Characterization of Anaphylatoxin Receptor Expression and C3a/C5a Functions in Anaphylatoxin Receptor Reporter Mice
Yves Laumonnier, Yves Laumonnier, Christian M. Karsten, Christian M. Karsten, Gabriele Köhl, Gabriele Köhl, Jörg Köhl, Jörg Köhl
Abstract
The anaphylatoxins (AT) C3a and C5a are effector molecules of C3 and C5 exerting multiple biologic functions through binding and activation of their cognate G protein−coupled receptors. C3a interacts with the C3a receptor (C3aR), whereas C5a and its primary degradation product C5a-desArg engage C5aR1 and C5aR2. In the past, analysis of AT expression has been hampered by cross reaction of antibodies designed to recognize the different AT receptors. Furthermore, assessment of effects mediated by cell-specific activation has been difficult. Here, floxed AT receptor reporter mice are described as tools to monitor AT receptor expression in cells and tissues and to study the functions of C3a and C5a by cell-specific deletion of their cognate AT receptors. © 2020 The Authors.
Basic Protocol 1 : Genotyping of floxed GFP-C5aR1 knockin mice
Support Protocol 1 : Genotyping of LysMcre-C5ar1-/- mice
Basic Protocol 2 : Genotyping of floxed tdTomato-C3aR and -tdTomato-C5aR2 knockin mice
Support Protocol 2 : Preparation of genomic DNA
Basic Protocol 3 : Determination of C5aR1, C5aR2, and C3aR expression using floxed AT receptor reporter mice
Support Protocol 3 : Determination of C3aR expression using a C3aR-specific antibody
Support Protocol 4 : Determination of C5aR1, C5aR2, and C3aR mRNA expression in floxed GFP-C5aR1, floxed tdTomato-C5aR2 or -tdTomato C3aR positive cells
Basic Protocol 4 : Analysis of C5aR1-driven ERK1/2 phosphorylation in GFP-C5aR1+ cells
Basic Protocol 5 : Assessment of C3aR functions in cells obtained from floxed tdTomato-C3aR knockin mice- Determination of C3aR internalization
Alternate Protocol : C3a-induced increase in intracellular Ca2+
Basic Protocol 6 : C5aR2-driven IFN-γ production from NK cells
Support Protocol 5 : Isolation of splenic NK cells by FACS
INTRODUCTION
In recent years, our view of the complement system as a guardian of the extracellular space has been markedly extended. Several reports have shown that complement is not activated systemically solely via the three canonical activation pathways, i.e., the classical, lectin, and alternative pathway, but by non-canonical pathways via specific proteases derived from pathogens, activated host cells, or the contact system (Hajishengallis, Reis, Mastellos, Ricklin, & Lambris, 2017). These newly discovered non-canonical activation pathways of complement, its unexpected contributions to cell homeostasis, metabolism, differentiation, and apoptosis, its crosstalk with several receptor classes and other cascade systems of innate immunity, as well as its impact on adaptive immune responses make the complement system more than ever an important field of research within immunology (Kolev, Le Friec, & Kemper, 2014; West, Kolev, & Kemper, 2018).
Many of these newly discovered functions can be attributed to the anaphylatoxins (ATs) C3a and C5a, which were originally considered mere proinflammatory molecules. Their pleiotropic functions are mainly mediated through activation of their G protein−coupled corresponding complement peptide receptors, i.e., C3aR for C3a and C5aR1 and C5aR2 for C5a. Until recently, AT receptor expression in professional and non-professional immune cells, as well as tissue-resident stroma cells, has been ill-defined, and is still controversial (Laumonnier, Karsten, & Köhl, 2017). To better understand the expression patterns of the AT receptors in health and disease, and to define the multiple roles of the ATs in the innate and adaptive immune networks, we have generated floxed AT receptor reporter mice that are now available to and widely used by the scientific community.
This article describes the use of C3aR and C5aR2 reporter mice with a tandem-dye (td)Tomato construct (Karsten et al., 2017; Quell et al., 2017), as well as the use of a C5aR1 knock-in mouse in which the Aequorea coerulescens green fluorescent protein [(Ac)GFP] was added to monitor C5aR1 expression (Karsten et al., 2015). These mice are available from the Köhl laboratory upon request. Basic Protocol 1 outlines the procedure to genotype GFP-C5aR1 knockin mice. Support Protocol 1 describes as an example the genotyping of mice with conditional deletion of C5aR1 in LysM-expressing cells after Cre-mediated targeting. Basic Protocol 2 describes the genotyping of tdTomato-C3aR and C5aR2 knock-in mice, with Support Protocol 2 detailing the preparation of the genomic DNA. Basic Protocol 3, together with two support protocols that describe the analysis of AT receptor protein (Support Protocol 3) and mRNA (Support Protocol 4) expression, outlines a flow cytometric approach to assess AT receptor expression in GFP-C5aR1+, tdTomato-C5aR2+, or tdTomato-C3aR+ cells. To link the expression of the AT receptor reporter constructs with AT receptor function, we provide three basic protocols to determine distinct functions of C5aR1 in GFP-C5aR1 knockin mice (Basic Protocol 4), C3aR in tdTomato-C3aR knockin mice (Basic Protocol 5), and C5aR2 in tdTomato-C5aR2 knockin mice (Basic Protocol 6). Finally, we have added Support Protocol 5 for the isolation of NK cells, which exclusively express C5aR2 but not C5aR1.
NOTE : All animals were used for organ removal according to protocols approved by local authorities of the Animal and Care and Use Committee (Ministerium für Landwirtschaft, Umwelt und Ländliche Räume, Kiel, Germany). All protocols using live animals must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of laboratory animals.
Basic Protocol 1: GENOTYPING OF FLOXED GFP-C5aR1 KNOCKIN MICE
The genes encoding for the three AT or complement peptide receptors are organized in a similar way, with a 5′-untranslated region (UTR), two exons, and a 3′-UTR after exon 2 (https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=5). This offers the opportunity to insert a fluorescent reporter sequence (tdTomato or GFP) before or at the beginning of exon 2.With the inserted construct, one locus of cross-over P1 (loxP) site is introduced at the 5′ end of the reporter sequence and another one after the coding region of the receptor before the 3′-UTR. According to the general design, the C5aR1 construct includes a GFP coding sequence ending with a stop codon, followed by an internal ribosomal entry site (IRES) placed upstream of exon 2 encoding for the C5ar1 gene, which is flanked by loxP sites at both ends. Between the loxP site at the 5′-UTR and the GFP reporter gene, a neomycin cassette with flippase recognition target (FRT) sites at both ends was included. During the generation of mice, the neomycin cassette was removed by breeding to a flippase (FLP) deleter mouse strain (Fig. 1).
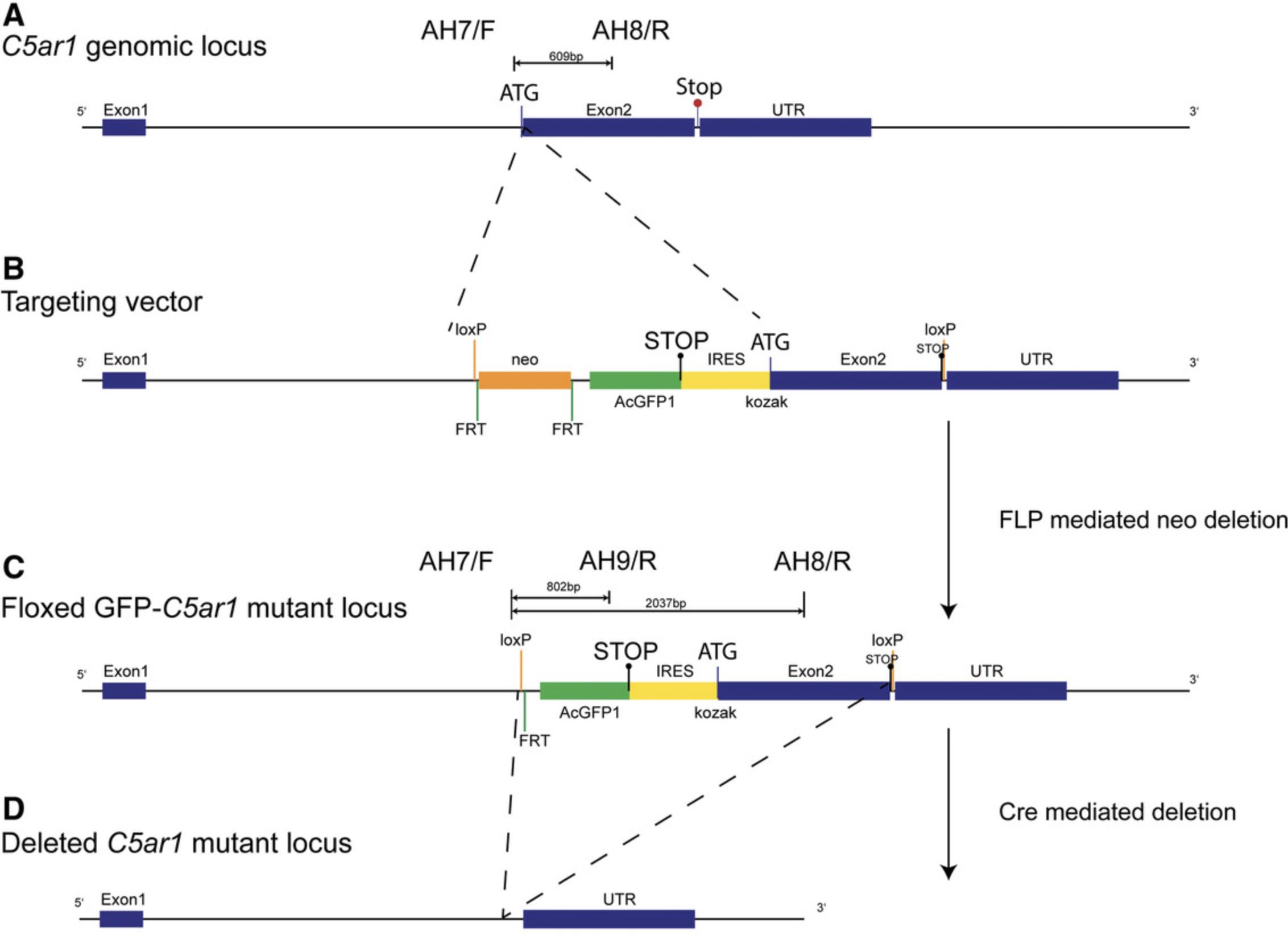
To determine the genotype of the AT receptor reporter mice, we have set up a PCR-based approach in which we detect the presence of the reporter sequence (GFP : PCR1) including the loxP sites and/or the AT receptor wild-type genes (C5ar1 : PCR 2), in particular in the case of target cell−specific AT receptor deletion after breeding of the AT receptor reporter mice to defined Cre mice (H. Kim, Kim, Im, & Fang, 2018; Song & Palmiter, 2018). In the latter case, the Cre mouse strain used needs to be genotyped in a separate approach, as outlined in Support Protocol 1.
Materials
- 100 µM primers for PCR 1 and 2 (Table 1; Eurofins Genomics/Thermo Fisher Scientific)
- Nuclease-free water (Life Technologies GmbH, cat. no. AM9937)
- 2× KAPA2G Fast Hot Start Genotyping Mix (Sigma Aldrich, cat. no. KK5621; manufactured by Roche)
- Template DNA: genomic DNA (Support Protocol 2) from floxed GFP-C5aR1 knockin mouse (Köhl laboratory)
- 1.5% SB agarose gel (Brody & Kern, 2004; also see Current Protocols article Voytas, 2001)
- Sodium borate (SB) buffer (see recipe)
- O'Gene Ruler DNA Ladder Mix (Thermo Fisher Scientific, cat. no. SM0333)
- GelRed working solution (see recipe)
| Use in PCR# | Primer/type/target | Sequence | bp | Design |
|---|---|---|---|---|
| 1 | AH7/F/C5ar1 | TAGAGTTGAGACTCAGAAAGACGG | 24 | Ozgene |
| 1 | AH9/R/GFP | GGGTGGACAGGTAGTGGTTATC | 22 | Ozgene |
| 1 | GK91/F/Tcrd | CAAATGTTGCTTGTCTGGTG | 20 | JAX LAb oIMR8744 |
| 1 | GK92/R/Tcrd | GTCAGTCGAGTGCACAGTTT | 20 | JAX LAb oIMR8745 |
| 2 | AH7/F/C5ar1 | TAGAGTTGAGACTCAGAAAGACGG | 24 | Ozgene |
| 2 | AH8/R/C5ar1 | GTACACGAAGGATGGAATGGTG | 22 | Ozgene |
| 2 | AH15/F/Il2 | CTAGGCCACAGAATTGAAAGATCT | 24 | Jax Lab oIMR7338 |
| 2 | AH16/R/Il2 | GTAGGTGGAAATTCTAGCATCATCC | 25 | Jax Lab oIMR7339 |
-
Abbreviations: bp, base pair; F, forward; GFP, green fluorescent protein; Il2, IL-2 precursor short; R, reverse; Tcrd, T cell receptor delta chain.
-
1.5-ml screw-cap microcentrifuge tube with O-ring (Sarstedt, cat. no. 72.692.405)
-
Microcentrifuge
-
0.2-ml 8-tube PCR strips without caps (BioRad (cat. no. TBS0201)
-
8-capstrips for PCR tubes (BioRad, cat. no. TBS0803)
-
Filtered pipet tips (10, 250, and 1000 µl; Sarstedt, cat. no. 701.116.210, 701.189.215, 701.186.210)
-
Microcentrifuge tubes (1.5-ml, Sarstedt, cat. no. 72.690)
-
PCR machine (BioRad C1000 or C1000 Touch Thermal Cycler)
-
Gel staining tray
-
UV transilluminator and camera
-
Additional reagents and equipment for agarose gel electrophoresis (Brody & Kern, 2004; also see Current Protocols article Voytas, 2001)
CAUTION : Remember the proper work safety requirements to protect eyes and skin against UV radiation. Boric acid belongs to the group of carcinogenic, mutagenic, reprotoxic (CMR) substances. Follow the national requirements for disposal and working safety.
NOTE : All chemicals need to be molecular grade (DNase-free). For all steps use filtered pipet tips only.
1.Prepare PCR1 primer mix by combining 20 µl each of primers AH7/AH9/GK91/GK92 (Table 1) from primer stock (100 µM) in a 1.5-ml screw-cap microcentrifuge tube with O-ring, and make up with 120 µl nuclease-free water to a final volume of 200 µl. Store at −20°C.
2.Prepare PCR2 primer mix by combining 20 µl each of primers AH7/AH8/AH15/AH16 (Table 1) from primer stock (100 µM) in a 1.5-ml screw-cap microcentrifuge tube with O-ring, and make up with 120 µl nuclease-free water to a final volume of 200 µl. Store at −20°C.
3.Prepare separate master mixes for PCR1 and PCR2 by pipetting the following reagents at the indicated volumes into a 1.5-ml microcentrifuge tube (per sample):
- 6.25 µl nuclease-free water
- 1.25 µl primer mix for PCR1 (step 1) or PCR2 (step 2)
- 12.5 µl 2× KAPA2G Fast Hot Start Genotyping Mix
- Mix and microcentrifuge briefly to bring mixture to bottom of tube.
The final concentration of each primer in PCR1 and 2 is 0.5 µM after adding the template.
4.Combine 5 µl template [genomic DNA (Support Protocol 2) from floxed GFP-C5aR1 knockin mouse diluted 1:10 with nuclease-free water] or nuclease-free water for control with 20 µl of the appropriate master mix in a 0.2-ml 8-tube PCR strip without caps, and run the following thermocycling conditions for both PCR1 and PCR2 after closing the tubes with the 8-capstrips for PCR tubes:
| Step no. | Temp. | Time | No. of cycles |
|---|---|---|---|
| 1 | 95°C | 180 s | 1 cycle |
| 2 | 95°C | 15 s | |
| 3 | 62°C | 15 s | |
| 4 | 72°C | 60 s | |
| 5 | As in 2, 3, and 4 | As in 2, 3, and 4 | 35 cycles (total) |
| 6 | 72°C | 120 s | 1 cycle |
| 7 | 15°C | Hold |
5.Prepare a 1.5% SB agarose gel as described in Brody & Kern (2004).
6.Run samples on 1.5% SB agarose gel using the following conditions: 30-40 min, 200 V, with a 7-cm running length. Include DNA markers. Load per lane: either 6 µl PCR reaction or 4 µl marker.
7.Stain the gel with GelRed working solution for 45 min in a tray.
8.Visualize the gel bands with UV light and document the result by photography.
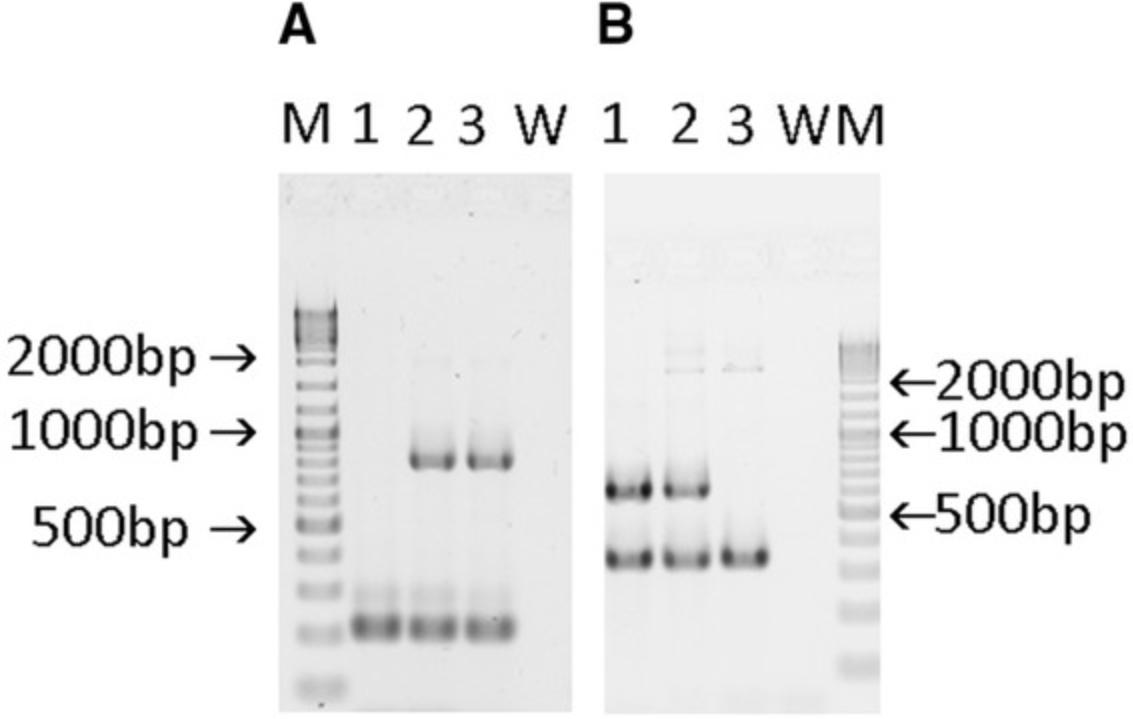
| Result PCR1 | Result PCR2 | |||
|---|---|---|---|---|
| Genotype | Specific:GFP | Internal control:Tcrd | Specific:C5ar1 | Internal control:Il2 |
| +/+ | - | 206 bp | 609 bp | 324 bp |
| flx/+ | 802 bp | 206 bp | 609 and 2037 bpa | 324 bp |
| flx/flx | 802 bp | 206 bp | 2037 bpa | 324 bp |
- a
The 2037 bp fragment is very faint and can even be missing.
-
Abbreviations: bp, base pair; GFP, green fluorescent protein; Il2, IL-2 precursor short; Tcrd, T cell receptor delta chain.
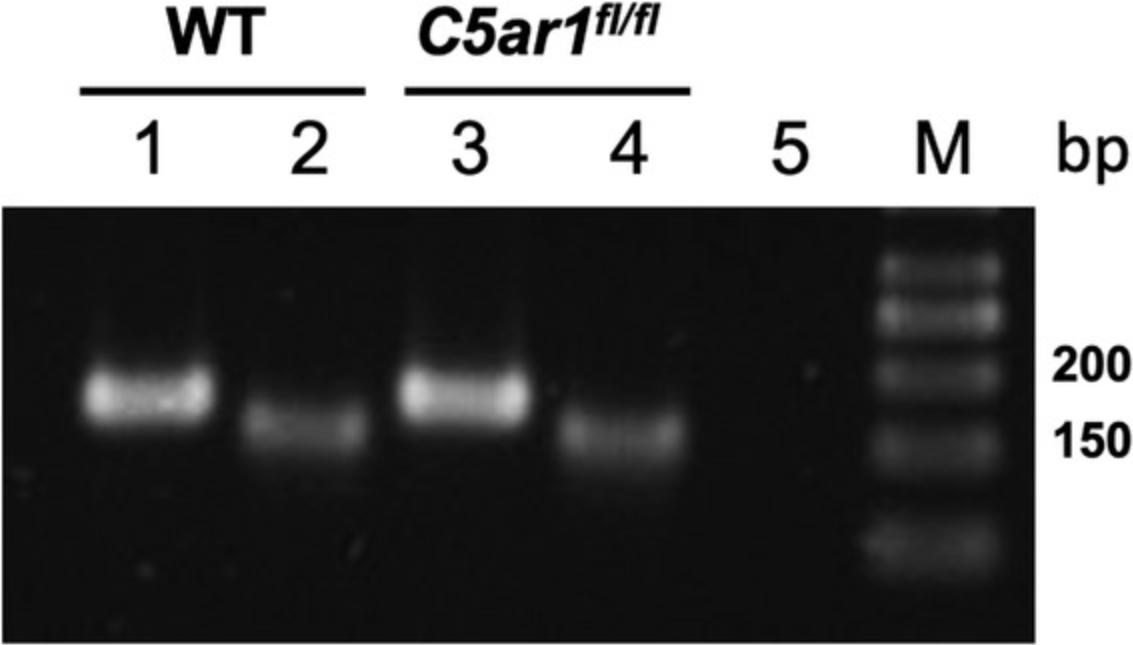
Support Protocol 1: GENOTYPING OF LysMcre-C5aR1 MICE
Breeding of the floxed GFP-C5aR1 knockin mice with strains that harbor a Cre recombinase will result in the targeted deletion of the GFP-C5aR1 cassette through excision at the two loxP sites (Fig. 1D). Similarly, the tdTomato-C3ar1 or C5ar2 genes will be excised in response to Cre (Figs. 4D and 5D). Here, we describe the genotyping of LysMcre-C5aR1 in which LysMcre mice (Clausen, Burkhardt, Reith, Renkawitz, & Forster, 1999) have been crossed with floxed GFP-C5aR1 knockin mice to specifically delete C5aR1 in granulocytes, monocytes, and macrophages (Karsten et al., 2015). Given that the C5ar1 gene is still expressed in many cell types, we have set up an analysis system that includes C5ar1 (Basic Protocol 1) and Lyz2 , the gene encoding for lysozyme M(lysM). The C5ar1 and Lyz2 genes are expressed on two different chromosomes, i.e., chromosomes 7 and 10. To delete C5aR1 in lysM-expressing cells, GFP-C5aR1fl/fl mice need to harbor at least one functional Cre allele under the control of the lysM promoter.
Materials
-
100 µM primers for PCR3 (Eurofins Genomics or Life Technologies GmbH)
-
Nuclease-free water (Life Technologies GmbH, cat. no. AM9937)
-
2× KAPA2G Fast Hot Start Genotyping Mix (Sigma Aldrich, cat. no. KK5621; manufactured by Roche)
-
Template DNA: genomic DNA (Support Protocol 2) from LysMcre-C5aR1 mouse
-
1.5% SB agarose gel (Brody & Kern, 2004; also see Current Protocols article Voytas, 2001)
-
Sodium borate (SB) buffer (see recipe)
-
O'Gene Ruler DNA Ladder Mix (Thermo Fisher Scientific, cat. no. SM0333)
-
GelRed working solution (see recipe)
-
1.5-ml screw-cap microcentrifuge tube with O-ring (Sarstedt, cat. no. 72.692.405)
-
Microcentrifuge
-
0.2-ml 8-tube PCR strips without caps (BioRad, cat. no. TBS0201)
-
8-capstrips for PCR tubes (BioRad, cat. no. TBS0803)
-
Filtered pipet tips (10, 250, and 1000 µl; Sarstedt, cat. no. 701.116.210, 701.189.215, 701.186.210)
-
Microcentrifuge tubes (1.5-ml, Sarstedt, cat. no. 72.690)
-
PCR machine (BioRad C1000 or C1000 Touch Thermal Cycler)
-
Gel staining tray
-
UV transilluminator and camera
-
Additional reagents and equipment for agarose gel electrophoresis (Brody & Kern, 2004; also see Current Protocols article Voytas, 2001)
CAUTION : Remember the proper work safety requirements to protect eyes and skin against UV radiation. Boric acid belongs to the group of carcinogenic, mutagenic, reprotoxic (CMR) substances. Follow the national requirements for disposal and working safety.
NOTE : All chemicals need to be molecular grade (DNase free). For all steps use filtered pipet tips only.
1.Prepare PCR3 primer mix to amplify Lyz2 by combining 20 µl of primers GK199/GK223/GK234 (Table 3) from primer stock (100 µM) in a 1.5-ml screw-cap microcentrifuge tube with O-ring and make up with 140 µl nuclease-free water to a final volume of 200 µl.
| Use in PCR# | Primer/type/target | Sequence | bp | Design |
|---|---|---|---|---|
| 3 | GK199/R/cre recombinase | CCCAGAAATGCCAGATTACG | 20 | Riken BRC 02302/3 = Jax Lab oIMR3066 |
| 3 | GK223/F/Lyz2 | GCATTGCAGACTAGCTAAAGGCAG | 24 | Riken BRC 02302/1 |
| 3 | GK224/R/Lyz2 | GTCGGCCAGGCTGACTCCATAG | 22 | Riken BRC 02302/2 |
-
Abbreviations: bp, base pair; F, forward; Lyz2, Lysozyme M; R, reverse.
2.Prepare a master mix for PCR3 by pipetting the following reagents and the indicated volumes into a 1.5-ml microcentrifuge tube (per sample):
- 1.25 µl nuclease-free water
- 1.25 µl primer mix for PCR3
- 12.5 µl 2× KAPA2G Fast Hot Start Genotyping Mix.
- Mix and microcentrifuge briefly to bring mixture to bottom of tube.
3.Combine 10 µl template [genomic DNA (Support Protocol 2) from floxed LysMCre-C5aR1 mouse diluted 1:10 with nuclease-free water] or nuclease-free water for control with 15 µl master mix in a 0.2-ml 8-tube PCR strip without caps and run the following thermocycling conditions for PCR3 after closing the tubes with the 8-capstrips for PCR tubes:
| Step no. | Temp. | Time | No. of cycles |
|---|---|---|---|
| 1 | 95°C | 180 s | 1 cycle |
| 2 | 95°C | 30 s | |
| 3 | 66.5°C | 30 s | |
| 4 | 72°C | 60 s | |
| 5 | As in 2, 3, and 4 | 35 cycles (total) | |
| 6 | 72°C | 120 s | 1 cycle |
| 7 | 15°C | Hold |
4.Run samples on 1.5% SB agarose gel (Brody & Kern, 2004) using the following conditions: 30-40 min, 200 V, with a 7-cm running length. Include DNA markers (O'Gene Ruler DNA Ladder Mix). Load per lane: either 6 µl PCR reaction or 4 µl marker.
5.Stain the gel with GelRed working solution for 45 min in a tray.
6.Visualize the gel bands with UV light and document the result by photography.
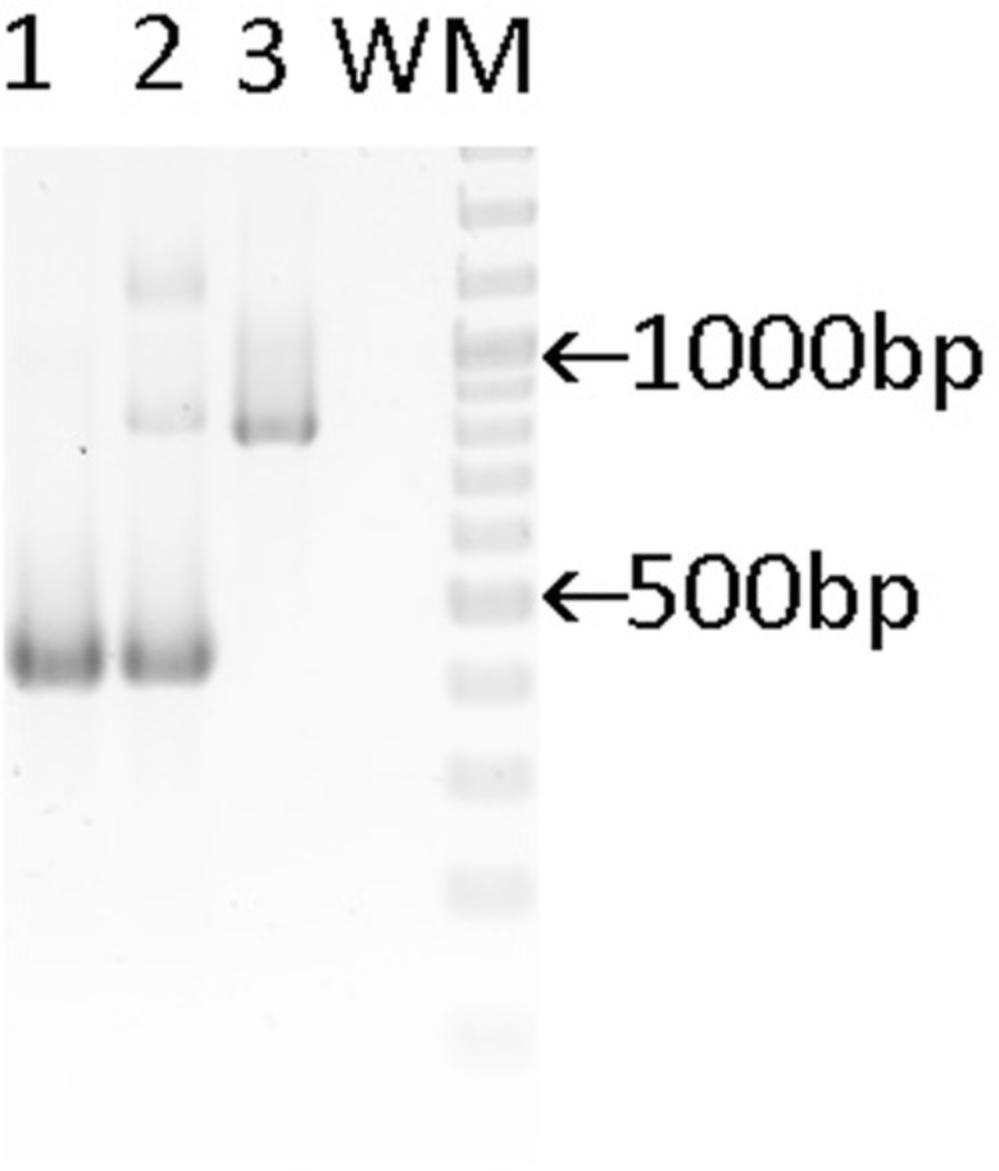
| Genotype | Result PCR3 |
|---|---|
| Lyz2 | |
| +/+ | 429 bp |
| cre/+ | 429 and 800 bp |
| cre/cre | 800 bp |
-
Abbreviations: bp, base pair; Lyz2, lysozyme M.
Basic Protocol 2: GENOTYPING OF FLOXED tdTomato-C3aR AND tdTomato-C5aR2 KNOCKIN MICE
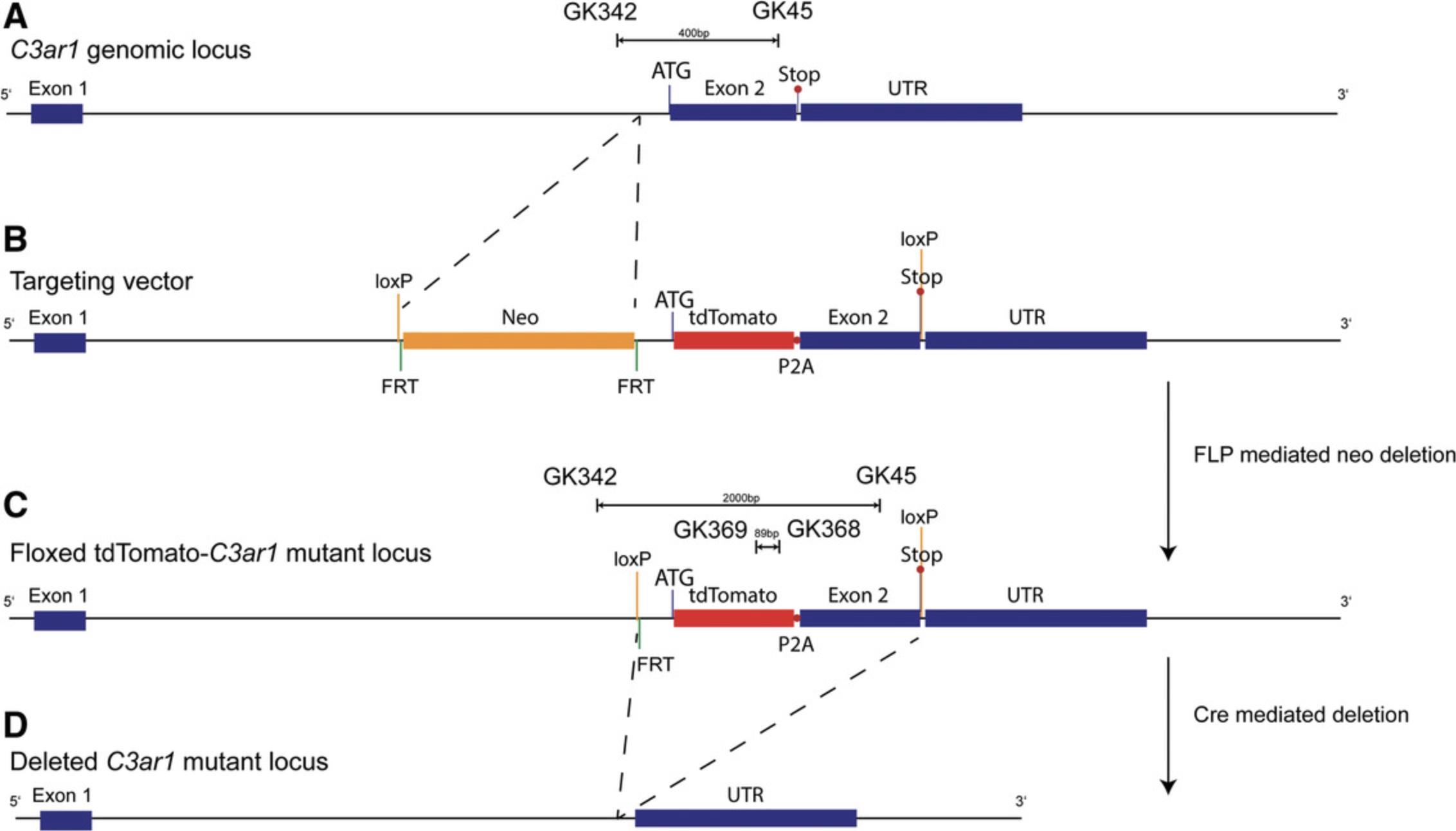
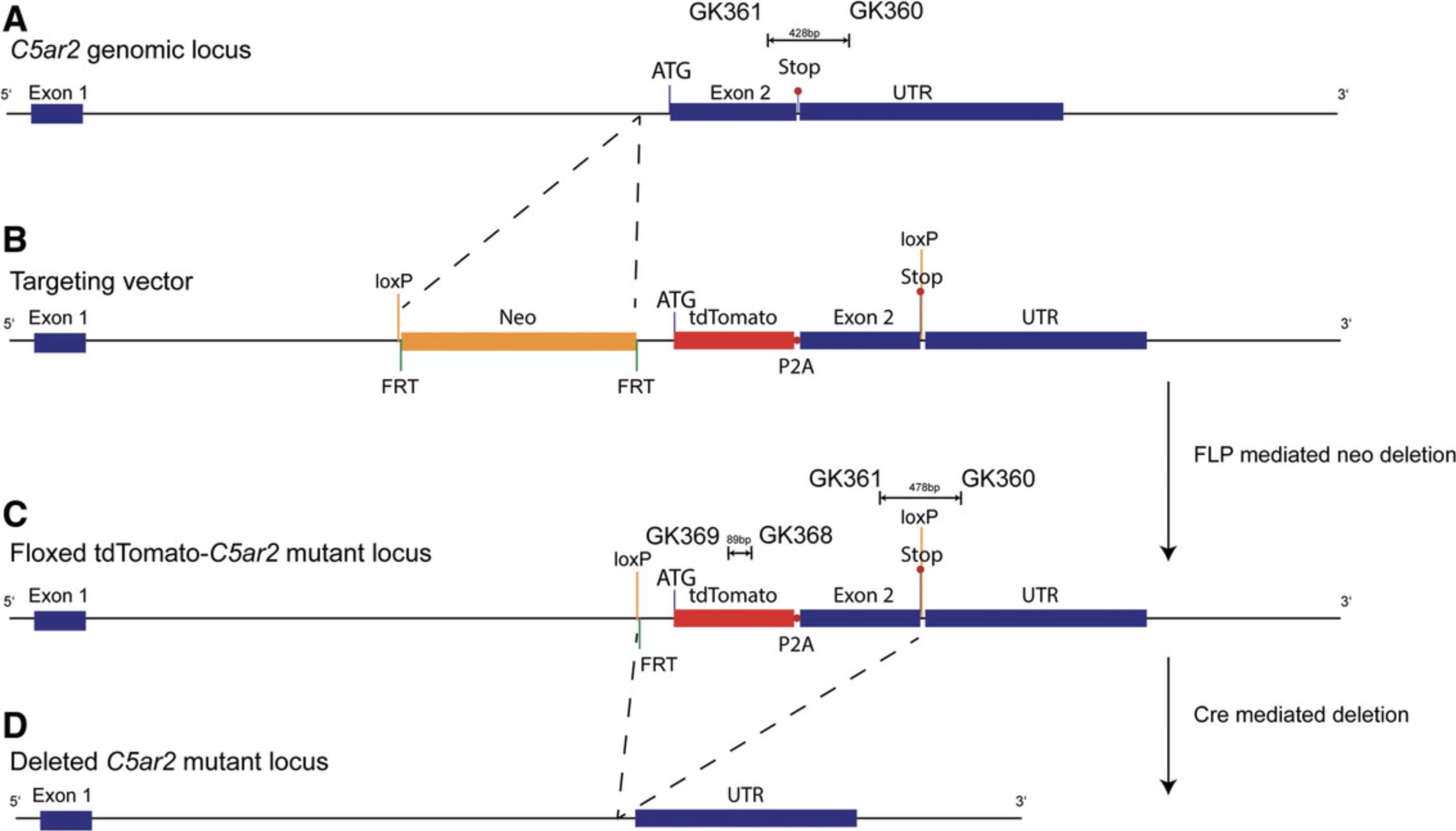
In addition to C5aR1, we have targeted the C3ar1 and C5ar2 genes to generate reporter mice that allow tracking of cellular C3aR and C5aR2 expression and cell-specific deletion. To provide a strong signal with a high stability, in particular upon standard fixation procedures, we used a tandem-dye (td)Tomato instead of GFP (de Felipe et al., 2006). Furthermore, although IRES is a widely used design to produce independent proteins from one mRNA, we and others have found that the translation of the coding sequence under the control of IRES is less efficient than the translation of the main ribosomal entry sequence (de Felipe et al., 2006; Karsten et al., 2015). As an alternative approach, we have used the short, self-cleaving peptide derived from porcine teschovirus-1 (P2A; Kim et al., 2011), which shows high cleavage efficiency and a stoichiometric expression of proteins flanking the 2A peptide (de Felipe et al., 2006; Kim et al., 2011). In the case of the C3ar1 gene, the resulting targeting construct encoded a 6.2-kb 5′ homology arm and a 3.0-kb 3′ homology arm flanking the floxed coding region of exon 2, in which tdTomato_P2A has been inserted in frame with the ATG of exon 2 (Fig. 4). Similarly, we have inserted the tdTomato sequence following the splice acceptor of exon 2 in-frame with the coding sequence of the C5ar2 gene (Fig. 5). In both cases, two loxP sites flank the constructs for potential excision of the C3ar1 or C5ar2 genes by Cre-mediated deletion.
Materials
-
100 µM primers for PCR4-6 (Eurofins Genomics or Life Technologies GmbH)
-
Nuclease-free water (Life Technologies GmbH, cat. no. AM9937)
-
2× KAPA2G Fast Hot Start Genotyping Mix (Sigma Aldrich, cat. no. KK5621; manufactured by Roche)
-
Template DNA: genomic DNA (Support Protocol 2) from floxed tdTomato-C3aR knockin mouse, or -tdTomato-C5aR2 knockin mouse
-
2.5% and 1% SB agarose gels (Brody & Kern, 2004; also see Current Protocols article Voytas, 2001)
-
Sodium borate (SB) buffer (see recipe)
-
O'Gene Ruler DNA Ladder Mix (Thermo Fisher Scientific, cat. no. SM0333)
-
O'Gene Ruler Ladder, 50 bp (Thermo Fisher Scientific, cat. no. SM 0371)
-
GelRed working solution (see recipe)
-
1.5-ml screw-cap microcentrifuge tube with O-ring (Sarstedt, cat. no. 72.692.405)
-
Microcentrifuge
-
0.2-ml 8-tube PCR strips without caps (BioRad (cat. no. TBS0201)
-
8-capstrips for PCR tubes (BioRad, cat. no. TBS0803)
-
Filtered pipet tips (10, 250, and 1000 µl; Sarstedt, cat. no. 701.116.210, 701.189.215, 701.186.210)
-
Microcentrifuge tubes (1.5-ml, Sarstedt, cat. no. 72.690)
-
PCR machine (BioRad C1000 or C1000 Touch Thermal Cycler)
-
Gel staining tray
-
UV transilluminator and camera
-
Additional reagents and equipment for agarose gel electrophoresis (Brody & Kern, 2004; also see Current Protocols article Voytas, 2001)
CAUTION : Remember the proper work safety requirements to protect eyes and skin against UV radiation. Boric acid belongs to the group of carcinogenic, mutagenic, reprotoxic (CMR) substances. Follow the national requirements for disposal and working safety.
NOTE : All chemicals need to be molecular grade (DNase free). For all steps use filtered pipet tips only.
1.Prepare PCR4 primer mix to amplify tdTomato by combining 20 µl of primers GK368/GK369/GK91/GK92 (Table 5) from primer stock (100 µM) in a 1.5-ml screw-cap microcentrifuge tube with O-ring and make up with 120 µl nuclease-free water to a final volume of 200 µl.
| Use in PCR# | Primer/type/target | Sequence | bp | Design |
|---|---|---|---|---|
| 4 | GK368/R/tdTomato | ATGACGGCCATGTTGTTGTC | 20 | Köhl lab |
| 4 | GK369/F/tdTomato | CACCACCTGTTCCTGGGG | 18 | Köhl lab |
| 4 | GK91/F/Tcrd | CAAATGTTGCTTGTCTGGTG | 20 | JAX LAb oIMR8744 |
| 4 | GK92/R/Tcrd | GTCAGTCGAGTGCACAGTTT | 20 | JAX LAb oIMR8745 |
| 5 | GK342/F/C3aR1 | AACAACAGAAGTAGGGAGGTGTAA | 24 | Köhl lab |
| 5 | GK45/R/C3ar1 | TCCCAATAGACAAGTGAGACCAA | 23 |
Harvard primerbank # 6753224a1 |
| 5 | GK190/R/Il2 | CCGTGCTTTCTCTCACATCC | 20 | Köhl lab |
| 5 | GK191/F/Il2 | CGATTACCTCAGTCCCCCTTTAC | 23 | Köhl lab |
| 6 | GK360/F/C5ar2 | TGTCAGCCCGGGACCTTTA | 19 | Köhl lab |
| 6 | GK361/R/C5ar2 | CTTATCACGTCCTGCGGGTAA | 21 | Köhl lab |
| 6 | GK91/F/Tcrd | CAAATGTTGCTTGTCTGGTG | 20 | JAX LAb oIMR8744 |
| 6 | GK92/R/Tcrd | GTCAGTCGAGTGCACAGTTT | 20 | JAX LAb oIMR8745 |
-
Abbreviations: bp, base pair; F, forward; Il2, IL-2 precursor long; R, reverse; Tcrd, T cell receptor delta chain.
2.Prepare PCR5 primer mix to amplify C3ar1 as in step 1 using 20 µl of primers GK342/GK45/GK190/GK191 (Table 5) analogous to step 1.
3.Prepare PCR6 primer mix to amplify C5ar2 as in step 1 using 20 µl of primers GK360/GK361/GK91/GK92 (Table 5) analogous to step 1.
4.Prepare master mixes for PCR 4, 5, and 6 separately by pipetting the following reagents at the indicated volumes into a 1.5-ml microcentrifuge tube (per sample):
- 6.25 µl nuclease-free water
- 1.25 µl primer mix (PCR4, 5 or 6)
- 12.5 µl 2× KAPA2G Fast Hot Start Genotyping Mix.
- Mix and microcentrifuge briefly to bring mixture to bottom of tube.
5.Combine 5 µl template [genomic DNA (Support Protocol 2) from floxed tdTomato-C3aR knockin mouse or -tdTomato-C5aR2 knockin mouse diluted 1:10 with nuclease-free water] or nuclease-free water for control with 20 µl of the respective master mix in a 0.2-ml 8-tube PCR strip without caps and run the following thermocycling conditions for PCR4 after closing the tubes with the 8-capstrips for PCR tubes:
| Step no. | Temp. | Time | No. of cycles |
|---|---|---|---|
| 1 | 95°C | 180 s | 1 cycle |
| 2 | 95°C | 15 s | |
| 3 | 67°C | 15 s | |
| 4 | 72°C | 10 s | |
| 5 | As in 2, 3, and 4 | 35 cycles (total) | |
| 6 | 72°C | 120 s | 1 cycle |
| 7 | 15°C | Hold |
6.Run samples on a 2.5% (PCR5 and 6) or an 1% (PCR4) SB agarose gel (Brody & Kern, 2004; also see Current Protocols article Voytas, 2001). Run SB agarose gels for 30-40 min at 200 V with a 7 cm running length. Include DNA markers (O'Gene Ruler DNA Ladder Mix for PCR 5, and O' Gene Ruler DNA Ladder 50 bp for PCR 4 and 6). Load per lane: either 6 µl PCR reaction or 4 µl marker.
7.Stain the gel with GelRed working solution for 45 min in a tray. Visualize the gel bands with UV light and document the result by photography.
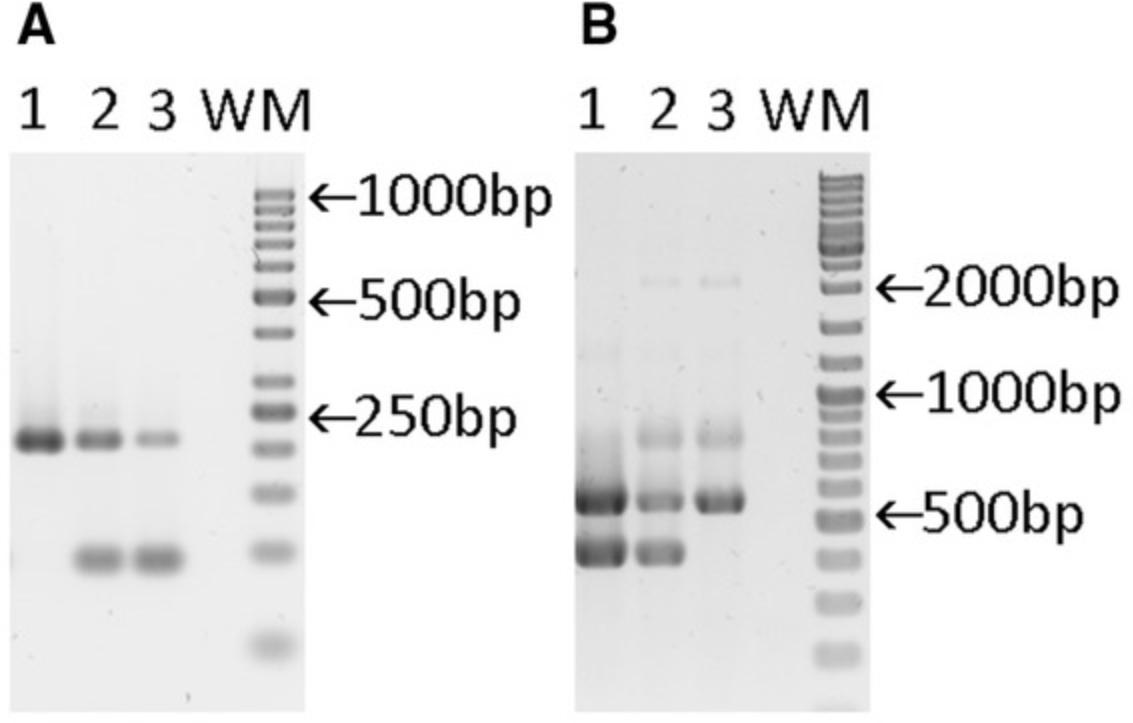
| Result PCR4 | Result PCR5 | |||
|---|---|---|---|---|
| Genotype | Specific:tdTomato | Internal control:Tcrd | Specific:C3ar1 | Internal control:Il2 |
| +/+ | - | 206 bp | 400 bp | 547 bp |
| flx/+ | 89 bp | 206 bp | 400 and 2000 bpa | 547 bp |
| flx/flx | 89 bp | 206 bp | 2000 bpa | 547 bp |
- a
The 2000 bp fragment is very faint and can be even missing.
-
Abbreviations: bp, base pair; Il2, IL-2 precursor long; Tcrd, T cell receptor delta chain.
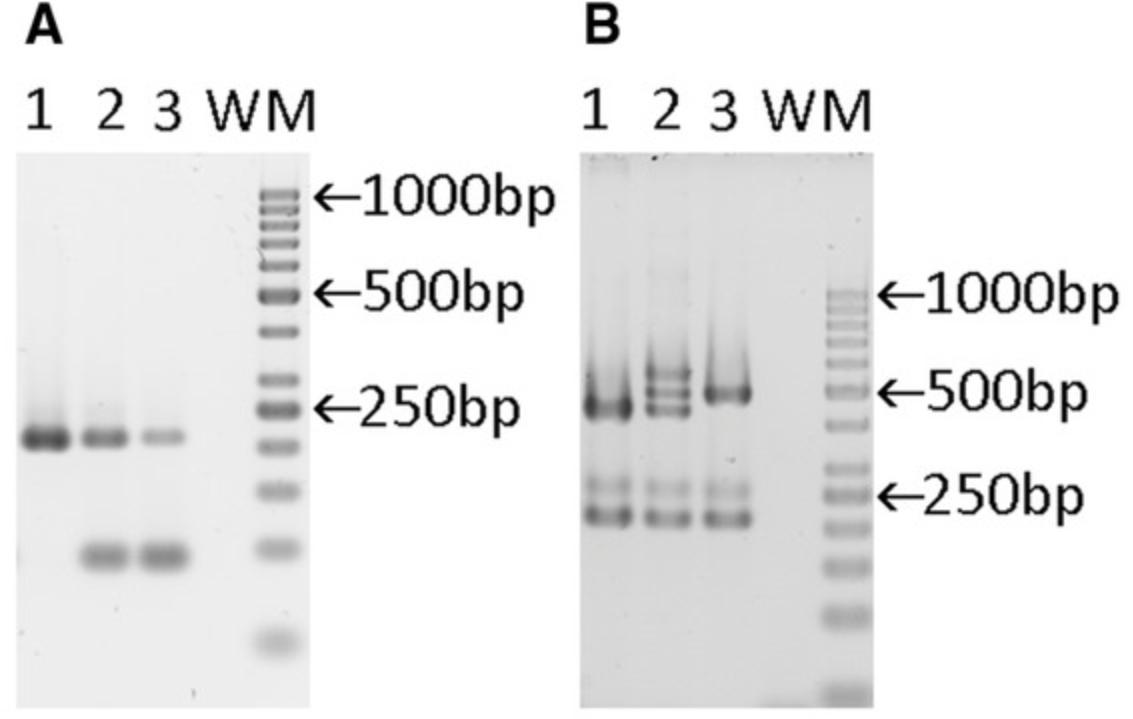
| Result PCR4 | Result PCR6 | |||
|---|---|---|---|---|
| Genotype | Specific:tdTomato | Internal control:Tcrd | Specific:C5ar2 | Internal control:Tcrd |
| +/+ | - | 206 bp | 428 bp | 206 bp |
| flx/+ | 89 bp | 206 bp | 428, 478, and 528 bpa | 206 bp |
| flx/flx | 89 bp | 206 bp | 478 bp | 206 bp |
- a
Heteroduplex formation.
-
Abbreviations: bp, base pair; Tcrd, T cell receptor delta chain.
Support Protocol 2: PREPARATION OF GENOMIC DNA
The prerequisite to amplify the different genomic regions of interest by the PCRs outlined in Basic Protocols 1 and 2 is the appropriate preparation of genomic DNA extracted from a piece of tissue. The most frequently used mouse tissues for genomic DNA preparation are from tail or ear. Our animal facility uses ear punch as the method of identification. Thus, we are using such ear punch biopsies for preparation of genomic DNA as outlined below. Several methods and kits are available to extract DNA from mouse tissue. We routinely use the KAPA Express Extract Kit, which results in high amounts of genomic DNA of good quality.
Materials
-
Ear punch biopsy (max. 2 mm2) from floxed GFP-C5aR1, floxed tdTomato-C5aR2, floxed tdTomato-C3aR knockin or LysMcre-C5ar1 mouse (Köhl laboratory)
-
Lysis master mix (see recipe)
-
UltraPure™ 1 M Tris·HCl, pH 8.0 (Thermo Fisher Scientific Invitrogen, cat. no. 15567027), autoclaved
-
BD Blunt Fill 18-G, 1 ½ in., 1.20 mm × 40 mm, red cannulas, single-use (BD Bioscience, cat. no. 305180)
-
Sterile 0.2-ml PCR tubes with lids (BioRad, cat. no. TFI0201)
-
PCR thermocycler or two independent heating blocks
-
Filtered pipet tips (size 10, 250, and 1000 µl; Sarstedt, cat. no. 701.116.210, 701.189.215, 701.186.210)
-
Microcentrifuge tubes (0.5 ml, 1.5 ml, Sarstedt, cat. no. 172.699, 72.690)
-
Microcentrifuge: e.g, Rotilabo Uni-fuge, Carl Roth GmbH + Co. KG)
NOTE : We recommend using a PCR workstation (e.g., Analytic Jena AG) to avoid cross contamination and a PCR cooler (e.g., Eppendorf AG) to secure the template quality.
1.Transfer biopsy using a Blunt Fill cannula from the tube that was used to collect the specimen from the animal into a 0.2-ml PCR tube with lid under sterile conditions.
2.Add 50 µl lysis master mix to the biopsy, vortex, and spin down in a microcentrifuge at room temperature.
3.Incubate 10 min at 75°C followed by 5 min at 95°C using a cycler or two independent heating blocks.
4.Vortex at full speed for 2-3 s.
5.Centrifuge samples 1 min at 2000 × g , room temperature.
6.Carefully transfer 40 µl supernatant to a 0.5-ml microcentrifuge tube containing 360 µl of 10 mM Tris·HCl (pH 8.0).
7.Use immediately or store at −20°C.
Basic Protocol 3: DETERMINATION OF C5aR1, C5aR2, and C3aR EXPRESSION USING FLOXED AT RECEPTOR REPORTER MICE
Floxed AT receptor reporter mice allow the assessment of AT receptor expression without the need for labeled ligands or antibodies. The GFP and tdTomato reporter genes that are under the control of the promoters of the C5ar1 , C5ar2 , or C3ar1 genes encode for strongly fluorescent proteins that can easily be detected by imaging technologies, including flow cytometry and fluorescence microscopy. As an example, we describe here a protocol to monitor the expression of tdTomato as a surrogate for C3aR expression in peritoneal macrophages. The same procedure can be used to delineate the expression of GFP as a surrogate for C5aR1 and tdTomato as a surrogate for C5aR2 expression, since peritoneal macrophages are known to strongly express all AT receptors (Karsten et al., 2015; Karsten et al., 2017; Quell et al., 2017). Importantly, the genetic association of a reporter gene to AT receptor genes can impact the trafficking of the AT receptors to the cell surface (Dunkelberger, Zhou, Miwa, & Song, 2012). Thus, it is important to also assess the surface expression of ATRs in GFP-C5aR1+, tdTomato-C5aR2+, or tdTomato-C3aR+ cells using AT-receptor-specific antibodies, and control for the specificity using the respective AT receptor−deficient mice. For this purpose, we provide Support Protocol 3, which determines C3aR expression using a C3aR-specific antibody. Finally, Support Protocol 4 outlines the assessment of AT receptor mRNA expression in GFP-C5aR1+, tdTomato-C5aR2+, or tdTomato-C3aR+ cells to verify the AT receptor expression using GFP-C5aR1 mice as an example.
Materials
-
Floxed GFP-C5aR1, floxed tdTomato-C5aR2, or tdTomato-C3aR knock-in mouse (Köhl laboratory); C57BL/6 or BALB/c wild-type mice (The Jackson Laboratory, stock no. 000664 or 000651)
-
Thioglycolate-elicited peritoneal macrophages (see Current Protocols article: Zhang, Goncalves, & Mosser, 2008])
-
Dulbecco's phosphate-buffered saline (D-PBS) without calcium or magnesium, pH 7.0-7.3 (Thermo Fisher Scientific, cat. no. 14190144)
-
Anti-CD16/32 (clone 93; Thermo Fisher Scientific, cat. no. 14-0161-82)
-
Anti-mouse F4/80 BrilliantViolet (BV)421 (Clone BM8; BioLegend, cat. no. 123131)
-
Anti-CD11b-BV510, (clone M1/70; BioLegend; cat. no. 101245)
-
D-PBS/BSA (see recipe)
-
75 × 12−mm polystyrene round-bottom FACS tubes (Sarstedt, cat. no. 50-809-212)
-
Refrigerated centrifuge
-
1.5-ml microcentrifuge tubes (Sarstedt, cat. no. 72.690.001)
-
Flow cytometer
-
Additional reagents and equipment for anesthesia (see Current Protocols article: Donovan & Brown, 2001) and euthanasia (see Current Protocols article: Donovan & Brown, 2006) of mice, and counting cells (see Current Protocols article: Strober, 2001)
1.Sacrifice mice by cervical dislocation (see Current Protocols article: Donovan & Brown, 2006) under anesthesia (see Current Protocols article: Donovan & Brown, 2001).
2.Isolate peritoneal cells from floxed GFP-C5aR1, -tdTomato-C5aR2, -tdTomato-C3aR, C57BL/6, or BALB/c wild-type mice as described in Current Protocols article Zhang et al. (2008).
3.Count cells (see Current Protocols article: Strober, 2001). Adjust the cells to a density of 1 × 107 cells/ml in D-PBS and transfer 100 µl of the cells to a 1.5-ml microcentrifuge tube.
4.Add 1 µl (0.5 µg diluted in D-PBS) of anti-CD16/32 antibody and incubate for 15 min at 4°C
5.Centrifuge the cell suspension 5 min at 500 × g , 4°C, in a refrigerated centrifuge.
6.Discard the supernatant by aspiration.
7.Break up the pellet by gently tapping the bottom of the tube.
8.Prepare an antibody master mix by diluting the anti-F4/80 antibody to a final concentration of 0.5 µg/ml and the anti-CD11b antibody to 0.25 µg/ml in 100 µl D-PBS/BSA.
9.Incubate the cells from each sample with 100 µl of the antibody master mix for 20 min at 4°C.
10.Add 1 ml D-PBS/BSA and centrifuge 5 min at 500 × g , 4°C, in a refrigerated centrifuge.
11.Discard the supernatant by careful aspiration.
12.Resuspend the cells in 300 µl D-PBS/BSA and transfer to a flow cytometer.
13.Plot FSC-H versus FSC-W and gate on singlet cells as shown in Figure 8A. Label the population “singlets.”
![Details are in the caption following the image tdTomato and C3aR expression in thioglycolate-elicited peritoneal macrophages. (A) Contour plot showing the gating strategy to identify thioglycolate-elicited peritoneal macrophages as CD11b<sup>+</sup> and F4/80<sup>+</sup> cells. Peritoneal macrophages stain positive for tdTomato-C3aR (middle panel) and C3aR (right panel) by flow cytometry. The solid black line depicts the signal from the tdTomato-C3aR knockin mice; the dashed line shows the signal obtained from C3ar1<sup>-/-</sup> mice; and the gray histogram shows the signal from wild-type Balb/c mice. (B) Contour plot of wild-type (left panel) and tdTomato-C3ar1<sup>fl/fl</sup> mice showing a tdTomato-C3aR signal only in tdTomato-C3ar1<sup>fl/fl</sup> mice. Part of this figure was originally published in Quell et al. (2017). Copyright © [2017] The American Association of Immunologists, Inc.](https://static.yanyin.tech/literature_test/cpim100-fig-0008-m.jpg)
14.Considering the singlet cells, plot CD11b (on the y axis) versus F4/80 (on the x axis). Gate on the CD11b+F4/80+ cells (Fig. 8A).
15.Record the tdTomato signal in the PE channel (also in case of tdTomato-C5aR2 knockin mice) or the GFP-signal in the FITC channel (in case of floxed GFP-C5aR1 knockin mice) (Fig. 8A).
Support Protocol 3: DETERMINATION OF C3aR EXPRESSION USING A C3aR-SPECIFIC ANTIBODY
The expression of the GFP or tdTomato AT receptor surrogate fluorochrome is not necessarily linked with surface or intracellular expression of the AT receptors. Although our previous results show a strong match of C5aR1 and C3aR surface expression with the GFP or tdTomato signal (Karsten et al., 2015; Quell et al., 2017), we strongly recommend also staining the GFP-C5aR1+, tdTomato-C3aR+, or tdTomato-C5aR2+ cells with AT receptor−specific antibodies to verify AT receptor surface expression. In the case of tdTomato-C5aR2+ cells, we have not yet been able to verify the surface expression with C5aR2-specific antibodies, as all antibodies that we have tested so far have also showed surface staining in C5aR2-deficient mice, suggesting cross-reactivity with other structures (Karsten et al., 2017). We strongly recommend using AT receptor−deficient mice to control for the specificity of a given AT receptor antibody and not to rely only on FMO or IgG isotype controls. Support Protocol 3 outlines an example of C3aR staining using an anti-C3aR antibody and an AF647-labeled secondary antibody, as no conjugated C3aR antibodies are commercially available yet. Alternatively, the anti-C3aR antibody might be conjugated with a fluorochrome by commercially available conjugation kits. For C5aR1 staining, several antibodies conjugated with different fluorochromes are available.
Materials
-
BALB/c wild-type (The Jackson Laboratory, stock no.000664), tdTomato-C3aR (Köhl laboratory), and C3ar1 -/- mice (The Jackson Laboratory, stock no. 005712)
-
Thioglycolate-elicited peritoneal macrophages (see Current Protocols article: Zhang et al., 2008)
-
D-PBS/20% FBS (see recipe)
-
Anti-C3aR antibody (clone 14D4; Hycult Biotech, cat. no. HM1123)
-
Anti−rat F(ab')2-AlexaFluor(AF)647 (clone ab150151; Abcam, cat. no. 150151)
-
Fc blocking buffer (see recipe)
-
Anti−mouse F4/80 BrilliantViolet (BV)421 (Clone BM8; BioLegend, cat. no. 123131)
-
Anti-CD11b-BV510, (clone M1/70; BioLegend, cat. no. 101245)
-
D-PBS/BSA (see recipe)
-
1.5-ml microcentrifuge tubes (Sarstedt, cat. no. 72.690.001)
-
Refrigerated centrifuge
-
75 × 12−mm polystyrene round-bottom FACS tubes (Sarstedt, cat. no. 50-809-212)
-
Flow cytometer
-
Additional reagents and equipment for anesthesia (see Current Protocols article: Donovan & Brown, 2001) and euthanasia (see Current Protocols article: Donovan & Brown, 2006) of mice, and counting cells (see Current Protocols article: Strober, 2001)
1.Sacrifice mice by cervical dislocation (see Current Protocols article: Donovan & Brown, 2006) under anesthesia (see Current Protocols article: Donovan & Brown, 2001).
2.Isolate peritoneal exudate cells from BALB/c wild-type, tdTomato-C3aR1, and C3ar1 -/- mice as described in the Current Protocols article Zhang et al. (2008).
3.Count cells (see Current Protocols article: Strober, 2001). Adjust the cells from each mouse strain to a density of 1 × 107 cells/ml in D-PBS/20% FBS buffer, transfer 100 µl of the cells into a 1.5-ml microcentrifuge tube, and incubate for 30 min at 4°C.
4.Add 2 µl (0.4 µg/ml) of the C3aR antibody to each tube and incubate for 30 min at 4°C.
5.Add 500 µl of D-PBS/20% FBS to each tube and centrifuge the cells 5 min at 500 × g , 4°C.
6.Discard the supernatants by aspiration.
7.Add 1 µl F(ab')2 antibody to 500 µl D-PBS/20% FBS per sample to prepare the F(ab')2 master mix.
8.Resuspend the cell pellet in each microcentrifuge tube with 50 µl of F(ab')2 master mix and incubate for 20 min at 4°C.
9.Add 500 µl D-PBS/20% FBS to each tube and centrifuge 5 min at 500 × g , 4°C, in a refrigerated centrifuge.
10.Discard the supernatant by aspiration and repeat this procedure once.
11.Resuspend the cells in each tube in 100 µl of Fc blocking buffer and incubate for 15 min at 4°C.
12.Prepare an antibody master mix by diluting the anti-F4/80 antibody to a final concentration of 0.5 µg/ml and the anti-CD11b antibody to 0.25 µg/ml in 100 µl of sample volume of D-PBS/BSA.
13.Centrifuge 30 sec at 21,000 × g , 4°C.
14.Discard the supernatant by aspiration.
15.Break up the pellet by tapping the bottom of the tube.
16.Add 100 µl of the CD11b/F4/80 antibody master mix to each tube and incubate the cells for 20 min at 4°C.
17.Add 500 µl D-PBS/BSA to each tube and centrifuge 30 sec at 21,000 × g , 4°C.
18.Discard the supernatant by aspiration.
19.Resuspend the cells for flow cytometric analysis in 300 µl D-PBS/BSA and analyze the sample with a flow cytometer according to the following steps.
20.Gate on the CD11b+F4/80+ cells as described in the Basic Protocol 3, steps 13 to 14 (Fig. 8A).
21.Record the tdTomato-C3aR signal in the PE channel and C3aR-AF647 emission in the APC channel (Fig. 8B).
Support Protocol 4: DETERMINATION OF C5aR1, C5aR2, AND C3aR mRNA EXPRESSION IN FLOXED GFP-C5aR1, -tdTomato-C5aR2, or -tdTomato C3aR−POSITIVE CELLS
In addition to the determination of AT receptor expression using the GFP or tdTomato signal of floxed AT receptor knockin mice or the signal from fluorochrome-labeled AT receptor antibodies, Support Protocol 4 describes an example of the analysis of AT receptor mRNA expression in peritoneal macrophages from floxed GFP-C5aR1 knockin mice. This procedure is useful to verify AT receptor expression when AT receptor antibody staining has revealed cross-reactivity with structures in AT receptor-deficient mice, as is frequently observed with anti-C5aR2 antibodies.
Materials
-
Floxed GFP-C5aR1, floxed tdTomato-C5aR2 or tdTomato-C3aR knockin mice (Köhl laboratory)
-
Thioglycolate-elicited peritoneal macrophages (see Current Protocols article: Zhang et al., 2008)
-
DMEM medium (Thermo Fisher Scientific Gibco, cat. no. 31885-023)
-
Dulbecco's phosphate-buffered saline (D-PBS) without calcium or magnesium, pH 7.0-7.3 (Thermo Fisher Scientific, cat. no. 14190144)
-
RNAeasy Mini Kit (Qiagen, cat. no. 74104)
-
Nuclease-free water (Thermo Fisher Scientific, cat. no. R0581)
-
DNase I buffer (10×) with Mg2+: 100 mM Tris·HCl (pH 7.5 at 25°C)/25 mM MgCl2/1 mM CaCl2 (part of the DNase I kit; Thermo Fisher Scientific, cat. no. EN0525)
-
DNase I (Thermo Fisher Scientific, cat. no. EN0525)
-
50 mM EDTA (part of DNase I kit; Thermo Fisher Scientific, cat. no. EN0525)
-
Oligo(dT)18 primer (Thermo Fisher Scientific, cat. no. S0132)
-
10× dNTP (Thermo Fisher Scientific, cat. no. R0191)
-
RNaseOUT recombinant RNase inhibitor (Thermo Fisher Scientific, cat. no. 10777019)
-
5× RT buffer: 250 mM Tris·HCl (pH 8.3 at 25°C)/250 mM KCl/20 mM MgCl2/50 mM DTT (part of the RevertAid kit; Thermo Fisher Scientific, cat. no. EP0441)
-
RevertAid reverse transcriptase (Thermo Fisher Scientific, cat. no. EP0441)
-
Primers for C5ar1, C5ar2, C3ar1 (100 µM; Eurofins Genomics or Thermo Fisher Scientific)
-
iQ SYBR Green Supermix (BioRad, cat. no. 1708880)
-
1.5% TBE agarose gel (see Current Protocols article: Voytas, 2001)
-
O'Gene Ruler Ladder, 50 bp (Thermo Fisher Scientific, cat. no. SM 0371)
-
24-well plates
-
Cell scraper (Sarstedt, cat. no. 83.1830)
-
1.5-ml microcentrifuge tubes (Sarstedt, cat. no. 72.690.001), autoclaved
-
Refrigerated centrifuge
-
NanoDrop Microvolume Spectrophotometer (Thermo Fisher Scientific, cat. no. ND-2000)
-
Two heat blocks
-
0.2-ml 8-microtube strip without caps: high-profile (BioRad, cat. no. TBS0201) or low-profile (BioRad, cat. no. TLS0801) depending on your thermocycler
-
0.2-ml optical flat PCR-tube 8-cap strips, ultraclear (Bio-Rad, cat. no. TCS0803)
-
1.5-ml screw-cap microcentrifuge tubes with O-rings (Sarstedt, cat. no. 72.692.405)
-
Thermal cycler
-
Additional reagents and equipment for anesthesia (see Current Protocols article: Donovan & Brown, 2001) and euthanasia (see Current Protocols article: Donovan & Brown, 2006) of mice, counting cells (see Current Protocols article: Strober, 2001), and agarose gel electrophoresis (see Current Protocols article: Voytas, 2001)
1.Sacrifice mice by cervical dislocation (see Current Protocols article: Donovan & Brown, 2006) under anesthesia (see Current Protocols article: Donovan & Brown, 2001).
2.Isolate peritoneal cells from floxed GFP-C5aR1, floxed tdTomato-C5aR2 or tdTomato-C3aR as well as C57BL/6 or BALB/c wild-type mice as described in the Current Protocols article Zhang et al. (2008).
3.Count cells (see Current Protocols article: Strober, 2001). Adjust the cells from each mouse strain to a density of 1 × 107 cells/ml in DMEM medium without serum, add 500 µl/well to adherent 24-well-plates, and incubate 3 hr.
4.Discard the supernatant with the non-adherent cells.
5.Carefully wash the plate with 1 ml D-PBS, to remove remaining non-adherent cells.
6.Add 1 ml D-PBS to each well.
7.Harvest the adherent cells using a cell scraper into 1 ml of D-PBS and transfer detached cells into a 1.5-ml microcentrifuge tube.
8.Wash cells three times, each time with 1 ml D-PBS by centrifuging 5 min at 500 × g , 4°C.
9.Discard the supernatant and isolate the RNA using the RNeasy Mini Kit according to the manufacturer's instructions.
10.Determine the concentration of isolated total RNA using a spectrophotometer (NanoDrop or equivalent).
11.Transfer up to 8 µl of total RNA into a fresh autoclaved 1.5-ml microcentrifuge tube
12.Add 1 µl of 10 × DNase I Buffer with Mg2+.
13.Add 1 µl of DNase I and mix by gently pipetting several times up and down.
14.Incubate for 30 min at 37°C in a heat block.
15.Add 1 µl of 50 mM EDTA, transfer to a heat block (65°C), and incubate for 10 min.
16.Spin down for 5 s at 21,000 × g , then add 1.25 µl oligo(dT) primer, 1.25 µl 10× dNTPs, 0.5 µl RNaseOUT, and 5.5 µl nuclease-free water.
17.Heat the sample for 5 min in a heat block (65°C).
18.Place the sample immediately on ice.
19.Spin down 5 s at 21,000 × g , add 5 µl of 5× RT buffer, and mix by tapping the bottom of the tube.
20.Remove 5 µl of the reaction and set aside.
21.Add 0.5 µl of reverse transcriptase to the remaining 19.5 µl.
22.Pipet gently up and down and incubate for 30 min in a heat block (50°C)
23.Heat for 5 min on a heat block (85°C).
24.Spin down 5 s at 21,000 × g.
25.Place 2 µl of cDNA, 2 µl of RT negative control (from step 20), or 2 µl nuclease-free water as PCR negative control in each of three different tubes of a 0.2-ml, 8-microtube strip.
26.Prepare PCR 7 (C5ar1), PCR 8 (C5ar2), and PCR9 (C3ar1) primer mixes by adding 20 µl of each forward and reverse primer combination (Table 8) from their respective primer stocks (100 µM) to a 1.5-ml micro tube and add 160 µl nuclease-free water for a final volume of 200 µl. Store at −20°C.
| Use in PCR# | Primer/type/target | Sequence | bp | Design |
|---|---|---|---|---|
| 7 | GK19/R/C5ar1 | CTGAGTAGAAGTCCTTATATGC | 22 | Köhl lab |
| 7 | GK18/F/C5ar1 | TTCCTGCTGGTGTTCAAG | 18 | Köhl lab |
| 8 | GK82/R/C5ar2 | GCCCCAGGAAGCCAAAGAGGA | 21 | (Atefi et al., 2011) |
| 8 | GK81/F/C5ar2 | CTGGGCCTCTTGCTGACTGTGC | 22 | (Atefi et al., 2011) |
| 9 | GK45/R/C3ar1 | TCCCAATAGACAAGTGAGACCAA | 20 | Harvard primerbank # 6753224a1 |
| 9 | GK44/F/C3ar1 | TCGATGCTGACACCAATTCAA | 20 | Harvard primerbank # 6753224a1 |
-
Abbreviations: bp, base pair; F, forward, R, reverse.
27.Prepare a master mix for these three samples by combining 36.5 µl of iQ SYBR Green Supermix with 3 µl each of C5ar1 (PCR7), C5ar2 (PCR8), or C3ar1 (PCR9) primer mix (step 26) and 28.5 µl nuclease-free water.
28.Add 23 µl of PCR master mix to each of the three tubes.
29.Place the microtube strip into a thermal cycler and run the following thermal cycling conditions:
| Step no. | Temp. | Time | No. of cycles |
|---|---|---|---|
| 1 | 25°C | 120 s | 1 cycle |
| 2 | 53°C | 600 s | |
| 3 | 95°C | 120 s | |
| 4 | 95°C | 15 s | |
| 5 | 55°C | 15 s | |
| 6 | 72°C | 15 s | |
| 7 | As in 4, 5, and 6 | As in 4, 5, and 6 | 35 cycles (total) |
| 8 | 72°C | 120 s | 1 cycle |
| 9 | 15°C | Hold |
30.Run samples on a 1.5% TBE agarose gel for 30-40 min at 200 V with a 7-cm running length. Include DNA markers (O'Gene Ruler DNA Ladder, 50 bp). Load per lane: 6 µl PCR reaction and 4 µl marker.
31.Visualize the gel bands with UV light and document the result by photography.
Basic Protocol 4: ANALYSIS OF C5aR1-DRIVEN ERK1/2 PHOSPHORYLATION IN GFP-C5aR1+ CELLS
C5aR1 is a G protein−coupled receptor (GPCR) that couples preferentially to pertussis toxin (PT)−sensitive G protein G a i2 or G a i3, but can also bind to the PT-insensitive G proteins G a 15 and G a 16 (Klos, Wende, Wareham, & Monk, 2013). Engagement of C5aR1 by its natural ligands C5a and C5a-desArg results in a series of signaling events that involve the activation of PI3K, Akt, and MAPK signaling, resulting in the phosphorylation of the MAPK ERK 1/2, in particular in bone marrow−derived neutrophils. Thus, phosphorylation of ERK1/2 can be used to assess the C5aR1 function in GFP-C5aR1+ cells. As an example, this protocol describes the analysis of C5a-driven ERK1/2 phosphorylation in GFP-C5aR1+ bone marrow−derived neutrophils.
Materials
-
Floxed GFP-C5aR1 knockin mice (Köhl laboratory)
-
Dulbecco's phosphate-buffered saline, no calcium, no magnesium (D-PBS; Thermo Fisher Scientific, cat. no. 14190144)
-
Recombinant human (rh) C5a, (Hycult Biotech, cat. no. HC2101)
-
37% formaldehyde (Sigma Aldrich, cat. no. 252549)
-
Methanol, anhydrous, 99.8%, (Sigma Aldrich, cat. no. 322415)
-
Anti-CD16/32 (clone 93; Thermo Fisher Scientific, cat. no. 14-0161-82)
-
Anti-p-ERK MAPK-APC, Thr202, Tyr204, (clone MILAN8R; Thermo Fisher Scientific, cat. no. 17-9109-42)
-
Anti-Ly6G-eF450 (clone 1A8; Thermo Fisher Scientific, cat. no. 48-9668-82)
-
FACS flow buffer (BD Bioscience, cat. no. 342003)
-
Low-retention tubes (0.5 and 1.5 ml, cat. no. 0030108094, 0030108116)
-
Low-retention pipet tips (10, 200, and 1000 µl; Eppendorf, cat. no. 0030072006, 0030072022, 0030072030)
-
Refrigerated centrifuge
-
Flow cytometer with lasers that can excite the APC and eF450 dyes [e.g., BD™ LSR II equipped with red (640 nm) and violet lasers (405 nm)]
-
Additional reagents and equipment for euthanasia of mice (see Current Protocols article: Donovan & Brown, 2006), isolation of bone marrow cells (see Current Protocols article Swamydas, Luo, Dorf, & Lionakis, 2015), counting cells (see Current Protocols article: Strober, 2001)
1.Sacrifice mice by cervical dislocation (see Current Protocols article: Donovan & Brown, 2006) under anesthesia (see Current Protocols article: Donovan & Brown, 2001).
2.Isolate bone marrow cells as described in the Current Protocols article Swamydas et al. (2015).
3.Count cells (see Current Protocols article: Strober, 2001). Adjust the number of bone marrow cells to 1.0 × 107cells/ml using D-PBS.
4.Add 100 µl of the bone marrow cells to a 0.5-ml low-retention tube and let the cells rest for 5 min at 37°C.
5.Add rhC5a at a final concentration of 10 nM using low-retention pipet tips.
6.Incubate the cells for 5 min at room temperature.
7.Quickly add 4 µl of 37% formaldehyde solution to the 100 µl cell suspension and place cells immediately on ice.
8.Keep the cells for 10 min on ice in the dark.
9.Centrifuge the cells 5 min at 500 × g , 4°C.
10.Aspirate the supernatant and break up the pellet by gently tapping the bottom.
11.Add 500 µl pre-cooled (−20°C) methanol and incubate for 10 min at −20°C to permeabilize the cells.
12.Centrifuge the cells 5 min at 500 × g , 4°C.
13.Aspirate the supernatant.
14.Prepare Fc blocking buffer by diluting anti-CD16/32 antibody to a final concentration of 100 µg/ml in D-PBS.
15.Carefully resuspend the cells in 100 µl Fc blocking buffer and incubate the cells for 15 min at 4°C.
16.Centrifuge the cells 5 min at 500 × g , 4°C.
17.Discard the supernatant.
18.Prepare an antibody master mix by diluting anti-pERK antibody to a final concentration of 10 µg/ml and anti-Ly6G antibody to 5 µg/ml in D-PBS.
19.Incubate the cells with 100 µl of the antibody master mix for 60 min at 4°C.
20.Centrifuge the cells 10 min at 500 × g , 4°C
21.Discard the supernatant.
22.Resuspend the cells in 250 µl FACS flow buffer. Analyze ERK1/2 phosphorylation using an appropriate flow cytometer equipped with lasers that can excite the APC and eF450 dyes [e.g., BD™ LSR II equipped with red (640 nm) and violet lasers (405 nm)].

ASSESSMENT OF C3aR FUNCTIONS IN CELLS OBTAINED FROM FLOXED tdTomato-C3aR KNOCKIN MICE
Engagement of C3aR by C3a on different human cell types including neutrophils, eosinophils, and endothelial cells results in activation of PT-sensitive and PT-insensitive G proteins, as well as a transient increase in intracellular Ca2+ which requires extracellular Ca2+ sources (Klos et al., 2013). Further, C3aR activation in mouse bone marrow−derived macrophages (Cui, Wu, Song, Chen, & Wan, 2019) has resulted in ERK1/2 phosphorylation. In contrast, we found only minor ERK1/2 phosphorylation in peritoneal macrophages that express high levels of C3aR at their surface (Quell et al., 2017), demonstrating that signaling pathways downstream of C3a do not always result in ERK1/2 phosphorylation. Here, we provide two assays that we have used to determine C3aR function in different cell types, i.e., the internalization of the receptor upon C3a ligation and the assessment of changes in intracellular Ca2+ concentration [Ca2+]i. These functions can be determined by flow cytometry and fluorescence microscopy.
Basic Protocol 5: Determination of C3aR Internalization
Materials
-
Floxed tdTomato-C3aR knockin mice (Köhl laboratory)
-
0.5 mg/ml human (h)C3a (Hycult Biotech, cat. no, HC2126)
-
Thioglycolate-elicited peritoneal macrophages (see Current Protocols article: Zhang et al., 2008)
-
Dulbecco's phosphate-buffered saline, no calcium, no magnesium (D-PBS; Thermo Fisher Scientific, cat. no. 14190144)
-
37% formaldehyde, (Sigma Aldrich, cat. no.252549)
-
D-PBS/20% FBS (see recipe)
-
Anti-C3aR antibody (clone 14D4; Hycult Biotech, cat. no. HM1123)
-
Anti−rat F(ab')2-AlexaFluor(AF)647 (clone ab150151; Abcam, cat. no. 150151)
-
Fc blocking buffer (see recipe)
-
Anti-F4/80-BV510 (clone BM8; BioLegend, cat. no,123135)
-
D-PBS/BSA (see recipe)
-
Low-retention tubes (0.5 and 1.5 ml; Eppendorf, cat. no. 0030108094, 0030108116)
-
Low-retention pipet tips (10, 200, and 1000 µl; Eppendorf, cat. no. 0030072006, 0030072022, 0030072030)
-
Heat block
-
1.5-ml microcentrifuge tubes (Sarstedt, cat. no. 72.690.001)
-
Flow cytometer
-
Additional reagents and equipment for anesthesia (see Current Protocols article: Donovan & Brown, 2001) and euthanasia (see Current Protocols article: Donovan & Brown, 2006) of mice, and counting cells (see Current Protocols article: Strober, 2001)
1.Sacrifice mice by cervical dislocation (see Current Protocols article: Donovan & Brown, 2006) under anesthesia (see Current Protocols article: Donovan & Brown, 2001).
2.Isolate peritoneal exudate cells from the peritoneum of BALB/c wild-type and tdTomato-C3aR knockin mice as described in the Current Protocols article Zhang et al. (2008).
3.Count cells (see Current Protocols article: Strober, 2001) Resuspend peritoneal cells from both mouse strains at a density of 1 × 106 cells/ml in D-PBS.
4.Distribute 100 µl of the cell suspension into four low-retention 1.5-ml microcentrifuge tubes.
5.Pre-incubate the cells for 5 min at 37°C in a heat block.
6.Prepare hC3a working solution by adding 1 µl of hC3a (0.45 mg/ml in D-PBS, pH 7.2) to a 1.5-ml low-retention tube containing 1 ml of D-PBS and vortex. From this solution, add 1 µl to 99 µl of D-PBS, resulting in a 0.5 μM hC3a working solution.
7.Add 2 µl of the hC3a working solution to three of the four low-retention 1.5-ml microcentrifuge tubes prepared in step 4 (10 nM final concentration). Add 2 µl D-PBS to one of the four low-retention 1.5-ml microcentrifuge tubes as a control.
8.Gently mix the cells by pipetting up and down and incubate one of three hC3a-stimulated tubes for 1 min, the second one for 3 min, and the third one for 9 min at 37°C in a heat block. Also, incubate the unstimulated control tube for 9 min at 37°C on the heat block.
9.Add 2 µl of 37% formaldehyde and immediately transfer the tubes on to a pre-cooled rack placed on ice and incubate on ice for 30 min.
10.Centrifuge the cells 30 s at 21,000 × g , 4°C.
11.Discard the supernatant by careful aspiration.
12.Resuspend the fixed cells in 100 µl D-PBS/20% FBS and incubate for 30 min at 4°C.
13.Perform steps 4 to 11 of Support Protocol 3.
14.Add F4/80-specific antibody to each microcentrifuge to to reach a final concentration of 0.5 µg/ml, and incubate the cells for 20 min at 4°C.
15.Add 500 µl D-PBS/BSA to each microcentrifuge tube and centrifuge 30 s at 21,000 × g , 4°C.
16.Discard the supernatant by aspiration.
17.Resuspend the cells in each tube in 300 µl of D-PBS/BSA and measure the samples on a flow cytometer.
![Details are in the caption following the image C3a drives rapid C3aR internalization and mobilization of intracellular calcium in peritoneal macrophages from wild-type and tdTomato-C3ar1<sup>fl/fl</sup> mice. (A) Comparison of C3aR surface expression in thioglycolate-elicited F4/80<sup>+</sup> peritoneal macrophages (gating shown in contour blot on the left) before as well as 1, 3, and 9 min after stimulation with 10 nM C3a at 37°C (graph on the right). Shown is the △MFI of C3aR staining normalized to the C3ar1<sup>-/-</sup> peritoneal macrophage equivalents. Values shown are the mean ± SEM; n = 5-6 per group. (B) Microscopic evaluation of the C3a-mediated change of [Ca<sup>2+</sup>]<sub>i</sub> in thioglycolate-elicited peritoneal macrophages from wild-type and tdTomato-C3ar1<sup>fl/fl</sup> mice. Adherent thioglycolate-elicited peritoneal macrophages were loaded with Fluo4-AM and challenged with C3a (37 nM). The images show the fluorescence emission in the FITC (Fluo4 emission) and PE channels (tdTomato), as well as from polarized light before and 6 s after C3a stimulation (×40 objective). Scale bar, 10 μm. Data are representative of three independent experiments. Originally published in Quell et al. (2017). Copyright © [2017] The American Association of Immunologists, Inc.](https://static.yanyin.tech/literature_test/cpim100-fig-0011-m.jpg)
Alternate Protocol: C3a-Induced Increase in Intracellular Ca2+
Additional Materials (also see Basic Protocol 5)
-
Complete RPMI medium (see recipe)
-
5 mM Fluo-4AM stock solution (see recipe)
-
hC3a working solution (see Basic Protocol 5, step 6)
-
Dulbecco's phosphate-buffered saline, calcium, magnesium (D-DBS, with calcium and magnesium; Thermo Fisher Scientific, cat. no. 14040133)
-
Biocoat fibronectin coverslips, 3 cm2 (BD Bioscience, cat. no. 354088)
-
6-well plates (Sarstedt, cat. no. 83.3920.300)
-
Cover glass forceps (Fine Science Tools, cat. no. 11073-10)
-
50-ml centrifuge tube (Sarstedt, cat. no. 62.547.254)
-
Confocal microscope with 40× objective
-
Camera
1.Sacrifice mice by cervical dislocation (see Current Protocols article: Donovan & Brown, 2006) under anesthesia (see Current Protocols article: Donovan & Brown, 2001).
2.Isolate peritoneal exudate cells from the peritoneum of BALB/c wild-type and tdTomato-C3aR knockin mice as described in the Current Protocols article Zhang et al. (2008).
3.Centrifuge the thioglycolate-elicited peritoneal cells 5 min at 350 × g , 4°C.
4.Remove supernatant and resuspend the cells at a density of 1 × 106 cells/ml with pre-warmed complete RPMI 1640 medium at 37°C.
5.Place 3-cm2 circular glass coverslips into wells of a 6-well plate.
6.Add 3 ml of the peritoneal cell suspension to each well and incubate for 2 hr at 37°C in an incubator.
7.Gently swirl the plates and then aspirate medium.
8.Add 3 ml D-PBS, mix, aspirate, and discard the supernatant.
9.Add 500 µl D-PBS to the glass coverslips.
10.Pipet 350 µl of D-PBS into a 0.5-ml microcentrifuge tube. Add 0.7 µl of 5 mM Fluo-4AM stock solution, resulting in a 10 µM working solution.
11.Remove the glass coverslip with forceps and transfer it into an unused well of the 6-well plate.
12.Place 300 µl of the Fluo-4AM working solution on top of the coverslip.
13.Keep the coverslip in the dark at room temperature for 30 min.
14.Take the coverslip out of the 6-well plate using forceps and discard the excess fluid by blotting it on a clean paper towel.
15.Transfer the coverslip into a 50-ml centrifuge tube filled with 40 ml of D-PBS.
16.Transfer the coverslip into a fresh well of the 6-well plate containing 5 ml D-PBS and incubate for 30 min at 37°C in the dark.
17.Take the coverslip out using the forceps and blot the excess fluid again with a paper towel
18.Insert the coverslip into the glass coverslip holder of an immunofluorescence or confocal microscope with a 40× objective, and add 463 µl D-PBS.
19.Set the focus using the polarized filter and analyze your sample in the FITC (Fluo-4AM) and PE channel (tdTomato).
20.Take a picture.
21.Carefully add 37 µl of the hC3a working solution (see step 6 of Basic Protocol 5) to the cells.
22.Record a second picture 6 s after the addition of hC3a.
23.Process and analyze images using the data analysis software of your microscope.
Basic Protocol 6: C5aR2-DRIVEN IFN-γ PRODUCTION FROM NK CELLS
C5aR2 expression frequently overlaps with C5aR1 expression, making it difficult to differentiate between C5aR1 and C5aR2 effects in response to C5a stimulation. One solution to this problem is to use the recently described specific C5aR2 agonist P32 (Croker et al., 2016). Another option is to use cell types that exclusively express C5aR2.Using floxed tdTomato-C5aR2 knockin mice, we uncovered that some naïve NK cells from spleen and blood show this property and express C5aR2 but not C5aR1 (Karsten et al., 2017). Also, naïve B cells from blood and spleen express C5aR2 but not C5aR1.Thus, NK cells are a perfect tool to study the function of C5aR2 independently of C5aR1. NK cells are innate immune cells that exert different effector functions. In addition to their cytotoxic effects critical for killing of tumor cells, they can produce a wide array of cytokines that play important roles in viral infections and the regulation of adaptive immune responses including IFN-γ, TNF-α, and IL-22 (see Current Protocols article: Zamora, Grossenbacher, Aguilar, & Murphy, 2015). In Basic Protocol 6, we therefore describe C5aR2-driven suppression of IL-12/IL-18-induced IFN-γ production using P32 as an example of C5aR2-mediated function in NK cells.
Materials
-
Floxed tdTomato-C5aR2 knockin mice (Köhl laboratory)
-
Splenic NK cells (see Support Protocol 5)
-
Complete RPMI medium (see recipe)
-
IL-12p70 (Peprotech, cat. no. 210-12)
-
IL-18 (BioLegend, cat. no. 767002)
-
Dulbecco's phosphate-buffered saline, no calcium, no magnesium (D-PBS; Thermo Fisher Scientific, cat. no. 14190144)
-
C5aR2-specific agonist P32 (T. M. Woodruff, University of Queensland, Brisbane, Australia) or recombinant human (rh)C5a (Hycult Biotech, cat. no. HC2101)
-
Duo-Set IFN-γ ELISA kit (R&D Systems, cat. no. DY485-05)
-
48-well plate (Sarstedt, cat. no. 83.3923.500)
-
CO2 incubator
-
Additional reagents and equipment for euthanasia of mice (see Current Protocols article: Donovan & Brown, 2006), counting cells (see Current Protocols article: Strober, 2001), and isolation of splenic NK cells (Support Protocol 5)
1.Sacrifice mice by cervical dislocation (see Current Protocols article: Donovan & Brown, 2006) under anesthesia (see Current Protocols article: Donovan & Brown, 2001).
2.Isolate splenic NK cells as described in Support Protocol 5.
3.Count cells (see Current Protocols article: Strober (2001). Adjust the cell number to 5 × 105 cells/ml in complete RPMI medium
4.Transfer 200 µl to a 48-well plate.
5.Prepare IL-12 and IL-18 working solutions by adding 1 µl of the IL-12p70 (10 µg/ml) to 9 µl D-PBS. Prepare IL-18 working solution by adding 1 µl of IL-18 (100 µg/ml) to 99 µl D-PBS.
6.Add 2 µl each of the IL-12 and IL-18 working solution (10 ng/ml each final concentration) or D-PBS (as control) to the cells.
7.Add 4 μl of the specific C5aR2 agonist P32 at 5 mM for a 100 µM final concentration, or an equal amount of D-PBS (as control) to the cells.
8.Incubate cells for 24 hr in a CO2 incubator at 37°C (5% CO2).
9.Collect the supernatant and measure IFN-γ concentration using a Duo-Set IFN-γ ELISA kit according to the manufacturer's instructions.
![Details are in the caption following the image C5aR2 controls IL12-/IL-18-induced IFN-γ production from NK cells: (A) IFN-γ production from sorted splenic NK cells of wild-type and C5ar2<sup>−/−</sup> mice 24 hr after stimulation with IL-12/IL-18. (B) IFN-γ production from sorted wild-type splenic NK cells 24 hr after stimulation with IL-12/IL-18 in the presence or the absence of the C5aR2 agonist P32. Shown is the relative decrease in IL-12/IL-18–mediated IFN-γ production in response to C5aR2 stimulation by the C5aR2 agonist P32. Originally published in Karsten et al. (2017). Copyright © [2017] The American Association of Immunologists, Inc.](https://static.yanyin.tech/literature_test/cpim100-fig-0012-m.jpg)
Support Protocol 5: ISOLATION OF SPLENIC NK CELLS BY FACS
Several methods have been published to isolate NK cells from spleen, some of which use selected mAbs to deplete contaminating cells such as T, B, dendritic cells, granulocytes, and monocytes by magnetic separation (see Current Protocols article: Pak-Wittel, Piersma, Plougastel, Poursine-Laurent, & Yokoyama, 2014). The advantage of magnetic isolation is that it is fast, easy to carry out, and cost-effective, and still results in a purity of up to 95%. However, to reach higher purities (up to 99%), we describe a protocol below for positive selection by FACS.
Materials
-
Floxed tdTomato-C5aR2 knockin mice (Köhl laboratory)
-
Dulbecco's phosphate-buffered saline (D-PBS) without calcium or magnesium, pH 7.0-7.3 (Thermo Fisher Scientific, cat. no. 14190144)
-
Red blood cell lysis (RBCL) buffer (see recipe)
-
Anti−mouse CD16/32 (Clone 93; Thermo Fisher Scientific, cat. no. 16-0161-82)
-
Live/Dead Staining Viability kit eFluor™ 780 for flow cytometry (Thermo Fisher Scientific, cat. no. 65-0865-18)
-
Anti-CD3e-PerCP-Cy5.5 (clone 145-2C11; BD Bioscience, cat. no. 551163)
-
Anti-NKp46-eFluor450 (clone 29A1.4; Thermo Fisher, cat. no. 48-3351-83)
-
Anti-NK1.1-AF700 (clone PK136; BioLegend, cat. no. 108730)
-
D-PBS/BSA (see recipe)
-
1.5-ml microcentrifuge tubes (Sarstedt, cat. no. 72.690.001)
-
Nylon cell strainer, 40-µm pore size (BD Bioscience, cat. no. 352340)
-
50-ml tube (Sarstedt AD & Co., cat. no. 62.547.254)
-
Refrigerated centrifuge
-
Cell sorter
-
Additional reagents and equipment for anesthesia (see Current Protocols article: Donovan & Brown, 2001) and euthanasia (see Current Protocols article: Donovan & Brown, 2006) of mice, removal of spleen (see Current Protocols article: Reeves, Reeves, & Chin, 2001), and counting cells (see Current Protocols article: Strober, 2001)
1.Sacrifice mice by cervical dislocation (see Current Protocols article: Donovan & Brown, 2006) under anesthesia (see Current Protocols article: Donovan & Brown, 2001).
2.Remove the spleen (see Current Protocols article: Reeves et al., 2001), transfer it into a 1.5-ml tube filled with ice-cold D-PBS, and store it on ice.
3.Pass the cells through a nylon cell strainer (40-µm pore size) into a 50-ml tube using a syringe plunger to obtain a single-cell suspension.
4.Add 15 ml of D-PBS to wash the cells and centrifuge 5 min at 400 × g , 4°C.
5.Add 3 ml of RBCL and incubate for 3 min at room temperature.
6.Add 37 ml of D-PBS and centrifuge cells 5 min at 400 × g , 4°C.
7.Resuspend pellet in 30 ml D-PBS.
8.Prepare a 10-µl aliquot to count the cell number.
9.Centrifuge the cells 5 min at 400 × g , 4°C.
10.Adjust the cell number to 1 × 108 cells/ml with D-PBS.
11.Add 3.33 µl of anti-CD16/CD32 antibody per ml of cells and incubate for 15 min at room temperature.
12.Prepare Live/Dead staining working solution (50 µM) by adding 10 µl Live/Dead stain from the kit to 10 µl D-PBS
13.Add 2 µl of the Live/Dead working solution per ml cell suspension.
14.Add 2.5 µl of each of the following antibodies per ml of cells:
- CD3e (1.25 µg/ml)
- NKp46 (0.5 µg/ml)
- NK1.1 (1.25 µg/ml).
15.Incubate cells for 15 min at room temperature in the dark.
16.Add 0.5 ml of D-PBS and centrifuge cells for 5 min at 400 × g , 4°C.
17.Resuspend cells in 350 µl D-PBS.
18.Run the cells on a cell sorter using the gating strategy outlined in Figure 13
![Details are in the caption following the image Gating strategy to purify NK cells from the spleen by FACS. (A) Contour plot showing the gate of spleen cells to exclude cell debris. (B-C) Contour plots showing the gates to exclude cell doublets. (D) Contour and dot plots showing Live/Dead staining to define living cells. (E) Contour and dot plots showing the gating strategy to purify NK cells as NK1.1<sup>+</sup>NKp46<sup>+</sup> cells (left) or NK1.1<sup>+</sup> CDe3<sup>-</sup> cells (right). Originally published in Karsten et al. (2017). Copyright © [2017] The American Association of Immunologists, Inc.](https://static.yanyin.tech/literature_test/cpim100-fig-0013-m.jpg)
19.Continue with Basic Protocol 6.
REAGENTS AND SOLUTIONS
NOTE: For all buffers use at least ultrapure water; for PCR related-buffers always use nuclease-free ultrapure water.
Complete RPMI 1640 medium
- Supplement RPMI 1640 medium (Gibco, cat. no. 42401042) medium with:
- 10% fetal bovine serum (FBS; PAA, cat. no. A15-043), heat inactivated 30 min at 56°C (stored in 50-ml aliquots)
- 100 U/ml penicillin, 100 µg/ml streptomycin (add from penicillin/streptomycin; Gibco, cat. no. 15-140-122)
- 2 mM L-glutamine (Thermo Fisher Scientific Gibco, cat. no. 25-030-081)
- Store up to 1 month at 4°C
D-PBS/BSA
Supplement 47.5 ml of D-PBS (Thermo Fisher Scientific, cat. no. 14190144) with 2.5 ml MACS BSA stock solution (20× concentrated, 10% solution; Milteny Biotec GmbH, cat. no. 130-091-376) in a 50-ml tube. Store up to 1 day at 4°C.
D-PBS/20% FBS
Supplement 40 ml of D-PBS (Thermo Fisher Scientific, cat. no. 14190144) with 10 ml fetal bovine serum (FBS; PAA, cat. no. A15-043; heat-inactivated 30 min at 56°C) in a 50-ml tube. Store up to 1 day at 4°C.
Fc block buffer
Dilute anti-CD16/32 antibody to a final concentration of 100 µg/ml in D-PBS/BSA (see recipe).
Fluo-4AM stock solution (5 mM)
Add 13.6 µl DMSO to a 50 µg vial of Fluo4AM (Thermo Fisher Scientific cat. no. F14201).
GelRed working solution
Add 150 µl 10,000× GelRed™ Nucleic Acid Stain (Biotrend, cat. no. 41003) to 500 ml SB working buffer (see recipe). Store up to 1 week at room temperature.
Lysis master mix
Combine 44 µl nuclease-free water (Life Technologies GmbH, cat. no. AM9937), 5 µl 10× KAPA Express Extract Buffer, and 1 µl 1 U KAPA Express Extract Enzyme from the KAPA Express Extract Kit (Sigma Aldrich, cat.no. KK7100; manufactured by Roche). Prepare immediately before use.
Red blood cell lysis (RBCL) buffer
Prepare in distilled H2O:
- 155 mM NH4Cl
- 10 mM KHCO3
- 0.1 mM EDTA
- pH 7.2
- Store up to 6 months at room temperature
Sodium borate (SB) buffer
Stock (200 mM, pH 8.5) : Dissolve 8 g NaOH per L distilled H2O and add solid H3BO3 to adjust pH to 8.5. Store up to 6 months at room temperature.
SB working buffer (10 mM; pH 8.5) : Dilute SB stock 1:20 with distilled H2O. Store up to 1 week at room temperature.
COMMENTARY
Background Information
The ATs were first identified more than 50 years ago (Cochrane & Muller-Eberhard, 1968). For many years, before the age of molecular biology, AT receptor expression was determined by biochemical methods through binding studies with radiolabeled ligands (Huey & Hugli, 1985). With the rise of molecular biology, human (Gerard & Gerard, 1991) and mouse C5aR1 (Gerard et al., 1992), human (Crass et al., 1996) and mouse C3aR (Hsu et al., 1997) and eventually, C5aR2, initially named C5L2 or GPR77 (Okinaga et al., 2003), were cloned, which allowed the genetic targeting of the AT receptors. Since then, considerable work has been done to develop new tools to track, delineate, and/or block the functions of AT receptors in mice, humans, and several other species including rats, guinea pigs, and even fish (trout) or amphibia (axolotl). Among the existing tools, the generation of AT receptor knock-out mouse strains has been one of the most important achievements for studying the role of AT receptors in steady state and under pathophysiological conditions. In the past decades, several laboratories have generated such mice, which are now broadly available and used by the scientific community. Another important advancement was the generation of AT receptor−specific antibodies that helped to identify AT receptor−expressing cells by immuno(histo)chemical methods or by flow cytometry and associated techniques (e.g., live stream imaging). However, controversial results have been obtained using antibodies to determine AT receptor expression, in particular by immunohistochemical methods (Drouin et al., 2001; Quell et al., 2017; Tschernig, Kiafard, Dibbert, Neumann, & Zwirner, 2007). Monitoring mRNA expression of AT receptors in the cells of interest in addition to antibody staining, either by northern blot or by PCR, can help to test the results obtained with antibody staining. However, mRNA expression of AT receptors does not necessarily match protein expression. Furthermore, the structure of the AT receptor genes, consisting of 2 exons with most of the coding sequence located in exon 2, strengthen the need for appropriate controls to prevent unspecific amplification of genomic DNA by PCR or recognition by radioactive probes via blot hybridization.
Recently, our laboratory has developed reporter mouse strains for C5aR1, C5aR2, and C3aR to overcome these inherent problems and ease expression studies of AT receptors. During the past few years, we generated floxed GFP-C5aR1- (Karsten et al., 2015), tdTomato-C3aR- (Quell et al., 2017), and tdTomato-C5aR2-knockin mice (Karsten et al., 2017), which allowed ourselves and more than 30 academic laboratories in the U.S., Europe, and Australia to re-evaluate AT receptor expression in many tissues under homeostatic and multiple disease conditions. Further, many conditional AT receptor knockout strains have already been generated or are currently being developed targeting several professional and non-professional immune cells. We expect that these mice will further broaden our understanding of the multiple roles of the AT receptors in allergy, autoimmunity, infection, cancer, metabolic disease, transplantation, ischemia reperfusion injury, hypertonia and several other diseases or disorders.
Critical Parameters
Working with anaphylatoxins
One critical issue in handling ATs is their tendency to bind to plastic surfaces such as polypropylene walls of microcentrifuge tubes due to their basic nature. To avoid absorption to plastic surfaces, use low-retention tubes as well as low-retention pipet tips whenever you use pure AT solutions. To achieve this, several companies provide special tubes with a highly polished surface. Further, it is important to avoid repeated freezing/thawing processes.
Analysis of C5aR1-driven ERK1/2 phosphorylation in GFP-C5aR1+ cells
For staining of intracellular proteins using methanol-mediated fixation and permeabilization, the centrifugation steps and the handling of cells after treatment are critical. Methanol treatment makes the cells very fragile and sensitive to centrifugal forces. Thus, handle cells with care during washing and in particular the centrifugation steps. Because methanol-treated cells change buoyancy, the pellet is very fragile, unstable and difficult to spin down. Do not exceed 500 × g in your centrifugation steps.
C3a-induced increase in intracellular Ca2+
Depending on the cell type, you have to consider use of D-PBS supplemented with Ca2+ when measuring C3a-induced increase in [Ca2+]i. In contrast to C5aR1 activation, which mobilizes intracellular Ca2+ stores of the endoplasmic reticulum (Norgauer et al., 1993), C3aR pathway activation requires uptake of extracellular calcium in human neutrophils (Norgauer et al., 1993). In some cells, C3aR activation drives the mobilization of intracellular calcium from intracellular stores as in transfected cell lines (Chao et al., 1999) or astrocytes (Sayah et al., 2003). When the C3aR signaling pathway has not been evaluated yet in the cell type under investigation, we recommend using D-PBS supplemented with Ca2+.
Troubleshooting
Determination of AT receptor expression using antibodies
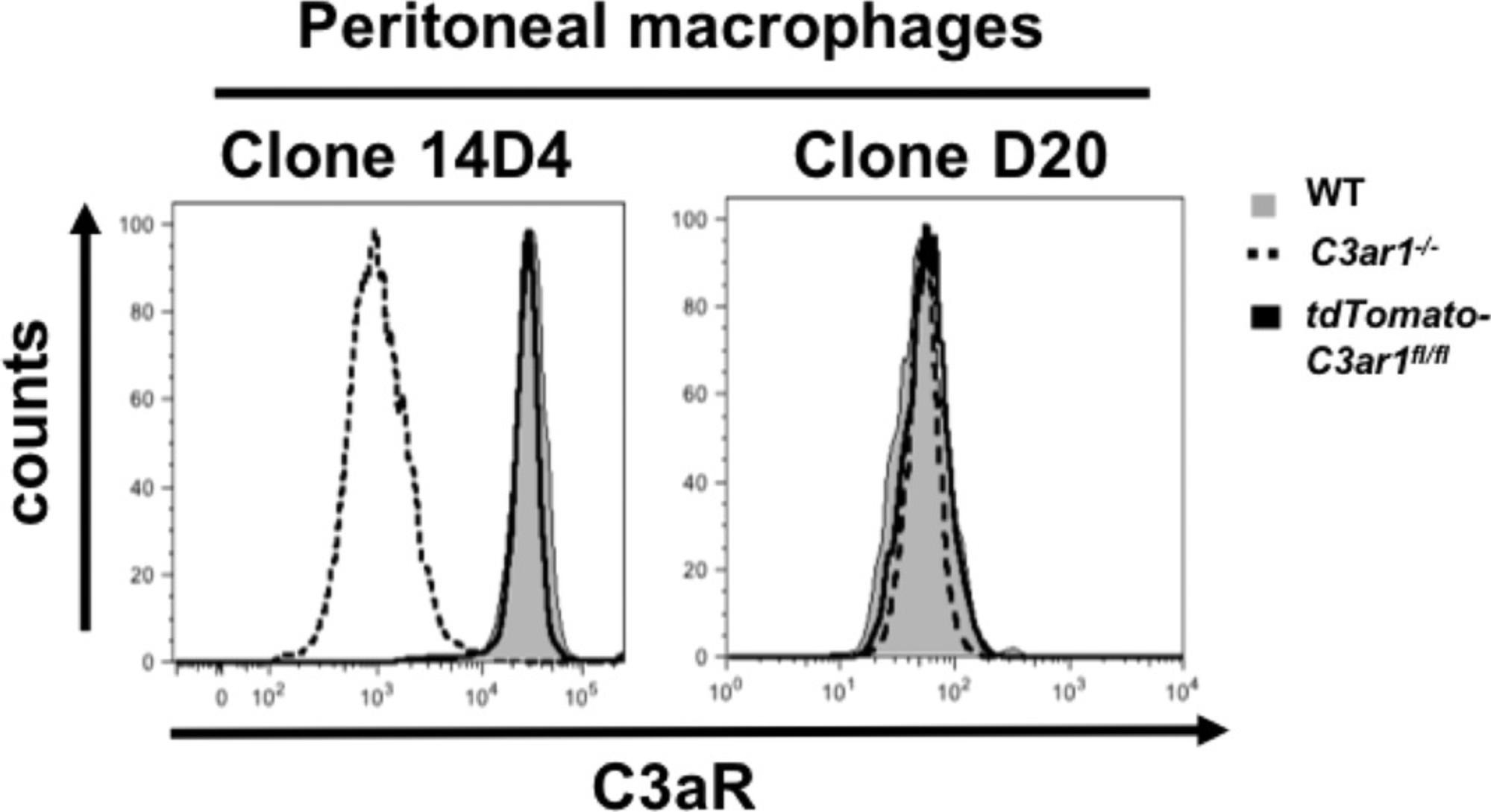
Several polyclonal and monoclonal antibodies have been reported that have been used to determine C5aR1, C5aR2, or C3aR expression in immune and tissue cells (Laumonnier et al., 2017). Unfortunately, many of the available AT receptor antibodies not only recognize the AT receptors, but can also bind additional structures and/or epitopes (Karsten et al., 2017). Thus, we recommend using AT receptor−deficient mice or additional PCR measurements to control for the specificity of a given AT receptor antibody staining, and not rely solely on fluorescence minus one (FMO) controls. In our hands, C5aR1 mAb 20/70 shows high specificity for C5aR1, and frequently matches results obtained with GFP-C5aR1 reporter mice. For C5aR2, mAb 468705 is widely used, but we found that this mAb not only stained bone marrow-derived neutrophils and macrophages as well as peritoneal macrophages from the tdTomato-C5ar2fl/fl mice, but also those from C5ar2-/- , suggesting that it cross reacts with other structures on these cells (Karsten et al., 2017). However, this antibody was found to specifically stain C5aR2 in endothelial cells (Miyabe, Miyabe, Mani, Mempel, & Luster, 2019), indicating that the specificity of this antibody is dependent on the cell type used. Regarding C3aR, we found that the anti-C3aR clone 14D4 matched the results obtained with floxed tdTomato-C3aR knock-in mice. In contrast, we found positive staining in different immune cells from C3ar1-/- mice using antibodies widely used in the past to stain C3aR, i.e., the D12, D20, and H300 clones (Quell et al., 2017) (Fig. 14).
Impact of fixatives on fluorochrome stability
Several fluorochromes (such as PE, PerCP), their respective tandem dyes (such as PE-Cy7 or PercP-Cy5.5), and to a certain extent APC, are sensitive to methanol treatment and lose their structural integrity and hence their capacity to emit light. Because of their instability, we do not recommend use of such dyes prior to methanol treatment but advise the reader to consider alternative fluorochromes for Basic Protocol 4. APC might be used when the expression of the target molecule/receptor is high.
Understanding Results
Genotyping of floxed AT receptor reporter mice
For good lab practice, include water controls, internal controls, i.e., primers targeting the wild-type gene in the PCR reaction and run reference lanes for wild-type, heterozygous, and homozygous genotypes with each experiment, as shown in the examples in Figures 2, 3, 6 and 7.
Strength of the reporter molecules
Due to the fact that the GFP protein emits quite strongly, the signal can be easily observed in cells expressing high levels of C5aR1 such as neutrophils using either homozygous or heterozygous C5aR1 reporter mice. The use of heterozygous mice should be preferred whenever the full functionality of the AT receptor is required, since we have shown previously that, although still functional, homozygous cells react somewhat more weakly to C5a than wild-type or heterozygous cells (Karsten et al., 2017). Interestingly, heterozygous animals can also be used to investigate the expression of C5aR1 in cells with high autofluorescence, such as alveolar macrophages (Fig. 15). Although very useful, a couple of things have to be considered regarding GFP as a reporter fluorochrome. It is well appreciated that it exerts high immunogenicity and cytotoxicity, and there is phototoxicity associated with the blue light used to excite GFP (Ansari et al., 2016). Also, GFP is sensitive to fixation by formaldehyde or ethanol dehydration (Jockusch, Voigt, & Eberhard, 2003). In contrast, the tdTomato protein is more stable in this regard (Shaner, Steinbach, & Tsien, 2005). It is well suited for immunofluorescence microscopy (Fig. 11B, [Morris, Klanke, Lang, Lim, & Crombleholme, 2010], [Quell et al., 2017]) without extensive optimization (Zhanmu et al., 2019). Furthermore, tdTomato emits more strongly than most other chromophores (Shaner et al., 2004; Shaner et al., 2005) making it a very interesting surrogate for cells with minor expression of the target molecule.
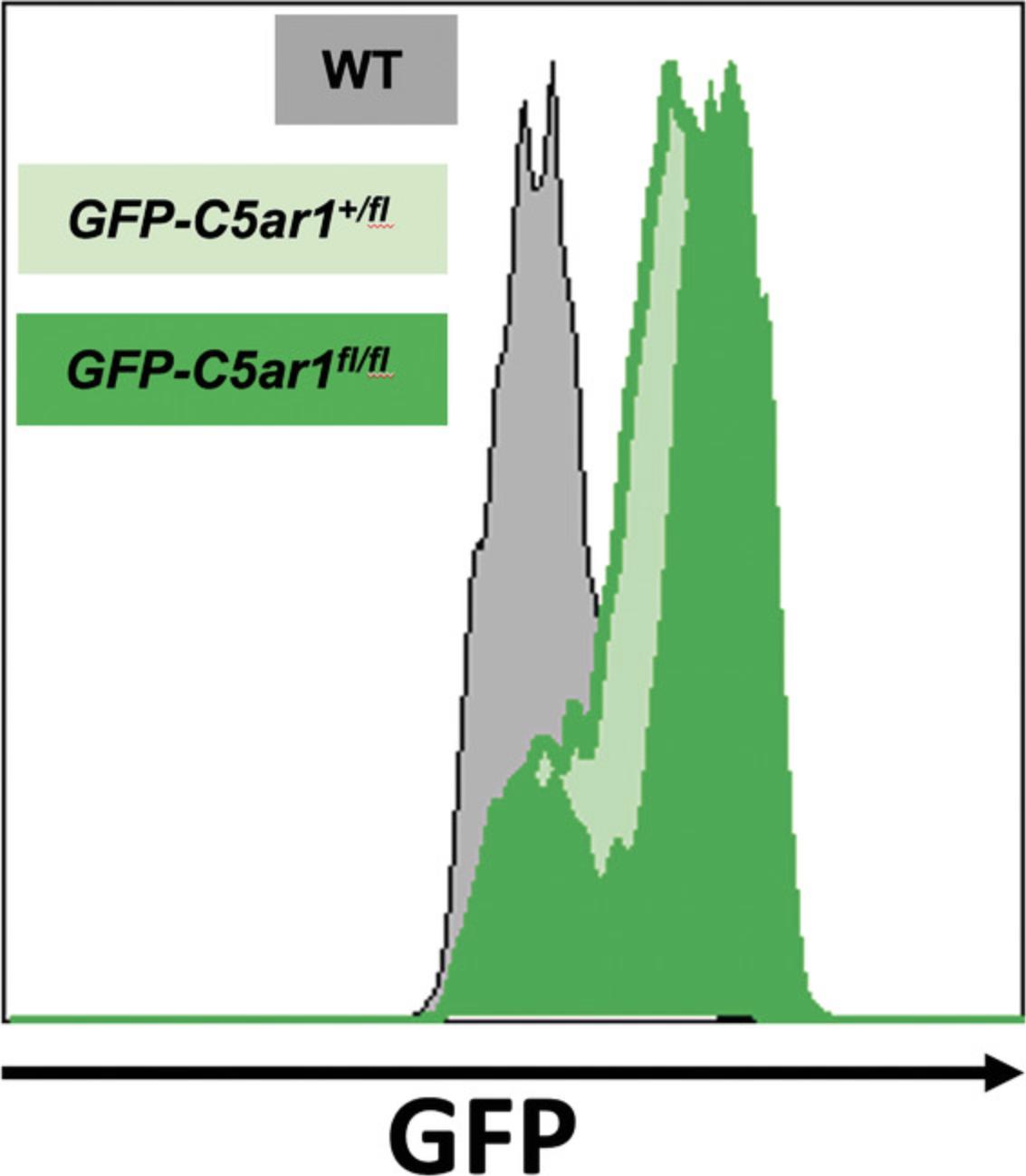
Flow cytometer settings used with AT receptor reporter mice
GFP is excited by the blue laser (488 nm) with an emission maximum at 509 nm. Therefore, GFP is typically detected in the FITC channel (530/25). Using a narrower-bandpass filter (510/12) that is closer to the emission maximum can improve the detection of dimly expressed GFP. Further, tdTomato is best excited by the yellow-green laser (561 nm), and is typically detected in the PE channel using a 585/15 bandpass filter. Consequently, antibodies labeled with fluorochromes that are detected in the FITC channel cannot be used with the GFP-C5aR1 reporter mouse, nor can antibodies labeled with fluorochromes detected in the PE channel with tdTomato-C3aR and tdTomato-C5aR2. Depending on the signal intensity of the fluorescent proteins, it is recommended to minimize the spillover into their detection channels. Autofluorescence from cells like macrophages, due to their high metabolic activity involving NADPH, might be a source of false positive signals in GFP and tdTomato detection channels. Therefore, when working with highly autofluorescent cells (like macrophages), fluorochromes emitting in the FITC or PE channel, but also emitting in the violet channels (BV421, 450, and 510), should be used with care.
Acknowledgments
Open access funding enabled and organized by Projekt DEAL. This work was supported by funds from the German Research Foundation (DFG) for the International Research Training Group (IRTG) 1911 (projects A1, A7, B1, and B2 to Y.L., C.M.K., and J.K.), as well as the EXC 306 and EXC 2167 to J.K. We thank Katharina Quell and Katharina Bröker for help in establishing the functional AT receptor assays and T. Vollbrandt for his comments regarding the flow cytometer settings.
Literature Cited
- Ansari, A. M., Ahmed, A. K., Matsangos, A. E., Lay, F., Born, L. J., Marti, G., & Sun, Z. (2016). Cellular GFP toxicity and immunogenicity: Potential confounders in in vivo cell tracking experiments. Stem Cell Reviews and Reports , 12, 553–559. doi: 10.1007/s12015-016-9670-8.
- Atefi, G., Zetoune, F. S., Herron, T. J., Jalife, J., Bosmann, M., Al-Aref, R., & Ward, P. A. (2011). Complement dependency of cardiomyocyte release of mediators during sepsis. FASEB Journal , 25, 2500–2508. doi: 10.1096/fj.11-183236.
- Brody, J. R., & Kern, S. E. (2004). Sodium boric acid: A Tris-free, cooler conductive medium for DNA electrophoresis. Biotechniques , 36, 214–216. doi: 10.2144/04362BM02.
- Chao, T. H., Ember, J. A., Wang, M., Bayon, Y., Hugli, T. E., & Ye, R. D. (1999). Role of the second extracellular loop of human C3a receptor in agonist binding and receptor function. Journal of Biological Chemistry , 274, 9721–9728. doi: 10.1074/jbc.274.14.9721.
- Clausen, B. E., Burkhardt, C., Reith, W., Renkawitz, R., & Forster, I. (1999). Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Research , 8, 265–277. doi: 10.1023/a:1008942828960.
- Cochrane, C. G., & Muller-Eberhard, H. J. (1968). The derivation of two distinct anaphylatoxin activities from the third and fifth components of human complement. Journal of Experimental Medicine , 127, 371–386. doi: 10.1084/jem.127.2.371.
- Crass, T., Raffetseder, U., Martin, U., Grove, M., Klos, A., Köhl, J., & Bautsch, W. (1996). Expression cloning of the human C3a anaphylatoxin receptor (C3aR) from differentiated U-937 cells. European Journal of Immunology , 26, 1944–1950. doi: 10.1002/eji.1830260840.
- Croker, D. E., Monk, P. N., Halai, R., Kaeslin, G., Schofield, Z., Wu, M. C., & Woodruff, T. M. (2016). Discovery of functionally selective C5aR2 ligands: Novel modulators of C5a signalling. Immunology and Cell Biology , 94, 787–795. doi: 10.1038/icb.2016.43.
- Cui, J., Wu, X., Song, Y., Chen, Y., & Wan, J. (2019). Complement C3 exacerbates renal interstitial fibrosis by facilitating the M1 macrophage phenotype in a mouse model of unilateral ureteral obstruction. American Journal of Physiology: Renal Physiology , 317, F1171–F1182. doi: 10.1152/ajprenal.00165.2019.
- Daley, J. M., Thomay, A. A., Connolly, M. D., Reichner, J. S., & Albina, J. E. (2008). Use of Ly6G- specific monoclonal antibody to deplete neutrophils in mice. Journal of Leukocyte Biology , 83, 64–70. doi: 10.1189/jlb.0407247.
- de Felipe, P., Luke, G. A., Hughes, L. E., Gani, D., Halpin, C., & Ryan, M. D. (2006). E unum pluribus: Multiple proteins from a self-processing polyprotein. Trends in Biotechnology , 24, 68–75. doi: 10.1016/j.tibtech.2005.12.006.
- Donovan, J., & Brown, P. (2001). Anesthesia. Current Protocols in Immunology , 27, 1.4.1-1.4.4. doi: 10.1002/0471142735.im0104s27.
- Donovan, J., & Brown, P. (2006). Euthanasia. Current Protocols in Immunology , 73, 1.8.1-1.8.5. doi: 10.1002/0471142735.im0108s73.
- Drouin, S. M., Kildsgaard, J., Haviland, J., Zabner, J., Jia, H. P., McCray, P. B., Jr., … Wetsel, R. A. (2001). Expression of the complement anaphylatoxin C3a and C5a receptors on bronchial epithelial and smooth muscle cells in models of sepsis and asthma. Journal of Immunology , 166, 2025–2032. doi: 10.4049/jimmunol.166.3.2025.
- Dunkelberger, J., Zhou, L., Miwa, T., & Song, W. C. (2012). C5aR expression in a novel GFP reporter gene knockin mouse: Implications for the mechanism of action of C5aR signaling in T cell immunity. Journal of Immunology , 188, 4032–4042. doi: 10.4049/jimmunol.1103141.
- Gerard, C., Bao, L., Orozco, O., Pearson, M., Kunz, D., & Gerard, N. P. (1992). Structural diversity in the extracellular faces of peptidergic G-protein-coupled receptors. Molecular cloning of the mouse C5a anaphylatoxin receptor. Journal of Immunology , 149, 2600–2606.
- Gerard, N. P., & Gerard, C. (1991). The chemotactic receptor for human C5a anaphylatoxin. Nature , 349, 614–617. doi: 10.1038/349614a0.
- Hajishengallis, G., Reis, E. S., Mastellos, D. C., Ricklin, D., & Lambris, J. D. (2017). Novel mechanisms and functions of complement. Nature Immunology , 18, 1288–1298. doi: 10.1038/ni.3858.
- Hsu, M. H., Ember, J. A., Wang, M., Prossnitz, E. R., Hugli, T. E., & Ye, R. D. (1997). Cloning and functional characterization of the mouse C3a anaphylatoxin receptor gene. Immunogenetics , 47, 64–72. doi: 10.1007/s002510050327.
- Huey, R., & Hugli, T. E. (1985). Characterization of a C5a receptor on human polymorphonuclear leukocytes (PMN). Journal of Immunology , 135, 2063–2068.
- Jockusch, H., Voigt, S., & Eberhard, D. (2003). Localization of GFP in frozen sections from unfixed mouse tissues: Immobilization of a highly soluble marker protein by formaldehyde vapor. Journal of Histochemistry and Cytochemistry , 51, 401–404. doi: 10.1177/002215540305100315.
- Karsten, C. M., Laumonnier, Y., Eurich, B., Ender, F., Broker, K., Roy, S., & Köhl, J. (2015). Monitoring and cell-specific deletion of C5aR1 using a novel floxed GFP-C5aR1 reporter knock-in mouse. Journal of Immunology , 194, 1841–1855. doi: 10.4049/jimmunol.1401401.
- Karsten, C. M., Wiese, A. V., Mey, F., Figge, J., Woodruff, T. M., Reuter, T., & Köhl, J. (2017). Monitoring C5aR2 expression using a floxed tdTomato-C5aR2 knock-in mouse. Journal of Immunology , 199, 3234–3248. doi: 10.4049/jimmunol.1700710.
- Kim, H., Kim, M., Im, S. K., & Fang, S. (2018). Mouse Cre-LoxP system: General principles to determine tissue-specific roles of target genes. Laboratory Animal Research , 34, 147–159. doi: 10.5625/lar.2018.34.4.147.
- Kim, J. H., Lee, S. R., Li, L. H., Park, H. J., Park, J. H., Lee, K. Y., & Choi, S. Y. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS One , 6, e18556. doi: 10.1371/journal.pone.0018556.
- Klos, A., Wende, E., Wareham, K. J., & Monk, P. N. (2013). International Union of Basic and Clinical Pharmacology. [corrected]. LXXXVII. Complement peptide C5a, C4a, and C3a receptors. Pharmacological Reviews , 65, 500–543. doi: 10.1124/pr.111.005223.
- Kolev, M., Le Friec, G., & Kemper, C. (2014). Complement—tapping into new sites and effector systems. Nature Reviews Immunology , 14, 811–820. doi: 10.1038/nri3761.
- Laumonnier, Y., Karsten, C. M., & Köhl, J. (2017). Novel insights into the expression pattern of anaphylatoxin receptors in mice and men. Molecular Immunology , 89, 44–58. doi: 10.1016/j.molimm.2017.05.019.
- Miyabe, Y., Miyabe, C., Mani, V., Mempel, T. R., & Luster, A. D. (2019). Atypical complement receptor C5aR2 transports C5a to initiate neutrophil adhesion and inflammation. Science Immunology , 4, eaav5951. doi: 10.1126/sciimmunol.aav5951.
- Morris, L. M., Klanke, C. A., Lang, S. A., Lim, F. Y., & Crombleholme, T. M. (2010). tdTomato and EGFP identification in histological sections: Insight and alternatives. Biotechnic & Histochemistry, 85, 379–387. doi: 10.3109/10520290903504753.
- Norgauer, J., Dobos, G., Kownatzki, E., Dahinden, C., Burger, R., Kupper, R., & Gierschik, P. (1993). Complement fragment C3a stimulates Ca2+ influx in neutrophils via a pertussis- toxin-sensitive G protein. European Journal of Biochemistry FEBS , 217, 289–294. doi: 10.1111/j.1432-1033.1993.tb18245.x.
- Okinaga, S., Slattery, D., Humbles, A., Zsengeller, Z., Morteau, O., Kinrade, M. B., & Gerard, C. (2003). C5L2, a nonsignaling C5A binding protein. Biochemistry , 42, 9406–9415. doi: 10.1021/bi034489v.
- Pak-Wittel, M. A., Piersma, S. J., Plougastel, B. F., Poursine-Laurent, J., & Yokoyama, W. M. (2014). Isolation of murine natural killer cells. Current Protocols in Immunology , 105(1), 3.22.1-3.22.9. doi: 10.1002/0471142735.im0322s105.
- Quell, K. M., Karsten, C. M., Kordowski, A., Almeida, L. N., Briukhovetska, D., Wiese, A. V., & Köhl, J. (2017). Monitoring C3aR expression using a floxed tdTomato-C3aR reporter knock-in mouse. Journal of Immunology , 199, 688–706. doi: 10.4049/jimmunol.1700318.
- Reeves, J. P., Reeves, P. A., & Chin, L. T. (2001). Survival surgery: Removal of the spleen or thymus. Current Protocols in Immunology , 2(1), 1.10.1–1.10.11. doi: 10.1002/0471142735.im0110s02.
- Sayah, S., Jauneau, A. C., Patte, C., Tonon, M. C., Vaudry, H., & Fontaine, M. (2003). Two different transduction pathways are activated by C3a and C5a anaphylatoxins on astrocytes. Brain Research Molecular Brain Research , 112, 53–60. doi: 10.1016/s0169-328x(03)00046-9.
- Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N., Palmer, A. E., & Tsien, R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology , 22, 1567–1572. doi: 10.1038/nbt1037.
- Shaner, N. C., Steinbach, P. A., & Tsien, R. Y. (2005). A guide to choosing fluorescent proteins. Nature Methods , 2, 905–909. doi: 10.1038/nmeth819.
- Song, A. J., & Palmiter, R. D. (2018). Detecting and avoiding problems when using the Cre-lox system. Trends in Genetics , 34, 333–340. doi: 10.1016/j.tig.2017.12.008.
- Strober, W. (2001). Monitoring cell growth. Current Protocols in Immunology , 21, 3A.1–A.3A.2. doi: 10.1002/0471142735.ima03as21.
- Swamydas, M., Luo, Y., Dorf, M. E., & Lionakis, M. S. (2015). Isolation of Mouse Neutrophils. Current Protocols in Immunology , 110(1), 3.20.1–3.20.15. doi: 10.1002/0471142735.im0320s110.
- Tschernig, T., Kiafard, Z., Dibbert, C., Neumann, D., & Zwirner, J. (2007). Use of monoclonal antibodies to assess expression of anaphylatoxin receptors in rat and murine models of lung inflammation. Experimental and Toxicologic Pathology , 58, 419–425. doi: 10.1016/j.etp.2007.03.004.
- Voytas, D. (2001). Agarose gel electrophoresis. Current Protocols in Molecular Biology , 51, 2.5A.1–2.5A.9. doi: 10.1002/0471142727.mb0205as51.
- West, E. E., Kolev, M., & Kemper, C. (2018). Complement and the regulation of T cell responses. Annual Review of Immunology , 36, 309–338. doi: 10.1146/annurev-immunol-042617-053245.
- Zamora, A. E., Grossenbacher, S. K., Aguilar, E. G., & Murphy, W. J. (2015). Models to study NK cell biology and possible clinical application. Current Protocols in Immunology , 110(1), 14.37.1–14.37.14. doi: 10.1002/0471142735.im1437s110.
- Zhang, X., Goncalves, R., & Mosser, D. M. (2008). The isolation and characterization of murine macrophages. Current Protocols in Immunology , 83(1), 14.1.1–14.1.14. doi: 10.1002/0471142735.im1401s83.
- Zhanmu, O., Zhao, P., Yang, Y., Yang, X., Gong, H., & Li, X. (2019). Maintenance of fluorescence during paraffin embedding of fluorescent protein-labeled specimens. Frontiers in Neuroscience , 13, 752. doi: 10.3389/fnins.2019.00752.
Citing Literature
Number of times cited according to CrossRef: 8
- Claudia Mickael, Mariah Jordan, Janelle N. Posey, Rubin M. Tuder, Eva S. Nozik, Joshua M. Thurman, Kurt R. Stenmark, Brian B. Graham, Cassidy A. Delaney, Activation of platelets and the complement system in mice with Schistosoma -induced pulmonary hypertension , American Journal of Physiology-Lung Cellular and Molecular Physiology, 10.1152/ajplung.00165.2024, 327 , 5, (L661-L668), (2024).
- Daniel L. Seiler, Katja H. Kähler, Marie Kleingarn, Christian D. Sadik, Katja Bieber, Jörg Köhl, Ralf J. Ludwig, Christian M. Karsten, The complement receptor C5aR2 regulates neutrophil activation and function contributing to neutrophil-driven epidermolysis bullosa acquisita, Frontiers in Immunology, 10.3389/fimmu.2023.1197709, 14 , (2023).
- Ranjit K. Sahu, Sandhya Xavier, Daniel Chauss, Luopin Wang, Claude Chew, Ronald Taylor, William B. Stallcup, Jennie Z. Ma, Majid Kazemian, Behdad Afzali, Jörg Köhl, Didier Portilla, Folic acid-mediated fibrosis is driven by C5a receptor 1-mediated activation of kidney myeloid cells, American Journal of Physiology-Renal Physiology, 10.1152/ajprenal.00404.2021, 322 , 6, (F597-F610), (2022).
- Lin Zhang, Wei Li, Minjie Gong, Zeyu Zhang, Xiaodong Xue, Jiarong Mao, Haibao Zhang, Siqi Li, Xiawan Liu, Feng Wu, Jingming Shi, Rongguo Fu, C‐reactive protein inhibits C3a/C3aR‐dependent podocyte autophagy in favor of diabetic kidney disease, The FASEB Journal, 10.1096/fj.202200198R, 36 , 6, (2022).
- Qi Sun, Yuhao Liu, Xiaojie Teng, Peng Luan, Xiaohua Teng, Xiujie Yin, Immunosuppression participated in complement activation-mediated inflammatory injury caused by 4-octylphenol via TLR7/IκBα/NF-κB pathway in common carp (Cyprinus carpio) gills, Aquatic Toxicology, 10.1016/j.aquatox.2022.106211, 249 , (106211), (2022).
- Henry David Mosquera-Daza, A look at the C3 and C5 proteins in the Covid-19, Actualidades Biológicas, 10.17533/udea.acbi.v43n115a05, 43 , 115, (1-9), (2021).
- Charles A. Warwick, Alex L. Keyes, Trent M. Woodruff, Yuriy M. Usachev, The complement cascade in the regulation of neuroinflammation, nociceptive sensitization, and pain, Journal of Biological Chemistry, 10.1016/j.jbc.2021.101085, 297 , 3, (101085), (2021).
- Qi Sun, Yuhao Liu, Xiaojie Teng, Peng Luan, Xiaohua Teng, Xiujie Yin, Immunosuppression Participated in Complement Activation-Mediated Inflammatory Injury Caused by 4-Octylphenol Via Tlr7/Iκbα/Nf-Κb Pathway in Common Carp (Cyprinus Carpio) Gills, SSRN Electronic Journal, 10.2139/ssrn.3993322.

