CRISPR/Cas9-Mediated Knockdown in LUHMES Cells: Nucleofection and Validation Protocol
Jason Waligorski, William J Buchser, Mallory Wright, Colin Kremitzki, Serena Elia, Graham Bachman, emanuel gerbi, Nicholas Tu, Lina Mohammed Ali
LUHMES
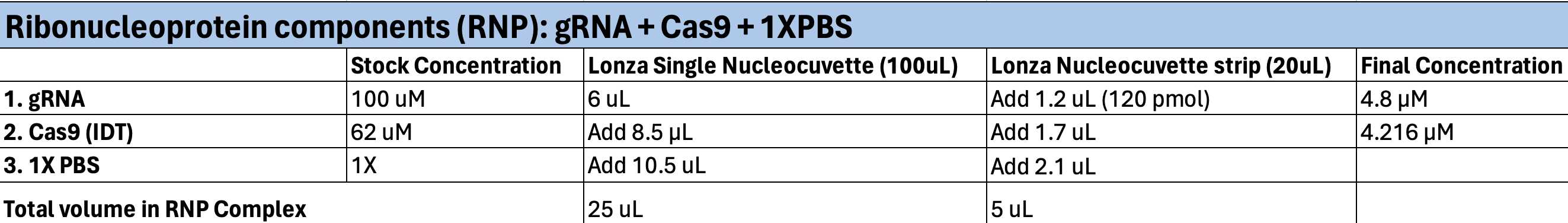
Ribonucleoprotein Complex (RNP)
Neurodegenerative disease models
Gene knockout
Neuroscience
CRISPR /Cas9
Gel electrophoresis
PCR
Abstract
Utilizing a CRISPR RNP complex and nucleofection, this protocol enables precise gene knockout. LUHMES cells, sourced from human fetal mesencephalic tissue, provide a valuable model for investigating dopaminergic dysfunction. They exhibit stimulus-induced dopamine release, pertinent electrophysiological traits, and distinctive dopaminergic markers, validating their phenotypic relevance.
LUHMES Cell Line: (ATCC Catalog Number: CRL-2927)
Nucleofection kit : P3 Primary Cell 4D-Nucleofector X Kit S Catalog #V4XP-3032
Steps
Nucleofection Protocol
Maintain cell confluency between 70–85% to optimize Nucleofection efficiencies; optimal results typically occur with cells in the logarithmic growth phase.
Coat a new 6-well plate freshly with poly-L-ornithine (50ug/mL) and fibronectin (2ug/mL) to facilitate LUHMES attachment.
Add 2mL of the LUHMES growth media to the 6-well plate and pre-incubate/equilibrate the plates in a humidified incubator set at 37°C with 5% CO2.
Rinse LUHMES with 5mL 1XPBS, then add 4mL for a T-75 flask, and incubate for 0h 3m 0s
Centrifuge1200rpm,0h 0m 0s for 0h 5m 0s, then carefully discard the supernatant
Re-suspend cells with5mL of 1X PBS and count cells.
After cell counting, distribute the cells into individual 15mL tubes according to the number of samples.
Add approximately 1 million cells per single cuvette and 500,000 cells per well in a nucleocuvette strip.
Centrifuge the samples a second time at a low speed90x g,0h 0m 0s for0h 10m 0s, ensuring minimal cell agitation before nucleofection.
Carefully re-suspend each cell pellet in room temperature 4D-Nucleofection Solution + supplement.
| A | B | C |
|---|---|---|
| Nucleofection supplement (P3) | 18 uL | 3.6 uL |
| Nucleofection solution (P3) | 82 uL | 16.4 uL |
| pmaxGFP Vector (0.5ug/uL) (optional positive control) | Add 4 uL | Add 1.2 uL |
Add cells to the cuvette, ensuring the sample covers the bottom of the cuvette. Gently tap to distribute evenly and avoid bubbles.
For each single nucleocuvette, add25µL of the RNP Complex, or for each well of a strip, add 5µL of the RNP complex.
Insert the nucleocuvettes into the nucleofector cuvette slot, ensuring they are properly aligned, then press start.
Following nucleofection, immediately add an additional500µL of pre-warmed media to each individual single cuvette or 50 uL to a curvette strip.
Maintain cell stability by avoiding any disturbance for at least0h 15m 0s at room temperature 2°Cor within an incubator
Transfer the cells to the previously coated flask or plate containing pre-warmed LUHMES growth media, and incubate.
A typical analysis time is 24-Hours post-nucleofection.
Nucleofection Validation
24 hours after nucleofection, rinse the cells using 5 mL 1X PBS, apply TE dissociation solution, allow a0h 3m 0sincubation period, transfer to a 15 mL tube, and then centrifuge1200rpm
Re-suspend the cells in 100µL 1X lysis buffer, gently triturating approximately 5 times to ensure efficient cell membrane disruption and DNA extraction.
Add cell suspension to a 1.5 mL snap cap tube
Add the 1.5 mL snap cap tube to a65°C heat block for 0h 15m 0s to disrupt cellular structures, denature proteins, and aid DNA extraction.
Follow with a second heat treatment at 95°C for0h 3m 0s
Add the Snap cap tube to fridge0h 3m 0s in 4°C
PCR Protocol
Utilize a nanodrop device to determine the nucleic acid concentration of your bulk sample.
Prepare the PCR reaction mix in a sterile microcentrifuge tube by combining the following components:
- Template DNA: The DNA you wish to amplify, need ~100 ng of DNA per
25µLreaction - Forward and Reverse Primers: Short DNA sequences that bind to the start and end of the target DNA region you want to copy.
- MyTaq Red Mix (Meridian Life Science Catalog # BIO-25047 ): A ready-to-use PCR master mix containing DNA polymerase, dNTPs, buffer, and a red dye.
- DMSO (Dimethyl Sulfoxide): Added to enhance PCR specificity and amplification of GC-rich templates
| A | B | C | D |
|---|---|---|---|
| Forward Primer (100uM) | 0.5 μM | 0.125 μL | 2.5 μL |
| Reverse Primer (100uM) | 0.5 μM | 0.125 μL | 2.5 μL |
| MyTaq (DNA Polymerase) | 2X | 12.5 | 250 uL |
| DMSO | 5% | 1.25 μL per reaction. | 25 μL |
| Water (ddH2O) | amount depends on the remaining volume after adding the other components | 9 uL | 180 uL |
| Template DNA | need ~100 ng per 25uL reaction | 2 uL | 2 uL |
| Total amount | 25 uL |
PCR1 components
Running an Agarose Gel for DNA Electrophoresis
Prepare Agarose Gel:
-
Measure
1gof agarose. -
Dissolve in
70mLof 0.5X TBE buffer in a beaker -
Microwave for
0h 1m 15sor until completely dissolved. -
Add SybrSafe dye at
1µLfor ever10mLof 0.5X TBE Buffer to the agarose solution for staining the DNA bands. -
Cool for about
0h 5m 0s
Pour Gel and Insert Combs:
-
Pour the cooled agarose into a gel tray.
-
Insert combs to create wells for loading samples.
-
Remove any bubbles in gel
-
Let the gel sit undisturbed for about
0h 30m 0sto solidify.
Prepare Samples:
-Remove combs
- Add
5µLof DNA ladder 100bp (Gold bio)
-Add 5µL of each DNA sample to the wells. Ensure proper labeling to keep track of the samples.
Run the Gel:
-Connect wires to matching color ports on the power supply (red to red, black to black)
-Set voltage to 170V, 400mA and run gel for 0h 25m 0s
Monitor gel progress as DNA fragments move through agarose: smaller ones faster, longer ones slower.
Once complete:
-
Carefully remove the gel from the casting tray and place it in a gel imaging system.
-
Visualize the DNA bands under UV light.
-
The Sybr Safe dye will fluoresce upon binding to DNA, allowing for the visualization of DNA fragments.
Analyze the gel to identify the presence of contaminants, estimate the size of amplified DNA fragments, and confirm whether they match the expected sizes based on primer design.
Last, perform PCR2 to incorporate barcodes and then submit the samples for next-generation sequencing.