A New Method of Bone Stromal Cell Characterization by Flow Cytometry
Chirayu Patel, Chirayu Patel, Lihong Shi, Lihong Shi, John F. Whitesides, John F. Whitesides, Brittni M. Foster, Brittni M. Foster, Roberto J. Fajardo, Roberto J. Fajardo, Ellen E. Quillen, Ellen E. Quillen, Bethany A. Kerr, Bethany A. Kerr
Abstract
The bone microenvironment cellular composition plays an essential role in bone health and is disrupted in bone pathologies, such as osteoporosis, osteoarthritis, and cancer. Flow cytometry protocols for hematopoietic stem cell lineages are well defined and well established. Additionally, a consensus for mesenchymal stem cell flow markers has been developed. However, flow cytometry markers for bone-residing cells—osteoblasts, osteoclasts, and osteocytes—have not been proposed. Here, we describe a novel partial digestion method to separate these cells from the bone matrix and present new markers for enumerating these cells by flow cytometry. We optimized bone digestion and analyzed markers across murine, nonhuman primate, and human bone. The isolation and staining protocols can be used with either cell sorting or flow cytometry. Our method allows for the enumeration and collection of hematopoietic and mesenchymal lineage cells in the bone microenvironment combined with bone-residing stromal cells. Thus, we have established a multi-fluorochrome bone marrow cell-typing methodology. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Partial digestion for murine long bone stromal cell isolation
Alternate Protocol 1 : Partial digestion for primate vertebrae stromal cell isolation
Alternate Protocol 2 : Murine vertebrae crushing for bone stromal cell isolation
Basic Protocol 2 : Staining of bone stromal cells
Support Protocol 1 : Fluorescence minus one control, isotype control, and antibody titration
Basic Protocol 3 : Cell sorting of bone stromal cells
Alternate Protocol 3 : Flow cytometry analysis of bone stromal cells
Support Protocol 2 : Preparing compensation beads
INTRODUCTION
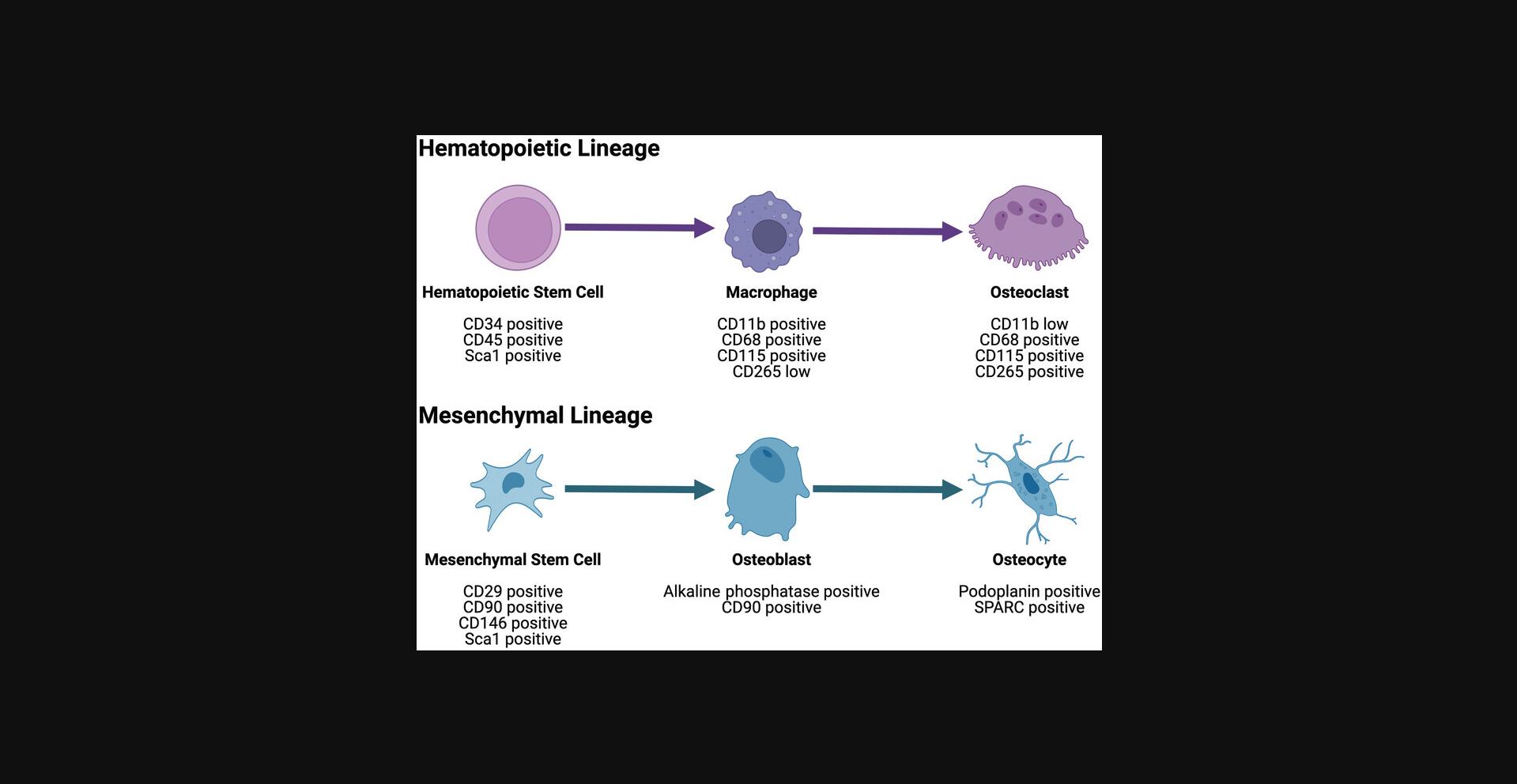
The composition of the bone microenvironment plays an important role in development and pathologies, with alterations in the marrow and stromal cell composition having profound effects on the bone structure. A balance between cells of the hematopoietic stem cell lineage, including osteoclasts, and mesenchymal stem cell lineage cells, including osteoblasts, is required for bone homeostasis. Flow cytometry and cell sorting are the gold standard methods for the enumeration and sorting of hematopoietic stem cell lineage cells. Additionally, a consensus for mesenchymal stem cell flow cytometry markers is established for these progenitor cells in the bone marrow (Dominici et al., 2006). However, flow cytometry markers for bone-residing stromal cells—osteoblasts, osteoclasts, and osteocytes—have not been established. This is partly due to the difficulty in removing these cells from the bone matrix for analysis. In Basic Protocol 1, an optimized partial collagenase digestion protocol to isolate bone marrow stromal cells is described. Further, in Alternate Protocols 1 and 2, isolation of bone marrow stromal cells by partial digestion of primate vertebrae or grinding from murine vertebrae is discussed. Basic Protocol 2 describes the staining of isolated bone marrow stromal cells to identify hematopoietic stem cells, mesenchymal stem cells, osteoblasts, osteoclasts, and osteocytes using the marker panel shown in Figure 1. Finally, in Basic Protocol 3 and Alternate Protocol 3, the analysis of bone stromal cells by cell sorting and flow cytometry are described. These protocols were optimized for use with common three laser cell sorters or two laser flow cytometers using the brightest and most stable fluorophores available for each antibody. This method allows for the enumeration and, using cell sorting, isolation of hematopoietic stem cells, mesenchymal stem cells, and bone-residing cells—osteoblasts, osteoclasts, and osteocytes—in a single experiment. This method will thus profile a majority of the bone microenvironment cells. It can be used for assessing the cellular composition of the bone microenvironment during development and examining the influence of various pathologies on the microenvironment.
STRATEGIC PLANNING
Choosing Analysis Method
Before starting experiments, determine whether flow cytometry (cell enumeration) or cell sorting (cell enumeration and separation) will provide the information needed for your experiments. If you need to perform downstream analyses on bone-residing stromal cells, cell sorting will be a better choice. Flow cytometry can be performed on fixed cells and has several potential storage/stoppage points. Cell sorting can only be completed on unfixed cells and must be completed within a few days of isolating cells. Check Time Considerations carefully to reserve time on a cell sorting machine.
Choice of Antibody Combinations
The selection of antibodies is essential to identify the proper cell population, especially for the rare bone-residing cells. For this protocol, multi-fluorophore panels were developed for two or three laser analysis machines using commercially available antibodies with brighter fluorophores assigned to rarer cell surface proteins. Markers for hematopoietic stem cells and mesenchymal stem cells are well defined (Boxall & Jones, 2012; Dominici et al., 2006). Due to fluorophore availability, not all the markers were tested in this protocol but could be used in multi-fluorophore panels with additional lasers for excitation. Antibody selection for bone-residing osteoclasts, osteoblasts, and osteocytes focused on surface marker expression. Osteoclast surface marker RANKL/CD254 (receptor activator of nuclear factor-κB ligand) or RANK/CD265 is associated with differentiation and activity of osteoclasts (Faust et al., 1999). Thus, CD265 or CD254 expression on cells expressing the macrophage surface markers CD68 and CD115 was postulated to represent osteoclasts or preosteoclast cells. Osteoblast surface proteins were proposed to be CD90/Thy-1 and alkaline phosphatase based on their roles in bone formation and bone matrix calcification (Picke et al., 2018; Rutkovskiy, Stensløkken, & Vaage, 2016; Trentz et al., 2010). Osteocyte markers SPARC (secreted protein acidic and rich in cysteine) and podoplanin (GP38) were derived from publications describing osteocyte ontogeny from osteoblasts (Bonewald, 2011; Zhu, Mackenzie, Millán, Farquharson, & MacRae, 2011). Markers were chosen based on their expression on the cell surface, their ability to differentiate between bone stromal cell populations, and the commercial availability of fluorophore-conjugated antibodies. When designing panels, look for antibodies in bright fluorophores with minimal overlap. Additional markers can be included as more fluorophore conjugations become available and for use with analyzers with additional excitation lasers.
Basic Protocol 1: PARTIAL DIGESTION FOR MURINE LONG BONE STROMAL CELL ISOLATION
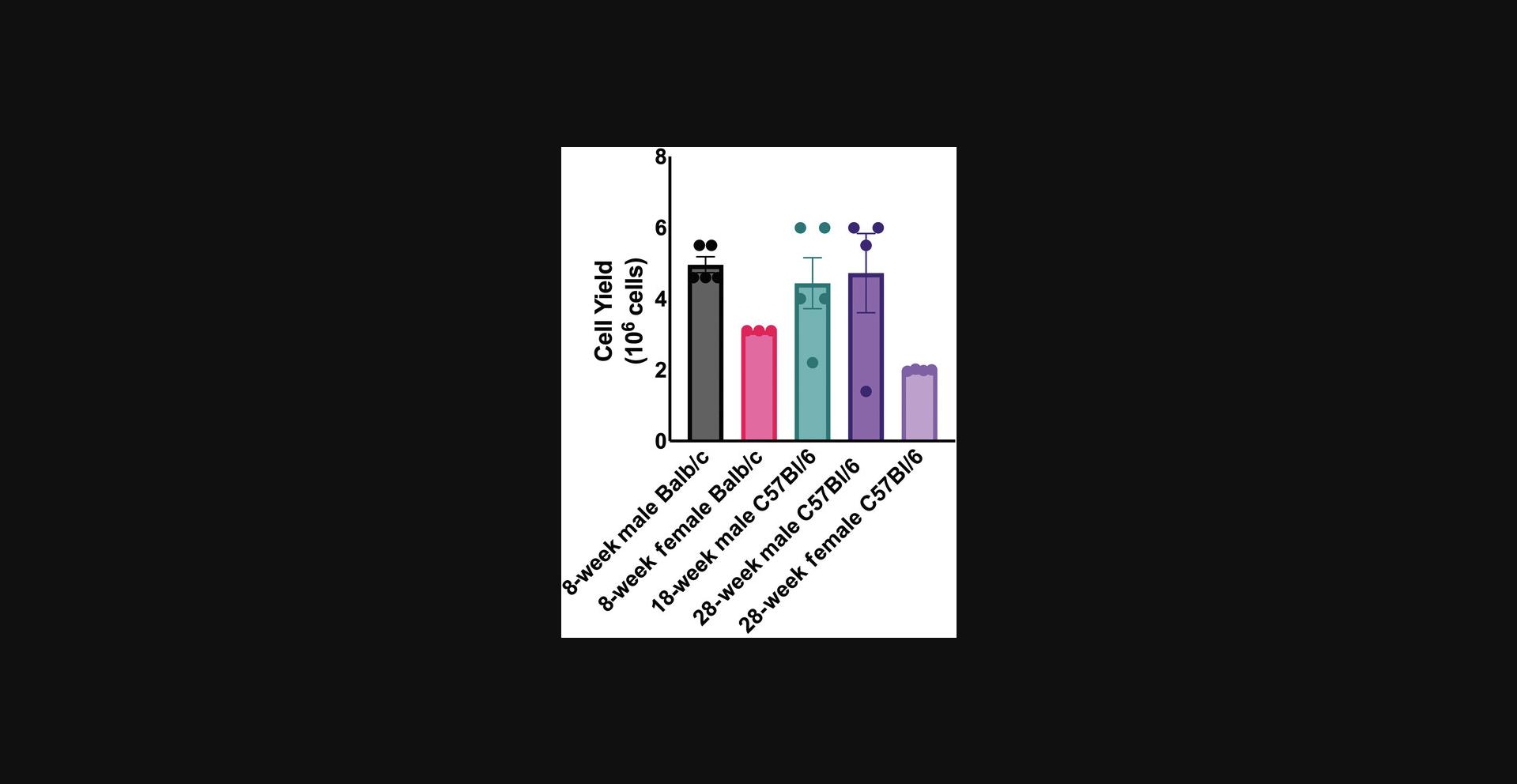
This protocol describes the partial digestion of murine long bones by collagenase I. While hindlimbs are often used, the procedure is the same for forelimbs, and these can be combined to maximize cell yield. This digestion method results in 3‒6 × 106 cells released from long bones per animal (see Fig. 2 and Table 1). Animal care and euthanasia should be performed under an institutionally approved protocol.
| Tissue | Live cell yield (107 cells) | Percent viable |
|---|---|---|
| Long bones | 11.9 | 85 |
| Vertebrae | 16.4 | 76 |
-
Spines, forelimbs, and hindlimbs were isolated from five 28-week-old male mice, and samples were pooled after digestion.
Materials
-
Mice of interest
-
1× phosphate-buffered saline (PBS) containing 100 U/ml penicillin and 100 µg/ml streptomycin (PBS+P/S)
-
1× PBS containing 1 mM EDTA
-
1 mg/ml collagenase I in PBS+P/S (e.g., Gibco, cat. no. 17-100-017)
-
Flow cytometry staining medium (see recipe)
-
1% methanol-free formaldehyde in PBS (diluted from 10% ultrapure, methanol-free formaldehyde; e.g., Polysciences Inc, cat. no. 04018-1)
-
Surgical scissors
-
10-ml syringe (e.g., Fisher Scientific, cat. no. 1482919C)
-
27.5-G needles (e.g., Medlab Gear, cat. no. BD305109)
-
Laminar chemical hood (e.g., Kewaunee Scientific Airflow Thin Wall Fume hood or equivalent)
-
Laminar biosafety cabinet (e.g., Baker Company SterilGard Hood Class II or equivalent)
-
100-mm dishes (e.g., Corning, cat. no. 353003)
-
No. 10 safety scalpel (e.g., Fisher Scientific, cat. no. SMS210)
-
50-ml conical tube
-
Forceps
-
Shaking incubator (e.g., Barnstead MaxQ 4000 or equivalent)
-
Vortex mixer
-
40-µm cell strainer (e.g., Denville, cat. no. 1006202)
-
Centrifuge (e.g., Eppendorf Eppendorf 5810)
NOTE : All protocols involving live animals must first be approved by an Institutional Animal Care and Use Committee (IACUC) or must follow local guidelines for the care and use of live animals.
Tissue dissection
1.Sacrifice mice according to an approved animal protocol.
2.Remove skin from the leg by cutting down the medial side and then removing skin with fingers to expose the entire limb musculature.
3.Remove hindlimb at hip joint, and place in PBS+P/S on ice.
4.In a biosafety or chemical hood with the light off, prepare a 10-ml syringe with PBS+P/S, and attach a 27.5-G needle.
5.Place bones in a 100-mm dish. Remove extra musculature and the foot with small surgical scissors and a scalpel using a 100-mm dish lid as a workspace. Cut off proximal and distal ends of the long bones (tibia, femur, humerus).
6.Insert syringe containing PBS with 1 mM EDTA into the marrow cavity, and flush diaphysis with 2 to 3 ml PBS with 1 mM EDTA into a 50-ml tube. Keep flushed bone marrow on ice.
7.Holding flushed bones with forceps, cut compact and trabecular bone into 1- to 3-mm chips with scissors.
Collagenase digestion
8.Transfer bone chips into a 50-ml conical tube containing 1 to 3 ml of 1 mg/ml collagenase I in PBS+P/S.
9.Incubate 30 to 60 min at 37°C in a shaking incubator at 200 rpm (0.04 × g).
10.Combine partially digested bone chips with bone marrow extract. Add PBS with 1 mM EDTA to a final volume of 30 ml.
11.Mix cell suspension vigorously by pipetting 20 times or vortexing.
12.Filter cell suspension through a 40-µm cell strainer into a new 50-ml conical tube.
13.Centrifuge cell suspension 5 min at 175 × g , room temperature.
14.Discard supernatant and resuspend in flow cytometry staining medium to an approximate concentration of 1 × 106 cells per ml. Proceed to Basic Protocol 2 to stain isolated cells.
Alternate Protocol 1: PARTIAL DIGESTION FOR PRIMATE VERTEBRAE STROMAL CELL ISOLATION
This protocol describes the isolation of bone marrow stromal cells from the vertebrae of nonhuman primates or humans. The protocol begins with the isolation from whole spines, but individual vertebrae can also be processed starting from step 6.While not tested, the trabecular regions from other primate bones could be used with this protocol with additional optimization of incubation times.
Materials
-
Human or nonhuman primate spine samples
-
3 mg/ml collagenase I in α Minimal Essential Medium (αMEM) containing 100 U/ml penicillin and 100 µg/ml streptomycin (αMEM+P/S)
-
Flow cytometry staining medium (see recipe)
-
Large plastic tube for thawing spines
-
Laminar biosafety cabinet (e.g., Baker Company SterilGard Hood Class II or equivalent)
-
No. 10 safety scalpel (e.g., Fisher Scientific, cat. no. SMS210)
-
Bone saw (e.g., Mopec 810 autopsy saw)
-
Dremel 300 with Dremel 409 cut-off wheel blades
-
Cut-resistant gloves
-
Safety goggles
-
Balance, for weighing samples
-
50-ml conical tube
-
Shaking incubator (e.g., Barnstead MaxQ 4000 or equivalent)
-
70-µm cell strainer (e.g., Fisher Scientific, cat. no. 08-771-2)
CAUTION : Tissue from some species of nonhuman primates are Biosafety Level 2 (BSL-2) pathogens owing to the risk of Herpes B virus. Follow all local guidelines and regulations for the use and handling of potentially pathogenic tissues.
NOTE : All protocols involving human samples must first be reviewed and approved by an Institutional Review Board (IRB) or must follow local guidelines for the use of human samples.
1.Retrieve wrapped spine samples from −20°C, and thaw for 36 hr in a tub at 4°C.
2.In a biosafety hood, unwrap spines and using a scalpel remove musculature and ligaments from one end of the spine until the intervertebral disc is revealed.
3.Using a bone saw, Dremel, or scalpel, remove vertebrae from the remainder of the spine.
4.Using a Dremel and a bone saw, remove spinal processes from the vertebrae.
5.Using a Dremel and a scalpel, remove intervertebral discs from both sides of the vertebrae.
6.Using a Dremel and a bone saw, cut a wedge into the vertebral body containing both cortical and trabecular bone and representing approximately one-sixth of the tissue mass. Further subdivide this wedge into 8 to 10 small pieces to expose as much surface area as possible for digestion.
7.Weigh samples and place in 50-ml conical tubes with 10 ml of 3 mg/ml collagenase I in αMEM+P/S.
8.Place sample in a 37°C shaking incubator overnight at 200 rpm (0.04 × g).
| Time | Initial weight (g) | Extracted cell concentration (105 cells/ml) |
|---|---|---|
| 4 hr | 2.88 | 2.19 |
| 6 hr | 2.87 | 3.17 |
| 8 hr | 2.13 | 7.06 |
| 22 hr | 2.76 | 7.94 |
- a
Archival 10-year-old female baboon vertebrae digested with 3 mg/ml collagenase I for the indicated times at 37°C with shaking.
| Animal pooled samplesa | Initial weight (g) | Extracted cell yield (107 cells) | Live cell yield (107 cells) | Percent viable |
|---|---|---|---|---|
| B1 | 21.927 | 94.7 | 30 | 31.7 |
| B2 | 5.934 | 4.87 | 1.04 | 21.4 |
| B3 | 6.790 | 2.74 | 0.985 | 35.9 |
- a
Each pool contains two female and one male baboon archival vertebrae with an average age of 19.6 years in each pool. Ages range from 18 to 22 years across all baboons in the sample.
| Subject pooled samplesa | Live cell yield (107 cells) | Percent viable |
|---|---|---|
| H1 | 2.42 | 21 |
| H2 | 10.8 | 37 |
| H3 | 6.1 | 27 |
- a
Each pool contains archival samples from 4 people, with 2 males and 2 females in pools H1 and H3. Ages across the samples range from 64 to 92 years. The pools for H1, H2, and H3 average 81, 89, and 80 years, respectively.
9.Strain sample through a 70-µm cell strainer, and centrifuge 3 to 5 min at 122 × g , room temperature.
10.Resuspend cells in 2 ml flow cytometry staining medium, and proceed to Basic Protocol 2 to stain isolated cells.
Alternate Protocol 2: MURINE VERTEBRAE GRINDING FOR BONE STROMAL CELL ISOLATION
This protocol is used to isolate bone marrow-residing stromal cells from murine spines. Compared with bone marrow isolation methods using femurs and tibias (Basic Protocol 1), this procedure allows for a much greater yield of cells (Table 1). When sacrificing mice, avoid performing cervical dislocation to prevent potential contamination of spinal bone marrow cells.
Additional Materials (also see Basic Protocol 1)
- PBS
- Bone scissors
- UV-sterilized mortar and pestle
1.Sacrifice mice according to an approved animal protocol.
2.Open peritoneal cavity, and remove organs to visualize the spine. Cut on both sides of the spine from the base of the tail to the neck, making sure to cut through attached ribs.
3.Use a scalpel or scissors to clean off any extra musculature and bones.
4.Remove ends of the spine near the brainstem and tail.
5.Cut remaining spine into three equal pieces using bone scissors.
6.Holding a spine piece vertically with forceps, use scissors to cut the spine open. Once open, move scissors into a horizontal position and open, spreading apart the sides of the spine to expose, and then remove spinal cord.
7.When the spinal cords are removed from all three pieces, place remaining spine into a mortar with 10 ml cold PBS. Use a pestle to crush pieces until the liquid becomes pink.
8.Pipette fluid from mortar through a 40-µm cell strainer into a 50-ml conical tube.
9.Repeat steps 7 and 8 until the PBS remains clear after crushing.
10.Centrifuge cell suspension 5 min at 274 × g , room temperature. 11.Discard supernatant and resuspend in flow cytometry staining medium to an approximate concentration of 1 × 106 cells per ml. Proceed to Basic Protocol 2 to stain isolated cells.
Basic Protocol 2: STAINING OF BONE STROMAL CELLS
This protocol describes the multi-fluorochrome staining for bone-residing stromal cells according to the schema in Figure 1.After staining, cells can be analyzed by cell sorting (Basic Protocol 3) or flow cytometry (Alternate Protocol 3).
Materials
-
Fresh or fixed cells (see Basic Protocol 1 or Alternate Protocols 1 or 2)
-
Trypan blue
-
PBS
-
Live/dead stain (e.g., Zombie Aqua; BioLegend, cat. no. 423102)
-
Flow cytometry staining medium (see recipe)
-
FcR blocking solution (e.g., BioLegend TruStain FcX PLUS, cat. no. 422302 [primate] or Miltenyi Biotec, cat. no. 130-092-575 [murine])
-
Antibodies of interest directly conjugated to fluorochromes
-
Cell sorting buffer (see recipe)
-
1% methanol-free formaldehyde in PBS
-
Automated cell counter and slides (e.g., BioRad TC20) or hemocytometer
-
Centrifuge
-
1.5-ml microcentrifuge tubes
-
Round-bottom 96-well plate (e.g., Falcon, cat. no. 3910)
-
Flow cytometry tubes (e.g., Fisher Scientific, cat. no. 02-681-385, Corning, cat. no. 352054, or equivalent)
-
Cell sorting tubes (e.g., Falcon, cat. no. 352052)
-
Aluminum foil
1.Bring volume of fresh or fixed cells to 10 ml with PBS+P/S and mix thoroughly. Take 10 µl sample, and combine with an equal volume of trypan blue. Count cells with an automated cell counter (or using hemocytometer).
2.Remove 1 × 106 cells per planned staining group, and centrifuge 3 to 5 min at 122 × g , room temperature.
| Antigen | Isotype control | Company | Cat. no. | RRID |
|---|---|---|---|---|
| Mesenchymal stem cell | ||||
| FITC Sca1 (Ly-6A/E) | FITC rat IgG2a, κ | BioLegend | 108106 | AB_313343 |
| PE CD146 | PE rat IgG2a, κ | BioLegend | 134704 | AB_2143527 |
| APC CD29 | APC hamster IgG | BioLegend | 102214 | AB_492831 |
| PerCP-Cy5.5 CD90 | PerCP-Cy5.5 rat IgG2b, κ | BioLegend | 105338 | AB_2571945 |
| Macrophage/osteoclast | ||||
| FITC CD11b | FITC rat IgG2b, κ | BioLegend | 101206 | AB_312789 |
| APC CD115 | APC rat IgG2a, κ | BioLegend | 135510 | AB_2085221 |
| PerCP-Cy5.5 CD68 | PerCP-Cy5.5 rat IgG2b, κ | BioLegend | 137010 | AB_2260046 |
| PE CD265 (RANK) | PE rat IgG2a, κ | BioLegend | 119806 | AB_2205353 |
| Osteoblast | ||||
| APC alkaline phosphatase | APC mouse IgG1, κ | R&D Systems | FAB1448A | AB_357039 |
| PerCP-Cy5.5 CD90 | PerCP-Cy5.5 rat IgG2b, κ | BioLegend | 105338 | AB_2571945 |
| Osteocyte | ||||
| PE podoplanin (GP38) | PE hamster IgG | BioLegend | 127408 | AB_2161929 |
| APC SPARC | APC rat IgG2b, κ | R&D Systems | IC942R | AB_2814696 |
| Hematopoietic stem cell | ||||
| PE CD34 | PE rat IgG2a, κ | BioLegend | 152204 | AB_2629648 |
| FITC Sca1 (Ly-6A/E) | FITC rat IgG2a, κ | BioLegend | 108106 | AB_313343 |
| Antigen | Company | Cat. no. | RRID |
|---|---|---|---|
| Mesenchymal stem cell | |||
| APC CD29 | BioLegend | 303018 | AB_2130080 |
| PE CD146 | BioLegend | 361006 | AB_2562981 |
| FITC CD90 | BioLegend | 328108 | AB_893429 |
| Macrophage/osteoclast | |||
| PE RANKL (CD254) | BioLegend | 347504 | AB_2256264 |
| FITC CD11b | BioLegend | 101206 | AB_312789 |
| PerCP-Cy5.5 CD68 | BioLegend | 333814 | AB_10683171 |
| APC CD115 | BioLegend | 347306 | AB_2562441 |
| Osteoblast | |||
| APC alkaline phosphatase | R&D Systems | FAB1448A | AB_357039 |
| FITC CD90 | BioLegend | 328108 | AB_893429 |
| Osteocyte | |||
| FITC SPARC | R&D Systems | IC941G | AB_2827438 |
| PE podoplanin (GP38) | BioLegend | 337004 | AB_1595457 |
| Hematopoietic stem cell | |||
| FITC CD34 | BioLegend | 343504 | AB_1731852 |
| APC CD45 | BioLegend | 304012 | AB_314400 |
3.Resuspend cells in PBS at 1 × 107 cells per ml.
4.For live/dead staining: Incubate cells with 1 µl live/dead staining solution per 1 ml cell solution for 20 min at room temperature. Wash cells and resuspend in flow cytometry staining medium at 1 × 107 cells per ml.
5.Block samples with 5 µl appropriate FcR blocking solution per 107 cells for 30 min on ice.
6.Aliquot 100 µl samples into each staining group, including unstained control, into 1.5-ml microcentrifuge tubes or 96-well plates.
7.Stain with primary antibody for 30 min on ice in the dark.
8.Wash twice in flow cytometry staining medium, centrifuging 5 min at 175 × g , room temperature.
9a. _For cell sorting (_Basic Protocol Basic Protocol 3 ) : Resuspend cells in a flow cytometry tube containing 100 µl cell sorting buffer.
9b. _For flow cytometry (_Alternate Protocol Alternate Protocol 3 ) : Resuspend sample in 100 µl of 1% methanol-free formaldehyde in PBS (1 × 106 cells per 100 µl) while mixing to prevent clumping, and move to flow cytometry tube for flow cytometry.
10.Store covered in foil at 4°C until ready to run cell sorting (Basic Protocol 3) or flow cytometry analysis (Alternate Protocol 3).
Support Protocol 1: FLUORESCENCE MINUS ONE CONTROL, ISOTYPE CONTROL, AND ANTIBODY TITRATION
This protocol describes controls that can be used to compensate for spectral overlap and to improve staining. Single color compensation controls and an unstained control are run with each experiment to determine autofluorescence. Antibody titration is performed to determine the optimal staining concentration of antibodies for high signal-to-noise peak expression while retaining identification of cell markers prior to collecting experimental data. FMO controls are conducted to assess the proper gating of positive cell populations due to the spread of the data along the log axis at the optimal antibody concentration for each multi-fluorophore staining group (Roederer, 2001). If there is a spectral spread between channels that is not compensated by the single color stained beads, then FMO controls must be run every time. FMO analysis should be performed on fixed and unfixed cells representative of the cells being analyzed in experiments. Isotype controls, while controversial (Herzenberg, Tung, Moore, Herzenberg, & Parks, 2006; Hulspas, O'Gorman, Wood, Gratama, & Sutherland, 2009; Maecker & Trotter, 2006), can be run for each experimental group (i.e., animal age, genotype, subject) to serve as controls for nonspecific staining, which can be a problem for rare cell populations. Isotype control antibodies must be at the same final concentration and fluorophore-to-protein ratio as the experimental antibody.
Additional Materials (also see Basic Protocol 2)
- Isotype antibodies
1.Follow Basic Protocol 2 to prepare and stain cells. Add groups for FMO analysis, isotype controls, and antibody titration.
2.For antibody titration, prepare antibody dilutions as described in Table 5 for murine samples and Table 6 for primate samples at 25%, 50%, 100%, and 200% of manufacturer-recommended volume.
3.For FMO controls, set up staining groups as shown in Table 7 for murine samples and Table 8 for primate samples.
| Antigen | FITC | PE | PerCP-Cy5.5 | APC | |
|---|---|---|---|---|---|
| Mesenchymal stem cells | |||||
| Sca1 (Ly-6A/E) FMO | ‒ | CD146 | CD90 | CD29 | |
| CD146 FMO | Sca1 | ‒ | CD90 | CD29 | |
| CD90 FMO | Sca1 | CD146 | ‒ | CD29 | |
| CD29 FMO | Sca1 | CD146 | CD90 | ‒ | |
| Macrophage/osteoclast | |||||
| CD11b FMO | ‒ | CD265 | CD68 | CD115 | |
| CD265 (RANK) FMO | CD11b | ‒ | CD68 | CD115 | |
| CD68 FMO | CD11b | CD265 | ‒ | CD115 | |
| CD115 FMO | CD11b | CD265 | CD68 | ‒ | |
| Osteoblast | |||||
| Alkaline phosphatase FMO | ‒ | ‒ | CD90 | ‒ | |
| CD90 FMO | ‒ | ‒ | ‒ | AlkPhos | |
| Osteocyte | |||||
| SPARC FMO | ‒ | GP38 | ‒ | ‒ | |
| Podoplanin (GP38) FMO | ‒ | ‒ | ‒ | SPARC | |
| Hematopoietic stem cells | |||||
| CD34 FMO | Sca1 | ‒ | ‒ | ‒ | |
| Sca1 (Ly-6A/E) FMO | ‒ | CD34 | ‒ | ‒ | |
-
AlkPhos, alkaline phosphatase; FMO, fluorescence minus one; SPARC, secreted protein acidic and rich in cysteine.
- a
FMO control panel was conducted to identify fluorochrome bleed over with cell-specific antibody mixes. Results indicated no fluorochrome bleed over into the FMO channel.
| Antigen | FITC | PE | PerCP-Cy5.5 | APC | |
|---|---|---|---|---|---|
| Mesenchymal stem cells | |||||
| CD146 FMO | CD90 | ‒ | ‒ | CD29 | |
| CD90 FMO | ‒ | CD146 | ‒ | CD29 | |
| CD29 FMO | CD90 | CD146 | ‒ | ‒ | |
| Macrophage/osteoclast | |||||
| CD11b FMO | ‒ | CD254 | CD68 | CD115 | |
| CD254 (RANKL) FMO | CD11b | ‒ | CD68 | CD115 | |
| CD68 FMO | CD11b | CD254 | ‒ | CD115 | |
| CD115 FMO | CD11b | CD254 | CD68 | ‒ | |
| Osteoblast | |||||
| Alkaline phosphatase FMO | ‒ | ‒ | CD90 | ‒ | |
| CD90 FMO | ‒ | ‒ | ‒ | AlkPhos | |
| Osteocyte | |||||
| SPARC FMO | ‒ | GP38 | ‒ | ‒ | |
| Podoplanin (GP38) FMO | ‒ | ‒ | ‒ | SPARC | |
| Hematopoietic stem cells | |||||
| CD34 FMO | ‒ | ‒ | ‒ | CD45 | |
| CD45 FMO | CD34 | ‒ | ‒ | ‒ | |
-
AlkPhos, alkaline phosphatase; FMO, fluorescence minus one; SPARC, secreted protein acidic and rich in cysteine.
- a
FMO control panel was conducted to identify fluorochrome bleed over with cell-specific antibody mixes. Results indicated no fluorochrome bleed over into the FMO channel.
4.For isotype controls, set up staining groups as shown in Table 9 for murine samples and Table 10 for primate samples.
| Isotype control | Company | Cat. no. | RRID |
|---|---|---|---|
| Mesenchymal stem cell | |||
| FITC rat IgG2a, κ | BioLegend | 400506 | AB_273619 |
| PE rat IgG2a, κ | BioLegend | 400508 | AB_326530 |
| APC hamster IgG | BioLegend | 402012 | AB_2814701 |
| PerPC-Cy5.5 rat IgG2b, κ | BioLegend | 400632 | AB_893677 |
| Macrophage/osteoclast | |||
| FITC rat IgG2b, κ | BioLegend | 400634 | AB_893662 |
| APC rat IgG2a, κ | BioLegend | 400512 | AB_2814702 |
| PerPC-Cy5.5 rat IgG2b, κ | BioLegend | 400632 | AB_893677 |
| PE rat IgG2a, κ | BioLegend | 400508 | AB_326530 |
| Osteoblast | |||
| APC rat IgG1, κ | BD Biosciences | 555751 | AB_398613 |
| PerCP-Cy5.5 rat IgG2b, κ | BioLegend | 400632 | AB_893677 |
| Osteocyte | |||
| PE hamster IgG | BioLegend | 400923 | AB_2814703 |
| APC rat IgG2b, κ | BioLegend | 400612 | AB_326556 |
| Hematopoietic stem cell | |||
| PE rat IgG2a, κ | BioLegend | 400508 | AB_326530 |
| FITC rat IgG2a, κ | BioLegend | 400506 | AB_273619 |
- a
Isotype control mixes were used as a negative control to determine the contribution of nonspecific background to staining and correct for nonspecific binding. Results indicated no nonspecific binding for cell identification schema.
| Isotype control | Company | Cat. no. | RRID | |
|---|---|---|---|---|
| Mesenchymal stem cell | ||||
| APC mouse IgG1, κ | BioLegend | 400120 | AB_2888687 | |
| PE mouse IgG1, κ | BioLegend | 400114 | AB_326435 | |
| FITC mouse IgG1, κ | BioLegend | 400110 | AB_2861401 | |
| Macrophage/osteoclast | ||||
| PE mouse IgG2b, κ | BioLegend | 400314 | NA | |
| FITC rat IgG2b, κ | BioLegend | 400634 | AB_893662 | |
| PerCP-Cy5.5 mouse IgG2b, κ | BioLegend | 400338 | NA | |
| APC rat IgG1, κ | BioLegend | 400412 | AB_326518 | |
| Osteoblast | ||||
| APC mouse IgG1, κ | BioLegend | 400120 | AB_2888687 | |
| FITC mouse IgG1, κ | BioLegend | 400110 | AB_2861401 | |
| Osteocyte | ||||
| FITC mouse IgG1, κ | BioLegend | 400110 | AB_2861401 | |
| PE rat IgG2a, κ | BioLegend | 400508 | AB_326530 | |
| Hematopoietic stem cell | ||||
| FITC mouse IgG1, κ | BioLegend | 400110 | AB_2861401 | |
| APC mouse IgG1, κ | BioLegend | 400120 | AB_2888687 | |
-
NA, not available.
- a
Isotype control mixes were utilized as a negative control to determine the contribution of nonspecific background to staining and correct for nonspecific binding. Results indicated no nonspecific binding for cell-identification schema.
Basic Protocol 3: CELL SORTING OF BONE STROMAL CELLS
This protocol describes the sorting of stained, isolated bone stromal cells for downstream analyses. At the end of the experiment, individual hematopoietic lineage and mesenchymal lineage cells are separated into tubes. The sorting report will include the number of cells isolated and percentages of stained cells.
Materials
-
Samples of interest (see Basic Protocol 2)
-
Stained composition beads (see Support Protocol 2)
-
Cell sorting buffer (see recipe)
-
Collection tubes
-
Fluorescence-activated cell sorter machine (e.g., Beckman Coulter Astrios EQ, BD FACSAria, or equivalent)
Cell sorting
1.Take samples and prepared compensation beads on ice to the cell sorting facility.
2.Prepare collection tubes with 2 ml cell sorting buffer.
3.Prepare side scatter (SSC) and forward scatter (FSC) gates based on unstained controls. Set photomultiplier tube (PMT) voltages according to procedures outlined in Maecker & Trotter (2006) ensuring that weakly stained cells will be separated from electronic noise and that all positive signals are on scale.
4.Run compensation beads and live/dead controls to generate a compensation matrix. Also, use beads to adjust FSC and SSC.
5.Run a single-stained bead sample to ensure positive peaks are on scale.
6.Rerun single-stained bead samples to perform compensation setup, and record files for compensation controls.
7.Set a gate around the SSC height (SSC-H) versus FSC height (FSC-H) scatter to remove debris.
8.Set a gate around the singlet population using FSC-H versus FSC area (FSC-A), and use it for further analysis (murine: Fig. 3A; primate: Fig. 4A).
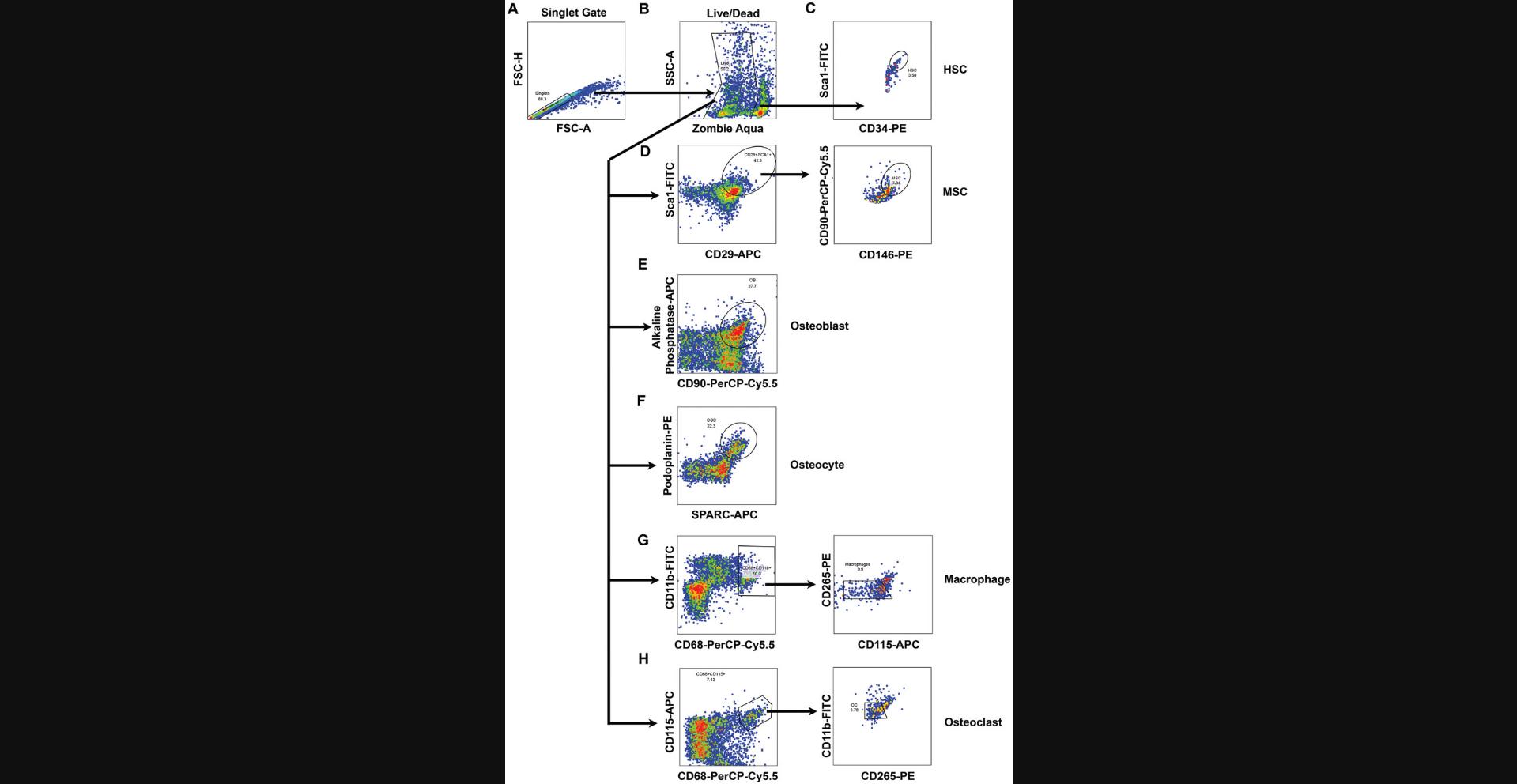
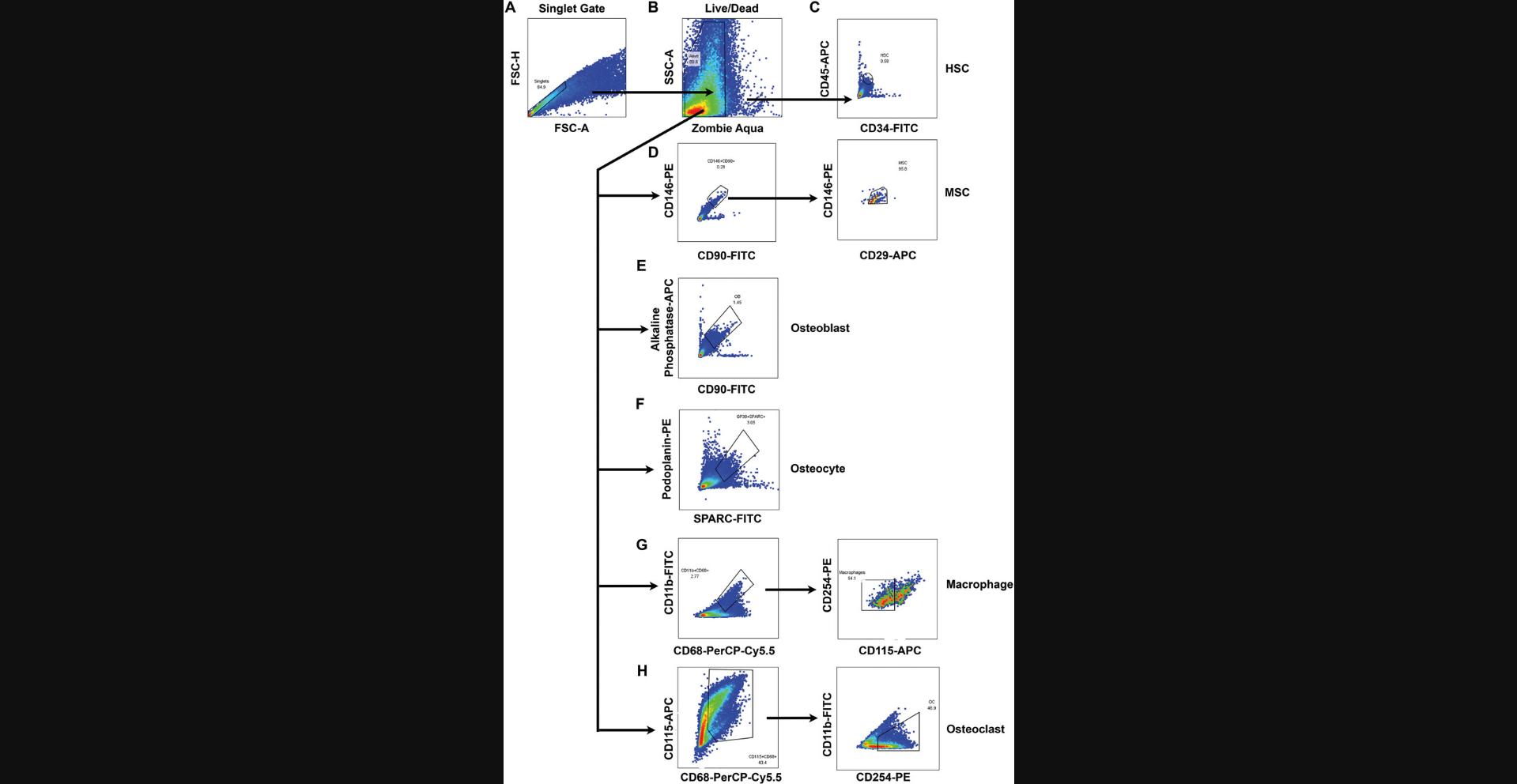
9.Gate live, Zombie Aqua (AmCyan channel) negative cells (Zombie Aqua area vs. SSC area [SSC-A]; murine: Fig. 3B; primate: Fig. 4B).
Data analysis
Hematopoietic stem cells
10a. Within the live, singlet population gated above, for murine samples, gate on the Sca1 high and CD34 high population (Fig. 3C). For primate samples, gate the CD45 high and CD34 high population (Fig. 4C).
Mesenchymal stem cells
10b. Within the live, singlet population gated above, for murine samples, gate on the Sca1 high and CD29 high population, and then gate the CD90 high and CD146 high population (Fig. 3D). For primate samples, within the live, singlet population gated above, gate on the CD146 high and CD90 high population, and then gate the CD146 high and CD29 high population (Fig. 4D).
Osteoblasts
10c. Within the live, singlet population gated above, gate on the alkaline phosphatase high and CD90 high population (murine: Fig. 3E; primate: Fig. 4E).
Osteocytes
10d. Within the live, singlet population gated above, gate on the SPARC high and podoplanin high population (murine: Fig. 3F; primate: Fig. 4F).
Macrophages
10e. Within the live, singlet population gated above, for murine samples, gate on the CD11b high and CD68 high population followed by the CD265 low and CD115 low population (Fig. 3G). For primate samples, gate on the CD11b high and CD68 high population followed by the CD254 low and CD115 low population (Fig. 4G).
Osteoclasts
10f. Within the live, singlet population gated above, for murine samples, gate on the CD68 high and CD115 high population followed by the CD11b low and CD265 high population (Fig. 3H). For primate samples, gate on the CD68 high and CD115 high population followed by the CD11b low and CD254 high population (Fig. 4H).
11.Run samples according to your sorter's manufacturer's protocol.
12.Place sorted samples on ice for downstream experiments.
13.Retain sorting reports with cell counts and percentage of stained cell data.
Alternate Protocol 3: FLOW CYTOMETRY ANALYSIS OF BONE STROMAL CELLS
This protocol describes the counting of stained, isolated bone-residing stromal cells by flow cytometry and data analysis to examine the percentages of stained cells in the total cell sample.
Additional Materials (also see Basic Protocol 3)
- Stained composition beads (see Support Protocol 2)
- Flow cytometer (e.g., BD FACS Canto II or equivalent)
- Flow cytometry analysis software (e.g., FlowJo v10.6.1 or equivalent)
Flow cytometry
1.Run compensation beads to generate a compensation matrix.
2.Generate unstained gating controls for SSC and FSC using unstained samples.
3.Using compensation beads, adjust FSC and SSC to visualize beads for each channel or fluorophore used and compensation samples.
4.Run a single-stained bead sample to ensure positive peaks are on scale.
5.Rerun single-stained bead samples to perform compensation setup, and record files for compensation controls. Set an FSC versus SSC gate to remove cell debris. Gate FSC-A versus FSC-A around the singlet population, and use it for further analysis.
6.Run samples according to flow cytometer manufacturer's protocol recording ≥100,000 events per sample. Export files for further analysis.
Data analysis
7.Open flow cytometry data in an appropriate gating software. Create a scatter gate of SSC-H versus FSC-H to remove any debris.
8.Use FSC-H versus FSC-A to create a singlets gate to remove doublets (murine: Fig. 3A; primate: Fig. 4A).
Hematopoietic stem cells
9a. Within the population gated above, for murine samples, gate on the Sca1 high and CD34 high population (Fig. 3C). For primate samples, within the population gated above, gate the CD45 high and CD34 high population (Fig. 4C).
Mesenchymal stem cells
9b. Within the population gated above, for murine samples, gate on the Sca1 high and CD29 high population, and then gate the CD90 high and CD146 high population (Fig. 3D). For primate samples, within the population gated above, gate on the CD146 high and CD90 high population, and then gate the CD146 high and CD29 high population (Fig. 4D).
Osteoblasts
9c. Within the population gated above, gate on the alkaline phosphatase high and CD90 high population (murine: Fig. 3E; primate: Fig. 4E).
Osteocytes
9d. Within the population gated above, gate on the SPARC high and podoplanin high population (murine: Fig. 3F; primate: Fig. 4F).
Macrophages
9e. Within the population gated above, for murine samples, gate on the CD11b high and CD68 high population followed by the CD265 low and CD115 low (Fig. 3G). For primate samples, gate on the CD11b high and CD68 high population followed by the CD254 low and CD115 low (Fig. 4G).
Osteoclasts
9f. Within the population gated above, for murine samples, gate on the CD68 high and CD115 high population followed by the CD11b low and CD265 high population (Fig. 3H). For primate samples, gate on the CD68 high and CD115 high population followed by the CD11b low and CD254 high population (Fig. 4H).
10.Calculate cell percentages or mean fluorescence intensities using analysis software, and graph to profile changes in the numbers of bone marrow-residing cells.
Support Protocol 2: PREPARING COMPENSATION BEADS
This protocol describes how to prepare compensation beads for cell sorting and flow cytometry. Compensation beads are used to set voltages and gating parameters while running samples to obtain accurate data across experiments. The use of compensation beads for this protocol is crucial as the cells being examined are rare within the sample. Further information on compensation beads in flow cytometry has been described by Perfetto, Ambrozak, Nguyen, Chattopadhyay, & Roederer (2006).
Materials
-
Compensation beads (e.g., UltraComp eBeads; Thermo Fisher Scientific, cat. no. 01-2222-41)
-
Antibodies of interest (see Basic Protocol 2 and Support Protocol 1)
-
Flow cytometry staining medium (see recipe)
-
1.5-ml microcentrifuge tube
-
96-well plate
-
Vortex mixer
-
Centrifuge
1.Prepare a tube or well for each fluorochrome.
2.Mix beads vigorously by inverting ≥10 times or pulse vortexing.
3.Add one drop of beads to each tube or well.
4.Add 2 µl of each antibody in 2 ml flow cytometry staining medium, and mix well by inversion or pulse vortexing.
5.Incubate at 4°C for 30 min in the dark.
6.Add 2 ml flow cytometry staining medium to each tube, and centrifuge 5 min at 400 × g , 4°C.
7.Remove supernatant and add 400 µl flow cytometry staining medium to each sample.
8.Vortex prior to running sample.
REAGENTS AND SOLUTIONS
Cell sorting buffer
- 500 ml PBS
- 100 µl 500 mM EDTA, pH 8.0 (e.g., Fisher Scientific, cat. no. S311)
- 1% (w/v) bovine serum albumin (e.g., Fisher Scientific, cat. no. BP1600)
- Store at 4°C for up to 12 months
Flow cytometry staining medium
- 500 ml Hank's buffered salt solution (e.g., Thermo Fisher Scientific, cat. no. 88284)
- 15 ml heat-inactivated fetal calf serum (3% [v/v] final; e.g., Gibco, cat. no. 21540079)
- 2 ml 500 mM EDTA, pH 8.0 (2 mM final; e.g., Sigma-Aldrich, cat. no. E134)
- 1 ml 1 M HEPES, pH 7.0 (10 mM final; e.g., Gibco, cat. no. 15-630-080)
- Store at 4°C for up to 12 months
Heat inactivate fetal calf serum for 30 min at 56°C before use.
COMMENTARY
Background Information
The bone marrow comprises two distinct stem cell lineages: hematopoietic and mesenchymal. Hematopoietic stem cells give rise to all blood cell types, macrophages, and osteoclasts. In contrast, mesenchymal stem cell differentiation pathways are responsible for generating stromal cells, osteoblasts, and osteocytes, which compose the bone marrow microenvironment (Laurenti & Göttgens, 2018). The bone-forming osteoblasts balance bone-resorbing osteoclasts to maintain bone homeostasis during development and in response to structural stressors. However, this balance is often dysregulated in disease states leading to an overabundance of bone in osteosclerosis and a loss of bone in osteoporosis. Additionally, the relative concentration of osteoclasts and osteoblasts has profound effects on the development of bone metastatic cancers, with enhanced osteoclasts resulting in tumor-induced osteolysis and increased osteoblast activity resulting in osteoblastic lesions (Jinnah, Zacks, Gwam, & Kerr, 2018). The progenitor cells also play a role in the homing and engraftment of cancer cells to the bone marrow. Thus, there is a need to characterize the composition of the bone microenvironment in healthy and disease states. There have been two major obstacles to characterizing bone marrow microenvironmental composition changes. Classification studies were (1) severely underpowered and (2) biased due to the lack of high-throughput methodologies, compounded with variance and lack of quantitation and reproducibility (Zeng & Sanes, 2017).
Thus, we sought to develop a multi-fluorophore cell identification panel to accurately identify cell-specific alterations of key bone marrow niche cell types based on markers proposed in the literature. Markers for hematopoietic stem cells and mesenchymal stem cells are already well defined (Boxall & Jones, 2012; Dominici et al., 2006). For hematopoietic stem cells, we chose CD34 and CD45 for primates, including Sca1 for mice, to identify hematopoietic progenitors capable of multiple differentiation lineages. Additional markers for hematopoietic stem cells could include CD117/c-kit and lineage negative (Lin & Goodell, 2011; Mayle, Luo, Jeong, & Goodell, 2013; Rundberg Nilsson, Bryder, & Pronk, 2013). CD117 was not included due to its high expression on mast cells and endothelial cells (Foster, Zaidi, Young, Mobley, & Kerr, 2018; Foster, Langsten, Mansour, Shi, & Kerr, 2021; Li et al., 2003). For mesenchymal stem cells, we chose the markers CD29, CD90, and CD146 for primates, with the addition of Sca1 for mice. CD29/integrin β1 and CD90/Thy1 are highly expressed by mesenchymal lineage cells (Morris, 2018; Shekaran et al., 2014). CD146 is expressed on mesenchymal stem cells in the bone marrow and peripheral tissues (Correa et al., 2016; Somoza, Correa, & Caplan, 2016). Additional markers for mesenchymal stem cells could include CD73 and CD105 (Dominici et al., 2006; Nery et al., 2013). CD105/endoglin is also associated with microvessel formation and endothelial cell differentiation (Dallas et al., 2008; Lee et al., 2012) and thus may also stain cells of the hematopoietic lineage. Therefore, CD105 is excluded from this panel. We focused on potential surface marker expression for bone-residing osteoclasts, osteoblasts, and osteocytes. Osteoclast surface marker CD265/RANK and CD254/RANKL are associated with differentiation and activity of osteoclasts (Faust et al., 1999). Thus, CD265 or CD254 expression on cells expressing the macrophage surface markers CD68 and CD115 was postulated to represent osteoclasts or preosteoclast cells. Osteoblast surface proteins were proposed to be CD90/Thy-1 and alkaline phosphatase based on their roles in bone formation and bone matrix calcification (Picke et al., 2018; Rutkovskiy et al., 2016; Trentz et al., 2010). Osteocyte markers SPARC and podoplanin (GP38) were derived from publications describing osteocyte ontogeny from osteoblasts (Bonewald, 2011; Zhu et al., 2011). Markers were chosen based on their expression on the cell surface, their ability to differentiate between bone stromal cell populations, and the commercial availability of fluorophore-conjugated antibodies. Antibodies were carefully selected with fluorophores to make a multi-fluorophore panel and have brighter fluorophores assigned to rarer cell surface proteins when possible. With this newly established multi-fluorochrome bone marrow cell-typing methodology, the cellular composition can be tracked during development and disease progression.
Critical Parameters
For best results, fresh tissues should be used for digestion. The protocols described were optimized using frozen tissues; however, cell viability after isolation is significantly decreased. If a high number of cells are required for downstream analysis after sorting, then additional samples would be needed if frozen, archival tissues are used. For cell staining, it is recommended that the antibody titration step be performed to optimize staining if any different antibodies or animal genotypes are tested. The inclusion of compensation beads in every experiment will improve the reproducibility of results between experiments. For flow cytometry, a high number of events is recommended for robust data, with ≥100,000 events being counted per sample.
Troubleshooting
See Table 11 for possible problems, causes, and solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Low signal/high noise | Contamination with B or T cells | Gate or sort B and T cells out of the target population by staining samples with PE/Cy7 or PE/Cy5 conjugated antibodies to CD3, CD4, CD19, and CD45 (Baumgarth & Roederer, 2000; McLaughlin et al., 2008; Perfetto, Chattopadhyay, & Roederer, 2004); remove cells from the analysis by gating, allowing for a higher signal to noise ratio |
| Depending on cytometer, use other fluorophores to create a “dump” channel where nontarget populations can be measured or sorted; include additional markers for B and T cells, if desired | ||
| Low cell isolation | Insufficient collagenase digestion | Incubate in collagenase I for longer times or at a higher concentration |
| Too small vessel size for tissue pieces to move freely | Cut tissues smaller to increase surface area exposed to the collagenase | |
| High autofluorescence | Fixation method; autofluorescence with formaldehyde fixation for hematopoietic lineage cells | Use another fixative to reduce autofluorescence |
| Loss of fluorescence signal | Fixation durationa | If longer storage is required or if signal degrades, perform fixation overnight, and centrifuge and resuspend in PBS for longer storage |
- a Fixation in 1% (v/v) formaldehyde should not affect fluorescence signal (including for PE and APC) if samples are run within 2 weeks.
Understanding Results
This protocol was developed to reliably isolate bone marrow‒derived cells for downstream fluorescence-activated cell-type identification and to isolate mineralized bone cells including osteoblasts, osteoclasts, and osteocytes. In initial optimization for murine long bones, digestions were conducted with 0.5, 1, 2, and 3 mg/ml collagenase I at 37°C for 0.5, 1, 2, and 4 hr to optimize cell yield and cell viability. The highest cell yield and cell viability were found at 1 mg/ml collagenase I digestion for 1 hr at 37°C. Yields are consistent across mouse age, sex, and genotype (Fig. 2). Fewer cells were isolated from female mice, although this is likely due to the smaller size of the starting material. Using this optimized procedure, 3‒10 × 106 cells are routinely recovered from each mouse. Additionally, protocols were developed and optimized to isolate cells from primate and murine vertebrae. Both procedures resulted in high numbers of cells recovered, as demonstrated in Tables 1, 2, 3, and 4. The number of cells recovered mainly indicates the efficacy of bone digestion. However, the length of digestion will also affect the viability of cells, and thus we have optimized the protocol to maximize cell number and viability.
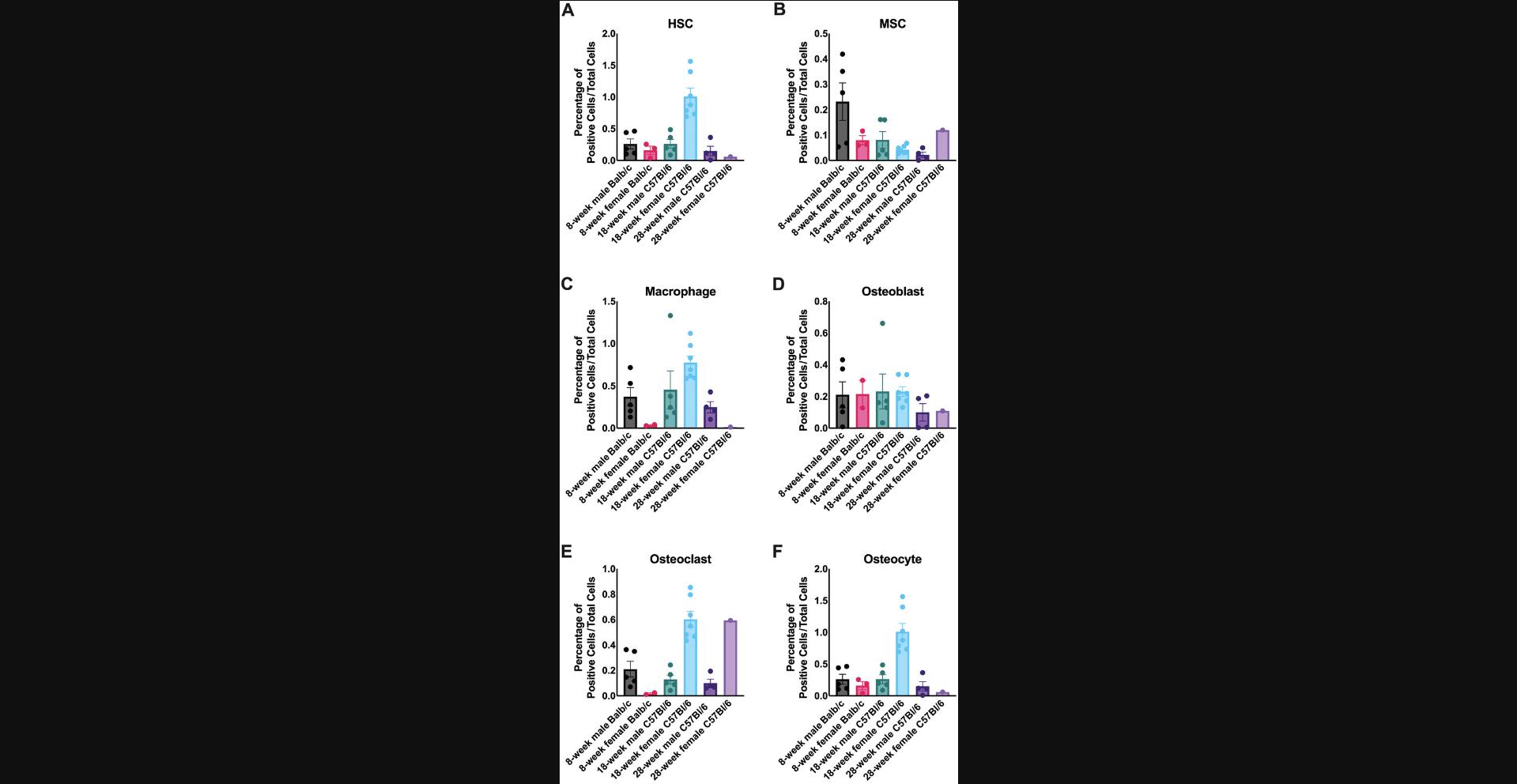
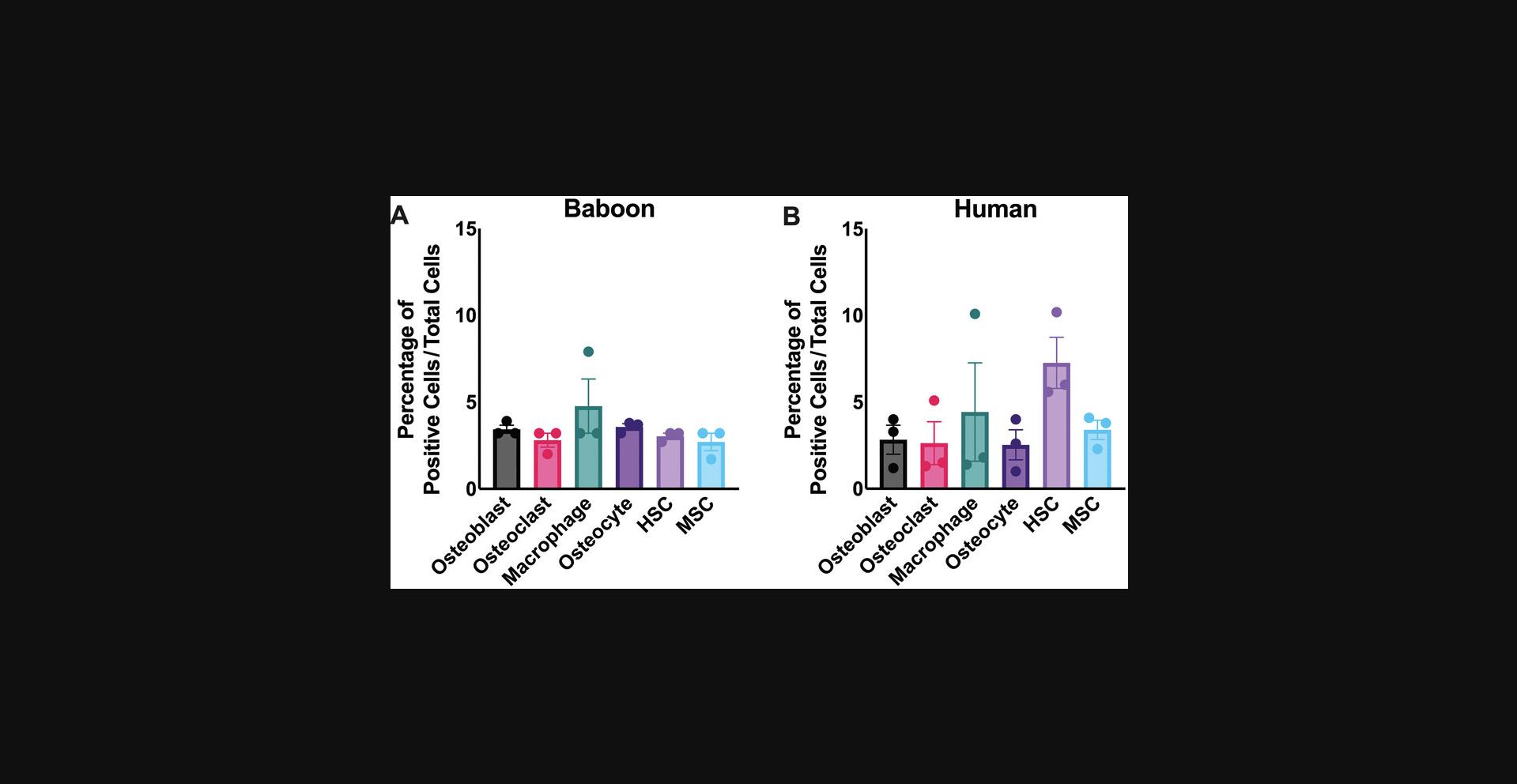
Using these freshly isolated samples, specific antibody panels for hematopoietic stem cells, mesenchymal stem cells, osteoblasts, osteoclasts, and osteocytes were created to identify bone marrow and stromal cells (Fig. 1). These cells were analyzed by either cell sorting or flow cytometry using the gating schemes shown in Figures 3 and 4. Using these analysis protocols, the numbers of progenitor cells vary with sex and age. Aged female mice demonstrate increased osteoclast numbers, whereas osteoblast numbers are only reduced in older mice (both male and female; Fig. 5). These data are consistent with known results of bone aging and sex differences. Between primates, humans, and baboons the cellular composition in the bone marrow and stromal compartment were consistent (Fig. 6). These data indicate that the results from nonhuman primates could be translatable to humans in future interventional studies. Either the total cell number or percentage of cells analyzed can be used to demonstrate cellular composition in the bone microenvironment. The cell count can be particularly important for downstream applications where the exact cell number is essential for reproducible results.
Time Considerations
Partial digestion for murine long bone stromal cell isolation in Basic Protocol 1 takes 1 to 3 hr, whereas partial digestion for primate vertebrae stromal cell isolation takes 57 to 58 hr. Murine vertebrae crushing for bone stromal cell isolation in Alternate Protocol 2 requires 1 to 3 hr. Staining of bone stromal cells in Basic Protocol 2 takes 4 to 5 hr, and preparing fluorescence minus one controls, isotype controls, and performing antibody titration in Support Protocol 1 takes 4 to 6 hr. Performing cell sorting of bone stromal cells (Basic Protocol 3) and flow cytometry analysis of bone stromal cells (Alternate Protocol 3) takes 4 to 8 hr for each protocol, and preparing compensation beads in Support Protocol 2 takes 45 min. Times will vary based on the number of samples.
Acknowledgments
The authors thank the Elsa U. Pardee Foundation and the Wake Forest School of Medicine Clinical and Translational Science Institute (NIH UL1 TR001420) for supporting this study. The flow cytometry core services were supported by a grant to the Wake Forest Baptist Comprehensive Cancer Center, NCI CCSG P30 CA012197. Care of animals in the SNPRC pedigree was supported by NIH grant P51 OD011133.
Author Contributions
Chirayu Patel : formal analysis, investigation, methodology, visualization, writing—original draft, review, and editing; Lihong Shi : formal analysis, visualization, writing—review and editing; John F. Whitesides : formal analysis, investigation, writing—review and editing; Brittni M. Foster : investigation, validation, writing—review and editing; Roberto J. Fajardo : resources, writing—review and editing; Ellen E. Quillen : conceptualization, funding acquisition, writing—review and editing; Bethany A. Kerr : conceptualization, funding acquisition, methodology, project administration, supervision, visualization, writing—original draft, review, and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data, tools, and material (or their source) that support the protocol are available from the corresponding author upon reasonable request.
Literature Cited
- Baumgarth, N., & Roederer, M. (2000). A practical approach to multicolor flow cytometry for immunophenotyping. Journal of Immunological Methods , 243, 77–97. doi: 10.1016/S0022-1759(00)00229-5.
- Bonewald, L. F. (2011). The amazing osteocyte. Journal of Bone and Mineral Research , 26, 229–238. doi: 10.1002/jbmr.320.
- Boxall, S. A., & Jones, E. (2012). Markers for characterization of bone marrow multipotential stromal cells. Stem Cells International , 2012, 975871. doi: 10.1155/2012/975871.
- Correa, D., Somoza, R. A., Lin, P., Schiemann, W. P., & Caplan, A. I. (2016). Mesenchymal stem cells regulate melanoma cancer cells extravasation to bone and liver at their perivascular niche. International Journal of Cancer , 138, 417–427. doi: 10.1002/ijc.29709.
- Dallas, N. A., Samuel, S., Xia, L., Fan, F., Gray, M. J., Lim, S. J., & Ellis, L. M. (2008). Endoglin (CD105): A marker of tumor vasculature and potential target for therapy. Clinical Cancer Research , 14, 1931–1937. doi: 10.1158/1078-0432.CCR-07-4478.
- Dominici, M., le Blanc, K., Mueller, I., Slaper-Cortenbach, I., Marini, F. C., Krause, D. S., … Horwitz, E. M. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy , 8, 315–317. doi: 10.1080/14653240600855905.
- Faust, J., Lacey, D. L., Hunt, P., Burgess, T. L., Scully, S., Van, G., … Shalhoub, G. (1999). Osteoclast markers accumulate on cells developing from human peripheral blood mononuclear precursors. Journal of Cellular Biochemistry , 72, 67–80. doi: 10.1002/(sici)1097-4644(19990101)72:1<67::aid-jcb8>3.0.co;2-a.
- Foster, B. M., Langsten, K. L., Mansour, A., Shi, L., & Kerr, B. A. (2021). Tissue distribution of stem cell factor in adults. Experimental and Molecular Pathology , 122, 104678. doi: 10.1016/j.yexmp.2021.104678.
- Foster, B., Zaidi, D., Young, T., Mobley, M., & Kerr, B. (2018). CD117/c-kit in cancer stem cell-mediated progression and therapeutic resistance. Biomedicines , 6, 31. doi: 10.3390/biomedicines6010031.
- Herzenberg, L. A., Tung, J., Moore, W. A., Herzenberg, L. A., & Parks, D. R. (2006). Interpreting flow cytometry data: A guide for the perplexed. Nature Immunology , 7, 681–685. doi: 10.1038/ni0706-681.
- Hulspas, R., O'Gorman, M. R. G., Wood, B. L., Gratama, J. W., & Sutherland, D. R. (2009). Considerations for the control of background fluorescence in clinical flow cytometry. Cytometry. Part B, Clinical Cytometry , 76, 355–364. doi: 10.1002/cyto.b.20485.
- Jinnah, A., Zacks, B., Gwam, C., & Kerr, B. (2018). Emerging and established models of bone metastasis. Cancers , 10, 176. doi: 10.3390/cancers10060176.
- Laurenti, E., & Göttgens, B. (2018). From haematopoietic stem cells to complex differentiation landscapes. Nature , 553, 418–426. doi: 10.1038/nature25022.
- Lee, N. Y., Golzio, C., Gatza, C. E., Sharma, A., Katsanis, N., & Blobe, G. C. (2012). Endoglin regulates PI3-kinase/Akt trafficking and signaling to alter endothelial capillary stability during angiogenesis. Molecular Biology of the Cell , 23, 2412–2423. doi: 10.1091/mbc.e11-12-0993.
- Li, T.-S., Hamano, K., Nishida, M., Hayashi, M., Ito, H., Mikamo, A., & Matsuzaki, M. (2003). CD117+ stem cells play a key role in therapeutic angiogenesis induced by bone marrow cell implantation. American Journal of Physiology. Heart and Circulatory Physiology , 285, H931–H937. doi: 10.1152/ajpheart.01146.2002.
- Lin, K. K., & Goodell, M. A. (2011). Detection of hematopoietic stem cells by flow cytometry. Methods in Cell Biology , 103, 21–30. doi: 10.1016/B978-0-12-385493-3.00002-4.
- Maecker, H. T., & Trotter, J. (2006). Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry. Part A , 69, 1037–1042. doi: 10.1002/cyto.a.20333.
- Mayle, A., Luo, M., Jeong, M., & Goodell, M. A. (2013). Flow cytometry analysis of murine hematopoietic stem cells. Cytometry. Part A , 83, 27–37. doi: 10.1002/cyto.a.22093.
- McLaughlin, B. E., Baumgarth, N., Bigos, M., Roederer, M., de Rosa, S. C., Altman, J. D., … Asmuth, D. M. (2008). Nine-color flow cytometry for accurate measurement of T cell subsets and cytokine responses. Part I: Panel design by an empiric approach. Cytometry, Part A , 73, 400–410. doi: 10.1002/cyto.a.20555.
- Morris, A. (2018). THY1 membrane glycoprotein linked to osteogenesis. Nature Reviews Endocrinology , 14, 564–564. doi: 10.1038/s41574-018-0086-1.
- Nery, A. A., Nascimento, I. C., Glaser, T., Bassaneze, V., Krieger, J. E., & Ulrich, H. (2013). Human mesenchymal stem cells: From immunophenotyping by flow cytometry to clinical applications. Cytometry. Part A , 83, 48–61. doi: 10.1002/cyto.a.22205.
- Perfetto, S. P., Ambrozak, D., Nguyen, R., Chattopadhyay, P., & Roederer, M. (2006). Quality assurance for polychromatic flow cytometry. Nature Protocols , 1, 1522–1530. doi: 10.1038/nprot.2006.250.
- Perfetto, S. P., Chattopadhyay, P. K., & Roederer, M. (2004). Seventeen-colour flow cytometry: Unravelling the immune system. Nature Reviews Immunology , 4, 648–655. doi: 10.1038/nri1416.
- Picke, A.-K., Campbell, G. M., Blüher, M., Krügel, U., Schmidt, F. N., Tsourdi, E., … Saalbach, A. (2018). Thy-1 (CD90) promotes bone formation and protects against obesity. Science Translational Medicine , 10, eaao6806. doi: 10.1126/scitranslmed.aao6806.
- Roederer, M. (2001). Spectral compensation for flow cytometry: Visualization artifacts, limitations, and caveats. Cytometry , 45, 194–205. doi: 10.1002/1097-0320(20011101)45:3%3c194::AID-CYTO1163%3e3.0.CO;2-C.
- Rundberg Nilsson, A., Bryder, D., & Pronk, C. J. H. (2013). Frequency determination of rare populations by flow cytometry: A hematopoietic stem cell perspective. Cytometry. Part A , 83, 721–727. doi: 10.1002/cyto.a.22324.
- Rutkovskiy, A., Stensløkken, K.-O., & Vaage, I. J. (2016). Osteoblast differentiation at a glance. Medical Science Monitor Basic Research , 22, 95–106. doi: 10.12659/MSMBR.901142.
- Shekaran, A., Shoemaker, J. T., Kavanaugh, T. E., Lin, A. S., LaPlaca, M. C., Fan, Y., … García, A. J. (2014). The effect of conditional inactivation of beta 1 integrins using twist 2 Cre, osterix Cre and osteocalcin Cre lines on skeletal phenotype. Bone , 68, 131–141. doi: 10.1016/j.bone.2014.08.008.
- Somoza, R. A., Correa, D., & Caplan, A. I. (2016). Roles for mesenchymal stem cells as medicinal signaling cells. Nature Protocols , 11. Retrieved from https://www.nature.com/nprot/posters.
- Trentz, O. A., Arikketh, D., Sentilnathan, V., Hemmi, S., Handschin, A. E., de Rosario, B., … Mohandas, P. V. A. (2010). Surface proteins and osteoblast markers: Characterization of human adipose tissue-derived osteogenic cells. European Journal of Trauma and Emergency Surgery , 36, 457–463. doi: 10.1007/s00068-010-0030-0.
- Zeng, H., & Sanes, J. R. (2017). Neuronal cell-type classification: Challenges, opportunities and the path forward. Nature Reviews Neuroscience , 18, 530–546. doi: 10.1038/nrn.2017.85.
- Zhu, D., Mackenzie, N. C. W., Millán, J. L., Farquharson, C., & MacRae, V. E. (2011). The appearance and modulation of osteocyte marker expression during calcification of vascular smooth muscle cells. PLoS One , 6, e19595. doi: 10.1371/journal.pone.0019595.
Internet Resources
Regularly updated list of phenotypic markers for progenitor cells of the hematopoietic lineage.
Regularly updated list of surface and secreted markers for osteoblasts and osteoclasts.
Recommended antibody titration protocol.
Resource describing how best to optimize multi-fluorophore flow cytometry staining.
Good overview of the principles of flow cytometry with detailed explanations on preparing controls and optimizing flow data.
Citing Literature
Number of times cited according to CrossRef: 4
- Zengxin Jiang, Guobin Qi, Xuecheng He, Yifan Yu, Yuting Cao, Changqing Zhang, Weiguo Zou, Hengfeng Yuan, Ferroptosis in Osteocytes as a Target for Protection Against Postmenopausal Osteoporosis, Advanced Science, 10.1002/advs.202307388, 11 , 12, (2024).
- Charles M. Bowlby, Devina Purmessur, Sushmitha S. Durgam, Equine peripheral blood CD14+ monocyte-derived macrophage in-vitro characteristics after GM-CSF pretreatment and LPS+IFN-γ or IL-4+IL-10 differentiation, Veterinary Immunology and Immunopathology, 10.1016/j.vetimm.2022.110534, 255 , (110534), (2023).
- O. Krasnova, I. Neganova, Assembling the Puzzle Pieces. Insights for in Vitro Bone Remodeling, Stem Cell Reviews and Reports, 10.1007/s12015-023-10558-6, (2023).
- Brittni M. Foster, Lihong Shi, Koran S. Harris, Chirayu Patel, Victoria E. Surratt, Kendall L. Langsten, Bethany A. Kerr, Bone Marrow-Derived Stem Cell Factor Regulates Prostate Cancer-Induced Shifts in Pre-Metastatic Niche Composition, Frontiers in Oncology, 10.3389/fonc.2022.855188, 12 , (2022).

