Cell Interaction by Multiplet sequencing (CIM-seq)
Nathanael Andrews, Jason T. Serviss, Natalie Geyer (Karolinska Institute Stockholm), Agneta B. Andersson, Ewa Dzwonkowska, Iva Šutevski, Rosan Heijboer, Ninib Baryawno (Karolinska Institute Stockholm), Marco Gerling, Martin Enge
Abstract
Single cell sequencing methods facilitate the study of tissues at high resolution, revealing rare cell types with varying transcriptomes or genomes, but so far have been lacking the capacity to investigate cell-cell interactions. Here, we introduce CIM-seq, an unsupervised and high-throughput method to analyze direct physical cell-cell interactions between every cell type in a given tissue. CIM-seq is based on RNA sequencing of incompletely dissociated cells, followed by computational deconvolution of these into their constituent cell types using machine learning. CIM-seq is broadly applicable to studies that aim to simultaneously investigate the constituent cell types and the global interaction profile in a specific tissue.
Before start
Before preparing cell lysis plates it is recommended to thoroughly clean all equipment with 70% EtOH and RNAse away to prevent contamination and avoid RNA degradation.
Steps
Prepare Lysis buffer
CIM-seq is compatible with both plate based and droplet based single cell RNA-seq methods. The following protocol is for Smart-seq2 based CIM-seq. For droplet based (10x), see the methods section of the manuscript and the vignettes in the Enge lab Github repository.
Prepare Lysis Buffer:
NOTE: Reagents are prepared on ice, working quickly. ERCC is stored in single-use aliquots at -80°C, thawed on ice and added last.
| A | B | C |
|---|---|---|
| Reagent: | Reagent concentration: | μl per reaction: |
| H20 | - | 1.31 |
| Inhibitor | 1 U/μl | 0.05 |
| ERCC (1:600 000) | - | 0.05 |
| Triton-X100 (10% solution) | 0.2% | 0.04 |
| 10mM dNTP | 2.5mM/each | 0.5 |
| 100uM dT | 2.5μM | 0.05 |
| Total | - | 2 |
Add 2µL lysis buffer mix to each well. Cover with appropriate lids. Spin down.
Snap freeze on dry ice . Store until use at -80°C
Sort cells
Sort single cells and multiplets (aggregates of multiple cells) into 2µL lysis buffer mix.
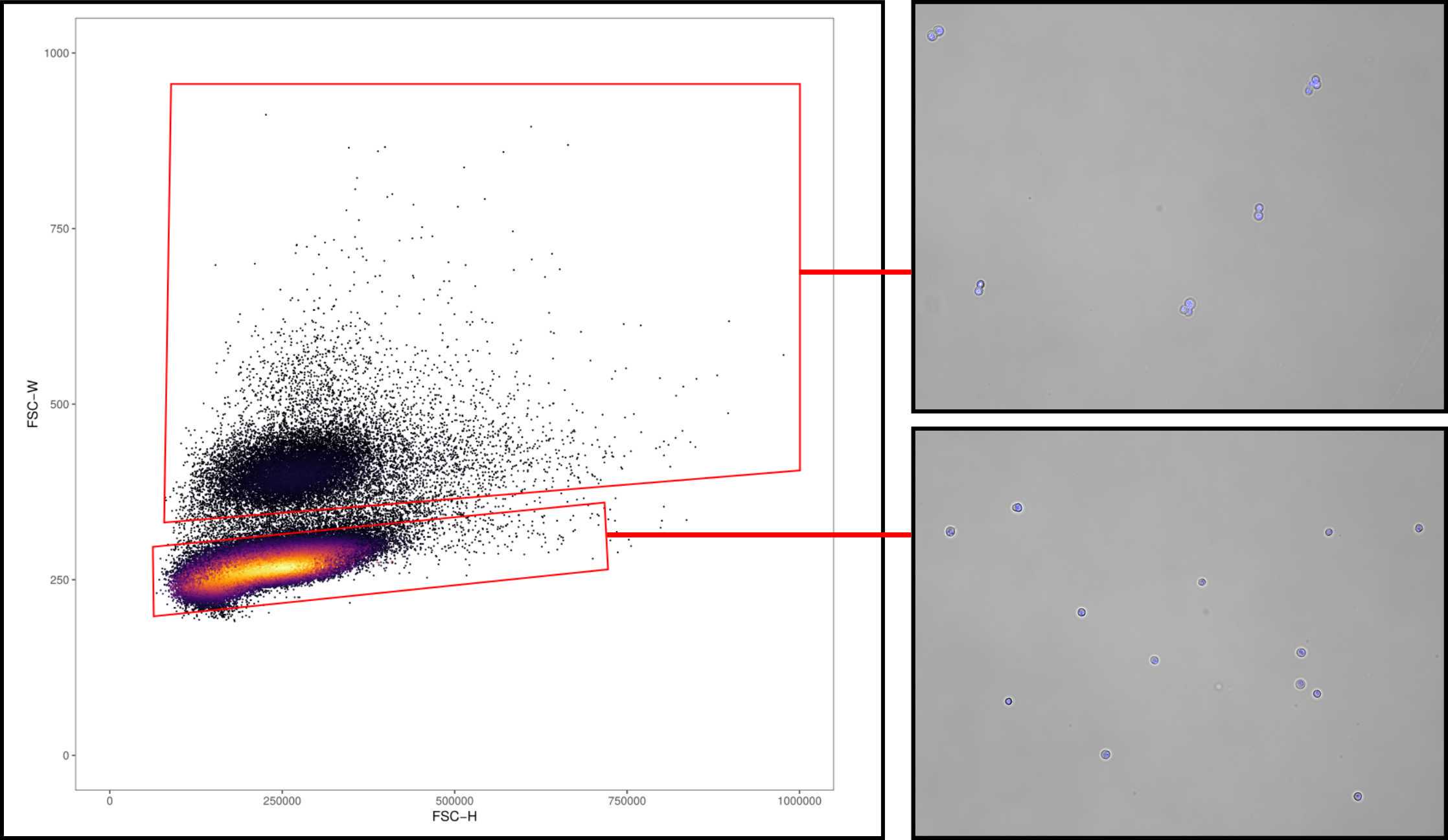
Multiplets can be discerned from singlets by gating on the basis of FSC-W (Forward scatter - Width) and FSC-H (Forward scatter - Height) (see Figure 1 ).
Following sort, immediately seal with appropriate seals (approved for -80C > 100C) and centrifuge at 2000x g,4°C.
Snap freeze on dry ice . Store until use at -80°C .
Reverse transcription and cDNA amplification
Primer annealing
Thaw plate. Spin down. Incubate in thermocycler at 72°C for 0h 3m 0s . Place on ice immediately.
Prepare RT master-mix
Made fresh.
| A | B | C |
|---|---|---|
| Reagent: | Reaction concentration: | Reagent volume: |
| SmartScribe | 15u/µl | 0.475 |
| RNase Inhibitor | 1.66u/µl | 0.125 |
| 5x First Strand buffer | 1X | 1 |
| DTT (100mM) | 8.33mM | 0.25 |
| Betaine (5M) [fridge] | 1.66M | 1 |
| MgCl2 (1M) [bench] | 10mM | 0.03 |
| TSO (100uM) | 1.66µM | 0.05 |
| H20 | - | 0.07 |
| Total | - | 3 |
Dispense 3µL per well.
Cover plate with new film and spin down.
Incubate in thermocycler
42°C 1h 30m 0s
70°C 0h 5m 0s
4°C hold
cDNA preamplification
Made fresh.
| A | B | C |
|---|---|---|
| Reagents | Reaction concentration: | Reagent volume: |
| H2O | - | 1.0688 |
| Kapa HiFi HotStart ReadyMix (2x) | 1X | 6.25 |
| IS_PCR primer (10uM) | 0.1µM | 0.125 |
| Lambda Exonuclease | 0,045u/µl | 0.05625 |
| Total | - | 7.5000 |
Dispense 7.5µL per well . Total reaction volume will be 12.5µl.
Spin down. Cover with new lid.
Incubate in thermocycler with the following program:
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Lambda exonuclease | 37ºC | 30 min | 1x |
| Initial denaturation | 95ºC | 3 min | 1x |
| Denaturation | 98ºC | 20 sec | 18-24x |
| Annealing | 67ºC | 15 sec | |
| Elongation | 72ºC | 4 min | |
| Final elongation | 72ºC | 5 min | 1x |
| 4C | Hold |
cDNA cleanup
We prepare SPRI-beads in 20% PEG-8000 solution as in: https://openwetware.org/wiki/SPRI_bead_mix#Ingredients_for_50_mL_2
Using 20% SPRI-bead solution:
-
Add 0.7x the reaction volume of SPRI beads per well. Mix well by pipetting. (i.e
8.75µLSPRI-bead solution for12.5µLreaction volume) -
Incubate
0h 5m 0sRoom temperature -
Place on magnetic stand for
0h 3m 0s -
Carefully remove supernatant
-
Add 40 µl 80% EtOH and incubate
0h 0m 30s -
Remove EtOH (without disturbing the beads)
-
Wash again with EtOH. Make sure to remove well.
-
Allow beads to air-dry for
0h 10m 0s-0h 15m 0s -
Remove plate from magnetic stand
-
Elute beads in
15µLEB or TE buffer. Mix well by pipetting -
Incubate
0h 5m 0sRoom temperature -
Place on magnetic plate for
0h 3m 0s -
Optional: Carefully remove supernatant to the elution plate
cDNA quantification
We measure concentration of random wells using Qubit HS dsDNA, adapted to a 96-well plate reader.
-
Add
97µLof 1X Qubit HS dsDNA solution to a flat-bottom, black plate -
Add
3µLof cDNA sample -
Add Standards (NOTE: We make a 8-step ladder from 0ng/µl --> 10ng/µl Qubit Standard DNA in TE buffer)
-
Read in plate reader using 485nM excitation/528nm emission
-
Calculate cDNA concentration
Make cDNA dilution plate
Dilute cDNA in water based on average concentration from Qubit measurements.
Target concentration 150pg per µl in 15µL.
cDNA tagmentation
Tn5 digestion
Tn5 is produced from psfTn5 (Addgene #79107), purified to ~3mg/ml and assembled with Illumina Tn5 adapters (see oligos) as in Picelli et al, 2014.
Prepare Tn5 master mix
| A | B | C |
|---|---|---|
| Reagent | Reaction conc. | µl per reaction |
| Nuclease free H2O | - | 1.05 |
| TAPS-PEG (50mM TAPS, 25mM MgCl2, 40% PEG-8000) | 10mM TAPS, 5mM MgCl2, 8% PEG-8000 | 0.5 |
| psfTn5, loaded with 50µM MEDS-A/B | 0.25 | |
| Total | 1.8 |
Dispense 1.8µL per well in a new plate(tagmentation plate)
Add 0.7µL cDNA (normalized to 150pg/µl)
Mix well by vortexing plate. Cover with new lid and spin down.
Incubate in thermocycler at 55°C``0h 10m 0s
Remove immediately and stop reaction by adding 1µL per well of 0.1% SDS.
Vortex, spin down and incubate 0h 7m 0s at 55Room temperature
cDNA library PCR and barcoding
Make PCR master-mix
| A | B |
|---|---|
| Reagents | µl per reaction |
| H2O | 13.25 |
| 5x buffer | 5 |
| dNTPs | 0.75 |
| KAPA | 0.5 |
| Total | 19.50 |
Dispense 19.5µL per well to tagmentation plate (containing 3.5µL sample after step 14)
Add primers/barcodes
2µL per well (from 384-well index plates, with 3.75µM/each forward/reverse primers; see oligos in materials ).
Total reaction volume is 25µL (3.5µL sample + 19.5µL PCR mix and 2µL primers).
Vortex. Spin down and cover. Incubate in thermocycler as following:
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Gap fill | 72ºC | 3 min | 1x |
| First denature | 95ºC | 30 sec | 1x |
| Denature | 95ºC | 15 sec | 12x |
| Denature | 67ºC | 30 sec | |
| Denature | 72ºC | 45 sec | |
| Final extension | 72ºC | 4 min | 1x |
| 4-10ºC | hold |
Pool 2.5µL from each well to an 1.5ml Eppendorf tube.
Library cleanup
-
Add 0.9x pooled library volume of SPRI-bead solution. Incubate for
0h 5m 0satRoom temperature. -
Place on magnetic rack for
0h 3m 0s. -
Remove supernatant without disturbing magnetic beads.
-
Add at least
1mL80% EtOH (fresh). Incubate for0h 0m 30s. -
Remove supernatant.
-
Repeat EtOH wash.
-
Air dry for
0h 10m 0s-0h 15m 0s. -
Re-suspend beads thoroughly in 100 µl EB or TE buffer.
-
Place eppendorf on magnetic rack for
0h 3m 0s. -
Transfer supernatant to new 1.5ml Eppendorf tube.
-
Repeat cleanup (from step 1-7) and elute in 30 µl EB or TE buffer.
12.(Optional) Place eppendorf on magnetic rack for 0h 3m 0s and transfer supernatant to new tube.
Data pre-processing
A series of pre-processing steps must be performed in order to generate a counts file:
- Trim reads, remove adapter sequences and align RNAseq data to reference genome using STAR:
- Remove duplicate reads using Picard:
Dimensionality reduction and classification
Once a counts file has been generated the data can be analyzed. CIM-seq requires four arguments in order to run:
- The raw counts data with gene IDs as rownames and sample IDs as
colnames.
- The ERCC spike-in counts data with gene IDs as rownames and sample
IDs as colnames.
-
The dimensionality reduced representation of the data.
-
A class for each of the individual singlets.
In order to generate the last two of these we recommend using the Seurat package in R, as CIM-seq is implemented in R as well. A number of tutorials for Seurat can be found on the Satijalab website:
CIM-seq
CIM-seq can be downloaded from:
https://github.com/EngeLab/CIMseq
Or installed directly in R using the devtools package:
devtools::install_github("EngeLab/CIMseq")
The CIM-seq vignette can be found at: