Elution, SDS-PAGE, and RNA Purification
Clémentine Delan-Forino, David Tollervey
RNA degradation
Protein–RNA interaction
RNA-binding sites
UV cross-linking
Yeast
Exosome
RNA processing
Abstract
The RNA exosome complex functions in both the accurate processing and rapid degradation of many classes of RNA in eukaryotes and Archaea. Functional and structural analyses indicate that RNA can either be threaded through the central channel of the exosome or more directly access the active sites of the ribonucleases Rrp44 and Rrp6, but in most cases, it remains unclear how many substrates follow each pathway in vivo. Here we describe the method for using an UV cross-linking technique termed CRAC to generate stringent, transcriptome-wide mapping of exosome–substrate interaction sites in vivo and at base-pair resolution.
We present a protocol for the identification of RNA interaction sites for the exosome, using UV cross-linking and analysis of c DNA (CRAC) [ 1 , 2 ]. A number of related protocols for the identification of sites of RNA–protein interaction have been reported, including HITS-CLIP, CLIP-Seq, iCLIP, eCLIP, and others [ 3 , 4 , 5 , 6 ]. These all exploit protein immunoprecipitation to isolate protein–RNA complexes. CRAC is distinguished by the inclusion of tandem affinity purification and denaturing purification, allowing greater stringency in the recovery of authentic RNA–protein interaction sites.
To allow CRAC analyses, strains are created that express a “bait” protein with a tripartite tag. This generally consists of His6, followed by a TEV-protease cleavage site, then two copies of the z-domain from Protein A (HTP). The tag is inserted at the C terminus of the endogenous gene within the chromosome. The fusion construct is the only version of the protein expressed and this is under the control of the endogenous promoter. Several alternative tags have been successfully used, including a version with N-terminal fusion to a tag consisting of 3× FLAG-PreSission protease (PP) cleavage site-His6 (FPH) [ 7 ]. This is a smaller construct and is suitable for use on proteins with structures that are incompatible with C-terminal tagging. An additional variant is the insertion of a PP site into a protein that is also HTP tagged. This allows the separation of different domains of multidomain proteins. Importantly, the intact protein is cross-linked in the living cell, with domain separation in vitro. This has been successfully applied to the exosome subunit Rrp44/Dis3 to specifically identify binding sites for the PIN endonuclease domain [ 8 ].
Briefly, during standard CRAC analyses, covalently linked protein–exosome complexes are generated in vivo by irradiation with UV-C (254 nm). This generates RNA radicals that rapidly react with proteins in direct contact with the affected nucleotide (zero length cross-linking). The cells are then lysed and complexes with the bait protein are purified using an IgG column. Protein–RNA complexes are specifically eluted by TEV cleavage of the fusion protein and cross-linked RNAs trimmed using RNase A/T1, leaving a protected “footprint” of the protein binding site on the RNA. Trimmed complexes are denatured using 6 M Guanidinium, immobilized on Ni-NTA affinity resin and washed under denaturing conditions to dissociate copurifying proteins and complexes. The subsequent enzymatic steps are all performed on-column, during which RNA 3′ and 5′ ends are prepared, labeled with 32P (to allow RNA–protein complexes to be followed during gel separation) and linkers ligated. Note, however, that alternatives to using 32P labeling have been reported (e.g., [ 6 ]). The linker-ligated, RNA–protein complexes are eluted from the Ni-NTA resin and size selected on a denaturing SDS-PAGE gel. Following elution, the bound RNA is released by degradation of the bait protein using treatment with Proteinase K. The recovered RNA fragments are identified by reverse transcription, PCR amplification and sequencing using an Illumina platform.
Relative to CLIP-related protocols, CRAC offers the advantages of stringent purification, that substantially reduces background, and on-bead linker ligation that simplifies separation of reaction constituents during successive enzymatic steps. It also avoids the necessity to generate high-affinity antibodies needed for immunoprecipitation. Potential disadvantages are that, despite their ubiquitous use in yeast studies, tagged constructs may not be fully functional. This can be partially mitigated by confirming the ability of the tagged protein to support normal cell growth and/or RNA processing, or by comparing the behavior of N- and C-terminal tagged constructs. Additionally, because linkers are ligated to the protein–RNA complex, a possible disadvantage is that UV-cross-linking of the RNA at, or near, the 5′ or 3′ end it may sterically hinder on-column (de)phosphorylation and/or linker ligation. With these caveats, CRAC has been successfully applied to >50 proteins in budding yeast, and in other systems ranging from pathogenic bacteria to viral infected mouse cells [ 7 , 9 ].
Before start
Appropriate negative controls and experimental replicates are required to determine the background signal and true positive binding sites. We routinely use the (untagged) yeast parental strain as a negative control, performing a minimum of two biological and technical replicates for each sample. It is commonly observed that technical replicates (even samples from the same culture) processed in two independent CRAC experiments show more differences than two biological replicates (independent cultures) processed together.
All steps should be performed wearing disposable gloves and materials should be free of DNase and RNase. Prior to each CRAC experiment, pipettes should be cleaned with DNAZap (ThermoFisher; AM9890) to avoid DNA contamination at the PCR step, followed by RNaseZAP (ThermoFisher; AM9890) treatment, and rinsed with deionized water. All the buffers should be prepared with deionized water and free of RNases; however, DEPC treatment is not normally essential. To minimize buffer contamination, adjust the pH by taking small aliquots for measurements. Filter-sterilize stock solutions following preparation, and store at 4 °C. Where required, add β-mercaptoethanol and protease inhibitors to the buffers shortly before use. Wash buffers should be prepared immediately before starting the CRAC experiment.
All steps must be carried out on ice, unless stated otherwise. For troubleshooting, it is a good idea to monitor the course of the experiment by retaining samples at points during the CRAC protocol. This allows potential problems with Protein–RNA purification steps to be identified. Three aliquots per sample are taken during the purification (Subheading 3.2.2 3.2.2 “Crude Lysate” and “IgG supernatant,” Subheading 3.2.3 3.2.3 “TEV Eluate”). These can be analyzed by Western blot.
Steps
Elution and Precipitation of Exosome Subunit–RNA Complexes
Spin out the void volume. Close the bottom of the column with the press-on stopper and add 200µL.
Incubate the resin On ice for 0h 5m 0s.
Collect the flow-through in a 1.5 ml microcentrifuge tube. Reclose the bottom of the column and repeat the elution with another 200µL. Use the Geiger counter to ensure that the elution flow-through is radioactive.
Collect the residual Elution buffer on the column by briefly spinning the column.
Pool the eluates into a single microcentrifuge tube. Add 40µg and 100µL. Vortex and incubate On ice for 1h 0m 0s.
Centrifuge at top speed for 0h 30m 0s at 4°C. Remove the supernatant (use the Geiger counter to ensure that the pellet has not been dislodged if the blue pellet is not visible).
Add 800µL to the pellet and centrifuge for 0h 20m 0s at 4°C. Pellets should be small and clear. If it is big and white, add additional acetone wash step and pipette up and down until the pellet has dissolved completely. Longer incubation with acetone on ice can also be considered.
Remove the supernatant and air-dry the pellet at room temperature. Pellets can be hard to resuspend; do not allow them to overdry as this can lead to loss of material during the resuspension step.
PAGE Separation and Transfer
Resuspend the pellet in 30µL (dilute 4× buffer in distilled water before use). Pipette along the wall of the tube very carefully and check with the Geiger counter to ensure that most of the material has been resuspended.
Heat the samples at 65°C for 0h 10m 0s.
Load the sample and SeeBlue2 ladder onto a NuPAGE 4–12% gradient gel. Run for 1h 0m 0s at maximum 150 V or until the dye reaches the bottom of the gel.
Transfer the protein–RNA complex to nitrocellulose using a wet transfer western blotting system, at 100 V for 1h 30m 0s in NuPage transfer buffer supplemented with 10% Methanol. To avoid overheating during transfer, the tank can be placed in ice.
Expose the membrane (wrapped in cling film or protected by a transparent plastic film) to a high-sensitivity X-ray film at -80°C. If samples are highly radioactive, a 0h 30m 0s–1h 0m 0s exposure time should be enough. Overnight exposure is often required for samples with weaker radioactive signal. Ensure that a chemiluminescent marker is included to realign membrane and film after developing.
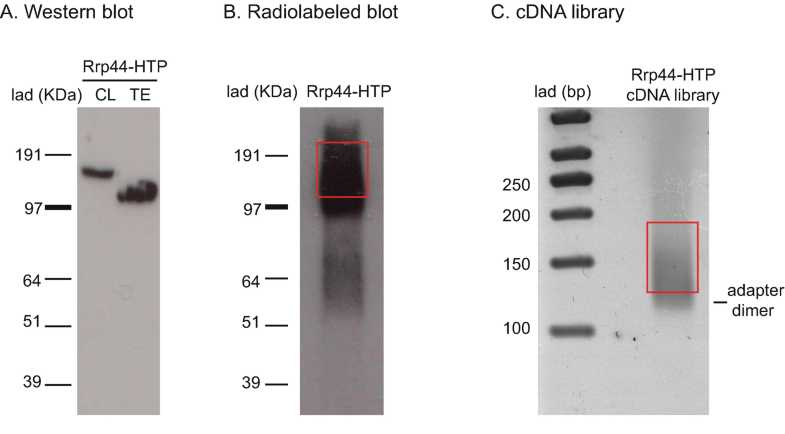
Develop the X-ray film and align it to the membrane using the chemiluminescent rulers. Cut out the smear corresponding to the size of the protein–RNA complex for all the samples. Cut at the same place in the negative control lane. Use clean scalpel for each sample. The first incision can be made in the middle of the band corresponding to the protein of interest plus the smear above to get most cross-link species. Once membrane fragments have been excised, they can be stored overnight (or longer) at -20°C or -80°C. An example of a radiolabeled blot for Rrp44-HTP is shown in Fig. 1b.
Recovery of Trimmed, Adapter-Ligated RNA
To digest away proteins, incubate the membrane slices with 400µL (50 mM Tris–HCl pH 7.8, 50 mM NaCl, 0.1% NP-40, 5 mM β-mercaptoethanol, 1% SDS, 5 mM EDTA). Add 100µg and incubate at 55°C for 2h 0m 0s with gentle mixing.
Add 50µL and 500µL. Vortex and centrifuge for 0h 5m 0s at Room temperature.
Transfer the aqueous phase to clean microcentrifuge tube and add 1mL and 20µg. Incubate at -80°C for 0h 30m 0s and centrifuge at 16000x g,4°C.
Wash the pellet with 500µL and centrifuge for 0h 20m 0s. Aspirate the supernatant and air dry.

