DOH Workshop Protocol Part 3: Library preparation for Rapid Sequencing DNA V14 Barcoding kit (SQK-RBK114.24) with Pronex modification
Natalie Ring, Bryan Wee, Vesa Qarkaxhija
Abstract
This protocol performs creation of Nanopore sequencing libraries for the MinION flow cell using the Rapid Barcoding Kit
Before start
Calculate the dilution of your starting genomic DNA from the extraction step to make up 10µL with a maximum concentration of20- 30 (200ng-300ng total DNA). (For example, if your DNA from the previous protocol was 100, you would only need 2µLof your sample, diluted in 8µLof NFW).1. Assign each sample to a Barcode (1-24) and note this down.
- Program the thermal cycler to incubate at
30°Cfor0h 2m 0sthen80°Cfor another0h 2m 0s. Do not start it yet.
Steps
DOH Workshop Protocol Part 3
Prepare one 0.2 ml thin-walled PCR tube for each sample from the previous step. Label the top of the tube with the barcode number.
In each 0.2 ml thin-walled PCR tubes:
Pipette an appropriate amount of your sample (1µL-10µL) and add NFW (1µL-10µL), if necessary, to get 200ng-300ng of DNA in a 10µLtotal volume.
1.5µL of your chosen Rapid Barcode (RB01-24). (1 barcode per sample).
Incubate the tubes in the thermal cycler (PCR machine) at 30°C for 0h 2m 0s then 80°C for another 0h 2m 0s.
Briefly place the tubes On ice to cool.
Only 1 person is required to carry out the following steps.
Resuspend the Pronex beads by vortexing for 0h 0m 10s or longer.
Incubate the sample at Room temperature for 0h 10m 0s.
To do while waiting
Take out Elution Buffer (EB) to thawOn ice.
Place the sample on a magnetic stand for 0h 2m 0suntil the solution becomes clear and the beads form a pellet on one side of the tube.
While leaving the tube on the magnet, carefully remove and discardsupernatant without disturbing the beads.
Wash 1: While still on the magnetic stand, carefully add 200µL of Pronex wash buffer without flushing directly onto the pellet. If 200µL is not enough to submerge the pellet, use more wash buffer.
Allow to incubate for 0h 0m 30s-0h 1m 0s .
While leaving the tube on the magnet, carefully remove and discard supernatant without disturbing the beads.
Wash 2: While still on the magnetic stand, carefully add 200µLof Pronex wash buffer without flushing directly onto the pellet. If 200µL is not enough to submerge the pellet, use more wash buffer.
Allow to incubate for 0h 0m 30s-0h 1m 0s .
While leaving the tube on the magnet, carefully remove and discard supernatant without disturbing the beads.
Remove the tube from the magnetic rack and add 15µL of elution buffer (included in the Nanopore Rapid Barcoding kit). Resuspend the beads by slowly pipetting or stirring with the pipette tip.
Incubate the samples at Room temperature for 0h 10m 0s to elute the DNA.
Return the tube to the magnetic stand for 0h 1m 0suntil the solution becomes clear and the beads form a pellet.
Store the Pronex beads in the fridge.
Take another 1µLof the elute from the tube on the magnetic stand for quantification on a fluorometer (Qubit or Quantus). The remaining beads can be kept in a closed tube On ice, for re-elution, if necessary.
In a new 1.5mL Eppendorf DNA LoBind tube, mix the following:
1.5µLRapid Adapter (RA)3.5µLAdapter Buffer (ADB)
Add 1µL of this RA + ADB mixture to the DNA.
Mix gently by flicking and spin down briefly (0h 0m 2s-0h 0m 3s).
Incubate this for0h 30m 0s atRoom temperature.
Preparing the flowcell
Remove the following Nanopore Rapid Kit (RBK-114.24) items from the -20°Cfreezer, spin down and store On ice.
- SB (Sequencing Buffer)
- LIB (Library Beads)
- FCT (Flow Cell Tether)
- FCF (Flow Cell Flush)
- Bovine Serum Albumin (BSA) at 50mg/ml
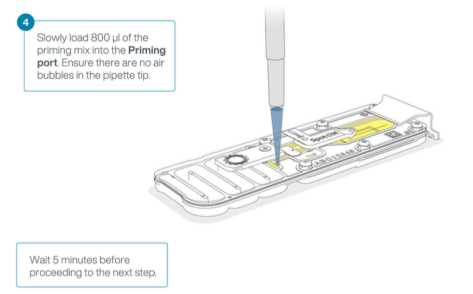
Prepare the flow cell Priming Mix in a fresh 1.5mL Eppendorf DNA LoBind. Mix by inverting the tube.
1170µLFCF5µLBSA (Bovine Serum Albumin)30µLFCT
Remove the flow cell you want to use and slide it under the metal clip in the Mk1B or Mk1C MinION. Press down firmly to ensure correct contact with the thermal and electrical connections.
- Mk1B: Plug in the MinION Mk1B to a laptop with Minknow software
- Mk1C: Turn on the MinION Mk1C.
Complete a flow cell check to assess the number of pores available on the flow cell.
Rotate the flow cell priming port cover clockwise to open the priming port.
After opening the priming port there will be a small air bubble under the cover that needs to be removed.
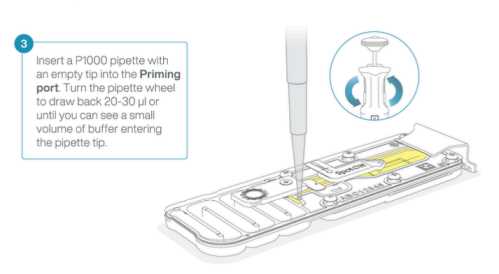
- Set a P1000 pipette to
200µL - Insert the tip into the priming port
- Turn the adjustment wheel slowly, pausing every few μls,until the pipette shows
220µL-230µLto draw a total of20µL-30µLout of the priming port, or until you can see a small volume of liquid entering the pipette tip.
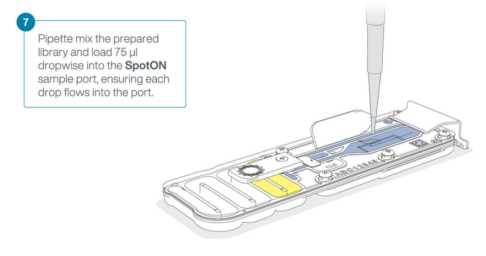
Mix the LIB (Library Beads) thoroughly by pipetting.
In a new 1.5 ml LoBind tube, mix the following to prepare your library:
37.5µLSB (Sequencing Buffer)25.5µLLIB (Library Beads), mixed immediately before use12µLof DNA library (your sample)
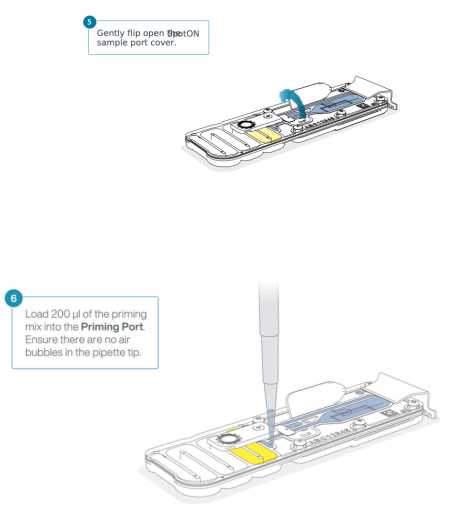
Gently open the SpotON sample port cover
Replace the SpotON sample port cover and gently press down to ensure the bung is in the port.
Rotate the Priming port cover back to close the Priming port.
Quickly cover the lid of the MinION to protect from light.
Start the run on the Minknow software interface.