromolecules interacting with losartan (‘3’ in Fig. 2).
6.If necessary, click the full-screen view button (‘4’ in Fig. 2) to view all rows and columns.
Basic Protocol 2: FINDING DRUG-LIKE COMPOUNDS SIMILAR TO A QUERY COMPOUND THROUGH 2-D SIMILARITY SEARCH
PubChem's search interface provides many features beyond the simple text search. For example, it supports a search by chemical structure. A chemical structure can be used as a query for various types of structure searches, including identity search, 2-D and 3-D similarity searches, and substructure and superstructure searches. The input chemical structure can be specified with a line notation (e.g., SMILES or InChI) or drawn using the PubChem Sketcher. If the input structure already exists in the PubChem Compound database, its CID can be used as a query. It is also possible to initiate a chemical structure search from one of the hit compounds returned from a previous search. More details about chemical structure searches in PubChem are outlined in the Background Information section of this article.
Another important feature of PubChem's search interface is that it provides filters that limit the search results to only those records with the desired attributes. Each data collection has a different set of filters. For example, the compound records can be filtered based on several molecular properties, such as molecular weight, hydrogen bond donor and acceptor counts, rotatable bond count, etc. The assay records can be filtered based on data sources and assay types (e.g., in vivo, in vitro, cell-based, biochemical, etc.). Taxonomy information can be used to filter the gene and protein records.
Basic Protocol 2, designed to demonstrate these two features (i.e., chemical structure search and filtering), aims to find drug-like chemicals that are structurally similar to a given chemical. In this protocol, the CID of the best hit compound returned from the text query losartan (in Basic Protocol 1) is used to specify the input chemical structure for a subsequent 2-D similarity search. The resulting compound list is further refined with filters to identify compounds that meet all criteria of Lipinski's rule of five (Lipinski, Lombardo, Dominy, & Feeney, 1997), which is a rule of thumb to evaluate drug-likeness of molecules. The refined compound list, along with the computed properties, is downloaded on a local computer.
Materials
- An up-to-date Web browser, such as Google Chrome, Microsoft Edge, Safari, or Firefox, is required for this protocol (and all other protocols in this article)
1.Repeat steps 1-2 of Basic Protocol 1 to search PubChem using losartan as a query.
2.Click the “Similar Structures Search” link at the bottom of the top panel that shows the best match (‘1’ in Fig. 3).
![Details are in the caption following the image Performing a similarity search using a hit compound returned from a previous search. Each hit compound is presented with links that allow the user to access commonly requested data or services relevant to the compound. Among them is the “Similar Structures Search” link (1). Clicking this link will invoke multiple structure searches [including 2-D similarity search (2)] using the compound as a query and present the search results. The user can rerun the 2-D similarity search with a different similarity threshold (3) and apply filters (4) to refine the hit compounds based on several molecular properties. The hit compound list can be downloaded using the “Download” button (5). The result for the 3-D similarity search can be viewed by clicking the “3D similarity” tab (indicated in the purple box).](https://static.yanyin.tech/literature_test/cpz1217-fig-0003-m.jpg)
3.If necessary, click the Settings button (‘3’ in Fig. 3) and adjust the similarity threshold to a desired value.
4.Click the “Filters” button (‘4’ in Fig. 3) and refine the hits to only drug-like compounds that satisfy Lipinski's rule of five.
1.A molecular weight less than 500 g/mol 2.No more than 5 hydrogen bond donors 3.No more than 10 hydrogen bond acceptors 4.An octanol-water partition coefficient (log P) that does not exceed 5.
Although PubChem has experimental log P values for more than 26,000 compounds, this corresponds to a very small fraction of the 100+ million compounds in PubChem, and it is not practical to use the experimental log P values as a filter to refine the search results. Therefore, for this purpose, PubChem uses computed log P values, called “XLogP” (Cheng et al., 2007). The XLogP values are available for more than 90% of compounds in PubChem (except for inorganic and organometallic compounds).
5.Click the “Download” button (‘5’ in Fig. 3) to save the hit list as a CSV file for further analysis.
Basic Protocol 3: FINDING COMPOUNDS SIMILAR TO A QUERY COMPOUND THROUGH 3-D SIMILARITY SEARCH
PubChem's search interface supports both 2-D and 3-D similarity searches. The molecular similarity methods used for the two similarity searches are complementary to each other. That is, one method can often recognize structural similarity that is unnoticed by the other approach. A brief overview of the underlying methods used in the 3-D similarity search is given in Background Information.
In Basic Protocol 3, a 3-D similarity search is performed to find the compounds structurally similar to losartan based on 3-D similarity scores, and the 3-D structures of the returned compounds are downloaded in a structure-data file (SDF) format. The downloaded SDF file can be opened in popular 3-D molecular viewers. Note that these 3-D structures are not experimentally determined, but computationally generated as described in detail in previous papers (Bolton, Kim, & Bryant, 2011a; Kim, Bolton, & Bryant, 2013).
Materials
- An up-to-date Web browser, such as Google Chrome, Microsoft Edge, Safari, or Firefox, is required for this protocol (and all other protocols in this article)
1.Repeat steps 1-2 of Basic Protocol 2 to perform a structure search with losartan as a query.
2.Click the “3D Similarity” tab to view the hit list for the 3-D similarity search (the purple box in Fig. 3).
1.Tier 1: Compounds with annotations, using up to ten conformers per compound 2.Tier 2: Compounds with patent links, using up to five conformers per compound 3.Tier 3: All remaining compounds, using up to three conformers per compound.
By default, a 3-D similarity search is performed against only the Tier 1 compounds (using up to ten conformers per compound). The search can be extended to the Tier 2 or Tier 3 compounds using the “SETTINGS” button (‘1’ in Fig. 4), but a smaller number of conformers per compound will be used.
Also, note that it is not possible to adjust the 3-D similarity search threshold, in contrast to the 2-D similarity search threshold (see Basic Protocol 2). During the 3-D similarity search, two compounds are considered to be similar if any conformer pair arising from them has a shape-Tanimoto (ST) score of ≥0.80 (or 80%) and a color-Tanimoto (CT) of ≥0.50 (or 50%) (Bolton, Kim, & Bryant, 2011b; Kim, Bolton, & Bryant, 2016). More information on the 3-D similarity method used in PubChem can be found in Background Information.
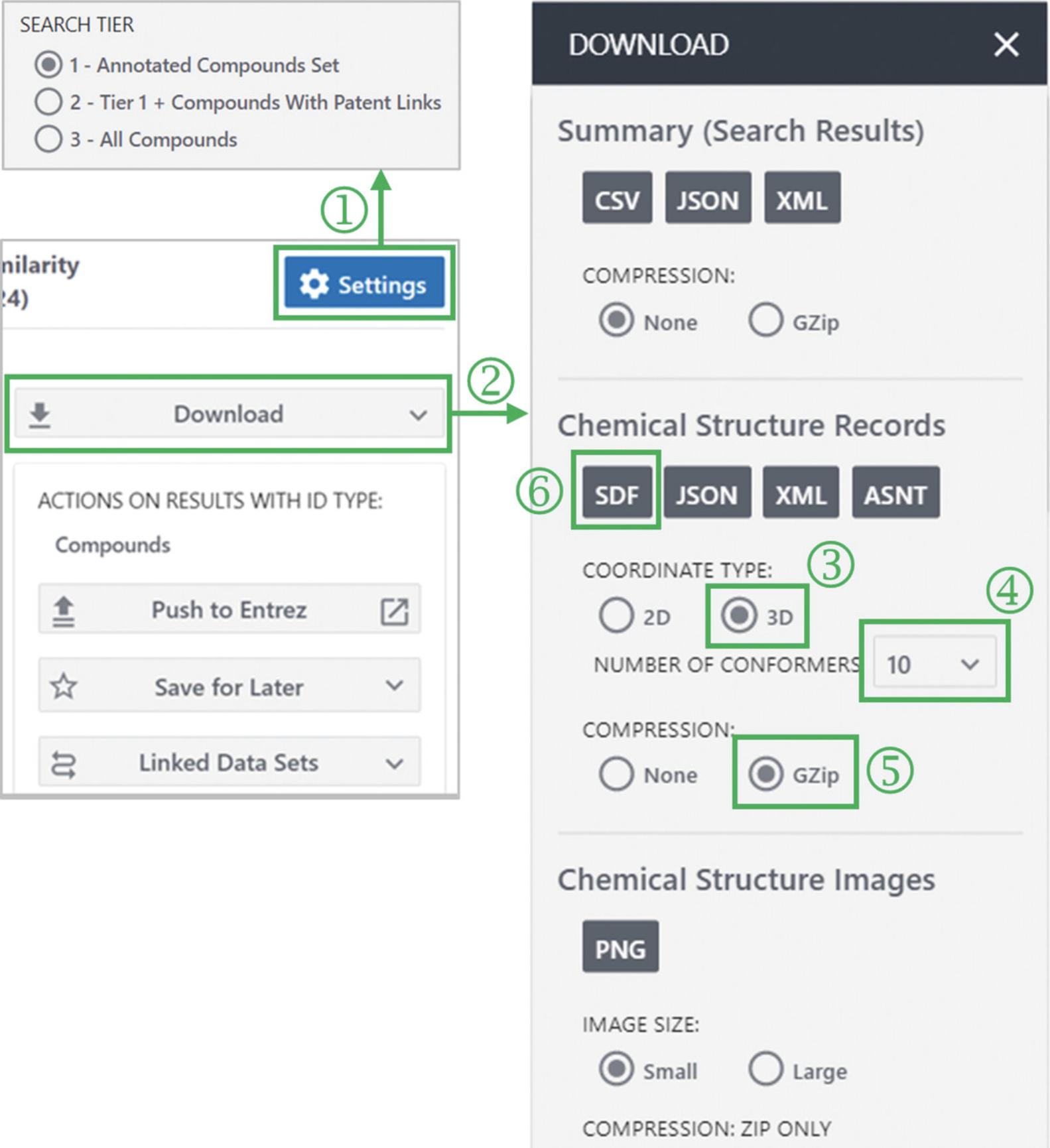
3.Click the “Download” button (‘2’ in Fig. 4).
4.To save the 3-D structures of hit compounds in an SDF format, select “3D” for the coordinate type, “10” for the number of conformers per compound, “gzip” for compression, and “SDF” for file format (‘3’ through ‘6’ in Fig. 4).
- Not too large (with ≤50 non-hydrogen atoms)
- Not too flexible (with ≤15 rotatable bonds)
- Has fewer than six undefined atom or bond stereocenters 4.Has only a single covalently bonded unit (i.e., not a salt or a mixture)
- Consists of only supported organic elements (H, C, N, O, F, Si, P, S, Cl, Br, and I)
- Contains only atom types recognized by the MMFF94s force field (Halgren, 1996 , 1996a b , 1999 ).
About 87% of compounds have computationally generated conformer models, and if a compound in the hit list does not have a conformer model, that compound will be ignored for download. While each of these conformer models contains up to 500 conformers, only up to 10 conformers per compound are accessible by the public. More detailed information on conformer generation in PubChem can be found in previous papers (Bolton et al., 2011a; Kim et al., 2013).
Support Protocol: COMPUTING SIMILARITY SCORES BETWEEN COMPOUNDS
Basic Protocols 2 and 3 demonstrate how to find compounds that are structurally similar to a query compound based on 2-D and 3-D similarity scores, respectively. However, the data returned from the similarity searches do not include the similarity scores between the query and the returned compounds. These scores can be used to sort the hit compounds and find higher-ranked compounds within the list. They can also be used to perform a cluster analysis to identify important structural patterns of the hit compounds.
In this Support Protocol, we download the 3-D similarity scores for the compounds returned from a 2-D similarity search (in Basic Protocol 2) using the PubChem Score Matrix Service (https://pubchem.ncbi.nlm.nih.gov/score_matrix). The PubChem Score Matrix Service computes 2-D and 3-D similarity scores between compounds in PubChem. This service takes a list of M compounds and another list of N compounds as an input, computes similarity scores for M × N compound pairs arising from the combination of the two lists, and returns the scores in a matrix form or in a list of CID-CID-score triples. When only one list (of M compounds) is provided as an input, similarity scores are computed for M(M+1)/2 unique CID pairs, arising from the combination of the M compounds.
Materials
- An up-to-date Web browser, such as Google Chrome, Microsoft Edge, Safari, or Firefox, is required for this protocol (and all other protocols in this article)
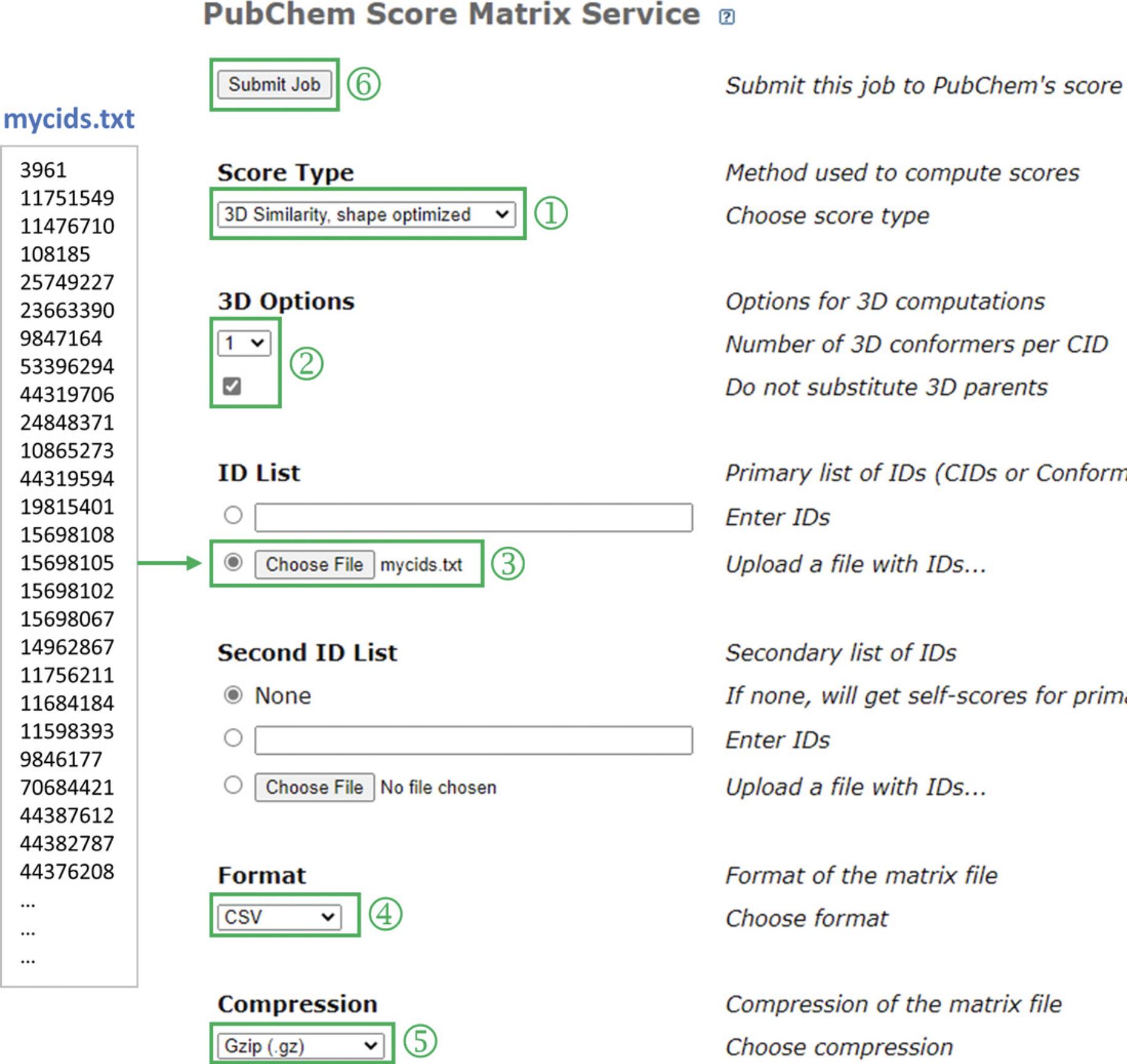
- In addition, this protocol requires a text file containing the CIDs of the hit compounds returned from Basic Protocol 2.This file can be generated from the CSV file downloaded in Basic Protocol 2. Open the CSV file in spreadsheet software (e.g., Microsoft Excel or Google Sheets). Copy the first column containing the CIDs (except for the column header), paste them into a text editor (e.g., Notepad on Windows PC and TextEdit on Mac), and save them as a text file. In this protocol, the file name is assumed to be mycids.txt. Double-check that the file has the same format as the mycids.txt file in Figure 5 (e.g., one CID for each line).
1.Go to the PubChem Score Matrix Service (https://pubchem.ncbi.nlm.nih.gov/score_matrix).
2.Select “3D Similarity, shape optimized” for the score type (‘1’ in Fig. 5).
3.Select “1 conformer per CID” and check the “Do not substitute 3D parents” box (‘2’ in Fig. 5).
4.Select the text file containing the input CID list (i.e., mycids.txt; ‘3’ in Fig. 5).
5.Select “CSV” for format and “gzip” for compression (‘4’ and ‘5’ in Fig. 5) and click “Submit Job (‘6’ in Fig. 5).
Basic Protocol 4: GETTING THE BIOACTIVITY DATA FOR THE HIT COMPOUNDS FROM SUBSTRUCTURE SEARCH
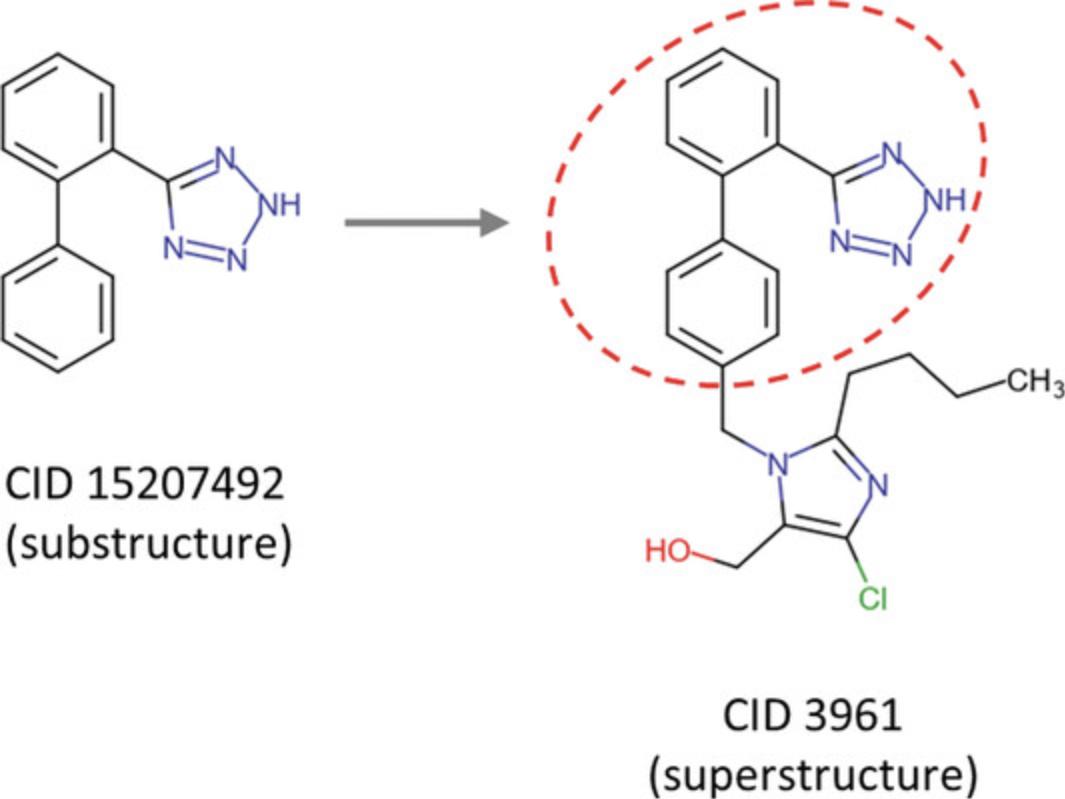
When a chemical structure pattern appears in a bigger chemical structure, the former is called a substructure and the latter is referred to as a superstructure (see Fig. 6). In this protocol, a substructure search is performed to find compounds with a given substructure, and their bioactivity data are downloaded on a local computer. The downloaded data can be used in further analysis by means of third-party software packages. This protocol uses two important features of PubChem's search interface, the PubChem Sketcher for structure input and the “Linked Data Sets” button for quick retrieval of linked data.
Previously, in Basic Protocols 2 and 3, a chemical name search (i.e., losartan as a query) was first performed to find the corresponding compound (CID 3961), which was used to specify the input chemical structure for a subsequent 2-D and 3-D similarity search. However, this approach cannot be used when the query structure does not exist in PubChem or when its name is unknown or ambiguous. In this case, the input structure can be provided by drawing it in the PubChem Sketcher.
This protocol also exemplifies the usefulness of linked data in PubChem. As mentioned in the Introduction, PubChem has multiple data collections. Some users often need to retrieve records in one data collection that are related to those in another data collection. For example, the present protocol retrieves bioactivity data (in BioAssay) associated with a list of chemicals (in Compound). This task can be done seamlessly with the “Linked Data Sets” button available on the search result page.
Materials
- An up-to-date Web browser, such as Google Chrome, Microsoft Edge, Safari, or Firefox, is required for this protocol (and all other protocols in this article)
1.Go to the PubChem homepage (https://pubchem.ncbi.nlm.nih.gov) and launch the PubChem Sketcher by clicking the “Draw Structure” button (‘1’ in Fig. 7).
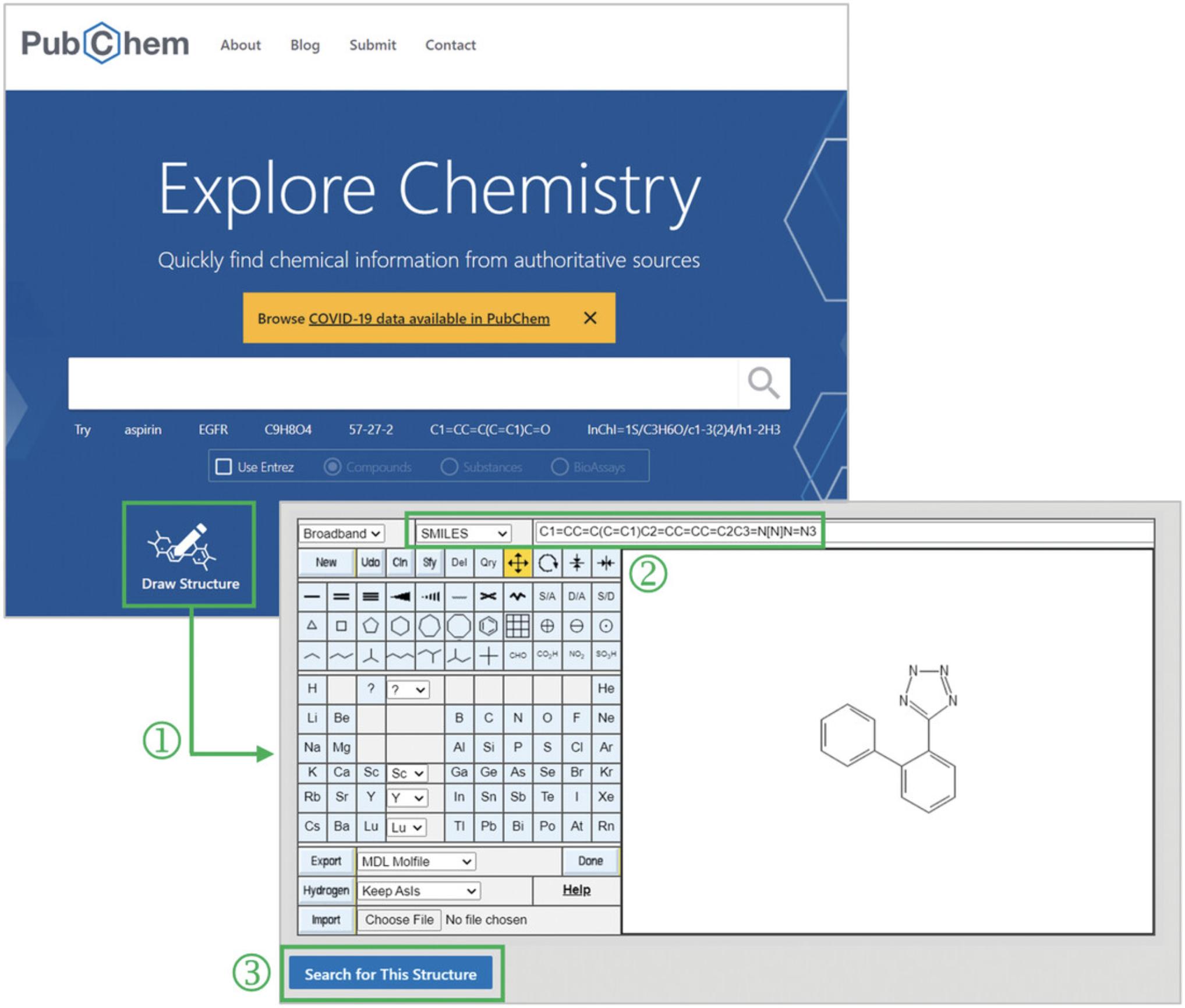
2.Draw the structure of 5-(2-phenylphenyl)-2H-tetrazole, by providing its SMILES string C1=CC=C(C=C1)C2=CC=CC=C2C3=N[N]N=N3 in the text box available at the top of the Sketcher (‘2’ in Fig. 7).
3.After drawing the input structure, click the “Search for This Structure” button (‘3’ in Fig. 7).
4.Click the “Substructure” tab to view the hit compounds from the substructure search (‘1’ in Fig. 8).
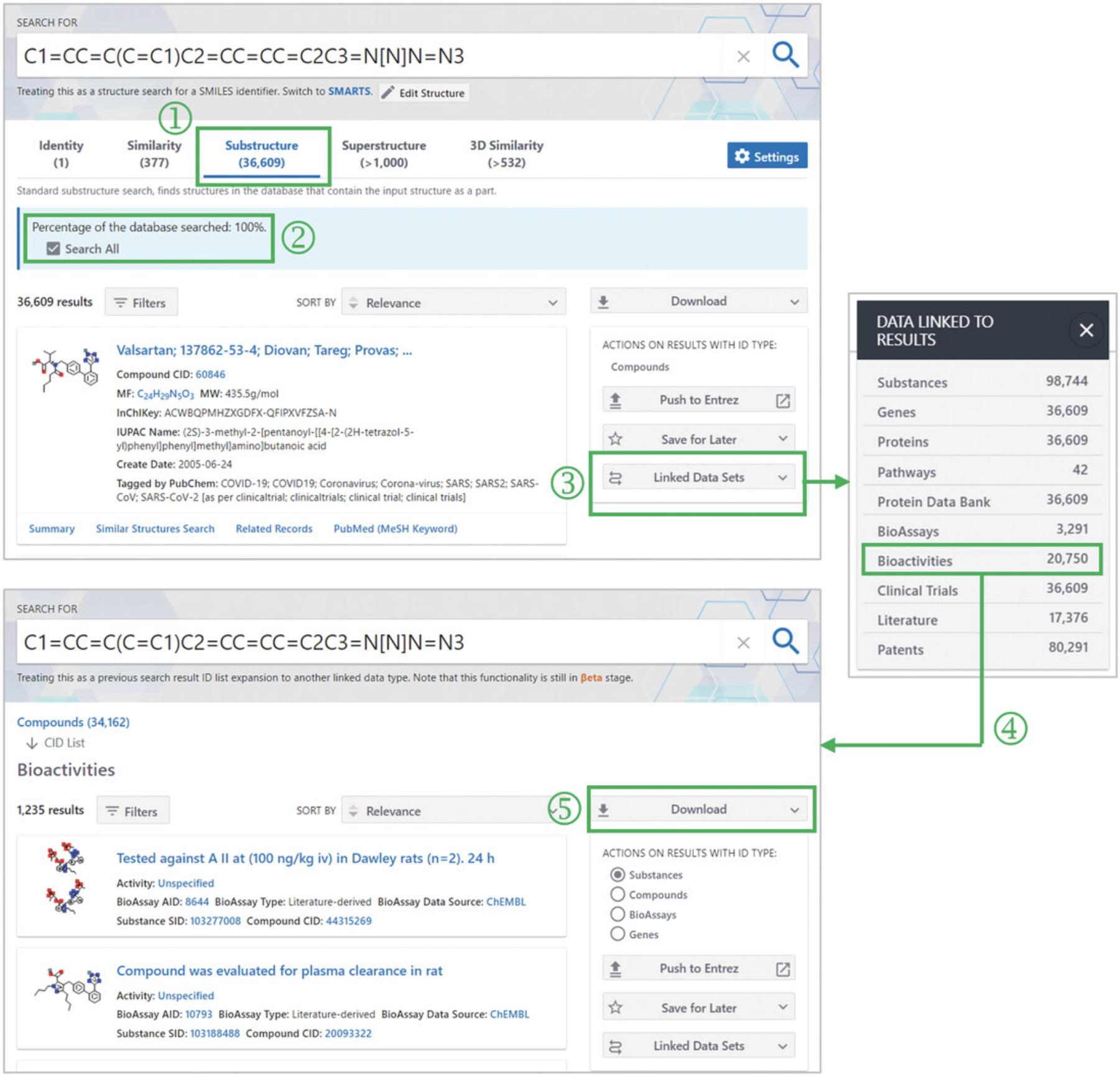
5.Check the “Search All” box (‘2’ in Fig. 8) to extend the search to all compounds in PubChem.
6.Click the “Linked Data Sets” button on the right column (‘3’ in Fig. 8) and select the “Bioactivities” link from the pop-up menu (‘4’ in Fig. 8).
7.Click the Download button to save the linked data on a local computer (‘5’ in Fig. 8).
Basic Protocol 5: FINDING DRUGS THAT TARGET A PARTICULAR GENE
While it is possible to retrieve all macromolecules interacting with a given chemical (as done in Basic Protocol 1), the user may want to find all chemicals that interact with a given gene or protein. This task can be done through the Summary page of a gene or protein record, which presents all PubChem data related to that macromolecule. It includes not only known drugs and tested chemicals, but also annotations collected from major gene or protein information resources.
Basic Protocol 5 aims to find all known drugs that interact with the gene encoding the human type-1 angiotensin II receptor, which is the target of losartan (see Basic Protocol 1). This protocol begins with a text search using the gene name as a query. Then, the resulting gene list is filtered based on taxons to identify the gene for humans. The Summary page of this gene contains lists of drugs targeting it, which are collected from DrugBank (Wishart et al., 2018), ChEMBL (Mendez et al., 2019), and IUPHAR/BPS Guide to PHARMACOLOGY (Armstrong et al., 2020). These lists can be downloaded on a local computer.
Materials
- An up-to-date Web browser, such as Google Chrome, Microsoft Edge, Safari, or Firefox, is required for this protocol (and all other protocols in this article)
1.Go to the PubChem homepage and perform a text search with type 1 angiotensin II receptor as a query (‘1’ in Fig. 9).
1.type 1 angiotensin II receptor 2.type-1 angiotensin II receptor 3.“type 1 angiotensin II receptor” (enclosed in double quotes) 4.“type-1 angiotensin II receptor” (enclosed in double quotes) 5.“angiotensin II receptor type 1” (enclosed in double quotes) 6.“angiotensin II receptor type-1” (enclosed in double quotes).
Among these examples, the first one is used as a query in Basic Protocol 5, as shown in Figure 9.It is interpreted as “type AND 1 AND angiotensin AND II AND receptor” and returns any records containing the five words. If the query needs to be interpreted as a phrase (e.g., “type 1 angiotensin II receptor”) to identify more specific hits, the query should be enclosed in double quotes. In this case, however, the search would miss records containing a phrase like “angiotensin II receptor type 1”.
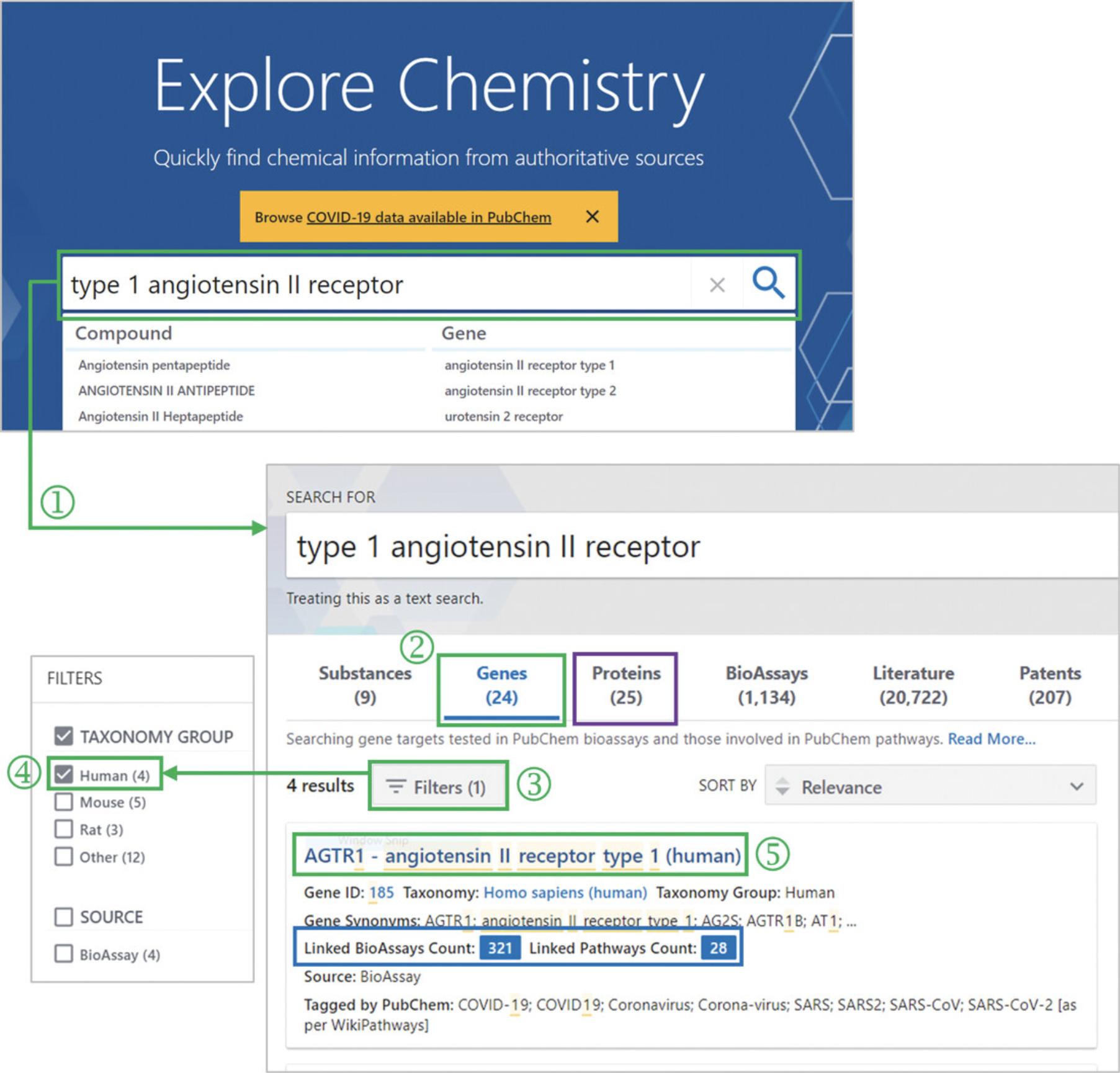
2.Select the “Genes” tab (‘2’ in Fig. 9) to display the search result from the gene collection.
3.Click the Filters button (‘3’ in Fig. 9) and select “Human” under the taxonomy group (‘4’ in Fig. 9).
4.Click the gene record for the human angiotensin II receptor type 1 (‘5’ in Fig. 9).
5.Use the Table of Contents (‘1’ in Fig. 10) in the right column to go to the DrugBank Drugs subsection (‘2’ in Fig. 10).
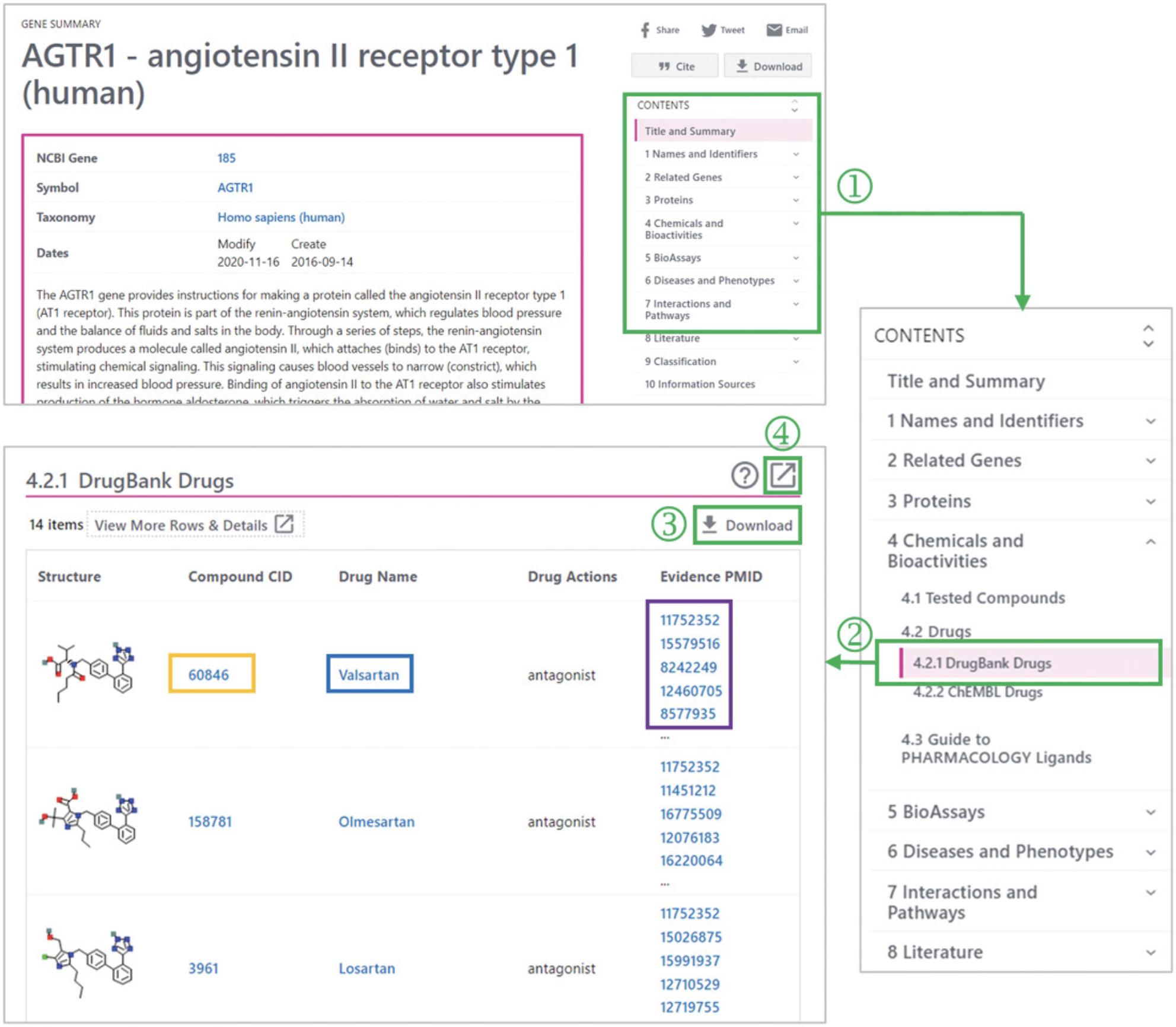
6.Click the Download button (‘3’ in Fig. 10) to download the data.
7.If necessary, click the “Full-view” button (‘4’ in Fig. 10) to get more detailed information.
8.Get the drug information from ChEMBL in a similar way to that described in steps 5 through 7.This information can be found in the “ChEMBL Drugs” section.
9.Get the drug information from Guide To PHARMACOLOGY in a similar way as described in steps 5 through 7. This information can be found in the “Guide to PHARMACOLOGY Ligands” section.
Basic Protocol 6: GETTING BIOACTIVITY DATA OF ALL CHEMICALS TESTED AGAINST A PROTEIN
Basic Protocol 6 is designed to demonstrate how to download the bioactivity data of all chemicals tested against a given protein and how to quickly access data for a protein orthologous to another protein, using the human type-1 angiotensin II receptor and its rat ortholog as an example. This protocol is similar to Basic Protocol 5, which downloads the list of drugs interacting with the gene encoding type-1 angiotensin II receptor. However, it should be kept in mind that a gene record in PubChem can be associated with multiple protein records, reflecting the fact that a gene can produce multiple protein sequences (e.g., isoforms or variants). Because bioassays archived in PubChem were performed typically against one of the multiple protein sequences that may arise from a single gene, the Summary pages of the different proteins from the same gene present different sets of bioactivity data. These data are merged together and presented on the Summary page of the encoding gene. Therefore, extra care should be taken when downloading the bioactivity data from the Summary page of a gene or protein.
Materials
- An up-to-date Web browser, such as Google Chrome, Microsoft Edge, Safari, or Firefox, is required for this protocol (and all other protocols in this article)
1.Go to the Protein Summary page of the human type-1 angiotensin II receptor. This can be done in a similar manner to steps 1 through 4 of Basic Protocol 5 (Fig. 9), except that the “Proteins” tab (the purple box in Fig. 9) should be clicked to access the hit protein records instead of the gene records.
2.Use the Table of Contents (‘1’ in Fig. 11) on the right column to go to the Tested Compounds subsection (‘2’ in Fig. 11).
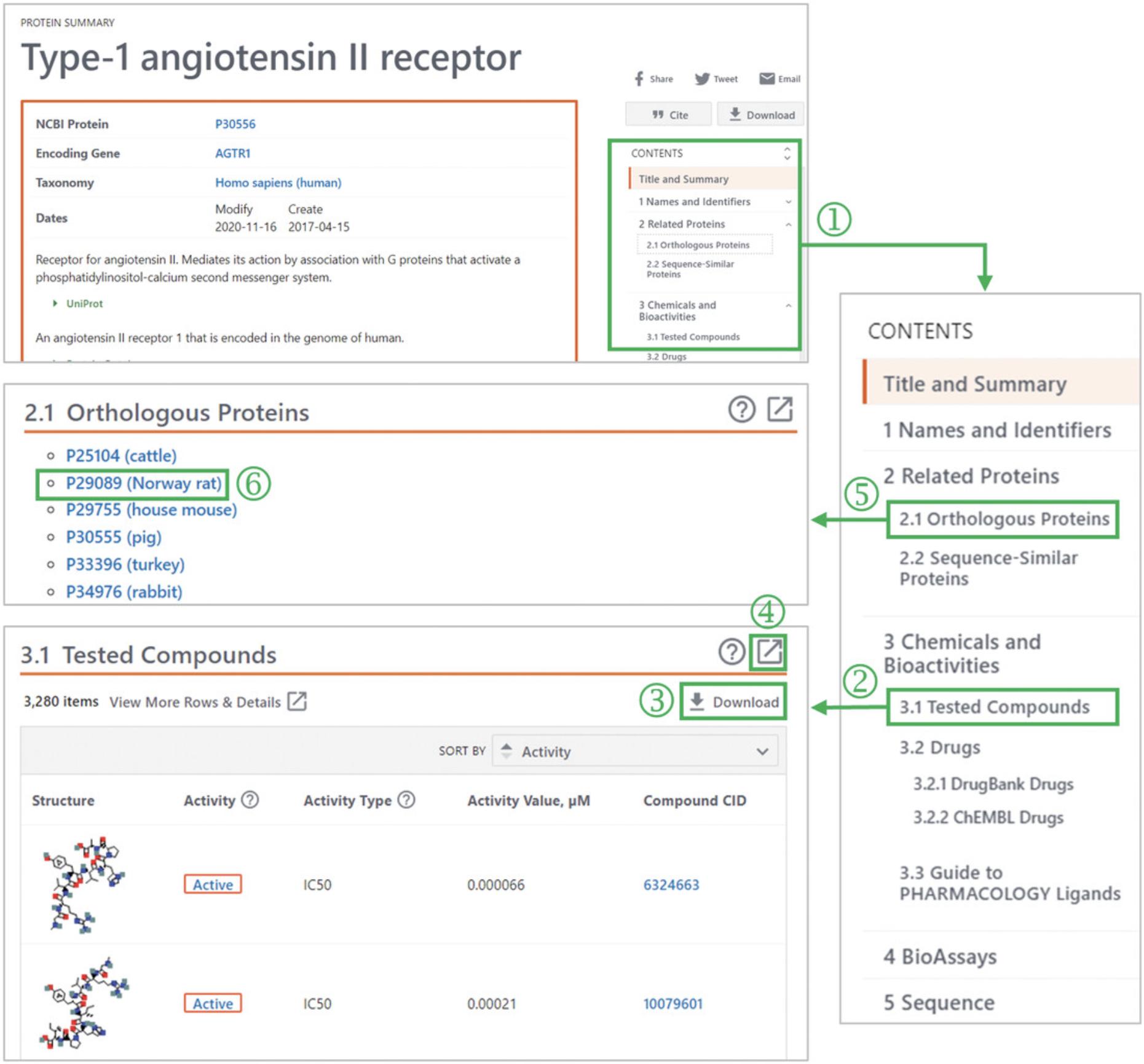
3.Download the list of the tested compounds with their bioactivity data against the target protein (‘3’ in Fig. 11).
4.If necessary, click the “Full-view button” (‘4’ in Fig. 11) to get more detailed information.
5.Go to the Orthologous Proteins section (‘5’ in Fig. 11) and click “P29089 (Norway rat)” (‘6’ in Fig. 11). This leads the user to the Summary page for the orthologous protein in rats.
6.Repeat steps 2 through 4 to download the list of the tested compounds and their bioactivity data for the rat type-1 angiotensin II receptor.
Basic Protocol 7: FINDING COMPOUNDS ANNOTATED WITH CLASSIFICATIONS OR ONTOLOGICAL TERMS
PubChem records are annotated with various classifications and ontological terms. For example, losartan (CID 3961) is annotated with three Medical Subject Headings (MeSH) terms, “Angiotensin II Type 1 Receptor Blockers”, “Antihypertensive Agents”, and “Anti-Arrhythmia Agents”, as shown at https://pubchem.ncbi.nlm.nih.gov/compound/3961#section=MeSH-Pharmacological-Classification.
PubChem users often want to access all records annotated with a particular term. This task can be done using the PubChem Classification Browser, which can be accessed from the PubChem homepage or via https://pubchem.ncbi.nlm.nih.gov/classification/.
The classification browser allows users to browse the distribution of PubChem records among nodes in the hierarchy of ontological terms and classifications and subset PubChem records annotated with the desired term.
In this protocol, the Classification Browser is used to retrieve chemicals with the same therapeutic uses as losartan, based on the MeSH annotations (that is, chemicals that are known as both antihypertensive and antiarrhythmic agents). This involves performing two independent searches (one for antihypertensive agents and the other for antiarrhythmic agents) and finding chemicals returned in both searches. PubChem users often need to perform a series of searches, followed by taking the intersection or union of the search results or identifying records returned from one search, but not from another. These tasks can be done in PubChem using Boolean operators (AND, OR, and NOT), as exemplified in this protocol.
Materials
- An up-to-date Web browser, such as Google Chrome, Microsoft Edge, Safari, or Firefox, is required for this protocol (and all other protocols in this article)
1.Go to the PubChem homepage and click the “Browse Data” icon below the search box (‘1’ in Fig. 12). This leads to the Classification Browser, which can also be accessed directly via https://pubchem.ncbi.nlm.nih.gov/classification/.
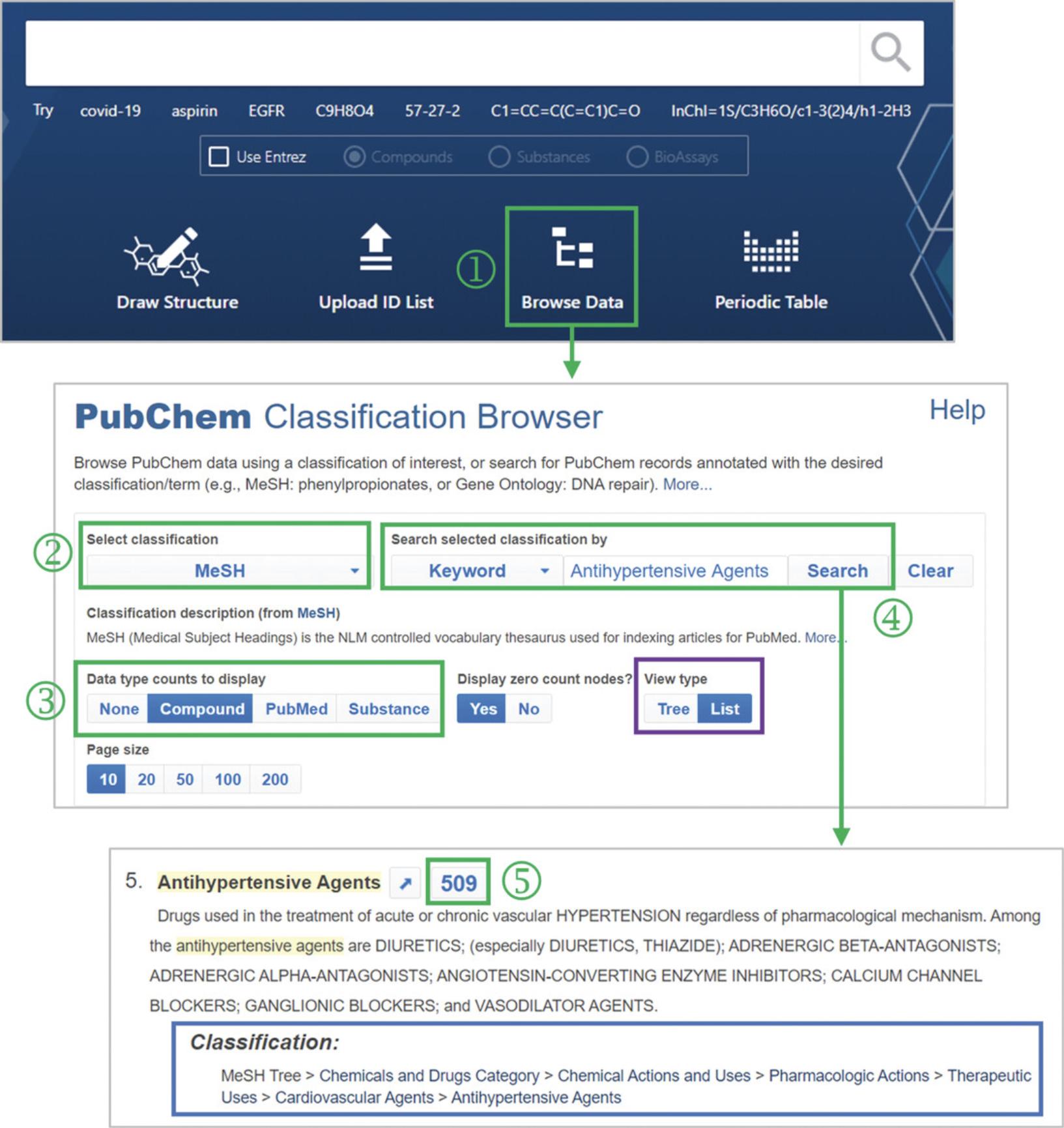
2.Select “MeSH” from the “Select classification” dropdown menu (‘2’ in Fig. 12).
1.Medical Subject Headings (see Internet Resources) 2._ChEBI Ontology (_Hastings et al. , 2016 ) 3._Gene Ontology (_Ashburner et al. , 2000; Carbon et al. , 2021 ) 4._Food and Drug Administration (FDA) Pharmacological Class (_FDA , 2021 ) 5._WIPO (World Intellectual Property Organization) International Patent Classification (_WIPO , 2021 ) 6._World Health Organization (WHO) Anatomical Therapeutic Chemical (ATC) classification system (_WHO , 2021 ) 7.PubChem Compound Table of Contents (TOC).
The PubChem Compound TOC is also available in the Classification Browser. This allows users to quickly identify and retrieve compounds that have a particular kind of annotation (e.g., those with solubility data, those with toxicological information, those which have been tested in a clinical trial, those mentioned in scientific articles or patent documents, etc.).
3.Select the “Compound” from the “Data type counts to display” menu (‘3’ in Fig. 12).
4.Type Antihypertensive Agents in the search box (‘4’ in Fig. 12).
5.From the returned hit list, find the “Antihypertensive Agents” node and click the record count for that node (‘5’ in Fig. 12).
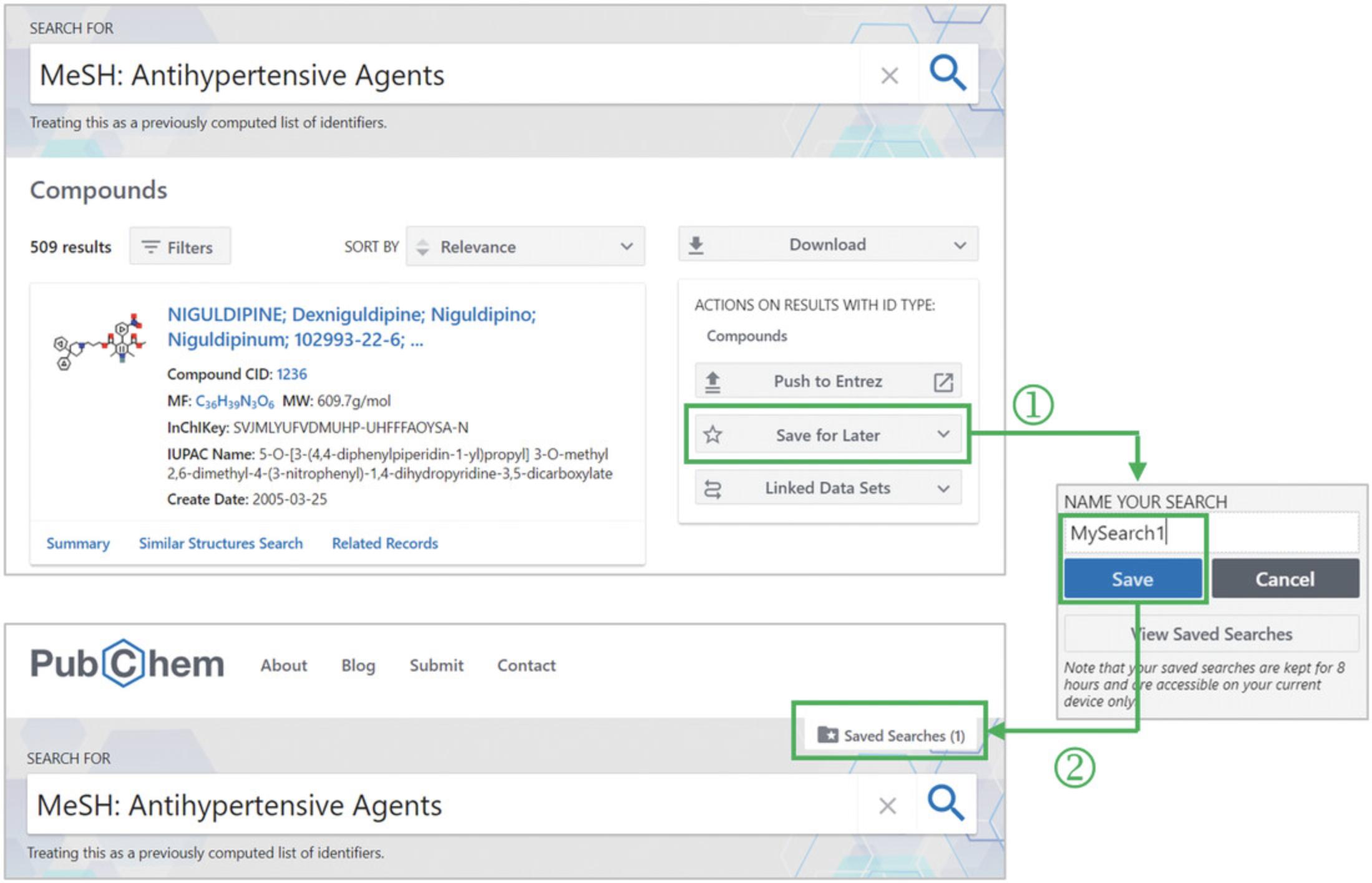
6.The previous step leads to a web page that shows compounds annotated as antihypertensives (Fig. 13). Save this list by clicking the “Save for Later” button available on the right column and providing an alias for that list (e.g., “MySearch1”) (‘1’ in Fig. 13). When the list is successfully saved, a new button “Saved Searched (1)” appears above the search box (‘2’ in Fig. 13).
7.Repeat steps 1 through 6 to retrieve the list of compounds annotated with the MeSH term “Anti-arrhythmia Agents” and save them as “MySearch2”.
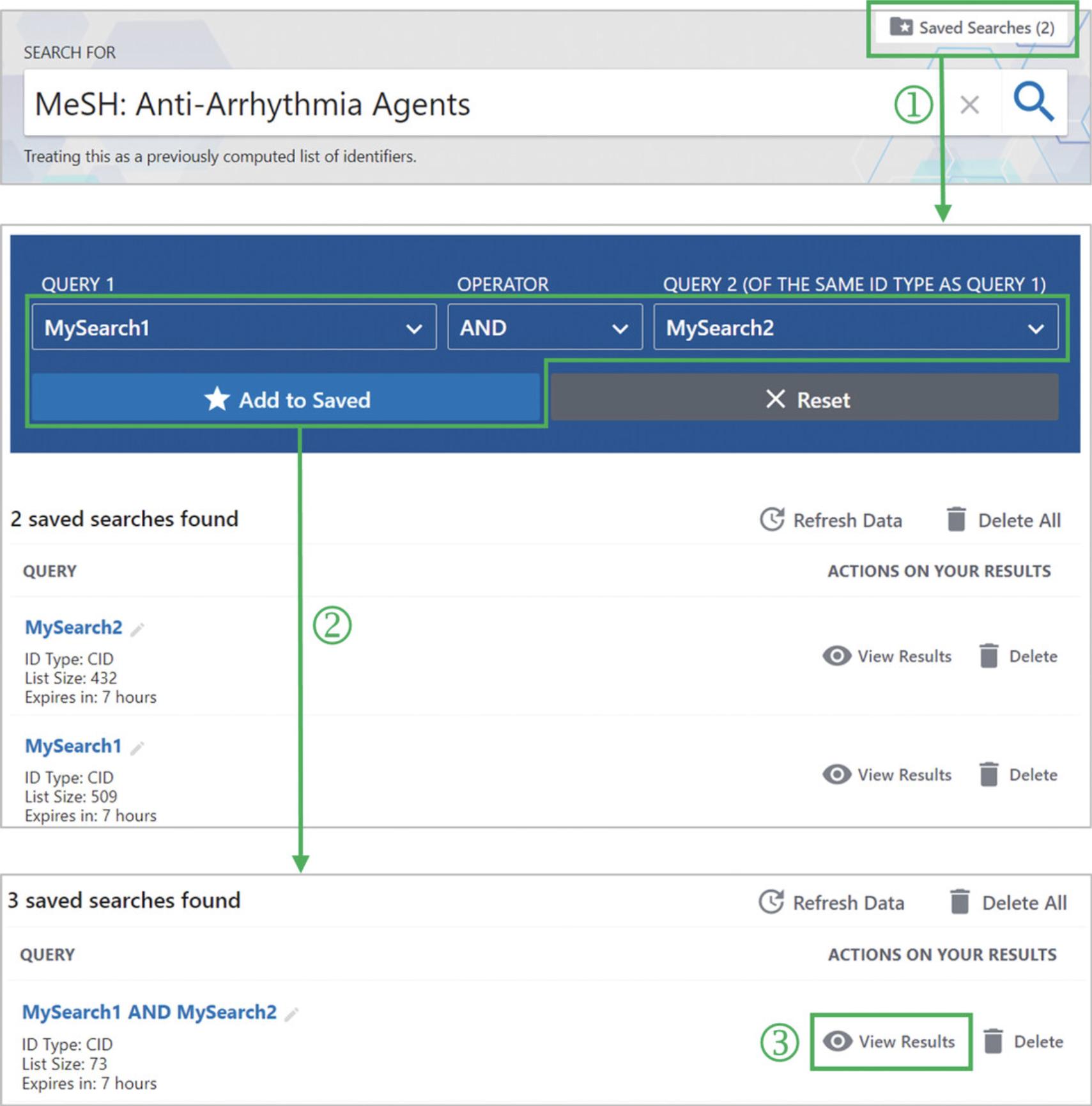
8.Click the “Saved Search (2)” button (‘1’ in Fig. 14). This launches a dialog box that enables users to perform advanced searches by combining results from previous searches using Boolean operators (AND, OR, and NOT).
9.Select the saved results, “MySearch1” and “MySearch2”, from the Query 1 and Query 2 dropdown menus and select “AND” from the Operator menu. Then, click the “Add to Saved” button (‘2’ in Fig. 14).
10.Click the “View Results” button to go to the web page that shows the resulting compound list (‘3’ in Fig. 14).
Basic Protocol 8: GETTING STEREOISOMERS AND ISOTOPOMERS OF A COMPOUND THROUGH IDENTITY SEARCH
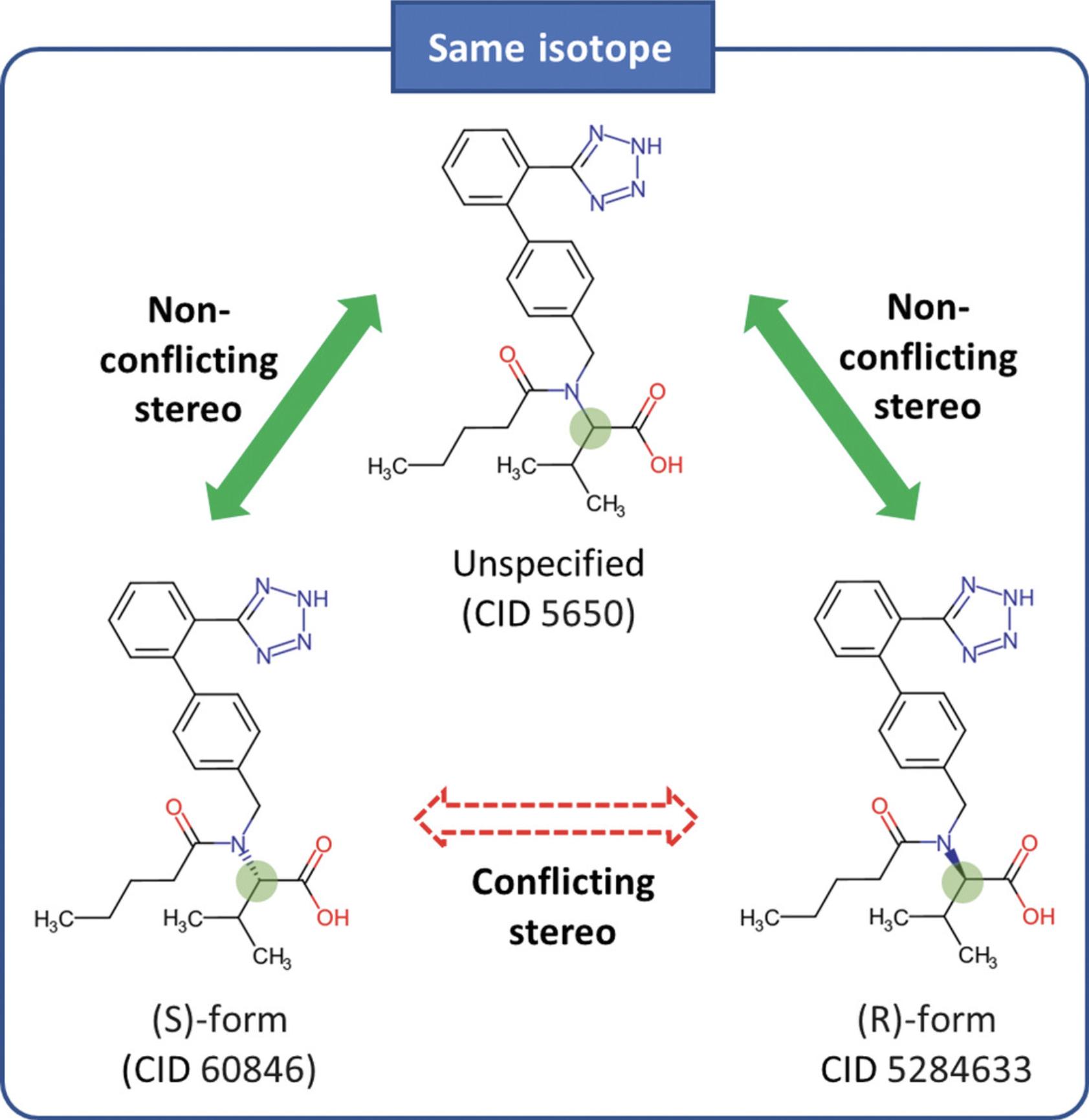
This protocol demonstrates how to find stereoisomers and isotopomers of a given compound, with valsartan (CID 60846) as an example. This task can be done using identity search, which is one of the structure search types supported by PubChem. An identity search returns compounds identical to the query molecule. While it may sound straightforward, the search results can vary, depending on what is meant by “identical” compounds. PubChem's identity search allows for some flexibility in the definition of chemical identity. By default, two molecules are considered identical if they have the same connectivity, isotopism, and stereochemistry [i.e., (R/S)-configuration and cis/trans-isomerism]. The user can change this behavior by choosing to ignore isotopism and/or stereochemistry. When stereochemistry is ignored, compounds with the same connectivity and isotopism, but with varying stereochemistry (i.e., stereoisomers), are returned. If isotopism is ignored, the identity search finds compounds with the same connectivity and stereochemistry, but with different isotopes (i.e., isotopomers). In this protocol, identity search is performed with different definitions of chemical identity to find stereoisomers and isotopomers of valsartan (CID 60846), which is a structural analog of losartan.
Materials
- An up-to-date Web browser, such as Google Chrome, Microsoft Edge, Safari, or Firefox, is required for this protocol (and all other protocols in this article)
1.Go to the PubChem homepage, type CID 60846 structure (‘1’ in Fig. 15), and hit the search button.
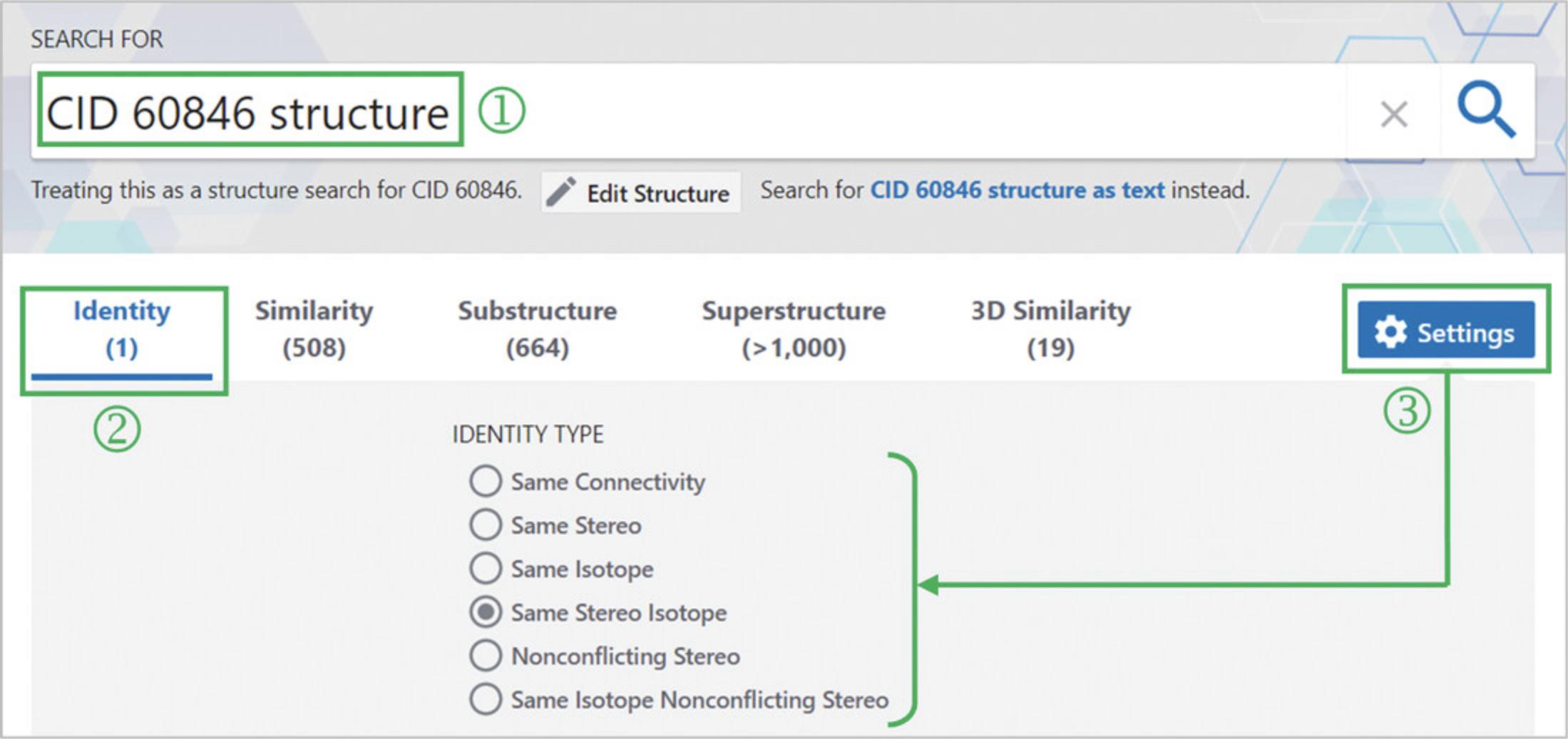
2.Click the “Identity” tab (‘2’ in Fig. 15) and the “Settings” button (‘3’ in Fig. 15).
3.Select the “Same Isotope” option to find stereoisomers of valsartan.
4.Download the returned stereoisomers in a CSV format.
5.Select the “Same Stereo” option to find stereoisomers of valsartan.
6.Download the returned isotopomers in a CSV format.
COMMENTARY
Background Information
PubChem as an archive and a knowledgebase
PubChem (https://pubchem.ncbi.nlm.nih.gov; Kim, 2016; Kim et al., 2019; Kim et al., 2021; Kim et al., 2016) is a popular chemical information resource that plays a dual role as a data repository (archive) and a knowledgebase. As a data repository, PubChem needs to archive various types of chemical information provided by individual data contributors. As a knowledgebase, it should provide the user with easy access to comprehensive chemical data from authoritative sources. These two demands are taken into account in data organization in PubChem. As mentioned previously, PubChem has multiple data collections, including Substance, Compound, BioAssay, Gene, Protein, Pathway, and Patent. Among them, Substance and BioAssay play a role as an archive. Substance stores chemical information provided by individual data sources, and BioAssay archives the description and test results of biological assay experiments. Compound is a knowledgebase that provides comprehensive information on unique chemical structures extracted from Substance. The other data collections (i.e., Gene, Protein, Pathway, and Patent) are also knowledgebases that provide information on chemicals associated with a specific gene, protein, pathway, and patent document, respectively.
Chemical structure search in PubChem
Beyond chemical name searches (Basic Protocols 1), PubChem allows the user to search by chemical structure. The input chemical structure can be provided using line notations like SMILES (Weininger, 1988, 1990; Weininger et al., 1989) and InChI (Heller et al., 2015), or drawn using the PubChem Sketcher (Ihlenfeldt, Bolton, & Bryant, 2009). If the input structure exists in the PubChem Compound database, its CID can also be used as a query. Alternatively, the structure of a hit compound from a previous search can be also be used, as demonstrated in Basic Protocols 2 and 3). Various types of structure searches are supported, including identity search, 2-D and 3-D similarity searches, and substructure/superstructure searches.
Identity search
Through identity search (Basic Protocol 8), the user can find compounds identical to a query compound. While it seems straightforward, the identity search can result in different hits, depending on the definition of “identical compounds.” For example, while isotopically labeled glucose (with 13C and 15N atoms) have the same chemical and biological properties as non-labeled one, they show different signals in nuclear magnetic resonance (NMR) or mass spectrometry (MS) experiments. Therefore, depending on the context, the two molecules may or may not be considered identical. PubChem's identity search allows the user to select one of several different contexts of “identity,” as demonstrated in Basic Protocol 8. By default, identity search returns compounds with the same connectivity, stereochemistry, and isotopism as the query molecule.
2-D and 3-D similarity search
Similarity search returns compounds structurally similar to a query molecule (Basic Protocols 2 and 3). Because molecular similarity is a subjective concept, which is not physically measurable, various similarity methods have been proposed to quantify it. The most widely used ones are 2-D similarity methods. In these approaches, the similarity between two molecules is evaluated by comparing their molecular fingerprints (binary fragment vectors encoding the 2-D structures of molecules) and computing a similarity score, which quantifies how similar the molecules are. This score can be computed using various metrics, but the Tanimoto coefficient is the most popular choice. In another group of methods, called 3-D similarity methods, 3-D structures of molecules are superimposed to find the “best” overlap between them. While 3-D similarity methods are much slower than 2-D similarity methods, they often recognize molecular similarity that is not readily detected by 2-D similarity methods. PubChem supports both 2-D and 3-D similarity searches. They usually give different lists of hit compounds, complementing each other. More detailed information on the 2-D and 3-D similarity methods used in PubChem is provided below.
Substructure and superstructure search
When a chemical structure occurs as a part of a bigger chemical structure, the former is called a substructure and the latter is referred to as a superstructure. For example, as shown in Figure 6, the structure of CID 15207492 (5-(2-phenylphenyl)-2H -tetrazole) occurs as a part of CID 3961. Therefore, CID 15207492 is a substructure of CID 3961.
In a substructure search, a substructure is provided as a query to find molecules that contain the substructure (that is, superstructures that contain the query substructure). On the contrary, superstructure search returns molecules that comprise or make up the provided superstructure query (that is, substructures that are contained in the query superstructure). PubChem supports both substructure and superstructure searches. It also provides flexible matching options that allow the user to specify how to deal with stereochemistry, isotopism, tautomerism, formal charges, aromatic bonds, and explicit hydrogens during the searches. Basic Protocol 4 demonstrates how to perform a substructure search using CID 15207942 as a query substructure.
2-D and 3-D molecular similarity assessment in PubChem
This section provides a brief overview of the 2-D and 3-D similarity methods used in PubChem and more detailed information on them is given elsewhere (Bolton et al., 2011; Kim et al., 2016; Kim, Bolton, & Bryant, 2011). PubChem evaluates 2-D molecular similarity using the PubChem substructure fingerprints (PubChem, 2009). They are 881-bit-long binary vectors, each bit of which represents the absence (0) or presence (1) of a particular structural characteristic found in a chemical structure, such as an element count, a type of ring system, atom pairing, and fragment patterns. The PubChem fingerprints are used to quantify 2-D similarity between two compounds, in conjunction with the Tanimoto coefficient, as shown in Equation 1 (Chen & Reynolds, 2002; Holliday et al., 2002; Holliday et al., 2003):
Tanimoto=NABNA+NB−NAB(1)
where NA and NB are the counts of bits set in the fingerprints representing molecules A and B, respectively, and NAB is the count of common bits set in both fingerprints. While a Tanimoto coefficient ranges from 0 (for no similarity between molecules) to 1 (for identical molecules, relative to the resolution of the substructure fingerprint).
On the other hand, 3-D similarity in PubChem is assessed using the Gaussian-shape overlay method of Grant and Pickup (Grant & Pickup, 1995, 1996, 1997; Grant, Gallardo, & Pickup, 1996), implemented in the Rapid Overlay of Chemical Structures (ROCS; Rush, Grant, Mosyak, & Nicholls, 2005). This method quantifies two aspects of 3-D similarity (i.e., shape similarity and feature similarity) between two conformers. The shape similarity is computed using the shape-Tanimoto (ST) (OpenEye Scientific Software, 2010a, 2010b), as shown in Equation 2:
ST=VABVAA+VBB−VAB(2)
where VAA and VBB are the self-overlap volumes of conformers A and B, respectively, and V AB is the overlap volume between conformers A and B. The feature similarity is evaluated using the color-Tanimoto (CT) (OpenEye Scientific Software, 2010a, 2010b), as shown in Equation 3:
CT=∑fVABf∑fVAAf+∑fVBBf−∑fVABf(3)
where the index “ f ” indicates any of six “fictitious” feature (color) atom types (hydrogen bond donors and acceptors, cations, anions, hydrophobes, and rings.), VAAf and VBBf are the self-overlap volumes of conformers A and B for feature atom type f , respectively, and VABf is the overlap volume between conformers A and B for feature atom type f. To consider the (steric) shape similarity and (chemical) feature similarity simultaneously, the combo-Tanimoto (ComboT) is used, as indicated in Equation 4:
ComboT=ST+CT(4)
Because both ST and CT scores range from 0 (for no similarity) to 1 (for identical molecules), by definition, the ComboT score can have a value from 0 to 2 (without normalization).
To find the best superposition between molecules, two approaches can be used: shape optimization and feature optimization. The shape-optimization approach finds the molecular superposition that maximizes the ST score and then computes the CT and ComboT scores at that superposition. On the other hand, the feature optimization approach considers the shape and feature simultaneously to find the best superposition.
It is noteworthy that the 3-D similarity comparison requires 3-D molecular structures (i.e., conformers) and that a molecule can have multiple conformers. Therefore, the 3-D similarity between two molecules is assessed by computing 3-D similarity scores for all possible conformer pairs arising from the combination of the conformers of the molecules, and selecting the highest score among them. For each compound in PubChem, a conformer model containing up to 500 diverse conformers is generated, among which up to 10 diverse conformers per compound are made accessible to the public and can also be used for 3-D similarity evaluation in PubChem (Bolton et al., 2011; Bolton et al., 2011a; Kim et al., 2013).
Critical Parameters and Troubleshooting
PubChem's search interface provides filters that allow users to refine hit records based on selected attributes. Each of the PubChem data collections has its own set of filters. For example, compound records can be filtered based on molecular properties (e.g., molecular weight, rotatable bond count, heavy atom count, hydrogen bond donor and acceptor counts, polar surface area, and XLogP) as well as the created date. The filters used on gene records include taxonomy groups (e.g., human, mouse, rat, etc.) and data source (e.g., BioAssay and Pathway). These filters help users to find information more specific to their needs.
Chemical structure searches in PubChem can be customized using various options available through the “Settings” button. It is worth mentioning that, because chemical structure searches are much more time-consuming than text (keyword) searches, they are set by default to stop when a thousand hit compounds have been returned. While the search can be extended beyond this 1000-hit limit (by checking the “Search All” box), only up to one million hits will be returned, at most. Therefore, a query structure should be specific enough not to exceed this limit.
The protocols in this article are designed to demonstrate the utility of PubChem, and can be readily modified and adopted for many other tasks. It is worth mentioning that these protocols are for interactive users who access PubChem data through web browsers (e.g., Google Chrome, Microsoft Edge, Safari, FireFox, etc.). When an interactive task needs to be repeated for a large number of PubChem records, it can likely be automated through PubChem's programmatic interfaces such as PUG-REST (Kim, Thiessen, Bolton, & Bryant, 2015; Kim, Thiessen, Cheng, Yu, & Bolton, 2018) and PUG-View (Kim et al., 2019). PubChem also supports the bulk download of its data through the PubChem FTP (file transfer protocol) site. Additional information about PubChem can be found in PubChemDocs (https://pubchemdocs.ncbi.nlm.nih.gov).
Understanding Results
PubChem contains a massive amount of data, collected from hundreds of data sources. Although PubChem makes every effort to ensure high data quality, inconsistency may be found in the data from different sources. For this reason, PubChem preserves information on the provenance of data (i.e., what source the data originated from), so that users can go to the original data source and find additional information that may help them to understand the data contained in PubChem.
Acknowledgements
This work was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health. The author would like to thank Dera Tompkins, NIH Library Editing Service, for reviewing the manuscript.
Author Contributions
Sunghwan Kim : conceptualization, methodology, visualization, writing original draft, writing review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
All PubChem data, tools, and services are provided to the public free of charge.
Literature Cited
- Armstrong, J. F., Faccenda, E., Harding, S. D., Pawson, A. J., Southan, C., Sharman, J. L. … Nc, I. (2020). The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: Extending immunopharmacology content and introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Research , 48(D1), D1006–D1021. doi: 10.1093/nar/gkz951.
- Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., … Gene Ontology, C. (2000). Gene Ontology: Tool for the unification of biology. Nature Genetics , 25(1), 25–29. doi: 10.1038/75556.
- Bolton, E. E., Chen, J., Kim, S., Han, L. Y., He, S. Q., Shi, W. Y. … Bryant, S. H. (2011). PubChem3D: A new resource for scientists. Journal of Cheminformatics , 3, 32. doi: 10.1186/1758-2946-3-32.
- Bolton, E. E., Kim, S., & Bryant, S. H. (2011a). PubChem3D: Conformer generation. Journal of Cheminformatics , 3, 4. doi: 10.1186/1758-2946-3-4.
- Bolton, E. E., Kim, S., & Bryant, S. H. (2011b). PubChem3D: Similar conformers. Journal of Cheminformatics , 3, 13. doi: 10.1186/1758-2946-3-13.
- Burley, S. K., Berman, H. M., Bhikadiya, C., Bi, C. X., Chen, L., Di Costanzo, L., … Zardecki, C. (2019). RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Research , 47(D1), D464–D474. doi: 10.1093/nar/gky1004.
- Carbon, S., Douglass, E., Good, B. M., Unni, D. R., Harris, N. L., & Mungall, C. J. … Gene Ontology, C. (2021) The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Research , 49(D1), D325–D334. doi: 10.1093/nar/gkaa1113.
- Chen, X., & Reynolds, C. H. (2002). Performance of similarity measures in 2D fragment-based similarity searching: Comparison of structural descriptors and similarity coefficients. Journal of Chemical Information and Computer Sciences , 42(6), 1407–1414. doi: 10.1021/ci025531g.
- Cheng, T., Zhao, Y., Li, X., Lin, F., Xu, Y., Zhang, X. … Lai, L. (2007). Computation of octanol−water partition coefficients by guiding an additive model with knowledge. Journal of Chemical Information and Modeling , 47(6), 2140–2148. doi: 10.1021/ci700257y.
- Davis, A. P., Grondin, C. J., Johnson, R. J., Sciaky, D., Wiegers, J., Wiegers, T. C., & Mattingly, C. J. (2021). Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Research , 49(D1), D1138–D1143. doi: 10.1093/nar/gkaa891.
- FDA. (2021). Pharmacologic class. Available at https://www.fda.gov/industry/structured-product-labeling-resources/pharmacologic-class.
- Freshour, S. L., Kiwala, S., Cotto, K. C., Coffman, A. C., McMichael, J. F., Song, J. J. … Wagner, A. H. (2021). Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Research , 49(D1), D1144–D1151. doi: 10.1093/nar/gkaa1084.
- Grant, J. A., Gallardo, M. A., & Pickup, B. T. (1996). A fast method of molecular shape comparison: A simple application of a Gaussian description of molecular shape. Journal of Computational Chemistry , 17(14), 1653–1666. doi: 10.1002/(sici)1096-987×(19961115)17:14<1653::Aid-jcc7>3.0.Co;2-k.
- Grant, J. A., & Pickup, B. T. (1995). A gaussian description of molecular shape. Journal of Physical Chemistry , 99(11), 3503–3510. doi: 10.1021/j100011a016.
- Grant, J. A., & Pickup, B. T. (1996). A gaussian description of molecular shape (vol 99, pg 3505, 1995). Journal of Physical Chemistry , 100(6), 2456–2456. doi: 10.1021/jp953707u doi: 10.1021/jp953707u.
- Grant, J. A., & Pickup, B. T. (1997). Gaussian shape methods. In W. F. Gunsteren, P. K. Weiner, & A. J. Wilkinson (Eds.), Computer simulation of biomolecular systems (pp. 150–176). Dordrecht: Kluwer Academic Publishers.
- Hähnke, V. D., Kim, S., & Bolton, E. E. (2018). PubChem chemical structure standardization. Journal of Cheminformatics , 10, 36. doi: 10.1186/s13321-018-0293-8.
- Halgren, T. A. (1996a). Merck molecular force field .1. Basis, form, scope, parameterization, and performance of MMFF94. Journal of Computational Chemistry , 17(5-6), 490–519. doi: 10.1002/(sici)1096-987×(199604)17:6<490::Aid-jcc1>3.3.Co;2-v.
- Halgren, T. A. (1996b). Merck molecular force field .2. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. Journal of Computational Chemistry , 17(5-6), 520–552. doi: 10.1002/(sici)1096-987×(199604)17:6<520::Aid-jcc2>3.3.Co;2-w.
- Halgren, T. A. (1999). MMFF VI. MMFF94s option for energy minimization studies. Journal of Computational Chemistry , 20(7), 720–729. doi: 10.1002/(sici)1096-987×(199905)20:7<720::Aid-jcc7>3.0.Co;2-x.
- Hastings, J., Owen, G., Dekker, A., Ennis, M., Kale, N., Muthukrishnan, V. … Steinbeck, C. (2016). ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Research , 44(D1), D1214–D1219. doi: 10.1093/nar/gkv1031.
- Heller, S. R., McNaught, A., Pletnev, I., Stein, S., & Tchekhovskoi, D. (2015). InChI, the IUPAC International Chemical Identifier. Journal of Cheminformatics , 7, 23. doi: 10.1186/s13321-015-0068-4.
- Holliday, J. D., Hu, C. Y., & Willett, P. (2002). Grouping of coefficients for the calculation of inter-molecular similarity and dissimilarity using 2D fragment bit-strings. Combinatorial Chemistry & High Throughput Screening, 5(2), 155–166. doi: 10.2174/1386207024607338.
- Holliday, J. D., Salim, N., Whittle, M., & Willett, P. (2003). Analysis and display of the size dependence of chemical similarity coefficients. Journal of Chemical Information and Computer Sciences , 43(3), 819–828. doi: 10.1021/ci034001x.
- Ihlenfeldt, W. D., Bolton, E. E., & Bryant, S. H. (2009). The PubChem chemical structure sketcher. Journal of Cheminformatics , 1, 20. doi: 10.1186/1758-2946-1-20.
- Kim, S. (2016). Getting the most out of PubChem for virtual screening. Expert Opinion on Drug Discovery , 11(9), 843–855. doi: 10.1080/17460441.2016.1216967.
- Kim, S., Bolton, E. E., & Bryant, S. H. (2011). PubChem3D: Biologically relevant 3-D similarity. Journal of Cheminformatics , 3, 26. doi: 10.1186/1758-2946-3-26.
- Kim, S., Bolton, E. E., & Bryant, S. H. (2013). PubChem3D: Conformer ensemble accuracy. Journal of Cheminformatics , 5, 1. doi: 10.1186/1758-2946-5-1.
- Kim, S., Bolton, E. E., & Bryant, S. H. (2016). Similar compounds versus similar conformers: Complementarity between PubChem 2-D and 3-D neighboring sets. Journal of Cheminformatics , 8, 62. doi: 10.1186/s13321-016-0163-1.
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S. … Bolton, E. E. (2019). PubChem 2019 update: Improved access to chemical data. Nucleic Acids Research , 47(D1), D1102–D1109. doi: 10.1093/nar/gky1033.
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S. … Bolton, E. E. (2021). PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Research , 49(D1), D1388–D1395. doi: 10.1093/nar/gkaa971.
- Kim, S., Thiessen, P. A., Bolton, E. E., & Bryant, S. H. (2015). PUG-SOAP and PUG-REST: Web services for programmatic access to chemical information in PubChem. Nucleic Acids Research , 43(W1), W605–W611. doi: 10.1093/nar/gkv396.
- Kim, S., Thiessen, P. A., Bolton, E. E., Chen, J., Fu, G., Gindulyte, A. … Bryant, S. H. (2016). PubChem Substance and Compound databases. Nucleic Acids Research , 44(D1), D1202–D1213. doi: 10.1093/nar/gkv951.
- Kim, S., Thiessen, P. A., Cheng, T., Zhang, J., Gindulyte, A. & Bolton, E. E. (2019). PUG-View: Programmatic access to chemical annotations integrated in PubChem. Journal of Cheminformatics , 11(1), 56. doi: 10.1186/s13321-019-0375-2.
- Kim, S., Thiessen, P. A., Cheng, T. J., Yu, B., & Bolton, E. E. (2018). An update on PUG-REST: RESTful interface for programmatic access to PubChem. Nucleic Acids Research , 46(W1), W563–W570. doi: 10.1093/nar/gky294.
- Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. (1997). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews , 23(1-3), 3–25. doi: 10.1016/s0169-409×(96)00423-1.
- Mendez, D., Gaulton, A., Bento, A. P., Chambers, J., De Veij, M., Felix, E. … Leach, A. R. (2019). ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Research , 47(D1), D930–D940. doi: 10.1093/nar/gky1075.
- OpenEye Scientific Software. (2010a). ROCS—Rapid Overlay of Chemical Structures. 3.1.0. Santa Fe, NM: OpenEye Scientific Software, Inc.
- OpenEye Scientific Software. (2010b). ShapeTK-C++. 1.8.0. Santa Fe, NM: OpenEye Scientific Software, Inc.
- PubChem. (2009). PubChem Substructure Fingerprint. (2/20/2021). Available at https://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.pdf.
- PubChem. (2014). What is the difference between a substance and a compound in PubChem? Available at http://go.usa.gov/x72qw.
- Rush, T. S., Grant, J. A., Mosyak, L., & Nicholls, A. (2005). A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction. Journal of Medicinal Chemistry , 48(5), 1489–1495. doi: 10.1021/jm040163o.
- Weininger, D. (1988). Smiles, a chemical language and information-system.1. Introduction to methodology and encoding rules. Journal of Chemical Information and Computer Sciences , 28(1), 31–36. doi: 10.1021/ci00057a005.
- Weininger, D. (1990). Smiles.3. depict−graphical depiction of chemical structures. Journal of Chemical Information and Computer Sciences , 30(3), 237–243. doi: 10.1021/ci00067a005.
- Weininger, D., Weininger, A., & Weininger, J. L. (1989). Smiles .2. algorithm for generation of unique smiles notation. Journal of Chemical Information and Computer Sciences , 29(2), 97–101. doi: 10.1021/ci00062a008.
- WHO. (2021). Anatomical Therapeutic Chemical (ATC) Classification. Available at https://www.who.int/tools/atc-ddd-toolkit/atc-classification.
- WIPO. (2021). International Patent Classification. Available at https://www.wipo.int/classifications/ipc/en/.
- Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R. … Wilson, M. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research , 46(D1), D1074–D1082. doi: 10.1093/nar/gkx1037.
Internet Resources
Daylight Chemical Information Systems Inc. SMARTS - A Language for Describing Molecular Patterns.
National Library of Medicine (NLM): Medical Subject Headings (2021).
Citing Literature
Number of times cited according to CrossRef: 24
- Jyoti Bashyal, Bimal Kumar Raut, Siddha Raj Upadhyaya, Kabita Sharma, Niranjan Parajuli, Exploration of Potent Human α‐Glucosidase Inhibitors Using In Silico Approaches: Molecular Docking, DFT, Molecular Dynamics Simulations, and MMPBSA, Journal of Chemistry, 10.1155/2024/2086167, 2024 , 1, (2024).
- Nasim Ahmed, Muhammad Abdul Bari, Partha Biswas, Sabbir Ahmed, Mohaimenul Islam Tareq, Shabana Bibi, A. H. M. Mazbah Uddin, Dhrubo Ahmed Khan, Mahmud Hasan, undefined Sohel, Nurul Islam, Norah A. Albekairi, Abdulrahman Alshammari, Nazmul Hasan, Amazon Plant‐Derived Compounds Suppressing Dengue NS5 Protein: Insights from Computational Drug Development and Network Pharmacology Approach, ChemistrySelect, 10.1002/slct.202303557, 9 , 10, (2024).
- Hongxiang Xu, Jiahua Cui, Yijun Cao, Lin Ma, Guixia Fan, Gen Huang, Kejia Ning, Jingzheng Wang, Yuntao Kang, Xin Sun, Jiushuai Deng, Shan Li, Construction of a Green-Comprehensive Evaluation System for Flotation Collectors, Processes, 10.3390/pr11051563, 11 , 5, (1563), (2023).
- Emmanuel Broni, Carolyn Ashley, Joseph Adams, Hammond Manu, Ebenezer Aikins, Mary Okom, Whelton A. Miller, Michael D. Wilson, Samuel K. Kwofie, Cheminformatics-Based Study Identifies Potential Ebola VP40 Inhibitors, International Journal of Molecular Sciences, 10.3390/ijms24076298, 24 , 7, (6298), (2023).
- Bryan Alejandro Espinosa-Rodriguez, Daniela Treviño-Almaguer, Pilar Carranza-Rosales, Monica Azucena Ramirez-Cabrera, Karla Ramirez-Estrada, Eder Ubaldo Arredondo-Espinoza, Luis Fernando Mendez-Lopez, Isaias Balderas-Renteria, Metformin May Alter the Metabolic Reprogramming in Cancer Cells by Disrupting the L-Arginine Metabolism: A Preliminary Computational Study, International Journal of Molecular Sciences, 10.3390/ijms24065316, 24 , 6, (5316), (2023).
- Jia‐yi Guo, Yong‐jun Wang, Si‐qi Li, Yu‐ping Wu, Molecular targets of metformin against ovarian cancer based on network pharmacology, Chemical Biology & Drug DesignChemical Biology & Drug DesignChemical Biology & Drug Design, 10.1111/cbdd.14234, 102 , 1, (88-100), (2023).
- K. Deepasree, Venugopal Subhashree, Molecular docking and dynamic simulation studies of terpenoid compounds against phosphatidylinositol-specific phospholipase C from Listeria monocytogenes, Informatics in Medicine Unlocked, 10.1016/j.imu.2023.101252, 39 , (101252), (2023).
- Manfred Hauben, Artificial Intelligence and Data Mining for the Pharmacovigilance of Drug–Drug Interactions, Clinical Therapeutics, 10.1016/j.clinthera.2023.01.002, 45 , 2, (117-133), (2023).
- Tong Wang, Daniel P. Russo, Dimitrios Bitounis, Philip Demokritou, Xuelian Jia, Heng Huang, Hao Zhu, Integrating structure annotation and machine learning approaches to develop graphene toxicity models, Carbon, 10.1016/j.carbon.2022.12.065, 204 , (484-494), (2023).
- Sunghwan Kim, Evan E. Bolton, PubChem: A Large‐Scale Public Chemical Database for Drug Discovery, Open Access Databases and Datasets for Drug Discovery, 10.1002/9783527830497.ch2, (39-66), (2023).
- Stephanie Holmgren, Chemical Toxicity Information Sources, Patty's Toxicology, 10.1002/0471125474.tox009.pub3, (1-33), (2023).
- Xiaoqing Chang, Yu-Mei Tan, David G. Allen, Shannon Bell, Paul C. Brown, Lauren Browning, Patricia Ceger, Jeffery Gearhart, Pertti J. Hakkinen, Shruti V. Kabadi, Nicole C. Kleinstreuer, Annie Lumen, Joanna Matheson, Alicia Paini, Heather A. Pangburn, Elijah J. Petersen, Emily N. Reinke, Alexandre J. S. Ribeiro, Nisha Sipes, Lisa M. Sweeney, John F. Wambaugh, Ronald Wange, Barbara A. Wetmore, Moiz Mumtaz, IVIVE: Facilitating the Use of In Vitro Toxicity Data in Risk Assessment and Decision Making, Toxics, 10.3390/toxics10050232, 10 , 5, (232), (2022).
- Sobika Bhandari, Bibek Raj Bhattarai, Ashma Adhikari, Babita Aryal, Asmita Shrestha, Niraj Aryal, Uttam Lamichhane, Ranjita Thapa, Bijaya B. Thapa, Ram Pramodh Yadav, Karan Khadayat, Achyut Adhikari, Bishnu P. Regmi, Niranjan Parajuli, Characterization of Streptomyces Species and Validation of Antimicrobial Activity of Their Metabolites through Molecular Docking, Processes, 10.3390/pr10102149, 10 , 10, (2149), (2022).
- Xiaowei Huang, Bian Wu, Fangxue Zhang, Fancheng Chen, Yong Zhang, Huizhi Guo, Hongtao Zhang, Epigenetic Biomarkers Screening of Non-Coding RNA and DNA Methylation Based on Peripheral Blood Monocytes in Smokers, Frontiers in Genetics, 10.3389/fgene.2022.766553, 13 , (2022).
- Qi Jin, Jie Li, Guang-Yao Chen, Zi-Yu Wu, Xiao-Yu Liu, Yi Liu, Lin Chen, Xin-Yi Wu, Yan Liu, Xin Zhao, Yue-Han Song, Network and Experimental Pharmacology to Decode the Action of Wendan Decoction Against Generalized Anxiety Disorder, Drug Design, Development and Therapy, 10.2147/DDDT.S367871, Volume 16 , (3297-3314), (2022).
- Rick Helmus, Bas van de Velde, Andrea M. Brunner, Thomas L. ter Laak, Annemarie P. van Wezel, Emma L. Schymanski, patRoon 2.0: Improved non-target analysis workflows including automated transformation product screening, Journal of Open Source Software, 10.21105/joss.04029, 7 , 71, (4029), (2022).
- Matthew R. Wilkinson, Uriel Martinez-Hernandez, Chick C. Wilson, Bernardo Castro-Dominguez, Images of chemical structures as molecular representations for deep learning, Journal of Materials Research, 10.1557/s43578-022-00628-9, 37 , 14, (2293-2303), (2022).
- Song Na, Li Ying, Cheng Jun, Xiong Ya, Zhang Suifeng, He Yuxi, Wang Jing, Lai Zonglang, Yang Xiaojun, Wu Yue, Study on the molecular mechanism of nightshade in the treatment of colon cancer, Bioengineered, 10.1080/21655979.2021.2016045, 13 , 1, (1575-1589), (2022).
- Yulong Wei, Ning Yu, Ziyuan Wang, Yiming Hao, Zongwei Wang, Zihui Yang, Jie Liu, Jing Wang, Analysis of the multi-physiological and functional mechanism of wheat alkylresorcinols based on reverse molecular docking and network pharmacology, Food & Function, 10.1039/D2FO01438F, (2022).
- Li Li, Shuang Dai, Jing-ya Liu, Wei Wu, Qian-xi Zhao, Xin Wang, Na Wang, Zhi-hong Xu, Antagonistic Effect and In Vitro Activity of Dauricine on Glucagon Receptor, Journal of Natural Products, 10.1021/acs.jnatprod.2c00446, 85 , 8, (2035-2043), (2022).
- Zhi-dan Gao, Hai-dong Yan, Ning-hua Wu, Qing Yao, Bin-bin Wan, Xiu-fen Liu, Zhen-wang Zhang, Qing-jie Chen, Cui-ping Huang, Mechanistic insights into the amelioration effects of lipopolysaccharide-induced acute lung injury by baicalein: An integrated systems pharmacology study and experimental validation, Pulmonary Pharmacology & Therapeutics, 10.1016/j.pupt.2022.102121, 73-74 , (102121), (2022).
- Sunghwan Kim, Tiejun Cheng, Siqian He, Paul A. Thiessen, Qingliang Li, Asta Gindulyte, Evan E. Bolton, PubChem Protein, Gene, Pathway, and Taxonomy Data Collections: Bridging Biology and Chemistry through Target-Centric Views of PubChem Data, Journal of Molecular Biology, 10.1016/j.jmb.2022.167514, 434 , 11, (167514), (2022).
- Maja A. Marinović, Edward T. Petri, Ljubica M. Grbović, Bojana R. Vasiljević, Suzana S. Jovanović‐Šanta, Sofija S. Bekić, Andjelka S. Ćelić, Investigation of the Potential of Bile Acid Methyl Esters as Inhibitors of Aldo‐keto Reductase 1C2: Insight from Molecular Docking, Virtual Screening, Experimental Assays and Molecular Dynamics, Molecular Informatics, 10.1002/minf.202100256, 41 , 10, (2022).
- Sho Nishimura, Kazune Nakamura, Miyako Yamamoto, Daichi Morita, Teruo Kuroda, Takanori Kumagai, Genome Sequence-Guided Finding of Lucensomycin Production by Streptomyces achromogenes Subsp. streptozoticus NBRC14001, Microorganisms, 10.3390/microorganisms10010037, 10 , 1, (37), (2021).

