Determining the Effects of Neddylation on Cullin-RING Ligase–Dependent Protein Ubiquitination
Kankan Wang, Kankan Wang, Xing Liu, Xing Liu
Abstract
As the largest family of ubiquitin (Ub) E3 ligases, cullin-RING ligases (CRLs) play crucial roles in various cellular processes, and their activities are tightly regulated by orchestrated mechanisms. Neddylation, the conjugation of a Ub-like protein NEDD8 to a target protein such as the cullin, represents a key regulatory mechanism for CRLs. Biochemical and structural studies of a few CRLs have revealed that cullin neddylation alters the CRL conformation and activates CRL-dependent protein ubiquitination. Here, using CUL2-RING ligase (CRL2) as an example, we describe our protocols for the preparation of recombinant CUL2 with or without NEDD8 conjugation, which is further used to quantitatively determine the effect of neddylation on CRL2-dependent protein ubiquitination in vitro. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Expression and purification of CUL2•RBX1 from Escherichia coli
Support Protocol : Further purification of CUL2•RBX1 with additional chromatography on an FPLC system
Basic Protocol 2 : Reconstitution of cullin neddylation for quantitative ubiquitination assay in vitro
INTRODUCTION
NEDD8 is a small protein that possesses high sequential and structural similarity to ubiquitin (Ub). As with protein ubiquitination, the conjugation of NEDD8 to substrates, referred to as neddylation, requires an enzyme cascade comprising NEDD8 E1 activating enzyme (NAE), NEDD8 E2 conjugating enzyme, and E3 NEDD8 ligase (Lydeard, Schulman, & Harper, 2013). Cullin (CUL) proteins are the best-known substrates for neddylation, serving as the catalytic core for cullin-RING ligases (CRLs) and mediating the ubiquitination of diverse proteins involved in various cellular pathways. A CRL becomes enzymatically active when an E2-conjugated Ub (E2∼Ub) is engaged by the RING protein RBX1 (or RBX2) bound to the C-terminus of cullin and when the protein substrate is engaged by one of the interchangeable substrate receptor modules that specifically binds to the N-terminus of cullin. Biochemical studies showed that cullin neddylation promotes the activity of the CUL1-RING ligase (CRL1) (Morimoto, Nishida, Honda, & Yasuda, 2000), and two distinct effects of neddylation on CRL1 activity were revealed (Saha & Deshaies, 2008). First, neddylation enhances Ub chain initiation by bringing the substrate and E2∼Ub into proximity. Second, neddylation promotes Ub chain elongation through increasing E2 recruitment. More recently, a structural study of CRL1 in complex with E2∼Ub further illustrated that the conjugation of NEDD8 to CUL1 induces protein-protein interactions within the CRL1•E2∼Ub complex, including the contacts between NEDD8 and E2 and between E2 and the substrate receptor. As a result, the CRL1 complex changes from an open conformation to a “locked” conformation and thereby accelerates the transfer of Ub from E2 to the substrate docked on the substrate receptor (Baek et al., 2020).
Although neddylation has been considered as a general mechanism for CRL activation, to what extent neddylation promotes the ubiquitination of a specific protein by a specific CRL remains largely unexplored. Given that diverse substrate receptor modules and substrate proteins are engaged by different CRLs, a robust quantitative assay is required to help investigate and evaluate the effects of neddylation on the activity of an individual CRL toward its substrate(s).
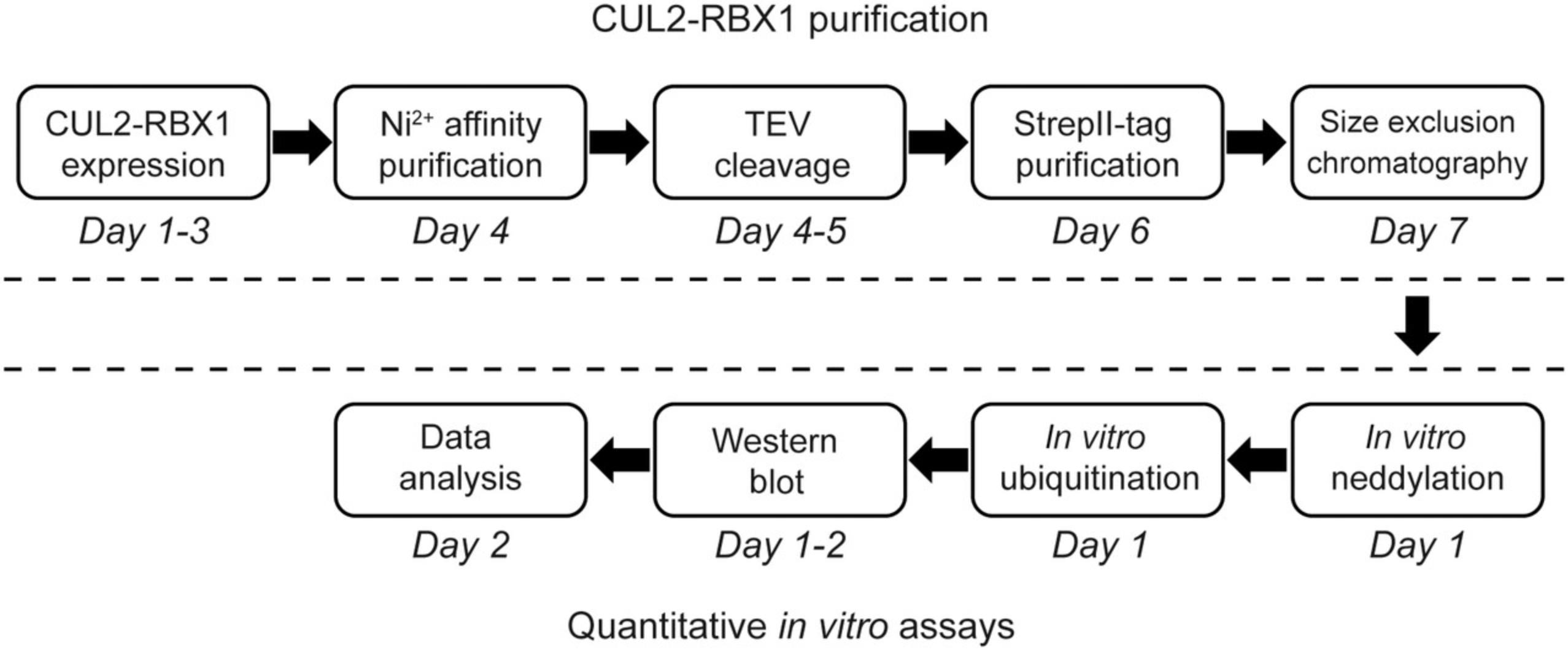
Here, we detail our protocols for the preparation of recombinant CUL2•RBX1 and neddylated CUL2•RBX1 (Basic Protocol 1 and Support Protocol) and the subsequent quantitative assay for CRL2-dependent protein ubiquitination (Basic Protocol 2) (Diaz, Li, Wang, & Liu, 2021; Wang, Reichermeier, & Liu, 2021). The protocols enable studies on the ubiquitination of a specific protein substrate with or without CUL2 neddylation, and the rate of substrate modification under each condition can be quantified for comparison. The workflow for these protocols is summarized in Figure 1, and it can be adapted to fit analyses using a different CRL and/or substrate of interest.
Basic Protocol 1: EXPRESSION AND PURIFICATION OF CUL2•RBX1 from Escherichia coli
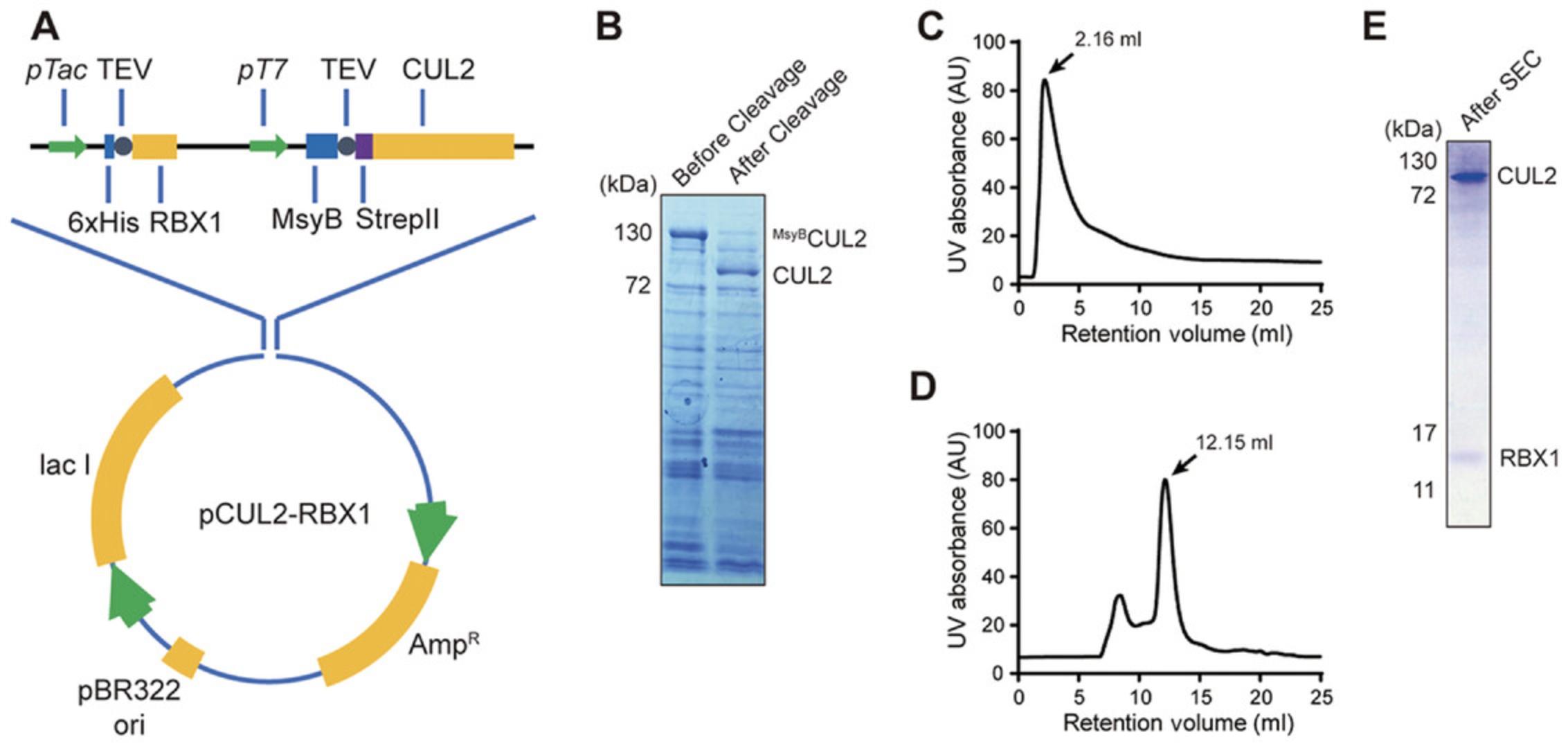
The enzymatic core of a CRL is a binary protein complex containing a cullin core and a RING domain protein (RBX1 or RBX2). To co-express the recombinant CUL•RBX protein complex in E. coli , the coding sequences of the two protein components can be incorporated either into one plasmid or separately into two plasmids. Using CUL2 as an example, this protocol describes the purification of recombinant CUL2•RBX1 complex using tandem affinity chromatography. The expression plasmid was derived from a pGEX-4T-2 vector (Cytiva, cat. no. 28-9545-50), with the 6×HisRBX1 coding sequence inserted after the Tac promoter, a T7 promoter sequence inserted after 6×HisRBX1, and the 6×HisMsyB-StrepIICUL2 coding sequence inserted after the T7 promoter. The hyper-acidic bacterial protein MsyB was fused to the N-terminus of CUL2 to improve protein solubility. A TEV protease cleavage site was inserted between the 6×His tag and RBX1 and between MsyB and StrepIICUL2.Figure 2A summarizes the structure of the CUL2•RBX1 expression plasmid.
Materials
-
BL21 (DE3) chemically competent E. coli cells (Thermo Fisher Scientific, cat. no. C600003)
-
CUL2•RBX1 expression plasmid (see Fig. 2A)
-
LB liquid medium (sterile; see recipe; make fresh), room temperature or 37°C
-
LB agar plates containing 100 µg/ml ampicillin (sterile; see recipe), 37°C
-
100 mg/ml ampicillin stock solution (sterile; see recipe)
-
0.8 M IPTG stock solution (see recipe)
-
Phosphate-buffered saline (PBS; Sigma-Aldrich, cat. no. P4417)
-
Ni-NTA lysis buffer (see recipe; make fresh), 4°C
-
Ni-NTA agarose beads (His-Tag Purification Resin, Roche, cat. no. COHISR-RO)
-
Ni-NTA wash buffer (see recipe)
-
Ni-NTA elution buffer (see recipe)
-
Bradford protein assay kit (Bio-Rad, cat. no. 5000202)
-
4× SDS sample buffer (see recipe)
-
Tobacco etch virus (TEV) protease (purified from Addgene plasmid #92414)
-
42°C water bath
-
37°C microbiological incubator
-
Microcentrifuge (Eppendorf 5425 or equivalent)
-
250-ml Erlenmeyer flask
-
2.8-L Fernbach culture flasks (Thermo Fisher Scientific, cat. no. 41052800)
-
Cell density meter (Biowave CO8000 or equivalent)
-
1000-ml centrifuge bottles (Thermo Fisher Scientific, cat. no. 3120-1000)
-
Refrigerated floor centrifuge (6-L capacity; Beckman Coulter J6-MI or equivalent)
-
15- and 50-ml conical centrifuge tubes (Falcon, cat. no. 352097, or equivalent, and Corning, cat. no. 352070, or equivalent)
-
Plastic beaker
-
Sonicator (Fisherbrand Model 505 or equivalent)
-
High-speed centrifuge tubes (Thermo Fisher Scientific, cat. no. 3139-0050)
-
Refrigerated high-speed centrifuge (Beckman Avanti J30I or equivalent), 4°C
-
Refrigerated benchtop centrifuge, 4°C (Eppendorf 5430 R or equivalent)
-
Refrigerated incubator shaker (New Brunswick I26R or equivalent), 4°C
-
Gravity flow columns (Bio-Rad, cat. no. 7321010)
-
Clamp stand
-
Two-way stopcock (Bio-Rad, cat. no. 7328102)
-
Additional reagents and equipment for SDS-PAGE (see Current Protocols article; Gallagher, 2012)
Transform E. coli cells with the CUL2•RBX1 expression construct
1.Thaw a tube of BL21 (DE3) chemically competent E. coli cells on ice.
2.Add 0.1 to 10 ng CUL2•RBX1 expression plasmid to competent cells and mix by gently flicking tube several times.
3.Incubate mixture on ice for 30 min.
4.Heat shock at 42°C in a water bath for 45 s.
5.Leave tube on ice for 5 min.
6.Add 900 µl LB liquid medium into mixture.
7.Place tube containing the cell mixture at 37°C in a microbiological incubator with vigorous shaking (200 to 250 rpm) for 60 min.
8.Centrifuge 2 min at 300 × g in a microcentrifuge to pellet cells.
9.Carefully discard supernatant and leave ∼100 µl supernatant in tube.
10.Resuspend cell pellet and spread suspended cells onto an LB agar plate containing 100 µg/ml ampicillin (or other appropriate antibiotic).
11.Incubate plate at 37°C overnight (typically ∼16 hr).
12.Check plate to make sure that a good amount of E. coli colonies show up on the plate.
Express the recombinant protein in E. coli
13.In a 250-ml Erlenmeyer flask, add 50 ml LB liquid medium and 50 µl of 100 mg/ml ampicillin stock solution. Inoculate a few colonies from LB agar plate into the LB medium and grow culture overnight at 37°C with vigorous shaking (200 to 250 rpm).
14.On the next day, add 1 ml of 100 mg/ml ampicillin stock solution into 1 L LB liquid medium in each of six 2.8-L Fernbach culture flasks and mix well by gently swirling for a few seconds.
15.Add overnight starter culture from step 13 into each 2.8-L culture flask to dilute the culture 200 times (5 ml culture per 1 L medium). Grow at 37°C with vigorous shaking (200 to 250 rpm).
16.Check optical density at 600 nm (OD600) of the culture periodically using a cell density meter to ensure that the OD600 does not go over 1.0.
17.When the OD600 is close to 1.0, stop shaker and cool down culture by transferring the flasks to a cold room. Then, set shaker to 16°C and allow it to cool.
18.After the culture and the shaker are cooled down, add 0.8 M IPTG stock solution to each flask to 0.2 mM to induce expression of the recombinant protein.
19.Incubate culture overnight at 16°C with vigorous shaking (200 to 250 rpm).
20.On the next day, transfer culture into 1000-ml centrifuge bottles and harvest E. coli cells by centrifugation for 30 min at 3000 × g using a refrigerated floor centrifuge. Discard supernatant and resuspend cells in PBS.
21.Transfer resuspended cells into 50-ml conical centrifuge tubes. Centrifuge 10 min at 3000 × g and discard supernatant.
Extract CUL2•RBX1 from E. coli cells using Ni2+ affinity chromatography
22.Add cold Ni-NTA lysis buffer to tubes containing bacteria cell pellets.
23.Sonicate pellets briefly to free the pellets from the bottom of the tubes.
24.Transfer pellets and lysis buffer from the two tubes into the same plastic beaker incubated in an ice-water bath.
25.Sonicate pellets in the beaker to lyse the cells thoroughly.
26.Transfer equal volumes of cell lysate into two high-speed centrifuge tubes. Centrifuge 30 min at 25,000 × g , 4°C, in a refrigerated high-speed centrifuge.
27.During the centrifugation period (step 26), prepare Ni-NTA agarose beads for protein extraction by first loading Ni-NTA agarose bead slurry into two 15-ml conical centrifuge tubes.
28.Centrifuge 2 min at 500 × g , 4°C, in a refrigerated benchtop centrifuge.
29.Discard supernatant and resuspend beads in two bed volumes of cold Ni-NTA lysis buffer.
30.Repeat steps 27 to 29 two more times. Then, transfer equal volumes of agarose bead slurry into two 50-ml conical centrifuge tubes.
31.After the centrifugation in step 26 is done, transfer supernatant into the tubes containing the Ni-NTA beads (prepared in step 30).
32.Incubate tubes at 4°C for ≥2 hr with mild agitation on a refrigerated incubator shaker.
33.Centrifuge as in step 28 and discard supernatant.
34.Resuspend beads with two bed volumes of Ni-NTA wash buffer.
35.Repeat steps 33 to 34 two more times to remove excess cell lysate from beads.
36.Set up an empty gravity flow column on a clamp stand and wash column with 10 ml Ni-NTA wash buffer.
37.Transfer all the bead slurry (from step 35) into the column. Use a two-way stopcock on bottom of the column to control the flow.
38.Wait for beads to settle and then let buffer flow out of the column.
39.Close stopcock and resuspend beads in two bed volumes of Ni-NTA elution buffer. Incubate for 10 min.
40.Open stopcock and collect eluate in a 50-ml conical centrifuge tube incubated on ice.
41.Repeat steps 39 and 40 three to four more times. Use a Bradford protein assay kit to determine if a fraction of the eluate contains a detectable amount of protein that is worth collecting.
42.Combine eluate from all collected fractions and estimate total amount (mg) of protein via the Bradford assay.
43.Mix 30 µl eluate with 10 µl of 4× SDS sample buffer. Store up to 6 months at –20°C.
44.Add TEV protease to eluate to remove the MsyB protein and the 6×His tag. Mix by gently inverting tube several times and incubate at 4°C overnight.
45.On the next day, mix 9 µl eluate with 3 µl of 4× SDS sample buffer. Analyze this sample together with the sample from step 43 using SDS-PAGE (see Current Protocols article; Gallagher, 2012) to assess the progress of the TEV cleavage. If the cleavage is not complete, add more TEV protease and incubate longer.
Support Protocol: FURTHER PURIFICATION OF CUL2•RBX1 WITH ADDITIONAL CHROMATOGRAPHY ON AN FPLC SYSTEM
In this protocol, the recombinant CUL2•RBX1 extracted by the Ni-NTA beads in Basic Protocol 1 is further purified by StrepII-tag affinity chromatography followed by size exclusion chromatography (SEC) on an FPLC system.
Materials
-
Deionized water
-
Buffer W (see recipe)
-
Biotin elution buffer (see recipe; make fresh)
-
CUL2•RBX1 solution (see Basic Protocol 1)
-
SEC buffer (see recipe; make fresh)
-
FPLC system (ÄKTA pure 25 or equivalent)
-
Strep-Tactin XT 4Flow cartridge, 1 ml (IBA Lifesciences, cat. no. 2-5024-001)
-
Centrifugal filter units (50-kDa cutoff; Millipore, cat. no. UFC905024 or UFC805024)
-
Microcentrifuge (Eppendorf 5425 or equivalent), 4°C
-
0.22-μm syringe filters (VWR, cat. no. 28145-477)
-
1- and 20-ml syringes (BD, cat. no. 301025 and 301031)
-
Fraction collector
-
SEC column (Superdex 200 Increase 10/300 GL column, Cytiva, cat. no. 28990944)
-
0.22-μm centrifugal filters (Millipore, cat. no. UFC30GVNB)
-
Syringe needles
-
UV spectrophotometer
-
Additional reagents and equipment for SDS-PAGE (see Current Protocols article; Gallagher, 2012)
-
NOTE : All buffers and water should be filtered and degassed using bottle-top filters (Thermo Fisher Scientific, cat. no. 5963320) before use on the FPLC system.
StrepII-tag affinity chromatography
1.Set up FPLC system:
-
Wash Lines A and B with deionized water.
-
Insert Line A in Buffer W and Line B in biotin elution buffer.
-
Install 10-ml sample loop on the injection valve.
2.Prepare FPLC system:
-
Set pre-column pressure alarm to 0.5 MPa.
-
Prime Line B with ≥15 ml biotin elution buffer.
-
Prime Line A with ≥15 ml Buffer W.
-
Wash sample loop with ≥20 ml Buffer W.
-
Attach Strep-Tactin XT 4Flow cartridge to the FPLC system. Pay close attention to joints and make sure that no leak occurs.
-
Wash cartridge with 10 ml Buffer W at 1 ml/min.
3.Concentrate CUL2•RBX1 solution to <10 ml using a centrifugal filter unit (50-kDa cutoff) by centrifuging at 5000 × g , 4°C, and checking the sample every 5 min.
4.Pass CUL2•RBX1 solution through a 0.22-μm syringe filter attached to a 20-ml syringe to remove precipitated protein or other small particles that can clog the column.
5.Inject filtered protein sample into the sample loop.
6.Load protein sample into the Strep-Tactin cartridge at 0.5 ml/min.
7.Wash column with Buffer W until the UV trace reaches the basal level.
8.Elute protein using 100% biotin elution buffer at 1 ml/min for ≤30 ml. Collect eluate as 1-ml fractions in the fraction collector.
9.Analyze fractions under the UV peak using SDS-PAGE (see Current Protocols article; Gallagher, 2012). Combine fractions containing CUL2•RBX1.
Size exclusion chromatography
10.Set up FPLC system:
-
Wash Lines A and B with deionized water.
-
Insert Line A in SEC buffer.
-
Install 1-ml sample loop on the injection valve.
11.Prepare FPLC system:
-
Set pre-column pressure alarm to 2.8 MPa.
-
In “inject” mode, prime Line A with ≥20 ml SEC buffer.
This will also wash the sample loop.
12.Adjust flow rate to 0.4 ml/min and connect SEC column. Make sure that no leak occurs.
13.In “inject” mode, equilibrate column with 40 ml Buffer W.
14.Concentrate protein sample from step 9 to ∼500 μl using a centrifugal filter unit (50-kDa cutoff) by centrifuging at 5000 × g , 4°C, and checking the sample every 5 min. Filter protein sample through a 0.22-μm centrifugal filter.
15.Load filtered protein sample into the sample loop using a 1-ml syringe with a needle. To avoid introducing air into the sample loop, insert syringe needle into the injection port in “inject” mode and then switch to “manual load” mode and load protein solution into the sample loop.
16.Run elution program at 0.4 ml/min for 30 ml. Collect eluate as 300-μl fractions in the fraction collector.
17.Analyze fractions under the UV peak(s) as in step 9.Pay close attention to fractions with expected retention volume (12 to 14 ml for CUL2•RBX1). Combine fractions containing the target protein and low levels of contaminants.
18.Using a UV spectrophotometer, determine concentration of the collected protein through obtaining the UV absorbance (at 280 nm) of the protein solution and the molar extinction coefficient (Ɛ) for the target protein (see Current Protocols article; Olson, 2016).
19.If needed, concentrate protein stock using a centrifugal filter unit to the desired concentration.
20.Make aliquots and store up to 1 year at −80°C.
Basic Protocol 2: RECONSTITUTION OF CULLIN NEDDYLATION FOR QUANTITATIVE UBIQUITINATION ASSAY IN VITRO
Using CUL2•RBX1 as an example, this protocol describes the conjugation of NEDD8 to CUL2 in vitro. The CUL2•RBX1 protein is further incorporated into quantitative ubiquitination assays to evaluate the effect of neddylation on the activity of CUL2•RBX1.
Materials
-
Distilled water
-
4× SDS sample buffer (see recipe)
-
Anti-FLAG antibody (Sigma-Aldrich, cat. no. F1804; 1:1000 dilution)
-
Anti-VHL antibody (Cell Signaling Technology, cat. no. 68547S; 1:1000 dilution)
-
Anti-CUL2 antibody (Thermo Fisher, cat. no. 51–1800; 1:1000 dilution)
-
1.5-ml microcentrifuge tubes
-
100°C heat block
-
ImageJ or equivalent software
-
Additional reagents and equipment for assembling neddylation reaction (see Table 1), “substrate + substrate receptor mixture” (see step 3 annotation), and “Ub mixture” (see Table 2), for SDS-PAGE (see Current Protocols article; Gallagher, 2012), and for western blot (see Current Protocols article; Gallagher, Winston, Fuller, & Hurrell, 2011)
| Component | Concentration |
|---|---|
| Reaction buffer (see recipe for 10×) | 1× |
| DTT (see recipe for 1 M) | 2 mM |
| ATP (see recipe for 200 mM) | 2 mM |
| NEDD8 (R&D Systems, cat. no. UL-812-500) | 2 μM |
| NAE (R&D Systems, cat. no. E-313-025) | 0.25 μM |
| UBC12 (R&D Systems, cat. no. E2-656-100) | 3 μM |
| CUL2•RBX1 (see Basic Protocol 1 and Support Protocol) | 0.2 μM |
| Component | Concentration |
|---|---|
| Reaction buffer (see recipe for 10×) | 2× |
| DTT (see recipe for 1 M) | 4 mM |
| ATP (see recipe for 200 mM) | 4 mM |
| Ub (R&D Systems, cat. no. U-100H-10M) | 120 μM |
| UBE1 (R&D Systems, cat. no. E-305-025) | 2 μM |
| CDC34 (R&D Systems, cat. no. E2-610-100) | 0.8 μM |
Reconstitute CUL2 neddylation in vitro
1.neddylation reaction by mixing all reagents at the concentrations listed in Table 1.Add distilled water to adjust to desired final volume before adding the protein stocks.
2.Incubate reaction mixture at room temperature for 1 hr.
Evaluate the effect of cullin neddylation on substrate ubiquitination in vitro
3.Prepare “substrate + substrate receptor mixture” at concentrations four times the final concentration in the ubiquitination reaction (step 5).
4.Prepare “Ub mixture” by mixing all reagents at the concentrations listed in Table 2.Add distilled water to adjust to desired final volume before adding the protein stocks.
5.Initiate ubiquitination reaction by mixing the solutions as described in Table 3 in a 1.5-ml microcentrifuge tube and incubating at room temperature.
| Volume | Component |
|---|---|
| 60 μl |
CUL2N8•RBX1 or CUL2•RBX1 or mock control (step 2) |
| 30 μl | Substrate + substrate receptor mixture (step 3) |
| 30 μl | Ub mixture (step 4) |
| Amount | Component |
|---|---|
| 1× | Reaction buffer |
| 2 mM | DTT |
| 2 mM | ATP |
| 0.5 μM | Ub E1 |
| 0.2 μM | Ub E2 (Cdc34) |
| 30 μM | Ub |
| 0.1 μM |
CUL2N8•RBX1 (or CUL2•RBX1 or mock control) |
| 0.1 μM | VHL•EloB•EloC |
| 0.1 μM | Substrate |
6.At each desired time point after the start of the reaction, withdraw 18 μl reaction mixture and mix with 6 μl of 4× SDS sample buffer to stop reaction. When samples from all time points have been collected, incubate them at 100°C for 5 min in a heat block.
7.Run all samples on an SDS-PAGE gel. Then, detect unmodified and ubiquitinated substrates by western blot (see Current Protocols article; Gallagher, Winston, Fuller, & Hurrell, 2011) with anti-FLAG antibody, anti-VHL antibody, and anti-CUL2 antibody.
8.Quantify intensity of the unmodified substrate bands from all time points using ImageJ or equivalent software (Davarinejad, 2015). Also quantify intensity of the VHL (loading control) bands for normalization purposes.
9.Fit normalized intensity of the unmodified substrate bands with a one-phase exponential decay model.
REAGENTS AND SOLUTIONS
Ampicillin stock solution, 100 mg/ml
- Dissolve 10 g ampicillin (Fisher Scientific, cat. no. BP1760-25) in 100 ml ultrapure H2O
- Sterilize by filtering through 0.22-μm syringe filter (VWR, cat. no. 28145-477)
- Aliquot and store ≤6 months at −20°C
ATP stock solution, 200 mM
- Dissolve 1.1 g adenosine 5′-triphosphate disodium salt hydrate (Chem-Impex International, cat. no. 00015) in 7 ml ultrapure H2O
- Adjust pH to 7.0 by adding 1 M KOH
- Ultrapure H2O to 10 ml
- Aliquot and store ≤3 months at –80°C
Biotin elution buffer
- 100 mM Tris·HCl, pH 8.0
- 300 mM NaCl
- 5% (v/v) glycerol
- 50 mM biotin (Enzo, cat. no. ALX-460-002)
- Adjust pH to 8.0 by adding 6 N HCl
- Prepare fresh immediately before use and store at 4°C
- Filter and degas by vacuum before use
Buffer W
- 100 mM Tris·HCl, pH 8.0
- 150 mM NaCl
- Store ≤1 month at 4°C
- Filter and degas by vacuum before use
DTT stock solution, 1 M
- Dissolve 1.54 g dithiothreitol (DTT; Fisher Scientific, cat. no. BP172-25) in 10 ml ultrapure H2O
- Aliquot and store ≤3 months at −20°C
IPTG stock solution, 0.8 M
- Dissolve 19 g isopropyl-β-D-thiogalactopyranoside (IPTG; Sigma-Aldrich, cat. no. I6758) in 100 ml ultrapure H2O
- Aliquot and store ≤1 year at −20°C
LB agar plates containing 100 µg/ml ampicillin
- Add 15 g agar (BD, cat. no. 214010) to 1 L LB liquid medium (see recipe)
- Sterilize by autoclaving
- Cool to ∼50°C
- Add 1 ml of 100 mg/ml ampicillin stock solution (see recipe) and mix well
- Pour into sterile 10-cm Petri dishes (Fisher Scientific, cat. no. FB0875712)
- Wait until agar is solidified and cool
- Store ≤1 month at 4°C
LB liquid medium
- Add one LB broth pre-buffered capsule (Fisher Scientific, cat. no. BP97315) per 1 L ultrapure H2O
- Sterilize by autoclaving
- Prepare fresh immediately before use
Ni-NTA lysis buffer
- Dissolve one tablet of EDTA-free Protease Inhibitor Cocktail (Roche, cat. no. COEDTAF-RO) in 50 ml Ni-NTA wash buffer (see recipe)
- Prepare fresh immediately before use and keep at 4°C
Ni-NTA elution buffer
- 50 mM HEPES, pH 7.5
- 150 mM NaCl
- 250 mM imidazole (Sigma-Aldrich, cat. no. 56750)
- Store ≤3 months at 4°C
Ni-NTA wash buffer
- 30 mM Tris·HCl, pH 7.5
- 200 mM NaCl (Sigma-Aldrich, cat. no. S7653)
- 10% (v/v) glycerol (Sigma-Aldrich, cat. no. G9012)
- 10 mM imidazole (Sigma-Aldrich, cat. no. 56750)
- Adjust pH to 7.5 by adding 6 N NaOH
- Store ≤3 months at 4°C
- Immediately before use, add 5 mM β-mercaptoethanol (Sigma-Aldrich, cat. no. M6250)
SDS sample buffer, 4×
- 240 mM Tris·HCl, pH 6.8
- 8% (w/v) SDS (Sigma-Aldrich, cat. no. 74255)
- 40% (v/v) glycerol
- 0.08% (w/v) bromophenol blue (Sigma-Aldrich, cat. no. B5525)
- Aliquot and store ≤6 months at –20°C
- Immediately before use, add 0.5 M β-mercaptoethanol (Sigma-Aldrich, cat. no. M6250)
SEC buffer
- 30 mM Tris·HCl, pH 7.5
- 100 mM NaCl
- 10% (v/v) glycerol
- 1 mM DTT (from 1 M stock; see recipe)
- Prepare fresh immediately before use and store at 4°C
- Filter and degas by vacuum before use
Reaction buffer, 10×
- 300 mM Tris·HCl, pH 7.5
- 50 mM MgCl2 (Sigma-Aldrich, cat. no. M8266)
- Store ≤3 months at 4°C
COMMENTARY
Background Information
NEDD8 is a highly conserved Ub-like protein in eukaryotic species. Structurally, NEDD8 is composed of a globular core and a C-terminal tail. NEDD8 is initially translated as a precursor containing a tail with the “GGLRQ” sequence. This tail is then proteolytically cleaved by Ub C-terminal hydrolase L3 (UCHL3) or deneddylase 1 (DEN1) to expose the Gly residues that are vital to the conjugation of NEDD8 to its substrates (Wada, Kito, Caskey, Yeh, & Kamitani, 1998; Wu et al., 2003). The processed NEDD8 is covalently conjugated to substrate proteins through concerted actions of the NEDD8 E1, E2, and E3. As the first step, NEDD8 forms a thioester bond with NAE (E1) via its C-terminal Gly. The human NAE complex contains two subunits, amyloid-β precursor protein binding protein 1 (APPBP1) and Ub-activating enzyme 3 (UBA3). NEDD8 binds the heterodimeric NAE complex with extensive contacts (Walden et al., 2003). In particular, a conserved adenylation domain on UBA3 and a catalytic cysteine domain on APPBP1 are crucial for the bipartite interface. The NAE-mediated activation of NEDD8 is an ATP-dependent process. First, NAE catalyzes the formation of the NEDD8-AMP intermediate that binds the adenylation domain of UBA3. The intermediate is then transferred to the catalytic Cys, generating a high-energy thioester bond. Following the activation, the thioester-linked NEDD8 is transferred to a conserved Cys on the NEDD8 E2 via a transthiolation reaction, forming the E2∼NEDD8. UBC12 (also known as UBE2M) and UBE2F are the two known NEDD8 E2s that exhibit distinct substrate specificities. UBC12 cooperates with RBX1 to neddylate CUL1/2/3/4, whereas UBE2F recognizes RBX2 for the neddylation of CUL5 (Huang et al., 2009). At the last step, E2∼NEDD8 interacts with the NEDD8 E3 to transfer the NEDD8 to the protein substrate. Evidence from biochemical and structural studies has suggested a dual E3 mechanism for cullin neddylation (Kurz et al., 2008). The RING domain protein RBX1 (or RBX2) functions as the main E3 that binds and activates the E2∼NEDD8. In addition, through binding both the cullin protein and the acetylated N-termini of NEDD8 E2s, defective-in-cullin-neddylation (DCN)-like (DCNL) proteins serve as co-E3s that shape the flexible RBX-bound E2∼NEDD8 to a catalytically favorable geometry (Monda et al., 2013). Taken together, the biochemical mechanisms of cullin neddylation indicate that an in vitro cullin neddylation reaction minimally requires NEDD8, NAE, UBC12/UBE2F, CUL•RBX1, and ATP. The neddylation co-E3 such as DCNL1 can be incorporated to accelerate the reaction or to study the kinetics of cullin neddylation.
The family of cullin proteins—including the canonical CUL1, CUL2, CUL3, CUL4a, CUL4b, and CUL5 and the non-canonical CUL7 and CUL9—are the best characterized neddylation substrates. Each type of cullin engages a unique adaptor (with the exception of CUL3) and a set of substrate receptor proteins through its N-terminus, forming a subfamily of CRLs (Petroski & Deshaies, 2005). Given that different CRLs comprise different subunits, it is not hard to imagine that structural differences exist across different CRL subfamilies and even within the same subfamily. Moreover, the same CRL can bind distinct substrates for ubiquitination, which adds another layer of conformational diversity. Therefore, although neddylation has been considered as a general mechanism for CRL activation, it remains important to quantitatively define the effects of neddylation on the ubiquitination activity of a specific CRL toward a given substrate.
The in vitro assays described here provide advantages over cell-based assays for studying protein neddyation and ubiquitination. Although the rate of protein ubiquitination in cells can be estimated by monitoring the remaining level of the target protein in a cycloheximide assay, the result can be affected by various factors other than ubiquitination or neddyation. In cells, the assembly of CRLs is dynamically regulated by multiple players to achieve timely and efficient ubiquitination of a changing pool of CRL substrates (Liu et al., 2018; Pierce et al., 2013; Reitsma et al., 2017; Wang, Deshaies, & Liu, 2020). After ubiquitination and before proteosome-dependent degradation, substrates can be reverted to the unmodified form by deubiquitinating enzymes (Reyes-Turcu, Ventii, & Wilkinson, 2009), resulting in a lower apparent rate of ubiquitination. Further, the CRL substrate may be ubiquitinated and/or degraded via CRL-independent pathways, making cell-based analyses nonspecific to the CRL of interest (Wang et al., 2021). Due to the complexity of the cellular environment, it is challenging to establish a cell-based assay to directly compare the activity of a specific CRL with and without cullin neddylation. Instead, reconstitution of the cullin neddylation and ubiquitination reactions in vitro allows the protein components and their concentrations to be precisely controlled for quantitative analyses. Thus, these in vitro assays provide important tools for studying cullin neddylation and CRL-dependent protein ubiquitination.
It is noteworthy that quite a few proteins besides cullins have been identified as neddylation substrates (Enchev, Schulman, & Peter, 2015). The complete neddylation enzymatic cascade for these substrates remains unclear, which hinders the development of corresponding neddylation assays in vitro. Thus, the important roles of neddylation in biological processes need to continue to be revealed through constant endeavors to fill knowledge gaps.
Critical Parameters
Strategy for expressing recombinant CUL•RBX1 in E. coli
Cullin proteins are routinely co-expressed with RBX1 (or RBX2 for CUL5), and the two subunits form a stable complex in E. coli. The MsyB protein fused to the N-terminus of CUL2 was designed to improve the solubility and folding of CUL2, and the same strategy can be used to help express other proteins with low solubility in bacterial cells. As an alternative strategy, “Split-n-Coexpress” can be used to co-express RBX1 with CUL that is split into two halves. This approach has been reported for CUL1•RBX1 (Li, Pavletich, Schulman, & Zheng, 2005), CUL2•RBX1 (Diaz et al., 2021), and CUL3•RBX1 (Zhuang et al., 2009).
Use of DCNL proteins
The activity of DCNLs relies on the N-terminal acetylation of the NEDD8 E2 enzymes (see Background Information above). Therefore, DCNLs should be used in a pair with the NEDD8 E2 expressed and purified from the baculovirus expression system.
Purification of full-length CRL substrate
Although chemically synthesized degron peptides are frequently used as the substrate in in vitro studies, it may be necessary to use full-length substrates for the ubiquitination assay. When enriching the epitope-tagged substrate protein from mammalian cells via affinity purification, the substrate receptor and/or Ub ligase co-immunoprecipitated with the substrate should be minimized. Using quantitative western blot, the concentration of the purified substrate can be determined, provided that a recombinant protein with the same epitope tag and a known concentration is included as the standard for quantification.
Troubleshooting
Please refer to Tables 5 and 6 for troubleshooting guidance for Basic Protocols 1 and 2, respectively.
| Problem | Possible cause | Solution |
|---|---|---|
| Poor expression in bacteria | Incorrect construct | Confirm the sequence of the construct |
| Incorrect bacterial strain used for transformation or the transformants are too old | Use the BL21 (DE3) strain to express genes driven by the T7 promoter. Use freshly transformed cells for expression. | |
| Inefficient induction | Use a new aliquot of IPTG stock solution or increase the IPTG concentration for induction | |
| Poor protein recovery | Protein precipitation | Increase the NaCl concentration in the lysis buffer (e.g., 500 mM) to help with protein solubility. Make sure β-mercaptoethanol is freshly added to the lysis and wash buffers. Include 5-10% glycerol in all buffers to help with protein solubility and stability. Keep the protein solution on ice or at 4°C as much as possible. |
| Insufficient binding to the Ni-NTA beads | Check the extraction buffer. If Ni-NTA beads from a different vendor are used, then increase the Tris·HCl concentration to 50-100 mM and adjust the pH to 8.0. | |
| Inefficient elution | Freshly prepare the elution buffer. Make sure the imidazole concentration is correct. | |
| Inefficient TEV cleavage after the first incubation period | Low TEV activity | Repeat the TEV cleavage and incubate the protein solution at 16°C |
| Problem | Possible cause | Solution |
|---|---|---|
| Low neddylation or ubiquitination efficiency | Reaction components have become inactive; for example, proteins are aggregated or denatured or the ATP has broken down | Use new aliquots of protein stocks or freshly prepared ATP stock solution |
| Insufficient reaction time | Incubate for a longer time | |
| Improper reaction temperature | Perform the reactions at or above room temperature (22-37°C) | |
| The unmodified substrates disappear much faster than the appearance of the ubiquitinated substrates | Ubiquitinated substrates located in the top portion of the SDS-PAGE gel have been removed | Gradient SDS-PAGE gels (e.g., 4-20%) are recommended for sample fractionation, and the whole gel should be preserved for downstream analyses |
| Unmodified substrates are cleaved by proteases present in the reaction mixture, which is more likely to occur when substrate proteins are immunoprecipitated from human cells | Include protease inhibitor in the reaction mixture. The protease inhibitor cocktail used in the Ni-NTA lysis buffer can also be used here, and the final concentration can be doubled. | |
| The unmodified substrates disappear over time in the mock control group | The reaction mixture is contaminated with proteases (see above) | Add protease inhibitor to the reaction mixture (see above) |
Understanding Results
In Basic Protocol 1 and the Support Protocol, the CUL2•RBX1 is purified sequentially by Ni-NTA affinity chromatography, removal of fusion proteins and tags by TEV cleavage, Strep II-tag affinity purification, and SEC. After the TEV cleavage, the MsyB protein fused to the N-terminus of CUL2 is removed, and a shift of the CUL2 band is expected on the SDS-PAGE gel (Fig. 2B). After the TEV cleavage is complete, CUL2•RBX1 is bound to a 1-ml Strep-Tactin cartridge on an ÄKTA FPLC system. Eluting CUL2•RBX1 from the cartridge using biotin elution buffer is very efficient, with the eluted protein peak lasting from ∼2 ml to ∼15 ml (Fig. 2C). It is very common for the elution peak to tail, and eluate in the tail region usually contains the target protein and should be collected. At this point, the CUL2•RBX1 should look very pure on the SDS-PAGE gel. Lastly, SEC is used to further separate the correctly folded CUL2•RBX1 from the aggregated population and to change the buffer for better protein storage. The retention volume of CUL2•RBX1 on the Superdex 200 Increase 10/300 GL column is ∼12 ml (Fig. 2D and E). Aggregated CUL2•RBX1 has a retention volume of ∼8 ml (void volume) and should not be collected. In terms of the protein yield, ∼0.2 mg CUL2•RBX1 per liter of bacterial culture can be expected.
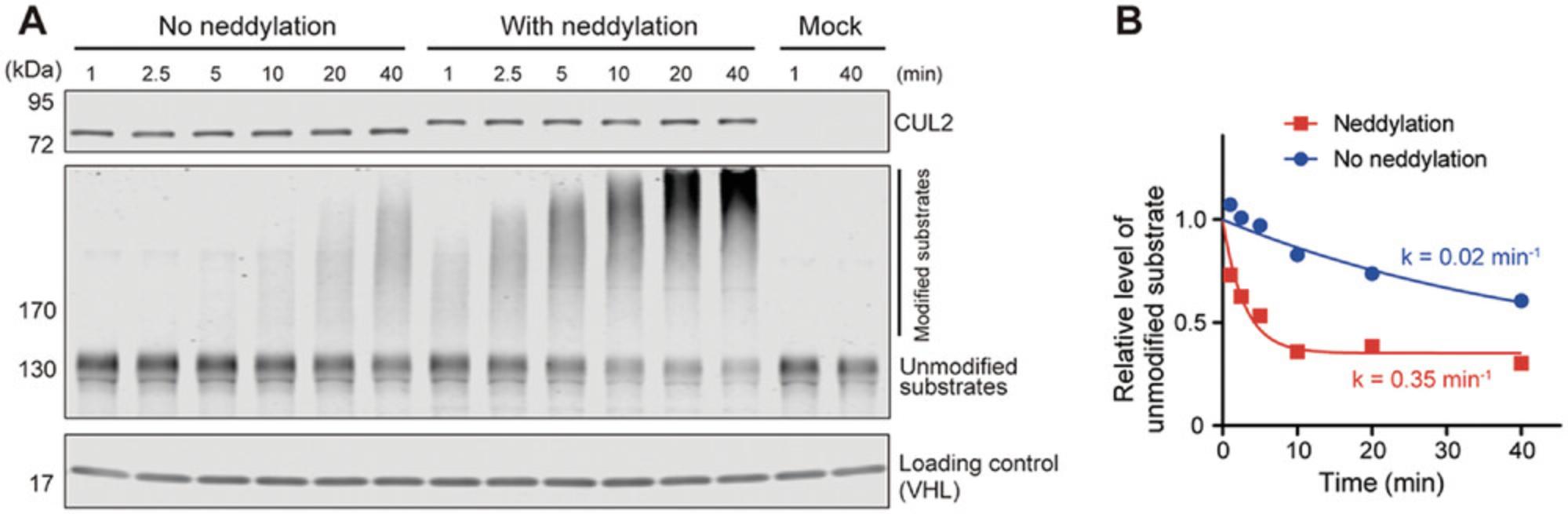
In Basic Protocol 2, CUL2 is conjugated with NEDD8 in vitro. Almost all CUL2 is converted to the neddylated form, and the CUL2 band is expected to shift upward in the western blot (Fig. 3A, upper blot). The neddylated CUL2•RBX1 is then used for time course–dependent substrate ubiquitination. Of note, a mock (negative) control group where no CUL2•RBX1 is added should be included and is expected to show no change in the level of unmodified substrate over time (Fig. 3A, last two lanes of the middle blot). Only with correct results obtained from the negative control group can the disappearance of the unmodified substrates in the presence of CUL2•RBX1 be interpreted as a result of CRL2-dependent ubiquitination (Fig. 3A, middle blot). To compare the rates at which substrates are ubiquitinated when different forms of CUL2•RBX1 are supplied, the remaining amounts of unmodified substrates at different time points should be fit with a one-phase exponential decay model. Intensities of the unmodified substrate bands are used for this quantification, and the values are normalized to account for variations in sample loading volumes. Because the same “substrate + substrate receptor” master mix is used for all samples, intensities of the substrate receptor bands are used for normalization (Fig. 3A, bottom blot). Finally, the normalized levels of unmodified substrates are used for regression analysis, and rates for substrate ubiquitination are quantitatively compared (Fig. 3B).
Time Considerations
Basic Protocol 1 (followed by the Support Protocol) can be completed in 7 days: plasmid transformation (∼2 hr) on Day 1, growth medium and starter culture preparation (∼2 hr) on Day 2, protein expression in E. coli (∼7 hr) on Day 3, bacterial cell collection (∼2 hr) on Day 4, Ni-NTA affinity purification (∼6 hr) on Day 4 (or another day), TEV cleavage and SDS-PAGE analysis (∼2 hr) on Day 5, StrepII-tag affinity purification and SDS-PAGE analysis (∼5 hr) on Day 6, and SEC (∼3 hr) on Day 7.
Basic Protocol 2 can be completed in 2 days: in vitro neddylation assay (2 hr) and in vitro ubiquitination assay (2 hr) on Day 1, sample analysis by SDS-PAGE (2 hr) on Day 1, and western blot on Days 1 and 2.
Acknowledgments
This work was supported by National Institutes of Health grant R35GM138016 (to X.L.) and an American Heart Association Career Development Award (to X.L.). This work was also supported by a start-up fund from Purdue University (to X.L.).
Author Contributions
Kankan Wang : Data curation, Formal analysis, Writing — original draft; Xing Liu : Funding acquisition, Supervision, Writing — review and editing.
Conflict of Interest
The authors declare that they have no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Literature Cited
- Adelmant, G., Garg, B. K., Tavares, M., Card, J. D., & Marto, J. A. (2019). Tandem affinity purification and mass spectrometry (TAP-MS) for the analysis of protein complexes. Current Protocols in Protein Science , 96(1), e84. doi: 10.1002/cpps.84.
- Baek, K., Krist, D. T., Prabu, J. R., Hill, S., Klugel, M., Neumaier, L. M., … Schulman, B. A. (2020). NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature , 578(7795), 461–466. doi: 10.1038/s41586-020-2000-y.
- Davarinejad, H. (2015). Quantifications of western blots with ImageJ. University of York. Retrieved from https://static.yanyin.tech/literature/current_protocol/10.1002/cpz1.401/attachments/ImageJ.pdf.
- Diaz, S., Li, L., Wang, K., & Liu, X. (2021). Expression and purification of functional recombinant CUL2*RBX1 from E. coli. Scientific Reports , 11(1), 11224. doi: 10.1038/s41598-021-90770-x.
- Enchev, R. I., Schulman, B. A., & Peter, M. (2015). Protein neddylation: Beyond cullin-RING ligases. Nature Reviews Molecular Cell Biology , 16(1), 30–44. doi: 10.1038/nrm3919.
- Gallagher, S. R. (2012). One-dimensional SDS gel electrophoresis of proteins. Current Protocols in Molecular Biology , 97, 10.2A.1–10.2A.44. doi: 10.1002/0471142727.mb1002as97.
- Gallagher, S., Winston, S. E., Fuller, S. A., & Hurrell, J. G. R. (2011). Immunoblotting and immunodetection. Current Protocols in Molecular Biology , 52, 10.8.1–10.8.28. doi: 10.1002/0471143030.cb0602s52.
- Huang, D. T., Ayrault, O., Hunt, H. W., Taherbhoy, A. M., Duda, D. M., Scott, D. C., … Schulman, B. A. (2009). E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Molecular Cell , 33(4), 483–495. doi: 10.1016/j.molcel.2009.01.011.
- Kurz, T., Chou, Y. C., Willems, A. R., Meyer-Schaller, N., Hecht, M. L., Tyers, M., … Sicheri, F. (2008). Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Molecular Cell , 29(1), 23–35. doi: 10.1016/j.molcel.2007.12.012.
- Li, T., Pavletich, N. P., Schulman, B. A., & Zheng, N. (2005). High-level expression and purification of recombinant SCF ubiquitin ligases. Methods in Enzymology , 398, 125–142. doi: 10.1016/s0076-6879(05)98012-9.
- Liu, X., Reitsma, J. M., Mamrosh, J. L., Zhang, Y., Straube, R., & Deshaies, R. J. (2018). Cand1-mediated adaptive exchange mechanism enables variation in F-box protein expression. Molecular Cell , 69(5), 773–786.e776. doi: 10.1016/j.molcel.2018.01.038.
- Lydeard, J. R., Schulman, B. A., & Harper, J. W. (2013). Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Reports , 14(12), 1050–1061. doi: 10.1038/embor.2013.173.
- Monda, J. K., Scott, D. C., Miller, D. J., Lydeard, J., King, D., Harper, J. W., … Schulman, B. A. (2013). Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structure , 21(1), 42–53. doi: 10.1016/j.str.2012.10.013.
- Morimoto, M., Nishida, T., Honda, R., & Yasuda, H. (2000). Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1). Biochemical and Biophysical Research Communications , 270(3), 1093–1096. doi: 10.1006/bbrc.2000.2576.
- Motulsky, H., & Christopoulos, A. (2004). Fitting models to biological data using linear and nonlinear regression: A practical guide to curve fitting. New York: Oxford University Press.
- Olson, B. (2016). Assays for determination of protein concentration. Current Protocols in Pharmacology , 73, A.3A.1–A.3A.32. doi: 10.1002/cpph.3.
- Petroski, M. D., & Deshaies, R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nature Reviews Molecular Cell Biology , 6(1), 9–20. doi: 10.1038/nrm1547.
- Pierce, N. W., Lee, J. E., Liu, X., Sweredoski, M. J., Graham, R. L., Larimore, E. A., … Deshaies, R. J. (2013). Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell , 153(1), 206–215. doi: 10.1016/j.cell.2013.02.024.
- Reitsma, J. M., Liu, X., Reichermeier, K. M., Moradian, A., Sweredoski, M. J., Hess, S., & Deshaies, R. J. (2017). Composition and regulation of the cellular repertoire of SCF ubiquitin ligases. Cell , 171(6), 1326–1339.e1314. doi: 10.1016/j.cell.2017.10.016.
- Reyes-Turcu, F. E., Ventii, K. H., & Wilkinson, K. D. (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual Review of Biochemistry , 78, 363–397. doi: 10.1146/annurev.biochem.78.082307.091526.
- Saha, A., & Deshaies, R. J. (2008). Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Molecular Cell , 32(1), 21–31. doi: 10.1016/j.molcel.2008.08.021.
- Wada, H., Kito, K., Caskey, L. S., Yeh, E. T., & Kamitani, T. (1998). Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochemical and Biophysical Research Communications , 251(3), 688–692. doi: 10.1006/bbrc.1998.9532.
- Walden, H., Podgorski, M. S., Huang, D. T., Miller, D. W., Howard, R. J., Minor, D. L., … Schulman, B. A. (2003). The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Molecular Cell , 12(6), 1427–1437. doi: 10.1016/s1097-2765(03)00452-0.
- Wang, K., Deshaies, R. J., & Liu, X. (2020). Assembly and regulation of CRL ubiquitin ligases. Advances in Experimental Medicine and Biology , 1217, 33–46. doi: 10.1007/978-981-15-1025-0_3.
- Wang, K., Reichermeier, K. M., & Liu, X. (2021). Quantitative analyses for effects of neddylation on CRL2(VHL) substrate ubiquitination and degradation. Protein Science , 30(11), 2338–2345. doi: 10.1002/pro.4176.
- Wu, K., Yamoah, K., Dolios, G., Gan-Erdene, T., Tan, P., Chen, A., … Pan, Z. Q. (2003). DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. Journal of Biological Chemistry , 278(31), 28882–28891. doi: 10.1074/jbc.M302888200.
- Zhuang, M., Calabrese, M. F., Liu, J., Waddell, M. B., Nourse, A., Hammel, M., … Schulman, B. A. (2009). Structures of SPOP-substrate complexes: Insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Molecular Cell , 36(1), 39–50. doi: 10.1016/j.molcel.2009.09.022.
Citing Literature
Number of times cited according to CrossRef: 1
- Lihong Li, Kankan Wang, Yun Zhou, Xing Liu, Review: A silent concert in developing plants: Dynamic assembly of cullin-RING ubiquitin ligases, Plant Science, 10.1016/j.plantsci.2023.111662, 330 , (111662), (2023).

