Current Strategies of Modeling Human Trophoblast Using Human Pluripotent Stem Cells in vitro
Virginia Chu Cheung, Virginia Chu Cheung, Tony Bui, Tony Bui, Francesca Soncin, Francesca Soncin, Tao Bai, Tao Bai, John A. Kessler, John A. Kessler, Mana M Parast, Mana M Parast, Mariko Horii, Mariko Horii
extravillous trophoblast
human trophoblast stem cell
human pluripotent stem cell
syncytiotrophoblast
trophoblast differentiation
Abstract
We previously established a trophoblast differentiation protocol from primed human pluripotent stem cells (PSC). To induce this lineage, we use a combination of Bone Morphogenetic Protein-4 (BMP4) and the WNT inhibitor IWP2. This protocol has enabled us to obtain a pure population of trophectoderm (TE)-like cells that could subsequently be terminally differentiated into syncytiotrophoblasts (STB) and extravillous trophoblasts (EVT). However, the resulting TE-like cells could only be terminally differentiated to a variable mixture of STB and EVT, with a bias toward the STB lineage. Recently, methods have been developed for derivation and culture of self-renewing human trophoblast stem cells (TSC) from human embryos and early gestation placental tissues. These primary TSCs were further able to differentiate into either STB or EVT with high efficiency using the lineage specific differentiation protocols. Based partly on these protocols, we have developed methods for establishing self-renewing TSC-like cells from PSC, and for efficient lineage-specific terminal differentiation. Here, we describe in detail the protocols to derive and maintain PSC-TSC, from both embryonic stem cells (ESC) and patient-derived induced pluripotent stem cells (iPSC), and their subsequent terminal differentiation to STB and EVT. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
This article was corrected on 14 April 2024. See the end of the full text for details.
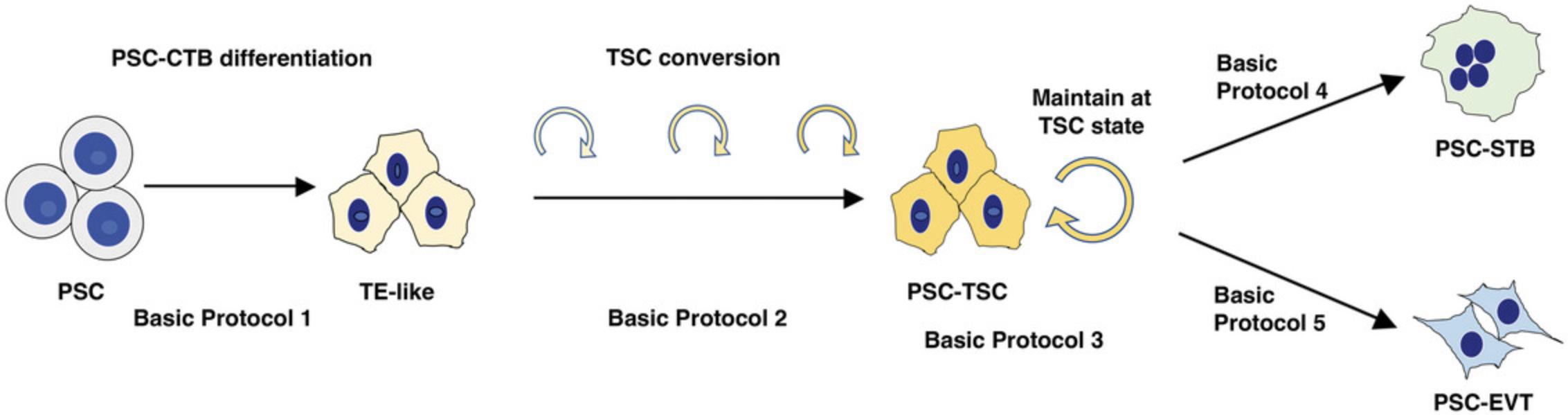
Basic Protocol 1 : Trophoblast Differentiation into TE-like Cells
Basic Protocol 2 : Conversion of PSC-Derived TE-like Cells to TSC
Basic Protocol 3 : Passaging PSC-Derived TSC in iCTB Complete Medium
Basic Protocol 4 : STB Differentiation from PSC-derived TSC
Basic Protocol 5 : EVT Differentiation from PSC-derived TSC
Support Protocol 1 : Geltrex-coated tissue culture plate preparation
Support Protocol 2 : Collagen IV-coated tissue culture plate preparation
Support Protocol 3 : Fibronectin-coated tissue culture plate preparation
INTRODUCTION
A significant portion of our current knowledge of the cellular and molecular mechanisms of trophoblast differentiation comes from the use of mouse trophoblast stem cells (TSC) (Tanaka et al., 1998) and tumor-derived or immortalized human trophoblast cell lines (Cerneus & van der Ende, 1991; Kohler & Bridson, 1971), due to the difficulties in deriving bona fide human trophoblast stem cells (TSC). However, the in-depth comparison of mouse and human placental development has revealed significant differences, particularly in early gestation (Blakeley et al., 2015; Soncin et al., 2018). Therefore, it is necessary to develop a model to enable studies of normal human trophoblast differentiation. Over the past two decades, multiple groups have shown that human pluripotent stem cells (PSC)—both embryonic (ESC) and induced pluripotent stem cells (iPSC)—can be differentiated into trophoblast and used to model both normal development and disease phenotypes (Aghajanova et al., 2012; Alici-Garipcan et al., 2020; Amita et al., 2013; Castel et al., 2020; Das et al., 2007; Erb et al., 2011; Horii et al., 2016; Horii et al., 2019; Horii et al., 2021; Kobayashi et al., 2022; Krendl et al., 2017; Kurek et al., 2015; Li et al., 2013; Sheridan et al., 2019; Sudheer et al., 2012; Telugu et al., 2013; Warmflash et al., 2014; Wu et al., 2008; Xu et al., 2002; Yabe et al., 2016; Yang et al., 2015). The use of patient-derived iPSC provides the added opportunity of exploring specific mutation-associated imprinting disorders in addition to genetic or epigenetic abnormalities associated with specific pregnancy and/or neonatal adverse outcomes (Alici-Garipcan et al., 2020; Horii et al., 2016; Horii et al., 2021; Sheridan et al., 2019). This is particularly important for studying placenta-based pregnancy disorders because they are known to be epigenetically affected by maternal lifestyle and the uterine environment (Apicella et al., 2019; Maccani & Maccani, 2015; Workalemahu et al., 2020). However, such previously established protocols result in a mixed population of terminally-differentiated syncytiotrophoblasts (STB) and extravillous trophoblasts (EVT), and are generally biased towards the STB lineage (Horii et al., 2019). The variability of a mixed culture is not ideal for determining the individual contribution of EVT and STB to a cellular or molecular phenotype, as characterization will be based on the balance of mixed culture in a dish. Further, the minor EVT population limits the types of assays we can use to further examine this cell type.
In 2018, Okae et al. established a method to derive and culture bona fide TSCs from human pre-implantation-stage embryos and early first trimester placenta (Okae et al., 2018). The in vitro maintenance and subsequent differentiation of TSC in a lineage-specific manner into EVT and STB has allowed the field to make tremendous progress towards understanding the molecular mechanisms of human trophoblast differentiation. However, the use of pre-implantation embryos and early trimester placentas holds multiple challenges due to both unknown pregnancy outcomes, and ethical, legal, and funding barriers associated with these tissues. Applying the Okae culture conditions to derive TSC from PSCs avoids the latter challenges. Importantly, iPSC-based models have the added benefit of known patient information, including pregnancy outcome and placental pathology.
Recently, new protocols for TSC derivation have been reported from both naïve (pre-implantation) and primed (post-implantation) pluripotent states (Cinkornpumin et al., 2020; Dong et al., 2020; Io et al., 2021; Jang et al., 2022; Li et al., 2019; Mischler et al., 2021; Soncin et al., 2022; Wei et al., 2021). TSC derived from both naïve and primed PSC have trophoblast characteristics, such as the capacity for terminal differentiation into multinucleated human chorionic gonadotropin (hCG)-secreting STB and invasive human leukocyte antigen-G+ (HLA-G+) EVT, with appropriate gene expression profiles, as well as E74 like ETS transcription factor 5 (ELF5) promoter hypomethylation and lack of expression of classical human leukocyte antigen (HLA) molecules (Io et al., 2021; Soncin et al., 2022). Naïve PSC-derived TSC also share one additional feature with early gestation trophoblast, which is high expression of chromosome 19 miRNA cluster (C19MC) genes (Io et al., 2021; Kobayashi et al., 2022). However, converting PSC to the naïve state is accompanied by a global loss of DNA methylation, which includes loss of imprinting throughout the genome (Theunissen et al., 2016). Differentiation of naïve PSC into TSC appears to lead to the re-emergence of at least some of the placenta-associated DNA methylation pattern (Cinkornpumin et al., 2020); however, the capacity of these cells for modeling trophoblast-associated defects underlying pregnancy disorders, particularly those associated with an epigenetic basis, remains unknown.
Here, we provide a detailed protocol for derivation of TSC from primed PSC, which is improved from the original version (Horii et al., 2019), and suited for modeling lineage-specific trophoblast differentiation (Soncin et al., 2022). This manuscript describes protocols for the induction of trophectoderm (TE)-like cells using a simple defined medium (Basic Protocol 1); conversion of these cells to TSC-like cells using “iCTB” medium, developed in the Kessler laboratory, for reprogramming of term CTB into TSC-like cells (Bai et al., 2021) (Basic Protocol 2); maintenance of PSC-derived TSC (PSC-TSC) (Basic Protocol 3); terminal differentiation of these PSC-TSC into STB (Basic Protocol 4); terminal differentiation of these PSC-TSC into EVT (Basic Protocol 5); Geltrex coated plate preparation (Support Protocol 1); Collagen IV coated plate preparation (Support Protocol 2); and Fibronectin coated plate preparation (Support Protocol 3) in a step-by-step method (Fig. 1).
CAUTION : All cell culture must be done in a suitable class II laminar flow hood to avoid contamination. All of the protocols described here require the proper use of aseptic technique.
CAUTION : Follow all appropriate guidelines and regulations for the use of human pluripotent stem cells at each individual institution/country (i.e., approval from an Embryonic Stem Cell Research Oversight (ESCRO) committee).
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals. Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol 1: TROPHOBLAST DIFFERENTIATION INTO TE-LIKE CELLS
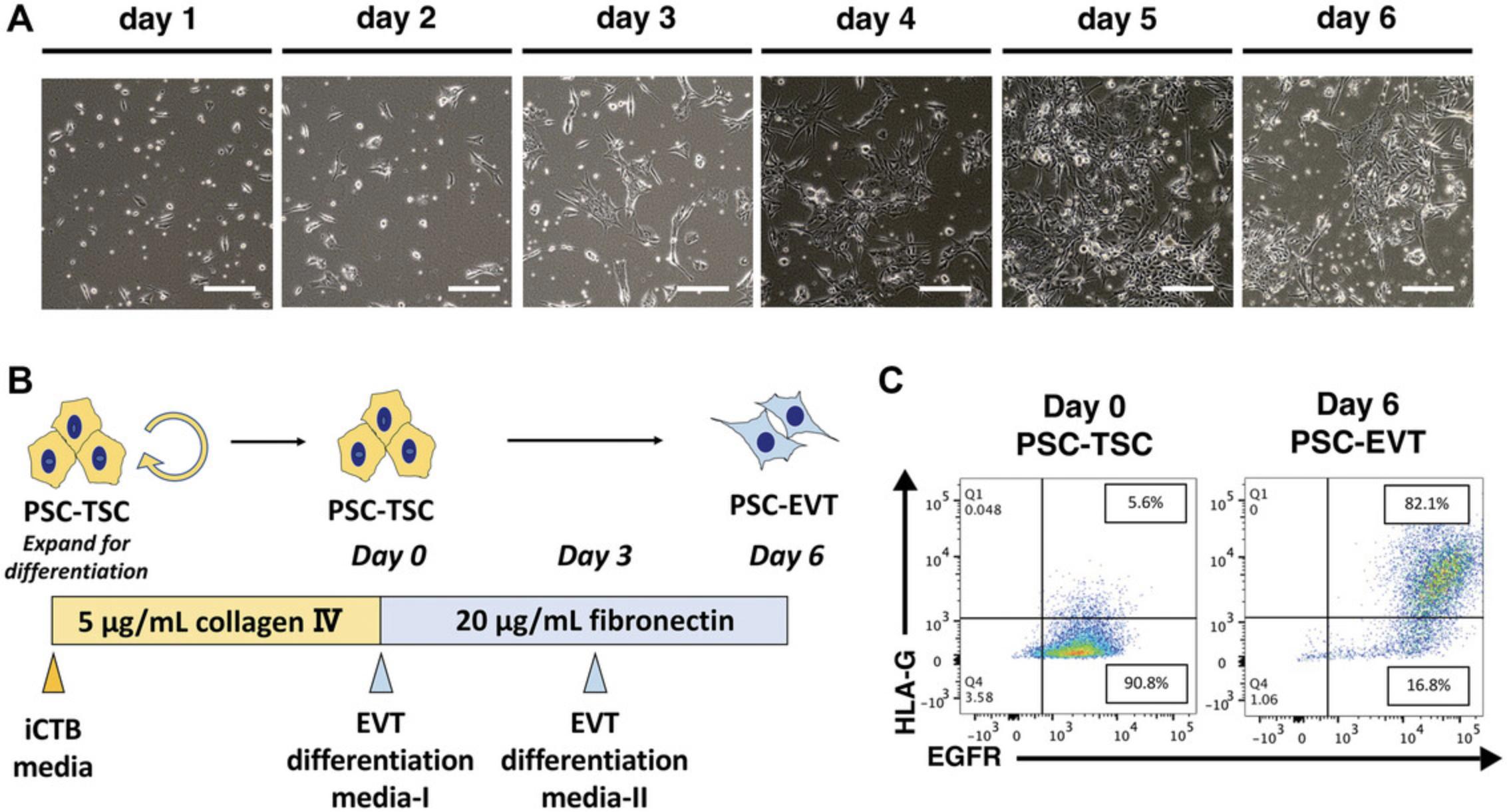
This protocol describes differentiation of PSCs into TE-like cells. Once the PSC are ready, it requires 5 days to make TE-like cells. This section is partially abstracted from Horii et al. (2019), which originally referred to the resulting cells as “CTB-like,” and expanded upon to provide a clear description of this protocol (see Background Information section below for details on the new designation of these as “TE-like” cells). Successful completion of this protocol will result in TE-like cells with a “cobblestone” morphology and over 80% EGFR positivity by flow cytometry.
Materials
-
PSC line(s)
- NOTE : We have used both hESC (including WA01/H1, WA09/H9) and several of our own iPSC lines (derived from human dermal fibroblast or human umbilical cord mesenchymal stem cells) in this protocol. Prior to differentiation, cells should be adapted to StemFlex media (ThermoFisher) or mTeSR Plus (STEMCELL Technologies), following the manufacturer's protocol.
-
StemFlex medium (ThermoFisher, cat. no. A3349401)
-
EDTA, 0.5 mM (see recipe)
-
TrypLE express (ThermoFisher, cat. no. 12604021)
-
Y27632 (see recipe)
-
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
-
Knockout serum replacement (KSR) (ThermoFisher, cat. no. 10828-028)
-
Trophoblast induction medium (see recipe and Table 8)
-
Geltrex coated 6-well tissue culture plates (see Support Protocol)
-
5-ml Serological pipettes
-
10-ml Serological pipettes
-
25-ml Serological pipettes
-
15-ml Conical tubes
-
50-ml Conical tubes
-
Hemocytometer or automated cell counter
-
Centrifuge (ThermoFisher, cat. no. 75004261 or equivalent)
-
Cell culture incubator
-
Flow cytometer and additional materials for cell staining and analysis as described in Horii et al, 2019
1.Use newly prepared Geltrex coated tissue culture plates (see Support Protocol 1). Culture PSCs in StemFlex until sub-confluent (∼80%). Cells are recommended to be passaged by clump passaging using a gentle dissociation buffer such as 0.5 mM EDTA every 2 to 3 days to expand the cells. Passaging details can also be found in Horii et al., 2019.
NOTE : It is essential to have good starting material. Ideal passage number for the use of ESC and iPSC are passage numbers below 50 for ESC, and passage number below 20 for iPSC. If ESC or iPSC are not growing well, consider thawing a new vial, or changing to new batches of StemFlex or Geltrex. For PSC medium, other regularly used medium (i.e., StemPro, E8, or mTeSR Plus) will also work for this protocol. We have found passaging at split ratios between 1:6 and 1:9 for H9 ESC, and 1:3 for our iPSC works well; however, splitting ratio varies between cell lines, so we recommend optimizing the split ratio beforehand.
2.On day -1, seed cells into a new Geltrex-coated 6-well plate (see Support Protocol 1). Pre-warm new Geltrex-coated plate (either a freshly-made or stored plate) at 37°C for at least 30 min. Aspirate the medium from sub-confluent well of PSCs and add 1 ml of room temperature TrypLE express for 3 min at 37°C to dissociate to single cells. Harvest cells, placing them in a conical tube. Wash wells with 2 to 5 ml of warm StemFlex media to collect as many cells as possible. Centrifuge 5 min at 200 × g , room temperature to pellet the PSC. Re-suspend and count the PSC.
3.Remove Geltrex from the well and seed 60,000 cells per well in StemFlex with 5 μM Y27632.Shake the plate gently by hand to spread the cells, allowing them to attach evenly on the plate (Fig. 2A).
NOTE : 60,000 cells per well of a 6-well plate is optimal for WA09/H9 ESC. However, seeding density will need to be optimized for each ESC/iPSC line. For our iPSC, we plate 100,000 cells per well of a 6-well plate. At these seeding densities, cells could grow up to roughly 1 million per well of a 6-well plate by day 4 of TE induction. It is very important to plate the cells sparsely at the beginning of the TE induction. If the PSC colonies are too big at the start, the center of the colony will not differentiate well. This is one reason to dissociate the cells using TrypLE express instead of EDTA. Daily cell culture work for TE induction should be done during a similar time in the day to ensure collection of samples and media changes are done every 24 hr. For subsequent conversion to TSC (Basic Protocol 2), you will need around 500,000 cells/well of a 6-well plate.
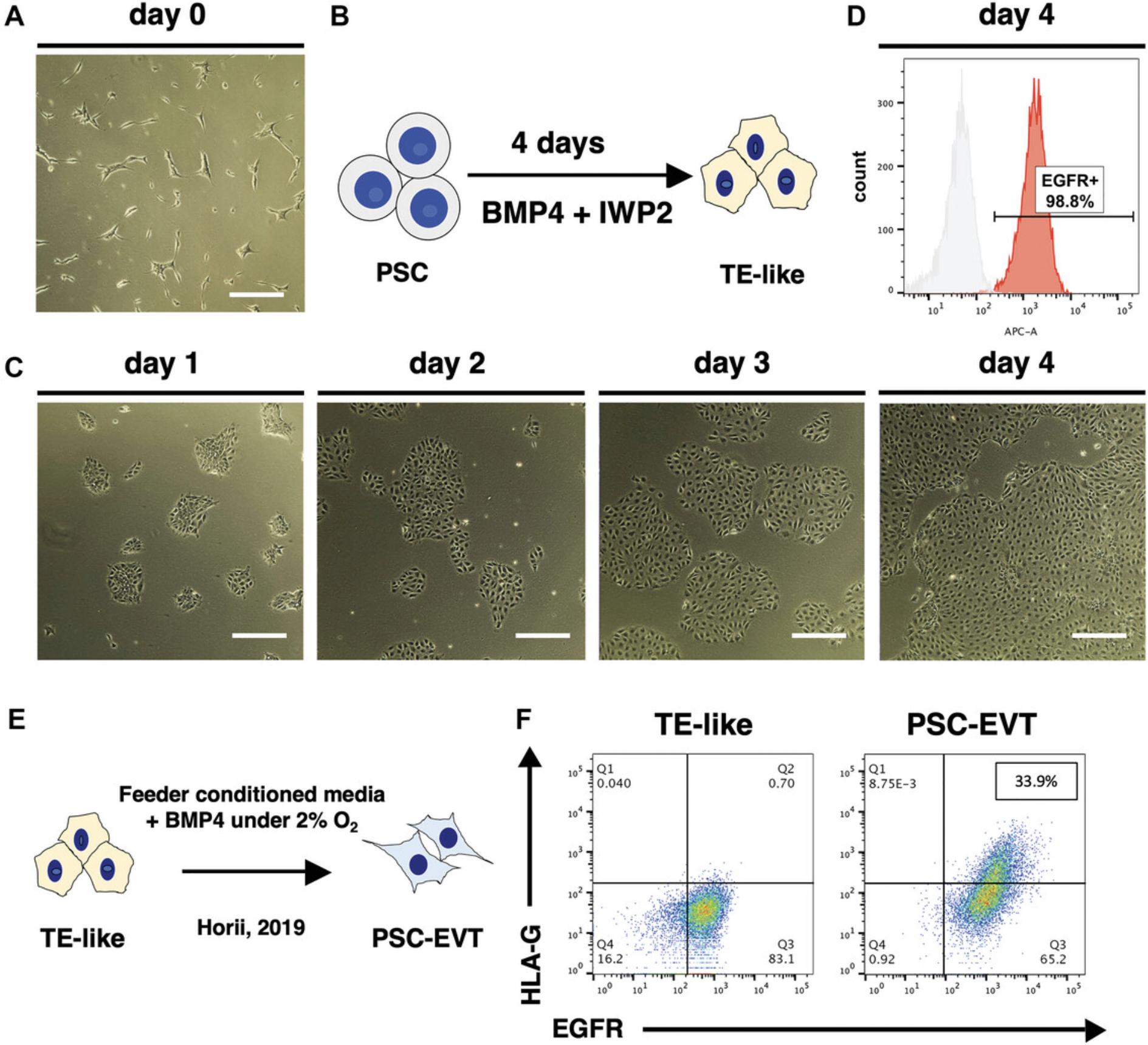
4.On day 0, check that the cells have attached evenly and formed small and sparse colonies (Fig. 2A). If the PSC morphology and density look good, change medium to Trophoblast Induction medium (Fig. 2B). Change the medium every day for 4 days. Cells will gradually acquire an epithelial (cobblestone-like) morphology (Fig. 2C).
5.On day 4, check cell surface EGFR expression using the flow cytometer. Using one well from the differentiation, remove the media and wash the cells with 1 ml of PBS. Aspirate PBS and add 1 ml of TrypLE express. Incubate at 37°C for 3 min. Add 1 ml of 10% Knockout Serum Replacement (KSR) in PBS onto the wells containing the TrypLE express, then wash the well and collect the cells in the TrypLE Express/10% KSR in PBS mixture in a 15 ml conical tube. Centrifuge 5 min at 200 × g , room temperature. Stain cells for EGFR expression following previously published protocol described in Horii et. al., 2019.
NOTE : Without the KSR, the cells will not form a dense pellet and there is a higher risk to lose the cells during aspiration of the supernatant.
6.If flow cytometric analysis shows over 80% of cells are EGFR+, proceed to the re-plating step in Basic Protocol 2. If the proportion of EGFR+ cells is less than 80%, change the medium on the remaining cells with Trophoblast Induction Medium and let the induction proceed for another day prior to measuring surface EGFR expression again by flow cytometry.
7.At the end of this step, successful induction should result in over 80% of the cells positive for surface EGFR expression by flow cytometry (Fig. 2D). Terminal differentiation of the TE-like cells generated by this protocol, previously referred to as CTB-like cells, using feeder-conditioned medium plus 10 ng/ml BMP4, as previously described by Horii et al., 2019, resulted in differentiation into mostly STB, with a small proportion (around 30%) of HLA-G+ EVT (Fig. 2E, F). EVT differentiation is significantly improved by adaptation to PSC-TSC (Basic Protocol 2) and subsequent use of Basic Protocol 5.
Basic Protocol 2: CONVERSION OF PSC-DERIVED TE-LIKE CELLS TO TSC
This protocol describes conversion of PSC-derived TE-like cells to TSC (PSC-TSC). Once the PSC-derived TE-like cells are confirmed to be over 80% EGFR positive by flow cytometry, proceed to Basic Protocol 2 to re-plate the TE-like cells onto Collagen IV-coated plate. Collagen IV-coated plates could be prepared on day 3 of the Basic Protocol 1.This step is essential to derive self-renewing PSC-TSC, and for subsequent successful terminal trophoblast differentiation.
Materials
-
TrypLE express (ThermoFisher, cat. no. 12604021)
-
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
-
iCTB complete medium (see recipe and Table 5)
-
Collagen IV-coated plate (see Support Protocol 2)
-
5-ml Serological pipettes
-
10-ml Serological pipettes
-
25-ml Serological pipettes
-
15-ml Conical tubes
-
50-ml Conical tubes
-
Hemocytometer or automated cell counter
-
Centrifuge (ThermoFisher, cat. no. 75004261 or equivalent)
-
Cell culture incubator
1.Before you begin, prepare the collagen IV-coated plates (see Support Protocol 2).
2.Aspirate media from the well containing PSC-derived TE from Basic Protocol 1 and add 1 ml TrypLE express to dissociate the cells to single cells. Incubate the plate at 37°C for 3 min.
3.Without removing TrypLE express, add 2 to 3 ml PBS to the well using a 5 ml pipette and collect the cells into a conical tube.
4.Centrifuge 5 min at 200 × g , room temperature. Aspirate the supernatant and re-suspend in iCTB complete medium to make a cell suspension of 250,000 cells per ml.
NOTE : If the cells do not form a dense pellet, add KSR for a final concentration of 10% to the cell suspension and centrifuge again.
5.Remove collagen IV from the well and wash once with PBS. Seed 500,000 cells per well in a 6-well plate, for a total volume of 2 ml per well. Agitate the plate to have the cells spread and attach evenly on the plate.
NOTE : Up to 1 million TE-like cells are generated in one well of a 6-well plate at the end of Basic Protocol 1. If there are 4 wells of a 6-well plate at the end of Basic Protocol 1, we recommend coating 4 to 6 wells of a 6-well plate with collagen IV.
6.Change iCTB medium every other day until the cells are confluent.
Basic Protocol 3: PASSAGING PSC-DERIVED TSC IN iCTB COMPLETE MEDIUM
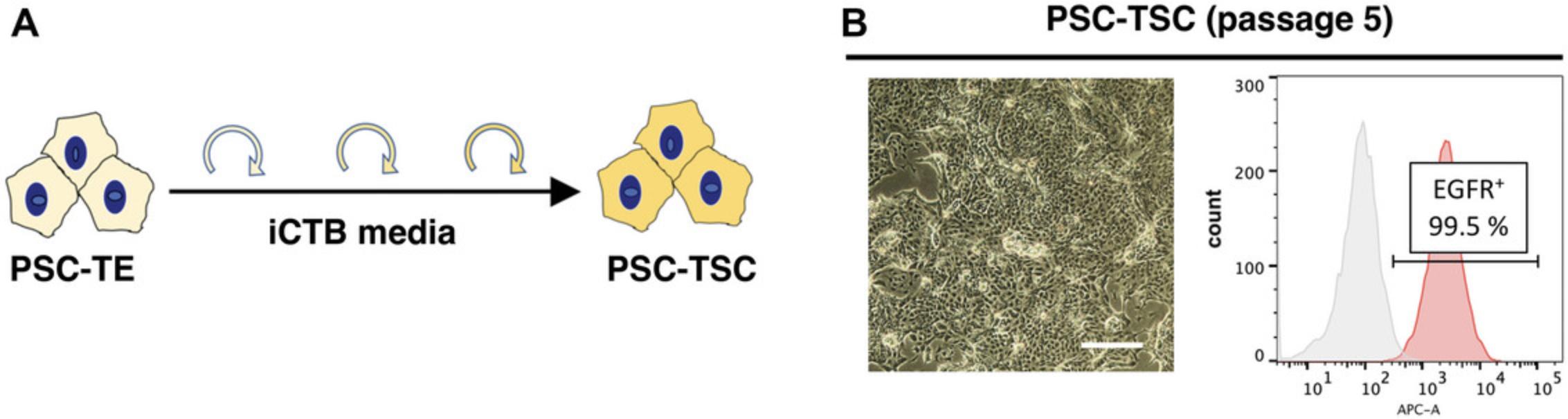
This protocol describes how to passage PSC-derived TSC. This step is essential to establish self-renewing PSC-TSC, and terminal trophoblast differentiation. Once the PSC-derived TE-like cells are replated onto a collagen IV-coated plate, EGFR expression level by flow cytometry will diminish during the first couple of passages. If completed successfully, EGFR expression will be regained and be expressed by over 90% of the cells by passage 5.This protocol typically takes up to 2 to 3 weeks to complete and will establish self-renewing PSC-TSC.
Materials
-
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
-
TrypLE express (ThermoFisher, cat. no. 12604021)
-
iCTB complete medium (see recipe and Table 5)
-
Collagen IV-coated plate (see Support Protocol 2)
-
5-ml Serological pipettes
-
10-ml Serological pipettes
-
25-ml Serological pipettes
-
15-ml Conical tubes
-
50-ml Conical tubes
-
Hemocytometer or automated cell counter
-
Centrifuge (ThermoFisher, cat. no. 75004261 or equivalent)
-
Cell culture incubator
1.Coat the appropriate number of wells with collagen IV before passaging (see Support Protocol 2). We recommend preparing enough wells to passage at a 1:3 split ratio. Therefore, for 1 well of PSC derived TSC, prepare 3 wells with collagen IV).
2.When the cells are 80% confluent, passage the cells at a 1:3 split ratio.
NOTE : Split ratio varies between the cell lines, so optimizing the split ratio beforehand is recommended. We typically use a 1:2 to 1:4 split ratio among different ESC- and iPSC-derived TSC lines.
3.Aspirate iCTB medium and add 1 ml of TrypLE express per well of a 6-well plate. Incubate at 37°C for 10 to 15 min.
4.Collect cells in 15 ml conical tube using PBS or iCTB basal medium.
5.Centrifuge 5 min at 200 × g , room temperature.
6.Aspirate the supernatant and then re-suspend cells in appropriate volume of iCTB complete medium for a split ratio of 1:3.
7.Aspirate collagen IV from the wells that are intended for plating and wash wells once with PBS.
8.Plate cells with appropriate split ratio in 2 ml medium for each well. Agitate the plate to allow the cells to spread out and attach evenly.
9.Passage cells every 2 to 3 days, or when 80% confluent.
10.Following 4 to 5 passages in this media (Fig. 3A), the cells should be over 90% EGFR+ when examined by flow cytometry (Fig. 3B). At this point, cells can be frozen down. After thawing, PSC-TSC can be passaged and differentiated in a lineage-specific manner into either STB or EVT.
NOTE : It takes about 1 week to prepare the cells for the initial TE induction (Basic Protocol 1) and an additional 2 to 3 weeks to complete the TSC derivation (Basic Protocols 2 and 3). During the adaptation phase into iCTB media, a drop in EGFR cell surface expression has been observed and is expected (as low as 10% EGFR+ cells). Over a few passages, cells will re-acquire EGFR expression as their morphology becomes more similar to primary TSC (Soncin et al., 2022). Conversion time to PSC-TSC varies between cell lines and is usually completed by passage 8 for most cell lines. After establishing PSC-derived TSC, cells can be passaged multiple times (we have successfully maintained them for up to 20 passages). Similar to primary TSC, it is important to passage PSC-derived TSC at sub-confluency (<80% confluency), rather than over-confluent, to avoid differentiation and acquisition of cell surface HLA-G. However, once a high proportion of cells acquire over 20% HLA-G, it is difficult to recover cells to the TSC stage (EGFR+, HLA-G-). In this case, thawing earlier passage cells or re-derivation of PSC-TSC is recommended.
Basic Protocol 4: STB DIFFERENTIATION FROM PSC-DERIVED TSC
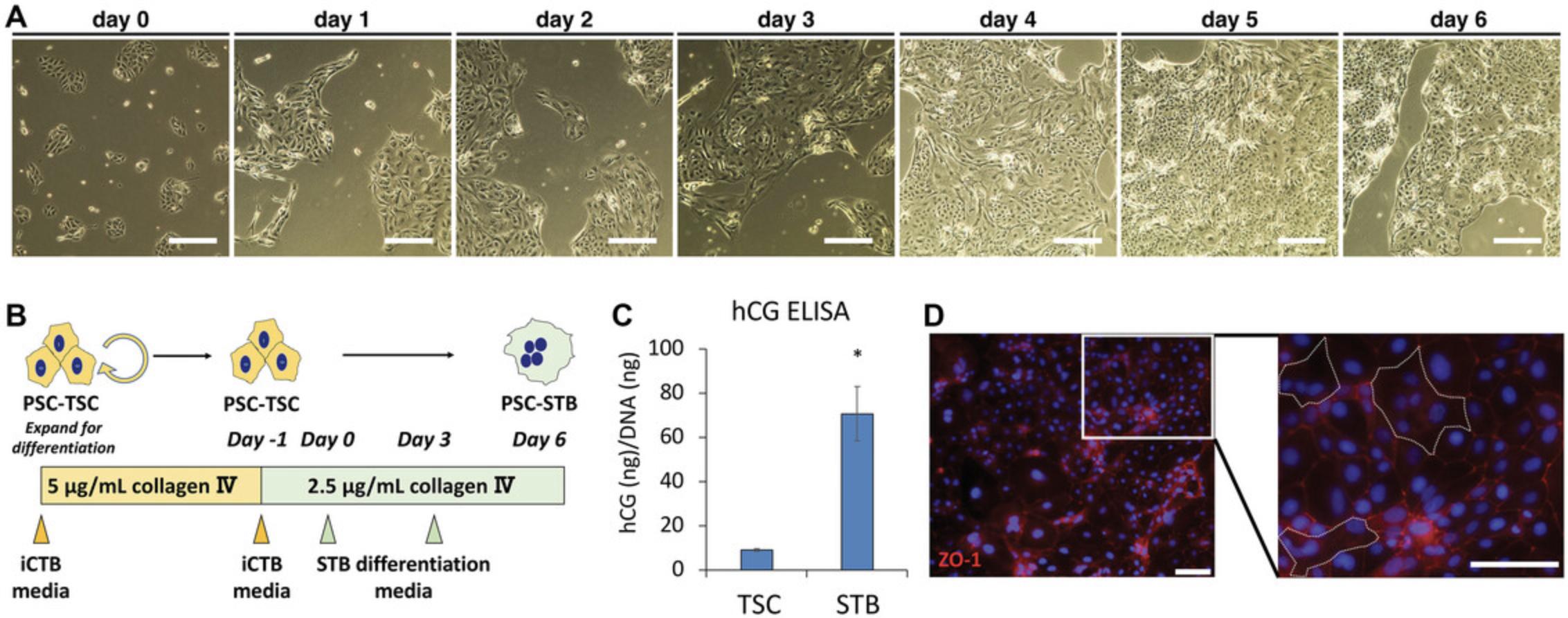
This protocol describes STB differentiation from PSC-derived TSC. Correct confluency (∼30%) of PSC-TSC on day 0 is essential for successful STB differentiation. If the cells are under- or over-confluent, they will detach from the plate before differentiation is complete. It will take around 8 days to complete the protocol (days -1 to day 6). If successful, resulting cells at day 6 of the protocol will exhibit STB-associated function, such as hCG secretion. Cellular assays such as ELISA for hCG and cell-cell fusion assays can be conducted on day 6.An example of positive results is shown in Figure 4C and D. The usage of these assays is described in detail in previous publications (Horii et al., 2016; Horii et al., 2021; Soncin et al., 2022).
Materials
- TrypLE express (ThermoFisher, cat. no. 12604021)
- PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
- iCTB complete medium (see recipe and Table 5)
- STB differentiation medium (see recipe and Table 6)
- Collagen IV-coated plate (see Support Protocol 2)
- 5-ml Serological pipettes
- 10-ml Serological pipettes
- 25-ml Serological pipettes
- 15-ml Conical tubes
- 50-ml Conical tubes
- Hemocytometer or automated cell counter
- Centrifuge (ThermoFisher, cat. no. 75004261 or equivalent)
- Cell culture incubator
1.Prepare plates coated with 2.5 μg/ml collagen IV in PBS (see Support Protocol 2).
2.Day -1: prior to differentiation, check the cells by flow cytometry to ensure the PSC-TSC population is over 80% EGFR+ and less than 20% HLA-G+.
3.Day -1: for STB differentiation, plate PSC-TSC at a density of 100,000 to 150,000 cells per well of a 6-well plate. Using PSC-TSC from Basic Protocol 3, roughly 300,000 cells are expected from one well of a 6-well plate.
4.Aspirate iCTB medium from the well that is going to be passaged, then add 1 ml of TrypLE express. Incubate at 37°C for 10 to 15 min.
5.Collect cells in 15 ml conical tube using PBS or iCTB basal medium. Count the cells.
6.Centrifuge 5 min at 200 × g , room temperature.
7.Aspirate the supernatant, then re-suspend cells in appropriate volume of iCTB complete medium.
8.Aspirate collagen IV solution from wells and wash with 1 ml of PBS.
9.Remove PBS from the well and plate 100,000 to 150,000 cells in 2 ml of iCTB complete medium into one well of a 6-well plate. Incubate cells for 1 to 2 days, until the cells are 30% confluent (see Fig. 4A).
10.Day 0: when the cells are 30% confluent (Fig. 4A), change medium to STB differentiation medium with 2 ml per well of a 6-well plate (Fig. 4B).
NOTE : Due to differences in growth rate, it may be required to optimize the seeding density of PSC-TSC to achieve 30% confluence at day 0. Alternatively, if the cells are less than 30%, wait until the PSC-TSC reach 30% of confluence before switching to STB differentiation media. This will avoid significant cell death throughout the differentiation and will perform better cell-cell fusion.
11.On day 3, change medium using freshly prepared STB differentiation medium (Fig. 4B).
12.On day 6, collect cells for analysis of STB function (Fig. 4B-D).
Basic Protocol 5: EVT DIFFERENTIATION FROM PSC-DERIVED TSC
This protocol describes EVT differentiation from PSC-TSC, which takes 8 days to complete (days -1 to day 6). It is important that the PSC-TSC are undifferentiated (over 80% EGFR+ and less than 20% HLA-G+) at the start of the protocol. If completed successfully, cells should be over 60% positive for EVT marker, HLA-G, at the end of day 6.This assay is also described in our previous publications (Horii et al., 2016; Horii et al., 2021; Soncin et al., 2022).
Materials
-
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
-
TrypLE express (ThermoFisher, cat. no. 12604021)
-
iCTB complete medium (see recipe and Table 5)
-
EVT differentiation medium-I (see recipe and Table 2)
-
EVT differentiation medium-II (see recipe and Table 3)
-
Fibronectin-coated plate (see Support Protocol 3)
-
5-ml Serological pipettes
-
10-ml Serological pipettes
-
25-ml Serological pipettes
-
15-ml Conical tubes
-
50-ml Conical tubes
-
Hemocytometer or automated cell counter
-
Centrifuge (ThermoFisher, cat. no. 75004261 or equivalent)
-
Cell culture incubator
1.Prepare plates coated with 20 μg/ml fibronectin in PBS (see Support Protocol 3).
2.Day -1 or 0: prior to differentiation, check the cells by flow cytometry to ensure the PSC-TSC population is over 80% EGFR+ and less than 20% HLA-G+.
3.Day 0: for EVT differentiation, plate cells at a density of 200,000 to 250,000 cells per well of 6-well plate. We expect to obtain about 300,000 PSC-TSC per confluent well of a 6-well plate at the end of Basic Protocol 3.
4.Thaw Matrigel on ice. Once Matrigel is thawed, prepare EVT differentiation medium-Ⅰ (see recipe) and allow medium to reach room temperature before use.
5.Aspirate iCTB medium, then add 1 ml of TrypLE express. Incubate at 37°C for 10 to 15 min.
6.Collect cells in 15 ml conical tube using PBS or iCTB basal medium. Count the cells.
7.Centrifuge 5 min at 200 × g , room temperature.
8.Aspirate supernatant, then re-suspend cells in EVT differentiation medium-Ⅰ.
9.Aspirate fibronectin solution from wells and wash with 1 ml of PBS per well.
10.Aspirate PBS and plate 200,000 to 250,000 cells in 2 ml of EVT differentiation medium-Ⅰ for each well of a 6-well plate.
11.On day 3, prepare EVT differentiation medium-II, and change medium (Fig. 5A, B).
12.On day 6, collect cells for further analysis (Fig. 5C).
Support Protocol 1: GELTREX-COATED TISSUE CULTURE PLATE PREPARATION
This protocol describes how to prepare Geltrex coated plates in a step-by-step manner for expansion of PSC and differentiation into TE like cells.
Materials
-
Geltrex (Thermo Fisher, cat. no. A14133-02)
-
DMEM/F-12, HEPES (Thermo Fisher, cat. no. 11330-032)
-
Tissue culture treated 6-well plates (Corning, cat. no. 3506)
-
5-ml Serological pipettes
-
10-ml Serological pipettes
-
25-ml Serological pipettes
-
15-ml Conical tubes
-
50-ml Conical tubes
-
Parafilm
-
Cell culture incubator
1.Thaw Geltrex on ice. Wait for 30 to 45 min until completely thawed (no ice remains). Prepare 20 ml of ice-cold DMEM/F-12, HEPES in a 50-ml conical tube and add 100 μl Geltrex to make a 1:200 dilution as recommended by the manufacturer. Mix gently.
2.Add medium/Geltrex mixture to the plate (2 ml per well for 6-well plate, and 1 ml per well for 12-well plate). Incubate the plate at 37°C for 30 min for immediate use. For preparation for later use, incubate for 2 hr in either room temperature or 37°C incubator. After the incubation, cover with Parafilm and store the plate at 4°C for up to 4 weeks. Do not use if the well is dry. When using the plate, simply aspirate the medium/Geltrex mixture and add stem cell culture media (i.e., StemFlex) containing cells.
Support Protocol 2: COLLAGEN IV-COATED TISSUE CULTURE PLATE PREPARATION
This protocol describes how to prepare Collagen IV-coated plates in a step-by-step manner for derivation and passaging of PSC-derived TSC as well as PSC-TSC differentiation into STB.
Materials
-
Collagen IV (MilliporeSigma, cat. no. C0543-1VL)
-
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
-
Tissue culture treated 6-well plates (Corning, cat. no. 3506)
-
5-ml Serological pipettes
-
10-ml Serological pipettes
-
25-ml Serological pipettes
-
15-ml Conical tubes
-
50-ml Conical tubes
-
Cell culture incubator
1.If collagen IV is frozen, thaw aliquot on ice prior to use. Once thawed, collagen IV can be stored in 4°C for up to 3 months, or according to the manufacturer's recommendation.
In a sterile environment, add 5 μl of 1 mg/ml collagen IV into 1 ml of PBS to make a 5 μg/ml solution for TSC derivation and maintenance (Basic Protocols 2 and 3). Mix gently.
2.Add diluted collagen IV mixture (5 μg/ml) to the plate (1 ml per well of a 6-well plate, 500 μl per well of a 12-well plate). Incubate the plate at 37°C for at least 2 to 3 hr at 37°C (optimal attachment occurs with overnight incubation at 37°C). The plate could be kept at 37°C up to a week. Do not use if the well is dry.
NOTE : Instead of 5 μl of 1 mg/ml collagen IV into 1 ml of PBS, add 2.5 μl of 1 mg/ml collagen IV into 1 ml of PBS to make a 2.5 μg/ml solution for STB differentiation (Basic Protocol 4).
Support Protocol 3: FIBRONECTIN-COATED TISSUE CULTURE PLATE PREPARATION
This protocol describes how to prepare fibronectin coated plates in a step-by-step manner for EVT differentiation of PSC-TSC.
Materials
-
Fibronectin (MilliporeSigma, cat. no. F1141)
-
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
-
Tissue culture created 6-well plates (Corning, cat. no. 3506)
-
5-ml Serological pipettes
-
10-ml Serological pipettes
-
25-ml Serological pipettes
-
15-ml Conical tubes
-
50-ml Conical tubes
-
Cell culture incubator
1.In a sterile environment, add 20 μl of fibronectin into 1 ml of PBS to make a 20 μg/ml solution. Mix gently.
2.Add fibronectin mixture to the plate (1 ml per well for 6-well plate,500 μl per well for 12-well plate). Incubate the plate at 37°C for 1 hr for immediate use. For preparation for use the next day, incubate the plate overnight at 37°C. Do not use if the well is dry. Rinse well with 1 ml of PBS prior use.
REAGENTS AND SOLUTIONS
Prepare all reagents and solutions in a sterile environment suitable for cell culture. Sterile filter as indicated.
A 83-01, 5 mM
A 83-01, 10 mg (Tocris, cat. no. TB2939-RMU
DMSO (MilliporeSigma, cat. no. D2650)
Add 4.74 ml DMSO to 10 mg A 83-01 to make 5 mM (w/v) stock. Make 80 μl aliquots. Store at −80°C for up to a month.
Basal trophoblast induction medium
DMEM/F-12 50/50 (Corning, cat. no. 10-092-CV)
ITS (see recipe)
L-ascorbic acid (MilliporeSigma, cat. no. A8960)
Sodium bicarbonate (NaHCO3), 7.5% solution (ThermoFisher, cat. no. 25080094)
BSA, 30% (w/v) solution (Gemini Bio-Products, cat. no. 700-110)
Prepare medium, referring to Table 1 for volumes and final concentrations. Sterile filter and sore at 4°C. Use within a week.
| Component | Stock concentration | Final concentration | Volume for 50 ml |
|---|---|---|---|
| DMEM/F-12 50/50 | – | – | 45.7 ml |
| ITS | 100× | 1× | 500 μl |
| L-ascorbic acid | 33.3 mg/ml | 64 μg/ml | 96 μl |
| NaHCO3 | 75 mg/ml (7.5%) | 543 μg/ml | 362 μl |
| BSA, 30% (w/v) solution | 30% | 2% | 3.3 ml |
| Component | Stock concentration | Final concentration | Volume for 12 ml |
|---|---|---|---|
| Terminal differentiation medium | – | – | 11.2 ml |
| Y-27632 | 10 mM | 2.5 μM | 3 μl |
| KSR | 100% | 4% | 480 μl |
| A 83-01 | 5 mM | 7.5 μM | 18 μl |
| NRG1 | 100 μg/ml | 100 ng/ml | 12 μl |
| Matrigel | 100% | 2% | 240 μl |
Bone morphogenetic protein 4 (BMP4), 10 μg/ml
BSA, 30% (Gemini Bio-Products, cat. no. 700-110)
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
BMP4, 50 ug (R&D Systems, cat. no. 314-BP)
Prepare 0.1% (w/v) BSA in PBS. To make a 10 μg/ml BMP4 solution, combine 5 ml of 0.1% (w/v) BSA in PBS solution to 50 μg of BMP4. Gently pipette up and down to reconstitute the protein. Thoroughly mix and prepare 100 μl aliquots. Store at −80°C for up to a year. Once thawed, keep in 4°C. Avoid freeze-thaw cycles.
CHIR99021, 3 mM
CHIR99021 (MilliporeSigma, cat. no. SML1046)
DMSO (MilliporeSigma, cat. no. D2650)
Add 3.58 ml DMSO to 5 mg CHIR99021 to prepare a 3 mM stock. Make 533 μl aliquots. Store at −80°C for up to a year.
Collagen IV, 1 mg/ml
HCl, 1 M (MilliporeSigma cat. no. H9892)
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
Collagen IV (MilliporeSigma, cat. no. C0543)
Dilute 1 M HCl with PBS to 0.05 M. Add 750 μl of 0.05 M HCl to 750 μg of collagen IV. Make 100 μl aliquots of 1 mg/ml collagen IV. Store at −80°C for up to a year. Once thawed, store at 4°C. Avoid freeze-thaw cycles.
EDTA, 0.5 mM
EDTA, 0.5 M (ThermoFisher, cat. no. 15575020)
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
Add 50 μl of 0.5 M EDTA to 50 ml PBS. Sterile filter and store at room temperature.
EVT differentiation medium-I
Terminal differentiation medium (see recipe)
Y-27632, 10 mM (Selleck Chemicals, cat. no. S1049)
Knockout serum replacement (KSR) (ThermoFisher, cat. no. 10828028)
A 83-01, 5 mM (Tocris, cat. no. 2939)
NRG1 100 μg/ml (Abcam, cat. no. ab50227)
Matrigel (Corning, cat. no. 354234)
Prepare EVT differentiation medium-I fresh prior to use. Matrigel will solidify and clump if added to warm media, so add ice-cold liquid Matrigel to ice-cold medium. The complete medium should be warmed up before feeding the cells. See Table 2 for volumes and concentrations.
EVT differentiation medium-II
Terminal differentiation medium (see recipe)
Y-27632, 10 mM (Selleck Chemicals, cat. no. S1049)
Knockout serum replacement (KSR) (ThermoFisher, cat. no. 10828028)
A 83-01, 5 mM (Tocris, cat. no. 2939)
Matrigel (Corning, cat. no. 354234)
Prepare EVT differentiation medium-II fresh prior to use. Matrigel will solidify and clump if added to warm media, so add ice-cold liquid Matrigel to ice-cold medium. The complete medium should be warmed up before feeding the cells. See Table 3 for volumes and concentrations.
| Component | Stock concentration | Final concentration | Volume for 12 ml |
|---|---|---|---|
| Terminal differentiation medium | – | – | 11.4 ml |
| Y-27632 | 10 mM | 2.5 μM | 3 μl |
| KSR | 100% | 4% | 480 μl |
| A 83-01 | 5 mM | 7.5 μM | 18 μl |
| Matrigel | 100% | 0.5% | 60 μl |
Forskolin, 10 mM
Forskolin, 10 mM (Selleck Chemicals, cat. no. S2449)
Aliquot the 10 mM forskolin in DMSO into 50 μl aliquots. If forskolin is in powder form, add appropriate amount of DMSO to dissolve the powder to a final concentration of 10 mM. Make 50 μl aliquots. Keep aliquots in −80°C for 2 years in liquid form. Keep powder stocks in −20°C for up to 3 years.
Geltrex, rowth factor-reduced
Geltrex, growth factor-reduced (ThermoFisher, cat. no. A1413302)
Thaw growth factor-reduced Geltrex on ice or overnight at 4°C. Using a cooled tip, make 100 μl aliquots on ice. We recommend putting the whole tip box in −20°C freezer for 5 min before use. Keep aliquots at −80°C for up to a year.
iCTB basal medium
Advanced DMEM/F-12 (ThermoFisher, cat. no. 12634028)
N-2 supplement, 100× (ThermoFisher, cat. no. 17502001)
B-27 supplement, 50× (ThermoFisher, cat. no. 17504001)
GlutaMax supplement, 100× (ThermoFisher, cat. no. 35050061)
1-Thioglycerol, 11.5 M (MilliporeSigma, cat. no. M6145)
BSA (30% (w/v) solution (Gemini Bio, cat. no. 700-110-100)
Knockout serum replacement (KSR) (ThermoFisher, cat. no. 10828028)
Combine ingredients according to volumes and concentrations in Table 4. Sterile filter. Use within 2 weeks.
| Component | Stock concentration | Final concentration | Volume for 250 ml |
|---|---|---|---|
| Advanced DMEM/F-12 | – | – | 237 ml |
| N-2 supplement | 100× | 1× | 2.5 ml |
| B-27 supplement | 50× | 1× | 5 ml |
| GlutaMax supplement | 100× | 1× | 2.5 ml |
| 1-Thioglycerol | 11.5 M | 150 μM | 3.3 μl |
| BSA | 30% | 0.05% | 417 μl |
| KSR | 100% | 1% | 2.5 ml |
iCTB complete medium
iCTB basal media (see recipe and Table 4)
TSC inhibitor cocktail, 1000× (see recipe and Table 9)
Y-27632, 10 mM (Selleck Chemicals, cat. no. S1049)
Valproic acid sodium salt, 300 mM, 50 mg/ml (MilliporeSigma, cat. no. 676380)
rhFGF2, 100 μg/ml (BioPioneer, cat. no. HRP-0011)
rhEGF, 100 μg/ml (R&D Systems, cat. no. 236-EG)
rhNoggin, 20 μg/ml (R&D Systems, cat. no. 6057-NG)
rhHGF, 100 μg/ml (STEMCELL Technologies, cat. no. 78019.2)
Prepare iCTB complete medium fresh prior to feeding. See Table 5 for volumes and concentrations.
| Component | Stock concentration | Final concentration | Volume for 12 ml |
|---|---|---|---|
| iCTB basal media (Table 4) | – | – | 11.9 ml |
| TSC inhibitor cocktail (Table 9) | 1000× | 1× | 12 μl |
| Y-27632 (Selleck Chemicals, cat. no. S1049) | 10 mM | 5 μM | 6 μl |
| Valproic acid sodium salt (MilliporeSigma, cat. no. 676380-5GM) | 50 mg/ml (300 mM) | 130 μg/ml (0.8 mM) | 31.4 μl |
| rhFGF2 | 100 μg/ml | 100 ng/ml | 12 μl |
| rhEGF | 100 μg/ml | 50 ng/ml | 6 μl |
| rhNoggin | 20 μg/ml | 20 ng/ml | 12 μl |
| rhHGF | 100 μg/ml | 50 ng/ml | 6 μl |
| Component | Stock concentration | Final concentration | Volume for 12 ml |
|---|---|---|---|
| Terminal differentiation medium | – | – | 11.5 ml |
| Y-27632 | 10 mM | 2.5 μM | 3 μl |
| KSR | 100% | 4% | 480 μl |
| Forskolin | 10 mM | 2 μM | 2.4 μl |
Insulin-transferrin-sodium selenite (ITS)
Insulin-transferrin-sodium selenite (ITS), 100× (MilliporeSigma, cat. no. I1884)
Water, sterile, cell culture-grade
Add 50 ml of sterile cell culture-grade water to the ITS vial. Mix well and make 1 ml aliquots. Store at −20°C for up to 1 year. Once thawed, store at 4°C. Avoid freeze-thaw cycles.
IWP-2
IWP-2 (Selleck Chemicals, cat. no. S7085)
Prepare a 2 mM IWP-2 stock solution in DMSO and make 100 μl aliquots. Store at −80°C for up to 2 years. Once thawed, store at 4°C. Avoid freeze-thaw cycles.
L-ascorbic acid 2-phosphate
L-ascorbic acid 2-phosphate (MilliporeSigma, cat. no. A8960)
Advanced DMEM/F-12 (ThermoFisher, cat. no. 12634028)
Add 6 ml of DMEM/F-12 to 200 mg of L-ascorbic acid to make 33.3 mg/ml L-ascorbic acid reagent. Sterile filter the solution and prepare 1 ml aliquots. Store at −20°C for up to 6 months. Once thawed, store at 4°C. Avoid freeze-thaw cycles.
Recombinant human epidermal growth factor (rhEGF)
Recombinant human epidermal growth factor (rhEGF) (R&D Systems cat. no. 236-EG)
BSA (Gemini Bio-Products, cat. no. 700-110)
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
First, prepare 0.1% (w/v) BSA in PBS solution. To make 100 μg/ml rhEGF, combine 10 ml of 0.1% (w/v) BSA in PBS solution to 1 mg of rhEGF. Gently pipette up and down to reconstitute the protein. Prepare 100 μl aliquots. Store at −80°C for up to 6 months. Once thawed, store at 4°C. Avoid freeze-thaw cycles.
Recombinant human basic fibroblast growth factor 2 (bFGF-2)
Recombinant human basic fibroblast growth factor 2 (FGF-2) (BioPioneer, cat. no. HRP-0011)
BSA (Gemini Bio-Products, cat. no. 700-110)
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
First, prepare 0.1% (w/v) BSA in PBS solution. To make 100 μg/ml bFGF-2, combine 500 μl of 0.1% (w/v) BSA in PBS solution to 50 μg of bFGF-2. Gently pipette up and down to reconstitute the protein and prepare 100 μl aliquots. Store at −20°C for up to 6 months. Once thawed, keep at 4°C. Avoid freeze-thaw cycles.
Recombinant human hepatocyte growth factor (rhHGF)
Recombinant human hepatocyte growth factor (rhHGF) (STEMCELL Technologies, cat. no. 78019.2)
PBS without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
To prepare 100 μg/ml rhHGF, combine 10 ml of sterile PBS to 1 mg of rhHGF. Gently pipette up and down to reconstitute protein and prepare 100 μl aliquots. Store at −80°C. Once thawed, store at 4°C. Avoid freeze-thaw cycles.
Recombinant human neuregulin 1 (NRG1)
Recombinant human neuregulin 1 (NRG1) (Abcam, cat. no. ab50227)
Water, sterile, cell culture-grade
Reconstitute 50 μg of recombinant human NRG1 with 500 μl of cell culture-grade water to make a concentration of 100 μg/ml. Prepare 50 μl aliquots and store at −80°C for 3 years.
Recombinant human noggin (rhNoggin)
Recombinant human noggin (rhNoggin) (R&D Systems, cat. no. 6057-NG)
BSA (Gemini Bio-Products, cat. no. 700-110)
PBS without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
First, prepare a 0.1% (w/v) BSA in PBS solution. To make 20 μg/ml rhNoggin, combine 5 ml of 0.1% (w/v) BSA in PBS solution to 100 μg of rhNoggin. Gently pipette up and down to reconstitute the protein and prepare 100 μl aliquots. Store at −80°C. Once thawed, store at 4°C. Avoid freeze-thaw cycles.
SB431542
SB431542 (MilliporeSigma, cat. no. 616464)
DMSO (MilliporeSigma, cat. no. D2650)
Thaw SB431542 at room temperature. Once thawed, spin down the sample to ensure no residual stock has remained on the wall of the tube. Dilute the vial to make 10 mM SB431542 (i.e., if the vial is at 100 mM [5 mg in 119 μl DMSO], add 1.071 ml DMSO). Mix well. Make 80 μl aliquots. Store at −80°C for up to a year.
STB differentiation medium
Terminal differentiation medium (see recipe and Table 7)
| Component | Stock concentration | Final concentration | Volume for 50 ml |
|---|---|---|---|
| DMEM/F-12 no HEPES | – | – | 48.9 ml |
| BSA 30% (w/v) solution | 30% | 0.3% | 500 μl |
| ITS-X | 100× | 1× | 500 μl |
| 2-Mercaptoethanol | 55 mM | 100 μM | 90 μl |
Y-27632, 10 mM (Selleck Chemicals, cat. no. S1049)
Knockout serum replacement (KSR) (ThermoFisher, cat. no. 10828028)
Forskolin, 10 mM (see recipe)
Prepare STB differentiation media fresh prior to feeding. See Table 6 for volumes and concentrations.
Terminal differentiation medium
DMEM/F-12 no HEPES (ThermoFisher cat. no. 11320-033)
BSA 30% (w/v) solution (Gemini Bio-Products, cat. no. 700-110-100)
ITS-X, 100× (ThermoFisher, cat. no. 51500056)
2-Mercaptoethanol (ThermoFisher, cat. no. 21985-023)
Combine ingredients using volumes and concentrations as listed in Table 7. Sterile filter and store at 4°C. Use within 2 weeks.
Trophoblast induction medium
Basal trophoblast induction medium (see recipe and Table 1)
BMP-4, 10 μg/ml (R&D Systems, cat. no. 314-BP-050)
IWP-2, 2 mM (Selleck Chemicals, cat. no. S7085)
Prepare trophoblast induction medium with a final concentration of 10 ng/ml BMP-4 (1:1000 from the stock solution) and 2 μM IWP-2 (1:1000 from the stock solution). 1.5 ml of media is required for one well of a 6-well plate. Prepare fresh. Filtering is not recommended. See Table 8 for volumes and concentrations.
| Component | Amount for 1 ml | Stock concentration | Final concentration |
|---|---|---|---|
| Basal trophoblast induction medium | 1 ml | – | – |
| BMP-4 | 1 μl | 10 μg/ml | 10 ng/ml |
| IWP-2 | 1 μl | 2 mM | 2 μM |
TSC inhibitor cocktail
CHIR99021, 3 mM (see recipe)
A 83-01, 5 mM (see recipe)
SB431542, 10 mM (see recipe)
DMSO (MilliporeSigma, cat. no. D2650)
Thaw CHIR99021, A 83-01, and SB431542 aliquots that were prepared as listed above. Combine ingredients according to Table 9 using volumes and concentrations as indicated. This will make 800 μl of the inhibitor cocktail. Mix well. Make 100 μl aliquots. Store at −80°C. Once thawed, store at 4°C. (H. Okae, personal communications).
| Component | Stock concentration | Final concentration | Volume for 800 μl |
|---|---|---|---|
| CHIR99021 | 3 mM | 2 mM | 533 μl |
| A 83-01 | 5 mM | 0.5 mM | 80 μl |
| SB431542 | 10 mM | 1 mM | 80 μl |
| DMSO | – | – | 107 μl |
Valproic acid sodium salt
Valproic acid sodium salt (MilliporeSigma, cat. no. 676380)
PBS, without calcium and magnesium (Corning, cat. no. 21-040-CV, or equivalent)
Add 20 ml sterile PBS to 1 g of valproic acid sodium salt. This will make a 50 mg/ml sodium valproate in PBS solution. Sterile filter the solution and make 500 µl aliquots. Store at −80°C. Once thawed, store at 4°C. Avoid freeze-thaw cycles.
Y-27632, 10 mM
Y-27632 (Selleck Chem, cat. no. S1049)
Cell culture grade water (Corning, cat. no. 25-055-CV)
Prepare a 10 mM stock solution and store in 100 μl aliquots. Store at −80°C for up to 2 years. Once thawed, store at 4°C. Avoid freeze-thaw cycles.
COMMENTARY
Background Information
Our initial work started with the establishment of a stepwise differentiation protocol of PSCs using BMP4 only, first into TE-like cells, which were previously described as CTB-like cells (Horii et al., 2016; Li et al., 2013). Recent single cell transcriptomic-based characterization found that these cells have a TE-like transcriptome signature (Soncin et al., 2022). We have therefore updated the terminology in this manuscript to reflect the TE-like nature of these cells. We previously terminally differentiated these TE-like cells directly into STB- and EVT-like cells (Fig. 6A), using PSC derived from both normal and diseased pregnancy (Horii et al., 2016). Specifically, we showed that, similar to primary CTB, hypoxia enhances differentiation of PSC-derived TE-like cells toward the EVT lineage. We also showed that T21-affected PSC recapitulate defects in STB differentiation, as seen in T21-affected placentas and primary CTB (Gerbaud et al., 2011), showing that PSC can be used to model trophoblast differentiation in normal and diseased settings. This protocol was further improved by the addition of IWP2 in the first step of the trophoblast differentiation protocol, which increased the purity of the TE-like population to over 80% (Fig. 6B). TE-like cells generated using this protocol could subsequently be differentiated by treatment with feeder-conditioned media (FCM) + BMP4. If differentiation was performed under 21% oxygen, most of the cells would become STB, while differentiation under 2% oxygen could increase the proportion of EVT (Horii et al., 2019).
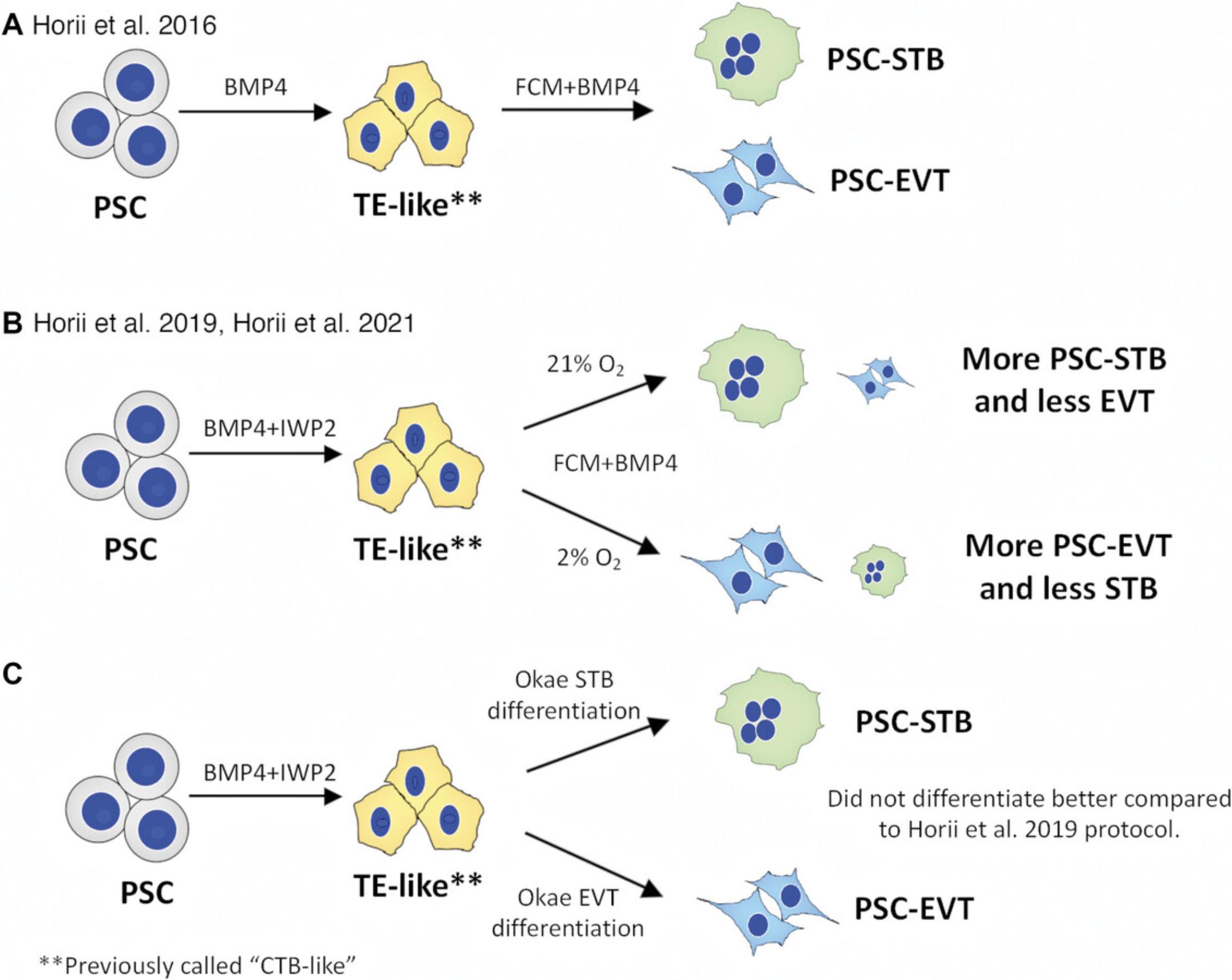
Using the 2019 improved protocol for trophoblast differentiation of PSCs, we established a “disease-in-a-dish” model to study molecular pathogenesis of the hypertensive disorder of pregnancy, called preeclampsia (Horii et al., 2019; Horii et al., 2021). However, a limitation of our study was the inefficient generation of EVT, which is a key cell type underlying PE pathophysiology. To overcome this limitation, we applied a modified media, named iCTB, developed in the Kessler lab for reprogramming of CTB from term placenta into TSC-like cells (Bai et al., 2021), to PSC-derived TE-like cells (Basic Protocol 1), to generate stable PSC-TSC (Basic Protocol 2). In our hands, the iCTB media is superior to the Okae TSC media for generation and maintenance of both primary and PSC-derived TSC (Bai et al., 2021; Soncin et al., 2022). Subsequent lineage-specific differentiation of PSC-TSC into EVT and STB lineages was further improved by adapting the medias and protocols established by Okae et al. (2018). In addition, we would also like to point out that we have attempted to directly perform Okae's STB and EVT differentiation protocol on the PSC-derived TE-like cells; however, this did not improve terminal differentiation of these cells, over and above the Horii et al. (2019) protocol (Fig. 6C). Therefore, TSC conversion of these TE-like cells (Basic Protocol 2) is required prior to terminal trophoblast differentiation (Fig. 1). These improvements are reflected in this updated protocol, and are currently employed among our three groups (Horii, Soncin, and Parast) to study trophoblast lineage specification, as well as preeclampsia-associated trophoblast phenotypes.
Both our protocol (using primed-PSC) and others’ (using naïve-PSC) include step-wise differentiation, starting with derivation of a TE-like cell, continuing to a TSC-like stage, and followed by terminal differentiation into EVT or STB through lineage-specific protocols (Io et al., 2021; Soncin et al., 2022). Our TE-like cells (from Basic Protocol 1) and naïve PSC-derived TE-like cells (Io et al., 2021) have a similar transcriptome profile and clustered together by RNAseq analysis (Soncin et al., 2022). Both protocols produce TSC, which have the ability to generate STB and EVT (with appropriate gene expression profiles), and show appropriate ELF5 promoter hypomethylation and lack of classical HLA (HLA-A and -B) expression (Io et al., 2021; Soncin et al., 2022). However, naïve-PSC derived TSC share one additional similarity to early gestation trophoblast, which is high expression of C19MC compared to primed PSC-derived TSC (Io et al., 2021; Soncin et al., 2022). The advantage of our protocol is that no additional step is required to convert primed-state PSC (regular hESC or reprogrammed iPSC) into naïve-PSC; therefore, there is no global loss of DNA methylation, including loss of imprinting throughout the genome (Theunissen et al., 2016). It is unclear whether this loss of DNA methylation which accompanies conversion to naïve pluripotency could negatively impact utility of patient-derived iPSC to model trophoblast development in the setting of placenta-based pregnancy disorders, such as pre-eclampsia, particularly as the latter is driven, at least in part, by environmental insults. The differences between TSC/trophoblast derived from naïve- and primed-PSC have recently been reviewed in depth (Morey et al., 2022; Roberts et al., 2022).
Critical Parameters
It is essential to have good starting material. If ESC or iPSC are not growing well, consider thawing a new vial, or changing to new batches of StemFlex or Geltrex. For PSC medium, other regularly-used media (i.e., StemPro, E8 or mTeSR Plus) besides StemFlex will also work for this protocol. Successful terminal differentiation to EVT requires undifferentiated (HLA-G <20%, EGFR >80%) PSC-TSC.
Cell attachment is improved if the collagen IV-coated plate is prepared a day before and kept in the incubator overnight. It is also acceptable to use the plate incubated for at least 2 to 3 hr on the day of plating but expect reduced cell attachment.
Troubleshooting
Following Basic Protocol 1 at day 4, TE-like cells express over 80% EGFR by flow cytometry. Do not proceed to Basic Protocol 2 if the cells show lower EGFR expression, but consider restarting the protocol and implementing the additional considerations listed in Table 10. Once the PSC-TSC are derived, consider expanding and freezing down for future experiments. After multiple passages, PSC-TSC may acquire HLA-G, and become difficult to recover as undifferentiated TSC. Cultures in which over 30% of the cells have acquired HLA-G by flow cytometry should be tossed, and younger passage cells should be thawed and expanded. All the reagents are used only from the listed manufactures; therefore, it will be necessary to test the differentiation efficiency if using reagents from different sources. A detailed troubleshooting guide is listed in Table 10.
| Problem | Possible cause(s) | Solution(s) |
|---|---|---|
| Poor attachment of the PSCs on Geltrex-coated plate | Matrix issues | Check matrix. Be sure to plate Geltrex in a cold environment. Keep on ice until plating, as it gels in 5-10 min at temperatures above 15°C. |
| No Y27632 | PSCs attach less efficiently when they are single cells. Include Y27632 when plating. | |
| PSCs detach from the plate | Matrix issues | Check matrix. Be sure to plate Geltrex in a cold environment. Keep on ice until plating, as it gels in 5-10 min at temperatures above 15°C. Heating the Geltrex could result in premature formation of gel. |
| Low EGFR at the induction of TE-like cells from PSC | Overseeding | Re-start the experiment. Seed at a lower concentration if necessary. |
| Cells are in big clusters | When treating cells with TrypLE, try to break up the cells into single cells. Use a P1000 to pipette up and down, harshly. Alternatively, incubate cells for a longer time to better dissociate the cells into single cells. | |
| Reagent issues | Check differentiation factors with a positive control to ensure that reagents are working. | |
| High passage number | Thaw earlier passage PSCs. | |
| Cells do not attach well on collagen IV coated plate | Matrix issue | Prepare plates and incubate overnight at 37°C. Longer incubation has shown better cell attachment. |
| PSC-derived TSC shows high HLA-G expression | High passage number | Thaw earlier passage PSC-derived TSC. |
| Cells grown to over-confluence | Thaw earlier passage PSC-derived TSC. | |
| Overgrown STB differentiation | High seeding density | Reduce the number of cells plated on day -1 to reach 30% confluency on day 0 before starting the differentiation. |
| Abbreviation | Definition |
|---|---|
| BMP4 | Bone morphogenetic protein 4 |
| BSA | Bovine serum albumin |
| CTB | Cytotrophoblast |
| EGF | Epidermal growth factor |
| EGFR | Epidermal growth factor receptor |
| ESC | Embryonic stem cells |
| EVT | Extravillous trophoblast |
| FCM | Feeder conditioned media |
| hCG | Human chorionic gonadotropin |
| HGF | Hepatocyte growth factor |
| HLA-G | Human leukocyte antigen G |
| iPSC | Induced pluripotent stem cells |
| ITS | Insulin-transferrin-selenium |
| KSR | Knockout Serum Replacement |
| NRG1 | Neuregulin 1 |
| PSC | Pluripotent stem cell |
| PSC-TSC | PSC-derived TSC |
| STB | Syncytiotrophoblast |
| TE | Trophectoderm |
| TSC | Trophoblast stem cells |
Understanding Results
The main part of this protocol uses TSC derivation and differentiation methods developed by Okae et al. (2018), and maintenance media (iCTB media) developed by Bai et al. (2021). Terminal differentiation protocols to STB and EVT were modified from Okae et al. (2018) (Morey et al., 2021; Soncin et al., 2022); in particular, coating tissue culture plates with fibronectin rather than collagen IV for EVT differentiation was adapted from primary EVT culture/differentiation protocols (Knofler & Pollheimer, 2013; Plessl et al., 2015; Wakeland et al., 2017), which is based on the decidual stromal composition of the EVT niche (Aplin et al., 1988; Earl et al., 1990). In the initial step of trophoblast induction (Basic Protocol 1), the colonies begin to show differentiation within a day, and by day 4, uniformly-flattened (epithelial) morphology appears throughout the culture (Fig. 2C). After 4 days of BMP4+IWP2 treatment, cells show over 80% EGFR positivity (Fig. 2D). This is important for the successful TSC derivation at the subsequent steps in Basic Protocol 2.
Using the previous protocol described by Horii et al. (2019), terminal differentiation of these TE-like cells (referred to as CTB-like cells in the original manuscript) using FCM plus 10 ng/ml BMP4 showed mixed differentiation into mostly STB, with a small proportion (less than 30%) of HLA-G+ EVT at the end of differentiation when cultured under 2% oxygen (Fig. 2E, F). However, after the development of TSC and lineage specific STB and EVT differentiation protocols (Okae et al., 2018), our group has applied the media to derive PSC-TSC from TE-like cells and was able to directly differentiate PSC-TSC to STB or EVT using lineage specific differentiation protocol (Fig. 1).
Notably, differentiation into EVT has had the greatest improvement. The PSC-TSC derived using Basic Protocols 1 to 3 (Fig. 3A, B), results in better yield of EVT showing over 60% HLA-G positive cells at the end of the Basic Protocol 5 (Fig. 5A-C) compared to 30% HLA-G positive EVT using our previous protocol (Fig. 2E, F). We can also specifically generate STB with high purity resulting in multinucleated cells with STB-associated gene expression (Soncin et al., 2022), high secretion of hCG (Fig. 4C), and cell-cell fusion (Fig. 4D). These assays are useful to study detailed mechanisms of trophoblast differentiation. A summary of abbreviations used can be found in Table 11.
Time Considerations
It takes about 1 week to expand and maintain the cells for the initial TE induction (Basic Protocol 1) and an additional 2 to 3 weeks to complete the TSC derivation (Basic Protocol 2). After establishing PSC-derived TSC, cells can be passaged multiple times (we have successfully maintained them for up to 20 passages). However, once a high proportion of cells acquire over 30% HLA-G, it is difficult to recover cells to the TSC stage (EGFR+, HLA-G-). In this case, thawing earlier passage cells may be required. For the STB differentiation (Basic Protocol 4), it takes about 8 days to complete the differentiation, and for EVT differentiation (Basic Protocol 5), it takes about 8 days to complete the differentiation. All time considerations include plate-coating steps.
Acknowledgments
Basic Protocol 1 was partially reproduced from Horii et al., 2019. Copyright 2019, with permission from John Wiley & Sons, Inc. This work was supported by funds from NIH/NICHD (R01-HD089537 to M.M.P., R01-HD096260 to F.S., K99/R00-HD091452 to M.H.). V.C.C. was supported through NIH (T32 HD007203 to the University of California, San Diego).
Author Contributions
Virginia Cheung : Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; visualization; writing original draft; writing review, and editing. Tony Bui : Conceptualization; data curation; investigation; methodology; writing review, and editing. Francesca Soncin : Conceptualization; methodology; funding acquisition; writing review, and editing. Tao Bai : Methodology; writing review, and editing. Jack Kessler : Methodology; writing review, and editing. Mana Parast : Conceptualization; funding acquisition; investigation; methodology; writing review, and editing. Mariko Horii : Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; visualization; writing original draft; writing review, and editing.
Conflict of Interest
M.M.P., F.S., and M.H. have filed a provisional patent for the protocol described. V.C.C., T.B., T.B., and J.A.K. declare no competing interests.
Open Research
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Literature Cited
- Aghajanova, L., Shen, S., Rojas, A. M., Fisher, S. J., Irwin, J. C., & Giudice, L. C. (2012). Comparative transcriptome analysis of human trophectoderm and embryonic stem cell-derived trophoblasts reveal key participants in early implantation. Biology of Reproduction , 86(1), 1–21. https://doi.org/10.1095/biolreprod.111.092775
- Alici-Garipcan, A., Özçimen, B., Süder, I., Ülker, V., Önder, T. T., & Özören, N. (2020). NLRP7 plays a functional role in regulating BMP4 signaling during differentiation of patient-derived trophoblasts. Cell Death & Disease, 11(8), 658. https://doi.org/10.1038/s41419-020-02884-1
- Amita, M., Adachi, K., Alexenko, A. P., Sinha, S., Schust, D. J., Schulz, L. C., Roberts, R. M., & Ezashi, T. (2013). Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proceedings of the National Academy of Sciences of the United States of America , 110(13), E1212–E1221. https://doi.org/10.1073/pnas.1303094110
- Apicella, C., Ruano, C. S. M., Méhats, C., Miralles, F., & Vaiman, D. (2019). The role of epigenetics in placental development and the etiology of preeclampsia. International Journal of Molecular Sciences , 20(11), 2837. https://doi.org/10.3390/ijms20112837
- Aplin, J. D., Charlton, A. K., & Ayad, S. (1988). An immunohistochemical study of human endometrial extracellular matrix during the menstrual cycle and first trimester of pregnancy. Cell and Tissue Research , 253(1), 231–240. https://doi.org/10.1007/BF00221758
- Bai, T., Peng, C.-Y., Aneas, I., Sakabe, N., Requena, D. F., Billstrand, C., Nobrega, M., Ober, C., Parast, M., & Kessler, J. A. (2021). Establishment of human induced trophoblast stem-like cells from term villous cytotrophoblasts. Stem Cell Research , 56, 102507. https://doi.org/10.1016/j.scr.2021.102507
- Blakeley, P., Fogarty, N. M. E., Del Valle, I., Wamaitha, S. E., Hu, T. X., Elder, K., Snell, P., Christie, L., Robson, P., & Niakan, K. K. (2015). Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development , 142(20), 3613. https://doi.org/10.1242/dev.131235
- Castel, G., Meistermann, D., Bretin, B., Firmin, J., Blin, J., Loubersac, S., Bruneau, A., Chevolleau, S., Kilens, S., Chariau, C., Gaignerie, A., Francheteau, Q., Kagawa, H., Charpentier, E., Flippe, L., François-Campion, V., Haider, S., Dietrich, B., Knöfler, M., … David, L. (2020). Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Reports , 33(8), 108419. https://doi.org/10.1016/j.celrep.2020.108419
- Cerneus, D. P., & van der Ende, A. (1991). Apical and basolateral transferrin receptors in polarized BeWo cells recycle through separate endosomes. Journal of Cell Biology , 114(6), 1149–1158. https://doi.org/10.1083/jcb.114.6.1149
- Cinkornpumin, J. K., Kwon, S. Y., Guo, Y., Hossain, I., Sirois, J., Russett, C. S., Tseng, H.-W., Okae, H., Arima, T., Duchaine, T. F., Liu, W., & Pastor, W. A. (2020). I human embryonic stem cells can give rise to cells with a trophoblast-like transcriptome and methylome. Stem Cell Reports , 15(1), 198–213. https://doi.org/10.1016/j.stemcr.2020.06.003
- Das, P., Ezashi, T., Schulz, L. C., Westfall, S. D., Livingston, K. A., & Roberts, R. M. (2007). Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Research , 1(1), 61–74. https://doi.org/10.1016/j.scr.2007.09.004
- Dong, C., Beltcheva, M., Gontarz, P., Zhang, B., Popli, P., Fischer, L. A., Khan, S. A., Park, K.-M., Yoon, E.-J., Xing, X., Kommagani, R., Wang, T., Solnica-Krezel, L., & Theunissen, T. W. (2020). Derivation of trophoblast stem cells Inaive human pluripotent stem cells. Elife , 9, e52504. https://doi.org/10.7554/eLife.52504
- Earl, U., Estlin, C., & Bulmer, J. (1990). Fibronectin and laminin in the early human placenta. Placenta , 11(3), 223–231. https://doi.org/10.1016/s0143-4004(05)80268-1
- Erb, T. M., Schneider, C., Mucko, S. E., Sanfilippo, J. S., Lowry, N. C., Desai, M. N., Mangoubi, R. S., Leuba, S. H., & Sammak, P. J. (2011). Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: The role of bone morphogenic protein 4 and histone deacetylases. Stem Cells and Development , 20(9), 1601–1614. https://doi.org/10.1089/scd.2010.0281
- Gerbaud, P., Pidoux, G., Guibourdenche, J., Pathirage, N., Costa, J. M., Badet, J., Frendo, J.-L., Murthi, P., & Evain-Brion, D. (2011). Mesenchymal activin-A overcomes defective human trisomy 21 trophoblast fusion. Endocrinology , 152(2), 5017–5028. https://doi.org/10.1210/en.2011-1193
- Horii, M., Li, Y., Wakeland, A. K., Pizzo, D. P., Nelson, K. K., Sabatini, K., Laurent, L. C., Liu, Y., & Parast, M. M. (2016). Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proceedings of the National Academy of Sciences of the United States of America , 113(27), E3882–E3891. https://doi.org/10.1073/pnas.1604747113
- Horii, M., Bui, T., Touma, O., Cho, H. Y., & Parast, M. M. (2019). An improved two-step protocol for trophoblast differentiation of human pluripotent stem cells. Current Protocols in Stem Cell Biology , 50(1), e96. https://doi.org/10.1002/cpsc.96
- Horii, M., Morey, R., Bui, T., Touma, O., Nelson, K. K., Cho, H.-Y., Rishik, H., Laurent, L. C., & Parast, M. M. (2021). Modeling preeclampsia using human induced pluripotent stem cells. Scientific Reports , 11(1), 5877. https://doi.org/10.1038/s41598-021-85230-5
- Io, S., Kabata, M., Iemura, Y., Semi, K., Morone, N., Minagawa, A., Wang, B., Okamoto, I., Nakamura, T., Kojima, Y., Iwatani, C., Tsuchiya, H., Kaswandy, B., Kondoh, E., Kaneko, S., Woltjen, K., Saitou, M., Yamamoto, T., Mandai, M., & Takashima, Y. (2021). Capturing human trophoblast developIwith naïve pluripotent stem cells in vitro. Cell Stem Cell , 28(6), 1023–1039.e13. https://doi.org/10.1016/j.stem.2021.03.013
- Jang, Y. J., Kim, M., Lee, B.-K., & Kim, J. (2022). Induction of human trophoblast stem-like cells from primed pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America , 119(20), e2115709119. https://doi.org/10.1073/pnas.2115709119
- Knöfler, M., & Pollheimer, J. (2013). Human placental trophoblast invasion and differentiation: A particular focus on Wnt signaling. Frontiers in Genetics , 4, 190. https://doi.org/10.3389/fgene.2013.00190
- Kobayashi, N., Okae, H., Hiura, H., Kubota, N., Kobayashi, E. H., Shibata, S., Oike, A., Hori, T., Kikutake, C., Hamada, H., Kaji, H., Suyama, M., Bortolin-Cavaillé, M.-L., Cavaillé, J., & Arima, T. (2022). The microRNA cluster C19MC confers differentiation potential into trophoblast lineages upon human pluripotent stem cells. Nature Communications , 13(1), 3071. https://doi.org/10.1038/s41467-022-30775-w
- Kohler, P. O., & Bridson, W. E. (1971). Isolation of hormone-producing clonal lines of human choriocarcinoma. Journal of Clinical Endocrinology and Metabolism , 32(5), 683–687. https://doi.org/10.1210/jcem-32-5-683
- Krendl, C., Shaposhnikov, D., Rishko, V., Ori, C., Ziegenhain, C., Sass, S., Simon, L., Müller, N. S., Straub, T., Brooks, K. E., Chavez, S. L., Enard, W., Theis, F. J., & Drukker, M. (2017). GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proceedings of the National Academy of Sciences of the United States of America , 114(45), E9579–E9588. https://doi.org/10.1073/pnas.1708341114
- Kurek, D., Neagu, A., Tastemel, M., Tüysüz, N., Lehmann, J., van de Werken, H. J. G., Philipsen, S., van der Linden, R., Maas, A., van Ijcken, W. F. J., Drukker, M., & ten Berge, D. (2015). Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Reports , 4(1), 114–128. https://doi.org/10.1016/j.stemcr.2014.11.007
- Li, Y., Moretto-Zita, M., Soncin, F., Wakeland, A., Wolfe, L., Leon-Garcia, S., Pandian, R., Pizzo, D., Cui, L., Nazor, K., Loring, J. F., Crum, C. P., Laurent, L. C., & Parast, M. M. (2013). BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a DeltaNp63+ cytotrophoblast stem cell state. Development , 140(19), 3965–3976. https://doi.org/10.1242/dev.092155
- Li, Z., Kurosawa, O., & Iwata, H. (2019). Establishment of human trophoblast stem cells from human induced pluripotent stem cell-derived cystic cells under micromesh culture. Stem Cell Res Ther , 10(1), 245. https://doi.org/10.1186/s13287-019-1339-1
- Maccani, J. Z. J., & MacCani, M. (2015). Altered placental DNA methylation patterns associated with maternal smoking: Current perspectives. Advances in Genomics and Genetics , 2015(5), 205–214. https://doi.org/10.2147/AGG.S61518
- Mischler, A., Karakis, V., Mahinthakumar, J., Carberry, C. K., San Miguel, A., Rager, J. E., Fry, R. C., & Rao, B. M. (2021). Two distinct trophectoderm lineage stem cells from human pluripotent stem cells. Journal of Biological Chemistry , 296, 100386. https://doi.org/10.1016/j.jbc.2021.100386
- Morey, R., Bui, T., Fisch, K. M., & Horii, M. (2022). Modeling placental development and disease using human pluripotent stem cells. Placenta , https://doi.org/10.1016/j.placenta.2022.10.011
- Morey, R., Farah, O., Kallol, S., Requena, D. F., Meads, M., Moretto-Zita, M., Soncin, F., Laurent, L. C., & Parast, M. M. (2021). Transcriptomic Drivers of Differentiation, Maturation, and Polyploidy in Human Extravillous Trophoblast. Frontiers in Cell and Developmental Biology , 9, 702046. https://doi.org/10.3389/fcell.2021.702046
- Okae, H., Toh, H., Sato, T., Hiura, H., Takahashi, S., Shirane, K., Kabayama, Y., Suyama, M., Sasaki, H., & Arima, T. (2018). Derivation of human trophoblast stem cells. Cell Stem Cell , 22(1), 50–63.e6. https://doi.org/10.1016/j.stem.2017.11.004
- Plessl, K., Haider, S., Fiala, C., Pollheimer, J., & Knöfler, M. (2015). Expression pattern and function of Notch2 in different subtypes of first trimester cytotrophoblast. Placenta , 36(4), 365–371. https://doi.org/10.1016/j.placenta.2015.01.009
- Roberts, R. M., Ezashi, T., Temple, J., Owen, J. R., Soncin, F., & Parast, M. M. (2022). The role of BMP4 signaling in trophoblast emergence from pluripotency. Cellular and Molecular Life Sciences , 79(8), 447. https://doi.org/10.1007/s00018-022-04478-w
- Sheridan, M. A., Yang, Y., Jain, A., Lyons, A. S., Yang, P., Brahmasani, S. R., Dai, A., Tian, Y., Ellersieck, M. R., Tuteja, G., Schust, D. J., Schulz, L. C., Ezashi, T., & Roberts, R. M. (2019). Early onset preeclampsia in a model for human placental trophoblast. Proceedings of the National Academy of Sciences of the United States of America , 116(10), 4336–4345. https://doi.org/10.1073/pnas.1816150116
- Soncin, F., Khater, M., To, C., Pizzo, D., Farah, O., Wakeland, A., Rajan, K. A. N., Nelson, K. K., Chang, C.-W., Moretto-Zita, M., Natale, D. R., Laurent, L. C., & Parast, M. M. (2018). Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development , 145(2), dev156273. https://doi.org/10.1242/dev.156273
- Soncin, F., Morey, R., Bui, T., Requena, D. F., Cheung, V. C., Kallol, S., Kittle, R., Jackson, M. G., Farah, O., Chousal, J., Meads, M., Pizzo, D., Horii, M., Fisch, K. M., & Parast, M. M. (2022). Derivation of functional trophoblast stem cells from primed human pluripotent stem cells. Stem Cell Reports , 17(6), 1303–1317. https://doi.org/10.1016/j.stemcr.2022.04.013
- Sudheer, S., Bhushan, R., Fauler, B., Lehrach, H., & Adjaye, J. (2012). FGF inhibition directs BMP4-mediated differentiation of human embryonic stem cells to syncytiotrophoblast. Stem Cells and Development , 21(16), 2987–3000. https://doi.org/10.1089/scd.2012.0099
- Tanaka, S., Kunath, T., Hadjantonakis, A.-K., Nagy, A., & Rossant, J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science , 282(5396), 2072–2075. https://doi.org/10.1126/science.282.5396.2072
- Telugu, B. P., Adachi, K., Schlitt, J. M., Ezashi, T., Schust, D. J., Roberts, R. M., & Schulz, L. C. (2013). Comparison of extravillous trophoblast cells derived from human embryonic stem cells and from first trimester human placentas. Placenta , 34(7), 536–543. https://doi.org/10.1016/j.placenta.2013.03.016
- Theunissen, T. W., Friedli, M., He, Y., Planet, E., O'neil, R. C., Markoulaki, S., Pontis, J., Wang, H., Iouranova, A., Imbeault, M., Duc, J., Cohen, M. A., Wert, K. J., Castanon, R., Zhang, Z., Huang, Y., Nery, J. R., Drotar, J., Lungjangwa, T., … Jaenisch, R. (2016). Molecular criteria foIining the naive human pluripotent state. Cell Stem Cell , 19(4), 502–515. https://doi.org/10.1016/j.stem.2016.06.011
- Wakeland, A. K., Soncin, F., Moretto-Zita, M., Chang, C.-W., Horii, M., Pizzo, D., Nelson, K. K., Laurent, L. C., & Parast, M. M. (2017). Hypoxia directs human extravillous trophoblast differentiation in a hypoxia-inducible factor-dependent manner. The American Journal of Pathology , 187(4), 767–780. https://doi.org/10.1016/j.ajpath.2016.11.018
- Warmflash, A., Sorre, B., Etoc, F., Siggia, E. D., & Brivanlou, A. H. (2014). A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nature Methods , 11(8), 847–854. https://doi.org/10.1038/nmeth.3016
- Wei, Y., Wang, T., Ma, L., Zhang, Y., Zhao, Y., Lye, K., Xiao, L., Chen, C., Wang, Z., Ma, Y., Zhou, X., Sun, F., Li, W., Dunk, C., Li, S., Nagy, A., Yu, Y., Pan, G., Lye, S. J., & Shan, Y. (2021). Efficient derivation of human trophoblast stem cells from primed pluripotent stem cells. Science Advances , 7(33), eabf4416. https://doi.org/10.1126/sciadv.abf4416
- Workalemahu, T., Ouidir, M., Shrestha, D., Wu, J., Grantz, K. L., & Tekola-Ayele, F. (2020). Differential DNA methylation in placenta associated with maternal blood pressure during pregnancy. Hypertension , 75(4), 1117–1124. https://doi.org/10.1161/HYPERTENSIONAHA.119.14509
- Wu, Z., Zhang, W., Chen, G., Cheng, L., Liao, J., Jia, N., Gao, Y., Dai, H., Yuan, J., Cheng, L., & Xiao, L. (2008). Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. Journal of Biological Chemistry , 283(36), 24991–25002. https://doi.org/10.1074/jbc.M803893200
- Xu, R.-H., Chen, X., Li, D. S., Li, R., Addicks, G. C., Glennon, C., Zwaka, T. P., & Thomson, J. A. (2002). BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nature Biotechnology , 20(12), 1261–1264. https://doi.org/10.1038/nbt761
- Yabe, S., Alexenko, A. P., Amita, M., Yang, Y., Schust, D. J., Sadovsky, Y., Ezashi, T., & Roberts, R. M. (2016). Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proceedings of the National Academy of Sciences of the United States of America , 113(19), E2598–E2607. https://doi.org/10.1073/pnas.1601630113
- Yang, Y., Adachi, K., Sheridan, M. A., Alexenko, A. P., Schust, D. J., Schulz, L. C., Ezashi, T., & Roberts, R. M. (2015). Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proceedings of the National Academy of Sciences of the United States of America , 112(18), E2337–2346. https://doi.org/10.1073/pnas.1504778112
Key References
- Soncin et al., 2022. See above.
- In this paper, we have shown four important points. First, the initial trophoblast induction of primed human PSCs (using BMP2+IWP2) resulted in cells showing a TE-like signature by transcriptome profile. Second, we showed that these TE-like cells can convert to TSC by applying iCTB media, which resulted in a transcriptome and ELF5 promotor DNA methylation profile similar to primary TSC. Third, primed PSC-derived TSC can further differentiate into STB or EVT by using both in vitro lineage-specific differentiation protocols and in vivo engraftment assay. Finally, PSC-derived TE and TSC showed similar transcriptome profile to naïve (pre-implantation state)-PSC derived TE and TSC. These data showed that primed human ESC/iPSC can in fact become functional TSC, and further give rise to STB or EVT through lineage-specific differentiation.
CORRECTIONS
In this publication, the following corrections have been made:
“Catalogue Numbers of Matrigel and 1-Thioglycerol” have been replaced with the correct numbers.
The current version online now includes these corrections and may be considered the authoritative version of record.
Citing Literature
Number of times cited according to CrossRef: 1
- Robert Morey, Tony Bui, Virginia Chu Cheung, Chen Dong, Joseph E. Zemke, Daniela Requena, Harneet Arora, Madeline G. Jackson, Donald Pizzo, Thorold W. Theunissen, Mariko Horii, iPSC-based modeling of preeclampsia identifies epigenetic defects in extravillous trophoblast differentiation, iScience, 10.1016/j.isci.2024.109569, 27 , 4, (109569), (2024).

