A Human Neuron/Astrocyte Co-culture to Model Seeded and Spontaneous Intraneuronal Tau Aggregation
Kevin Llewelyn Batenburg, Kevin Llewelyn Batenburg, Susan Karijn Rohde, Susan Karijn Rohde, Paulien Cornelissen-Steijger, Paulien Cornelissen-Steijger, Nicole Breeuwsma, Nicole Breeuwsma, Vivi Majella Heine, Vivi Majella Heine, Wiep Scheper, Wiep Scheper
Abstract
Communication and contact between neurons and astrocytes is important for proper brain physiology. How neuron/astrocyte crosstalk is affected by intraneuronal tau aggregation in neurodegenerative tauopathies is largely elusive. Human induced pluripotent stem cell (iPSC)-derived neurons provide the opportunity to model tau pathology in a translationally relevant in vitro context. However, current iPSC models inefficiently develop tau aggregates, and co-culture models of tau pathology have thus far utilized rodent astrocytes. In this article, we describe the co-culture of human iPSC-derived neurons with primary human astrocytes in a 96-well format compatible with high-content microscopy. By lentiviral overexpression of different mutated tau variants, this protocol can be flexibly adapted for the efficient induction of seeded or spontaneous tau aggregation. We used this novel co-culture model to identify cell type–specific disease mechanisms and to provide proof of concept for intervention by antisense therapy. These results show that this human co-culture model provides a highly translational tool for target discovery and drug development for human tauopathies. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Human neuron/astrocyte co-culture for seeded and spontaneous intraneuronal tau aggregation
Support Protocol 1 : Human induced pluripotent stem cell culture
Support Protocol 2 : Human primary astrocyte culture
INTRODUCTION
Intraneuronal tau aggregation is the major pathological hallmark of neurodegenerative tauopathies and affects neurons as well as astrocytes. Despite its strong correlation with neurodegeneration, the mechanism by which tau aggregation leads to neurodegeneration is largely elusive, partly due to the limited translational value of current rodent-derived cellular models. Neurons differentiated from human induced pluripotent stem cells (iPSCs) hold a promise of better translational models of tau pathology (Ehrlich et al., 2015; García-León et al., 2018; Iovino et al., 2015; Verheyen et al., 2015). However, their broad implementation in the study of tau-mediated neurodegeneration currently entails technical challenges, including lengthy neuronal differentiation with variable efficiency. In addition, cell clustering and/or detachment from the culture plates is commonly associated with prolonged culturing of iPSC-derived neurons and may complicate downstream microscopy analyses (Kuijlaars et al., 2016).
Inefficient tau aggregation has also been a major challenge to study the cell autonomous effects in iPSC-derived neurons. Importantly, also astrocytes contribute to tau pathogenesis in a cell nonautonomous manner (Leyns & Holtzman, 2017; Reid et al., 2020; Sidoryk-Węgrzynowicz & Struzyńska, 2019; Sidoryk-Wegrzynowicz et al., 2017). Thus far, astrocytes are often used in co-cultures as a mere supporting cell type for maturation into functional human iPSC-derived neurons (Johnson et al., 2007; Muratore et al., 2014; Taga et al., 2019; Tang et al., 2013). Likely because the maturation of human iPSC-derived neurons is generally more challenging with human compared to rodent astrocytes (Rhee et al., 2019), few systems employ human iPSC-derived (Hedegaard et al., 2020) or primary human astrocytes (Kuijlaars et al., 2016), and none have yet been reported in the context of intraneuronal tau aggregation. Remarkable species-specific differences exist between human and rodent astrocytes; these differences include that human astrocytes are larger and more morphologically complex, differentially respond to immune triggers, and display greater susceptibility to oxidative stress (Hodge et al., 2019; Kelley et al., 2018; Oberheim et al., 2009; Tarassishin et al., 2014; Zhang et al., 2016). Due to these differences, the role of astrocytes in human tauopathies is currently not well understood. Altogether, this emphasizes the need to establish a sufficiently stable human co-culture that includes human astrocytes, in which intraneuronal tau aggregation can be efficiently introduced.
Here we provide a protocol to generate microwell co-cultures of human iPSC-derived neurons and primary human astrocytes. We show that Geltrex coating greatly prevented cell clustering and detachment. This system can be employed to model seeded tau aggregation by overexpression of tau containing the P301L mutation (FTDtau1) and by subsequent treatment with tau preformed fibrils (PFFs). In addition, overexpression of tau containing the P301L and S320F mutations (FTDtau1+2) leads to spontaneous tau aggregation in the absence of seeds. Finally, we report the supporting procedures for culturing the human iPSC line and human primary astrocytes used in this study. The presented human co-culture system was used to identify disease mechanisms in neurons and astrocytes in a quantitative, cell-specific manner by high-content microscopy (Batenburg et al., 2022), thus representing a highly translational tool to identify and validate novel therapeutic targets and interventions.
CAUTION : To ensure cultures are kept sterile, cells and media should be handled in a class 2 biosafety cabinet using standard aseptic technique.
CAUTION : Reagent-impermeable protective gloves should be worn when handling reagents, cells, viral particles, and PFFs.
NOTE : For dilutions, the dilution factor (i.e., 1:2 for 50% [v/v]) is used unless otherwise stated.
NOTE : All protocols involving humans or human study material should be reviewed and approved by an institutional review board or independent ethics committee. Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol: HUMAN NEURON/ASTROCYTE CO-CULTURE FOR SEEDED AND SPONTANEOUS INTRANEURONAL TAU AGGREGATION
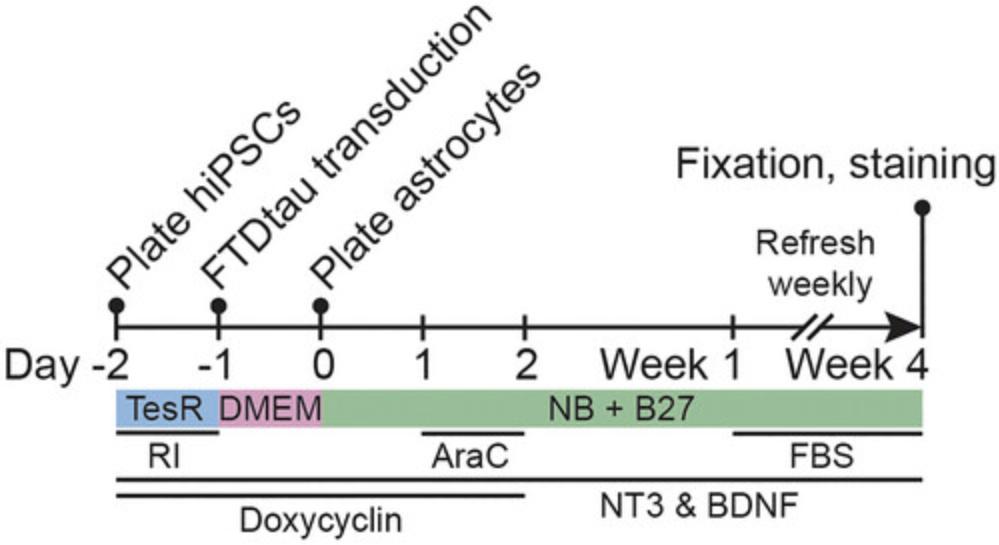
This protocol describes the generation of a human neuron/astrocyte co-culture to model seeded and spontaneous intraneuronal tau aggregation. A visual representation of the co-culture protocol is shown in Figure 1.Before the experiment, human iPSCs with a stably integrated, doxycycline-inducible neurogenin 2 (Ngn2) transgene are cultured as described in Support Protocol 1, and human primary astrocytes are cultured as described in Support Protocol 2.At day –2, human iPSCs are plated in TesR-E8 medium in the presence of a Rho kinase (ROCK) inhibitor (RI). After 1 day, the medium is switched to Dulbecco's modified Eagle medium (DMEM), and iPSC-derived neural progenitors are virally transduced for FTDtau variants. At day 0, human primary astrocytes are plated on top of iPSC-derived neural progenitors in neurobasal medium with neurotrophic factors. Arabinosylcytosine (AraC) treatment at day 1 inhibits cell proliferation. Doxycycline supplementation until day 2 drives Ngn2 transgene expression, and neurotrophin 3 (NT3) and brain-derived neurotrophic factor (BDNF) supplementation supports neuronal differentiation. From week 1, medium is refreshed weekly and supplemented with fetal bovine serum (FBS) to support astrocyte viability until fixation and analysis at week 4.
Materials
- Geltrex (see recipe)
- DMEM/F12 (e.g., Life Technologies, cat. no. 11320033)
- TesR-E8 medium (see recipe)
- 10 μg/ml NT3 (see recipe)
- 20 μg/ml BDNF (see recipe)
- 40 mg/ml doxycycline (see recipe)
- RI (see recipe)
- Near-confluent plate of human iPSCs with stably integrated doxycycline-inducible reverse tetracycline (rtTA)/Ngn2 (see Support Protocol 1; generated as described in Frega et al., 2017) or EBiSC line BIOni010-C-13 (see Support Protocol 1 and Schmid et al., 2021; e.g., European Collection of Authenticated Cell Cultures, cat. no. 66540561)
- Gentle cell dissociation reagent (e.g., StemCell Technologies, cat. no. 07174)
- Trypan blue (e.g., Life Technologies, cat. no. 15250061)
- DMEM (see recipes)
- Viral particles encoding fluorescently tagged 2N4R human tau (microtubule-associated protein tau [MAPT]) containing the P301L (FTDtau1) or P301L+S320F (FTDtau1+2) mutations (prepared as described in Moro et al., 2021; Naldini et al., 1996)
- Accutase (e.g., Sigma-Aldrich, cat. no. SF006)
- Neurobasal medium with or without FBS (see recipes)
- Confluent plate of human primary astrocytes (see Support Protocol 2; e.g., ScienCell, cat. no. 1800)
- Phosphate-buffered saline (PBS), sterile or filtered (see recipe)
- 4 mM AraC (see recipe)
- Tau PFFs (e.g., Stressmarq, cat. no. SPR-330, or prepared as described in Peeraer et al., 2015; Wiersma et al., 2019)
- 0.1 M sodium acetate (e.g., Sigma-Aldrich, cat. no. 32319)
- 4% (w/v) paraformaldehyde (PFA; e.g., Electron Microscopy Sciences, cat. no. 15710-S)
- Methanol (MeOH), ice-cold (e.g., Millipore Sigma, cat. no. 106012)
- Blocking buffer (see recipe)
- Primary antibodies (see Table 1)
- Alexa Fluor–conjugated secondary antibodies, 1:1000 dilution (e.g., Invitrogen)
- 50 mg/ml 4′,6-diamidino-2-phenylindole (DAPI; see recipe)
-
- 96-well plates, black (e.g., Greiner, cat. no. 655090)
- 37°C, 20% O2, 5% CO2 cell culture incubator
- Parafilm M
- 1.5-ml microcentrifuge tube (e.g., Eppendorf, cat. no. 0030108051)
- 15-ml conical tubes
- Tabletop centrifuge with rotor equipped for 1.5- and 15-ml tubes
- Hemocytometer (e.g., Bürker chamber; NanoEntek, cat. no. DHC-B01)
- Aluminum foil
- Probe sonicator
| Antigen | Supplier, cat. no. | RRID | Species | Dilution |
|---|---|---|---|---|
| CD44 | DSHB, H4C4 | AB_528147 | Mouse | 1:100 |
| GFAP | Dako, Z0334 | AB_10013382 | Rabbit | 1:1000 |
| MAP2 | Millipore Sigma, AB5543 | AB_571049 | Chicken | 1:5000 |
| MC1 | Kind gift of Peter Daviesa | – | Mouse | 1:500 |
| pTau (AT100) | Thermo Fisher Scientific, MN1060 | AB_223652 | Mouse | 1:200 |
| pTau (AT8) | Thermo Fisher Scientific, MN1020 | AB_223647 | Mouse | 1:200 |
| PSD95 | Abcam, AB2723 | AB_303248 | Mouse | 1:200 |
| SYP1 | Synaptic Systems, 101004 | AB_1210382 | Guinea pig | 1:1000 |
| vGLUT2 | Synaptic Systems, 135402 | AB_2187539 | Rabbit | 1:1000 |
-
DSHB, Developmental Studies Hybridoma Bank; GFAP, glial fibrillary acidic protein; MAP2, microtubule-associated protein-2; PSD95, postsynaptic density protein 95; pTau, phosphorylated tau; SYP1, synaptophysin 1; vGLUT2, vesicular glutamate transporter 2.
-
aContact antibodies@alzforum.org.
CAUTION : Viral transduction should be carried out in a class 2 biosafety cabinet to avoid viral contamination. Prepare and use safety equipment according to local guidelines.
CAUTION : All steps for PFA and MeOH fixation should be carried out in a chemical fume hood to prevent exposure to volatile vapors.
Coat 96-well plates with Geltrex
We observed that Geltrex greatly prevented cell clustering and culture detachment, which may complicate the culture of human iPSC-derived neurons on surfaces coated with poly-L-ornithine and mouse laminin (Kuijlaars et al., 2016; Fig. 2).
![Details are in the caption following the image Geltrex-coating increases culture integrity compared with coating with poly-L-ornithine (PLO) and laminin. Representative confocal images of neurons plated on 96-well plates coated with Geltrex (1:100 in DMEM/F12, left) or PLO and mouse laminin (PLO+laminin; 20 μg/ml in phosphate-buffered saline; right) and co-cultured with human primary astrocytes for 4 weeks. Immunostaining was performed for neurons (microtubule-associated protein-2 [MAP2]), and cell nuclei were visualized using DAPI. Merged fluorescence channels are shown for MAP2 (grayscale) and DAPI (blue).](https://static.yanyin.tech/literature_test/cpz1900-fig-0002-m.jpg)
1.Thaw Geltrex aliquot at 4°C, and dilute 1:50 with cold DMEM/F12.
2.Add 100 μl diluted Geltrex per well to the inner 32 wells of a 96-well plate.
3.Incubate for 4 hr at 37°C.
4.Use immediately (continue to step 5), or seal with parafilm and incubate 1 to 3 days at 4°C.
Start neuronal induction of Ngn2-iPSCs (day –2)
Human iPSCs should lack obvious signs of differentiation. See Figure 3, left panel, for a representative image of a human iPSC colony before detachment.
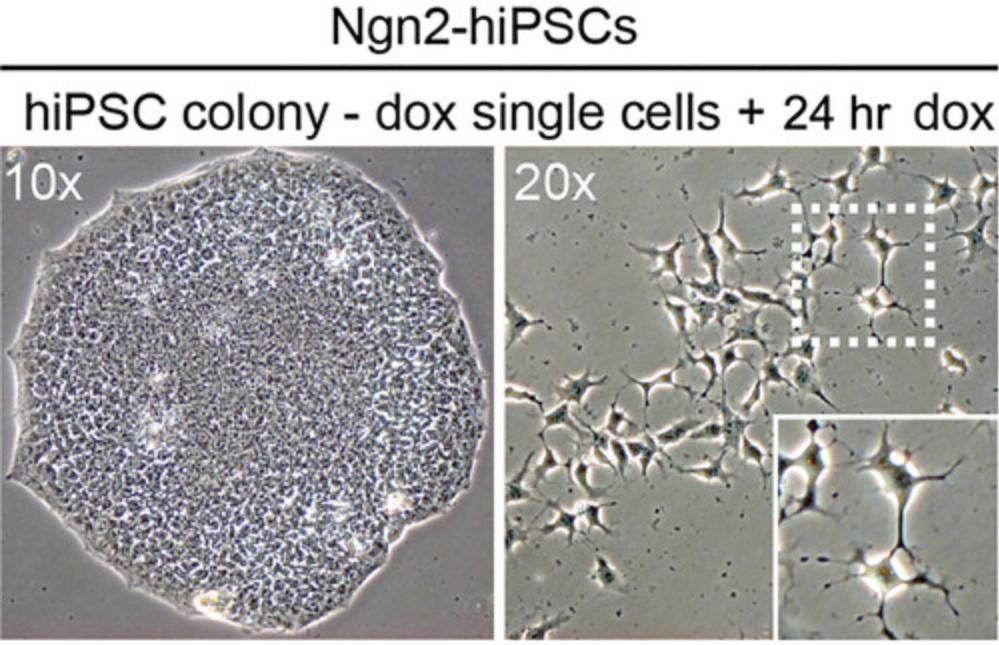
Human iPSCs should harbor a stably integrated doxycycline-inducible rtTA/Ngn2 transgene cassette. We used a previously generated clonal human iPSC line stably expressing rtTA/Ngn2 (Frega et al., 2017) that was maintained as previously described (Batenburg et al., 2022, 2023). For culture procedures for human iPSCs, see Support Protocol 1.
Human iPSCs are cultured in a 6-well plate. One near-confluent well of iPSCs in a 6-well plate gives rise to ∼4–7 × 105 cells. For co-culture, 1.3 × 105 iPSCs are needed for 32 wells in a 96-well plate. Therefore, to generate three to five co-culture plates you need one near-confluent well of iPSCs.
5.Let 96-well plate come to room temperature (RT). In the meantime, proceed to step 6.
6.Prepare TesR-E8 medium containing 10 ng/ml NT3, 10 ng/ml BDNF, 2 µg/ml doxycycline, and 10 µM RI. Prepare a total of 5 ml for dissociation of one well of iPSCs (prewarmed), 1 ml for resuspension of one well of iPSCs (prewarmed), and 7 ml for plating 32 wells in a 96-well plate (RT). Also prepare a sterile 1.5-ml microcentrifuge tube with 10 μl trypan blue.
7.Aspirate spent medium of human iPSCs with a suction needle or 5-ml serological pipette.
8.Gently add 1000 μl gentle cell dissociation reagent per well of a 6-well plate. Return plate to 37°C humidified incubator, 20% O2 and 5% CO2. Incubate for 7 to 9 min until gaps appear between cells at the border and center of the colonies.
9.Gently aspirate gentle cell dissociation reagent with a 1000-μl pipette.
10.Detach colonies by flushing with 1 ml prewarmed TesR-E8 medium (prepared in step 6). Gently make single cells by pipetting up and down with a 1000-μl pipette against the border of the well. Avoid bubble formation.
11.Collect cell suspension into a 15-ml tube containing the remaining 4 ml prewarmed TesR-E8 medium. Centrifuge 7 min at 340 × g , RT.
12.Aspirate supernatant with a suction needle or 10-ml serological pipette. Resuspend pellet in 1 ml prewarmed TesR-E8 medium.
13.Mix 10 μl cell suspension in 10 μl trypan blue, and count viable human iPSCs in a hemocytometer following the manufacturer's instruction. Dilute human iPSCs in TesR-E8 medium at RT to a density of 4000 iPSCs per 200 μl medium.
14.Remove Geltrex solution from the 96-well plate, and add 200 μl cell suspension per well using a multichannel pipette.
15.Cover 96-well plate with aluminum foil, and incubate for 15 min on a bench at RT.
16.Incubate plate overnight in a 37°C humidified incubator, 20% O2 and 5% CO2.
Transduce neural progenitors with FTDtau variants (day –1)
Compare different batches of viral vectors in order to ensure consistent transduction efficiencies across independent experiments. Since all viral vectors used in this study contain an enhanced green fluorescent protein (eGFP) tag, we recommend comparing the fluorescence intensity of different concentrations (2, 4, 6, and 8 μl/ml recommended) of newly prepared viral vector with previous viral vectors or a reference vector of known titer. This can be performed in cell culture systems of reduced complexity (e.g., rodent primary neurons or neuronal cell lines). Typical viral titers obtained with the method described in Moro et al. (2021) and Naldini et al. (1996) range from 5 × 108 to 1 × 109 infectious units/ml.
Before proceeding, confirm the formation of neurite-like cellular protrusions after 24 hr of doxycycline-induced Ngn2 expression (Fig. 3, right panel). If absent, start a new experiment, and make sure that doxycycline is present from day –2.
17.For 32 wells of a 96-well plate, prepare 8 ml DMEM containing 10 ng/ml NT3, 10 ng/ml BDNF, and 2 µg/ml doxycycline. Prewarm in water bath. With this medium, prepare viral dilutions (200 μl per well).
18.Remove spent medium using a multichannel pipette, and add 200 μl of each viral preparation per well.
19.Incubate plate for 24 hr in a 37°C humidified incubator, 20% O2 and 5% CO2.
Wash viral particles, plate primary human astrocytes on top of iPSC-derived neuronal progenitors (day 0)
Human primary astrocytes should not be overconfluent, grown under nutrition-limited conditions (yellowish medium), or lack obvious signs of cell death. We used primary human fetal astrocytes that were cultured according to distributor's instructions and as previously described (Batenburg et al., 2022, 2023). For the culture procedures of human primary astrocytes, see Support Protocol 2.
Human astrocytes are cultured in a 6-well plate. One confluent well of astrocytes in a 6-well plate gives rise to ∼0.8–1 × 106 cells. For co-culture, 1.3 × 105 astrocytes are needed for 32 wells in a 96-well plate. Therefore, to generate six to eight co-culture plates, you need one confluent well of astrocytes.
20.Let Accutase come to RT.
21.Prepare neurobasal medium containing 10 ng/ml NT3, 10 ng/ml BDNF, and 2 µg/ml doxycycline. Prepare a total of 7 ml for washing Ngn2 precursors (prewarmed), 5 min for dissociating astrocytes (prewarmed), 1 ml for resuspending astrocytes (prewarmed), and 7 ml for plating astrocytes in 32 wells of a 96-well plate (RT). Prepare 1 ml PBS for washing one well of astrocytes in a 6-well plate (prewarmed) and 13 ml PBS for filling outer wells of the 96-well plate (RT). Also prepare a sterile 1.5-ml microcentrifuge tube with 10 μl trypan blue.
22.Aspirate all medium containing viral particles from the 96-well plate using a multichannel pipette.
23.Add 200 μl freshly prepared neurobasal medium per well. Incubate for 1 hr in a 37°C humidified incubator, 20% O2 and 5% CO2. In the meantime, proceed to step 24.
24.Aspirate all spent medium from human primary astrocytes using a suction needle or 5-ml serological pipette. Wash once with 1 ml prewarmed, sterile PBS.
25.Gently add 1000 μl RT Accutase per well of a 6-well plate. Return plate to a 37°C humidified incubator, 20% O2 and 5% CO2.
26.Detach astrocytes by flushing with Accutase. Gently make single cells by pipetting up and down with a 1000-μl pipette against the border of the well. Avoid bubble formation.
27.Collect cell suspension into a 15-ml tube with 5 ml prewarmed neurobasal medium to neutralize the Accutase. Centrifuge 7 min at 340 × g , RT. In the meantime, proceed to step 28.
28.Using a multichannel pipette, add 200 μl sterile PBS at RT to the surrounding 64 wells in the 96-well plate to prevent evaporation of medium. Cover plate with aluminum foil, and carefully place plate on a bench to come to RT. In the meantime, proceed to step 29.
29.After centrifugation, aspirate supernatant with a suction needle or 10-ml serological pipette. Resuspend pellet in 1 ml prewarmed neurobasal medium.
30.Mix 10 μl cell suspension in 10 μl trypan blue, and count viable astrocytes in a hemocytometer following the manufacturer's instruction. Dilute astrocytes in neurobasal medium at RT to a density of 4000 astrocytes per 200 μl.
31.Aspirate medium from the 96-well plate using a multichannel pipette. Add 200 μl cell suspension per well using a multichannel pipette.
32.Cover 96-well plate with aluminum foil, and incubate for 15 min on bench at RT.
33.Incubate plate overnight in a 37°C humidified incubator, 20% O2 and 5% CO2.
Inhibit cell proliferation by AraC treatment (day 1)
34.For 32 wells of a 96-well plate, freshly prepare 3.5 ml neurobasal medium containing 10 ng/ml NT3, 10 ng/ml BDNF, 2 µg/ml doxycycline, and 4 µM AraC. Prewarm in water bath.
35.Using a multichannel pipette, remove 100 μl medium per well from all wells without touching the culture surface, and add 100 μl freshly prepared neurobasal medium per well.
36.Incubate plate overnight in a 37°C humidified incubator, 20% O2 and 5% CO2.
Refresh medium 2 days after co-culture (day 2)
37.For 32 wells of a 96-well plate, freshly prepare 3.5 ml neurobasal medium containing 10 ng/ml NT3 and 10 ng/ml BDNF. Prewarm in water bath.
38.Using a multichannel pipette, remove 100 μl medium per well from all wells without touching the culture surface, and add 100 μl freshly prepared neurobasal medium per well.
39.Incubate plate overnight in a 37°C humidified incubator, 20% O2 and 5% CO2.
Refresh medium on a weekly basis (week 1 until week 4)
Optional seeding of FTDtau1 is performed at day 14 (week 2; see step 43).
40.Starting from week 1 of co-culture, refresh half the medium once a week (i.e., at days 7, 14, and 21 of co-culture). To do so, freshly prepare and prewarm 3.5 ml neurobasal medium containing FBS, 10 ng/ml NT3, and 10 ng/ml BDNF per 32 wells of a 96-well plate.
41.Remove 90 μl spent medium, and add 100 μl neurobasal medium per well using a multichannel pipette.
42.Incubate plate in a 37°C humidified incubator, 20% O2 and 5% CO2, until next medium refreshment.
Optional: add preformed fibrils at week 2 for seeded aggregation of FTDtau1
This step is optional, and PFFs are only added to induce seeded aggregation of FTDtau1. For spontaneous aggregation of FTDtau1+2, continue with medium refreshment until week 4.
In this section, human K18-truncated FTDtau1 PFFs that were previously generated using heparin and diluted in sodium acetate were used (Wiersma et al., 2019).
In a similar system, it was previously demonstrated that seeding is optimal after 2 weeks culture compared with seeding at week 1 or 3 (Verheyen et al., 2015).
Lipofectamine-3000 (e.g., Invitrogen, cat. no. L3000001) may be used to enhance seeding capacity of PFFs.
CAUTION : PFFs are hazardous and may thus require special decontamination and disposal procedures that are subject to local guidelines. We recommend handling PFFs in a laminar flow hood, discarding all PFF-exposed consumables in a biohazard waste bin, and cleaning PFF-exposed surfaces with 1% (w/v) sodium dodecyl sulfate in MilliQ water (see also Fenyi et al., 2018).
43.Freshly prepare 3.5 ml neurobasal medium containing FBS, 10 ng/ml NT3, and 10 ng/ml BDNF per 32 wells of a 96-well plate. Prewarm in water bath. With this medium, dilute PFFs to 150 nM (100 μl per well). Dilute an equal volume of buffer (e.g., 0.1 M sodium acetate) in medium to be used as a vehicle control.
44.Sonicate PFFs and vehicle control with a probe sonicator (25 pulses at 50% power for 24 to 30 μm peak to peak, with 3 s rest every fifth pulse).
45.Remove 90 μl spent medium with a multichannel pipette. To each well, add 100 μl PFF- or vehicle-containing medium using a 200-μl pipette.
46.Incubate plate in a 37°C humidified incubator, 20% O2 and 5% CO2, until next medium refreshment.
Fix co-cultures with PFA or MeOH at week 4
MeOH fixation can be used to remove soluble proteins, including tau (Guo et al., 2016; Wiersma et al., 2019). As a result, only aggregated tau is detected by subsequent (immuno)fluorescent staining. PFA fixation, on the other hand, preserves both soluble and insoluble (i.e., aggregated) tau.
Wrap the plate in aluminum foil during each incubation step to prevent photobleaching of fluorescently tagged FTDtau variants.
47a. Fix co-cultures with PFA:
- a. Remove 100 μl medium per well from all wells without touching the culture surface, and gently add 100 μl of 4% (w/v) PFA (2% final) per well using a multichannel pipette. Incubate for 20 min at RT.
We observed that adding PFA to an equal volume of culture medium prevents culture detachment.
- b. Remove all medium/PFA, and add 100 μl of 4% (w/v) PFA per well using a multichannel pipette. Incubate for 20 min at RT.
- c. Remove all PFA, and gently add 200 μl filtered PBS per well using a multichannel pipette. Incubate for 10 min at RT.
If needed, parafilm the plate, and store in fridge until further use.
- d. Proceed to step 48 for immunostaining.
47b. Fix co-cultures with MeOH:
- a. Prepare filtered PBS. Put 96-well plate on ice for 5 min.
- b. Using a multichannel pipette, remove all medium without touching the culture surface. Gently add 200 μl ice-cold MeOH per well with a 1000-μl pipette.
Use prewetting and reverse pipetting techniques to prevent MeOH dripping, and increase dispensing speed.
- c. Incubate plate for 20 min at –20°C.
- d. Remove all MeOH, and gently add 200 μl filtered PBS per well using a multichannel pipette. Incubate for 10 min at RT.
- e. Proceed to step 48 for immunostaining.
Long-term storage at 4°C is not recommended for MeOH-fixed cultures because this increases the likelihood of culture detachment.
Perform immunostaining of co-cultures
Wrap the plate in aluminum foil during each incubation step to prevent photobleaching of fluorescently tagged FTDtau variants.
48.Prepare blocking buffer and filtered PBS. If applicable, bring plates to RT.
49.Remove PBS from all wells, and add 50 μl blocking buffer per well using a multichannel pipette. Seal lid with parafilm, and incubate for 3 hr at RT.
50.Dilute primary antibodies in blocking buffer, and centrifuge 5 min at 16,400 × g , RT. See Table 1 for antibody dilutions.
51.Remove blocking buffer, and gently add 50 μl primary antibody solution to each well using a 200-μl pipette. Incubate for 1 hr at RT and then overnight at 4°C.
52.Gently remove primary antibody solution, and gently wash wells once (for MeOH-fixed cultures) or twice (for PFA-fixed cultures) with 200 μl filtered PBS per well. Incubate 10 min at RT.
53.Dilute secondary antibodies 1:1000 in blocking buffer, and centrifuge 5 min at 16,400 × g , RT. Wrap tubes containing diluted secondary antibodies in aluminum foil to prevent photobleaching.
54.Aspirate PBS and gently add 50 μl secondary antibody solution to each well using a 200-μl pipette. Seal lid with parafilm, and incubate for 3 hr at RT.
55.Dilute DAPI to 2.5 µg/ml in filtered PBS, and centrifuge 5 min at 16,400 × g , RT. Proceed to step 56a for PFA-fixed cultures or 56b for MeOH-fixed cultures.
56.Gently aspirate all secondary antibody solution using a multichannel or 200-μl pipette. Then stain with DAPI:
- a. For PFA-fixed cultures : Wash once with 200 μl filtered PBS per well. Incubate for 10 min at RT. Add 200 μl of 2.5 μg/ml DAPI to all wells. Incubate for 10 min at RT to stain cell nuclei.
- b. For MeOH-fixed cultures : Add 200 μl of 2.5 μg/ml DAPI to all wells. Incubate for 10 min at RT to stain cell nuclei.
To prevent culture detachment, separate washing of antibodies is omitted but is instead performed during incubation with DAPI.
57.Remove DAPI solution and add 200 μl filtered PBS per well. Seal lid with parafilm, and cover plate in aluminum foil to prevent photobleaching. Incubate for ≥10 min at RT before proceeding to microscopy.
Support Protocol 1: HUMAN INDUCED PLURIPOTENT STEM CELL CULTURE
This protocol describes the thawing, medium refreshment, passaging, and cryopreservation of human iPSCs harboring a stably integrated, doxycycline-inducible Ngn2 transgene. Here we use a clonal human iPSC line stably expressing rtTA/Ngn2 that has been previously generated, as specified in the procedures described in Frega et al. (2017). Cryopreserved human iPSCs are thawed and cultured feeder-free on vitronectin-coated plates in TeSR-E8 medium. Medium is changed daily, and colonies are passaged once or twice a week until passage number 25.
Materials
- 250 μg/ml vitronectin (e.g., StemCell Technologies, cat. no. 07180)
- Dilution buffer (e.g., StemCell Technologies, cat. no. 07183)
- TesR-E8 medium (see recipe)
- RI (see recipe)
- 50 mg/ml geneticin (G418; e.g., Sigma-Aldrich, cat. no. A1720)
- 1 mg/ml puromycin (e.g., Sigma-Aldrich, cat. no. P8833)
- DMEM/F12 (e.g., Life Technologies, cat. no. 11320033)
- Cryopreserved stock of human iPSCs with stably integrated doxycycline-inducible rtTA/Ngn2 (generated as described in Frega et al., 2017) or EBiSC line BIOni010-C-13 (e.g., European Collection of Authenticated Cell Cultures, cat. no. 66540561)
- Gentle cell dissociation reagent (e.g., StemCell Technologies, cat. no. 07174)
- Dimethyl sulfoxide (DMSO; e.g., Sigma-Aldrich, cat. no. D2650)
-
- 6-well plate (e.g., VWR, cat. no. 734-2777)
- Parafilm M
- 37°C, 20% O2, 5% CO2 cell culture incubator
- Water bath
- 15-ml conical tubes
- Tabletop centrifuge with rotor equipped for 1.5- and 15-ml tubes
- Freezing container (e.g., Thermo Fisher Scientific, cat. no. 5100-0001)
- Cryovials (e.g., Sarstedt cat. no. 72.380)
- Liquid nitrogen cell storage container or −150°C deep freezer
Coat 6-well plate with vitronectin
1.Thaw vitronectin aliquot at 4°C, and dilute 1:25 with dilution buffer to prepare vitronectin coating solution.
2.Add 1 ml of 10 μg/ml vitronectin coating solution per well of a 6-well plate.
3.Seal plate with parafilm, and incubate for ≥1 hr at RT.
4.Use immediately (continue to step 5), or store at 4°C for up to 1 week.
Thaw iPSCs
Human iPSCs should harbor a stably integrated doxycycline-inducible rtTA/Ngn2 transgene cassette. Here we use a clonal human iPSC line stably expressing rtTA/Ngn2 that has been previously generated according to the procedures described in Frega et al. (2017). Clonal selection of this line is performed using G418 and puromycin. Alternatively, the EBiSC line BIOni010-C-13 (generated as described in Schmid et al., 2021) or equivalent may also be used, but G418 and puromycin should then be omitted from the medium in all subsequent steps.
We recommend using iPSCs that were cryopreserved below passage number 10.
5.Let 6-well plate come to RT. In the meantime, proceed to step 6.
6.Prepare TesR-E8 medium containing 10 µM RI, 50 µg/ml G418, and 0.5 µg/ml puromycin. Prepare a total of 1.5 ml per well of a 6-well plate for plating and 1 ml for resuspension (prewarmed). Also prepare 5 ml DMEM/F12 for centrifugation (prewarmed).
7.Remove vitronectin coating solution from the wells in the 6-well plate, and wash wells once with 1 ml dilution buffer per well.
8.Remove dilution buffer, and add 1.5 ml TesR-E8 medium containing 10 µM RI, 50 µg/ml G418, and 0.5 µg/ml puromycin per well using a 1000-μl pipette or serological pipette. Transfer plate to a 37°C humidified incubator, 5% O2 and 5% CO2.
9.Remove cryopreserved iPSCs from the liquid nitrogen storage container or −150°C freezer. Quickly thaw cryovial in a water bath by gently shaking the cryovial continuously until only a small frozen cell pellet remains.
10.Gently aspirate iPSC suspension from the cryovial with a 1000-μl pipette, and transfer to an empty 15-ml centrifuge tube.
11.Slowly, drop by drop, add 5 ml prewarmed DMEM/F12 to the cell suspension. Gently rock 15-ml centrifuge tube containing the cell suspension while adding medium to minimize osmotic shock to the cells.
12.Centrifuge cell suspension 5 min at 340 × g , RT.
13.After centrifugation, aspirate supernatant with a suction needle or 10-ml serological pipette. Gently resuspend pellet in 1 ml prewarmed TesR-E8 medium containing 10 µM RI, 50 µg/ml G418, and 0.5 µg/ml puromycin using a 1000-μl pipette.
14.Add cell mixture drop-wise using a 1000-μl pipette to the vitronectin-coated 6-well plate containing TesR-E8 medium with 10 µM RI, 50 µg/ml G418, and 0.5 µg/ml puromycin.
15.Transfer plate to a 37°C humidified incubator, 5% O2 and 5% CO2. In the incubator, move plate in several back-and-forth and side-to-side motions to evenly distribute cells across the surface of the wells. Do not disturb plate for 24 hr.
16.The next day, aspirate spent medium of human iPSCs with a suction needle or 5-ml serological pipette. Add 2 ml TesR-E8 medium containing 50 µg/ml G418 and 0.5 µg/ml puromycin per well of a 6-well plate. Transfer plate to a 37°C humidified incubator, 5% O2 and 5% CO2.
17.Perform daily medium changes using TesR-E8 medium (see step 16), and visually inspect cultures to monitor growth of colonies. Proceed to passaging iPSCs (step 18) once cells reach 70% to 80% confluence.
Passage iPSCs
18.Before passaging iPSCs, coat a 6-well plate with vitronectin (see step 1).
19.Prepare TesR-E8 medium containing 10 µM RI, 50 µg/ml G418, and 0.5 µg/ml puromycin. Prepare 2 ml per well of a 6-well plate for plating and 1 ml for dissociating iPSCs from one well of a 6-well plate (prewarmed).
20.Remove vitronectin coating solution from the wells in the 6-well plate, and wash wells once with 1 ml dilution buffer per well.
21.Remove dilution buffer, and add 2 ml TesR-E8 medium containing 10 µM RI, 50 µg/ml G418, and 0.5 µg/ml puromycin per well using a 1000-μl pipette or serological pipette. Transfer plate to a 37°C humidified incubator, 5% O2 and 5% CO2.
22.Aspirate spent medium of human iPSCs with a suction needle or 5-ml serological pipette.
23.Gently add 1 ml gentle cell dissociation reagent per well of a 6-well plate. Return plate to a 37°C humidified incubator, 5% O2 and 5% CO2. Incubate for 4 to 6 min until gaps appear between cells at the border of the colonies.
24.Gently aspirate gentle cell dissociation reagent using a 1000-μl pipette. Gently add 1 ml TesR-E8 medium containing 10 µM RI, 50 µg/ml G418, and 0.5 µg/ml puromycin per well of iPSCs using a 1000-μl pipette. Tap plate until colonies begin to come loose.
25.Transfer detached cell aggregates to a 15-ml conical tube with a 1000-μl pipette.
26.Carefully pipette cell aggregate mixture up and down to break up aggregates as needed.
27.Using a 1000-μl pipette, plate cell aggregate mixture at desired density drop-wise on the vitronectin-coated 6-well plate containing TesR-E8 medium with 10 µM RI, 50 µg/ml G418, and 0.5 µg/ml puromycin.
28.Transfer plate to a 37°C humidified incubator, 5% O2 and 5% CO2. In the incubator, move plate in several back-and-forth and side-to-side motions to evenly distribute cells across the surface of the wells. Do not disturb plate for 24 hr.
29.The next day, aspirate spent medium of human iPSCs with a suction needle or 5-ml serological pipette. Add 2 ml TesR-E8 medium containing 50 µg/ml G418 and 0.5 µg/ml puromycin per well of a 6-well plate. Transfer plate to a 37°C humidified incubator, 5% O2 and 5% CO2.
30.Perform daily medium changes using TesR-E8 medium (see step 29), and visually inspect cultures to monitor growth of colonies. Passage iPSCs, cryopreserve, or start co-culturing with human primary astrocytes (Basic Protocol) once cells reach 70% to 80% confluence.
Cryopreserve iPSCs
We recommend cryopreserving iPSCs at a low passage number (<10) for stock-keeping.
31.Cool freezing container to 4°C.
32.Per well of iPSCs in a 6-well plate, prepare 1 ml TesR-E8 medium containing 10 µM RI and 10% (v/v) DMSO. Cool to 4°C.
33.Aspirate spent medium of human iPSCs with a suction needle or 5-ml serological pipette.
34.Gently add 1 ml gentle cell dissociation reagent per well of a 6-well plate. Return plate to a 37°C humidified incubator, 5% O2 and 5% CO2. Incubate for 4 to 6 min until gaps appear between cells at the border of the colonies.
35.Gently aspirate gentle cell dissociation reagent using a 1000-μl pipette. Gently add 1 ml prechilled TesR-E8 medium containing 10 µM RI and 10% (v/v) DMSO per well of iPSCs using a 1000-μl pipette. Tap plate until colonies begin to come loose.
36.Transfer 1 ml detached cell aggregates to an appropriately labelled cryovial using a 1000-μl pipette.
37.Immediately place cryovial(s) into a prechilled freezing container, and transfer freezing container to a –80°C freezer.
38.The next day, transfer cryovials to a liquid nitrogen cell storage container or −150°C deep freezer.
Support Protocol 2: CULTURE HUMAN PRIMARY ASTROCYTES
This protocol describes the thawing, medium refreshment, passaging, and cryopreservation of human primary astrocytes. Here we use primary human fetal astrocytes. Astrocytes are cultured in astrocyte medium on Geltrex-coated plates, and half the medium is refreshed twice a week. Astrocytes are passaged once a week by enzymatic dissociation with Accutase. Cultures are kept at 37°C with 20% O2 and 5% CO2 and used until passage number seven.
Materials
- Geltrex (see recipe)
- DMEM/F12 (e.g., Life Technologies, cat. no. 11320033)
- Astrocyte medium (see recipe)
- Cryopreserved stock of human primary astrocytes (e.g., ScienCell cat. no. 1800)
- Accutase (e.g., Sigma-Aldrich, cat. no. SF006)
- Trypan blue (e.g., Life Technologies, cat. no. 15250061)
- DMSO (e.g., Sigma-Aldrich, cat. no. D2650)
-
- 6-well plate (e.g., VWR, cat. no. 734-2323)
- 37°C, 20% O2, 5% CO2 cell culture incubator
- Parafilm M
- Water bath
- 15-ml conical tubes
- Tabletop centrifuge with rotor equipped for 1.5- and 15-ml tubes
- 1.5-ml microcentrifuge tubes (e.g., Eppendorf, cat. no. 0030108051)
- Hemocytometer (e.g., Bürker chamber; NanoEntek, cat. no. DHC-B01)
- Freezing container (e.g., Thermo Fisher Scientific, cat. no. 5100-0001)
- Cryovials (e.g., Sarstedt, cat. no. 72.380)
- Liquid nitrogen cell storage container or −150°C deep freezer
Coat 6-well plate with Geltrex
1.Thaw Geltrex aliquot at 4°C, and dilute 1:50 with cold DMEM/F12.
2.Add 1 ml Geltrex coating solution per well of a 6-well plate.
3.Incubate for 4 hr at 37°C.
4.Use immediately (continue to step 5), or seal with parafilm and store at 4°C for up to 1 week.
Thaw astrocytes
We recommend using astrocytes that were cryopreserved below passage number five.
5.Let 6-well plate come to RT. In the meantime, proceed to step 6.
6.Prepare astrocyte medium. Per cryovial prepare 1.5 ml per well of a 6-well plate for plating and 1 ml for resuspension (prewarmed). Also prepare 5 ml DMEM/F12 for centrifugation (prewarmed).
7.Remove Geltrex coating solution from the wells in the 6-well plate, and add 1.5 ml astrocyte medium per well using a 1000-μl pipette or serological pipette. Transfer plate to a 37°C humidified incubator, 20% O2 and 5% CO2.
8.Remove cryopreserved astrocytes from the liquid nitrogen storage container or −150°C freezer. Quickly thaw cryovial in a water bath by gently shaking the cryovial continuously until only a small frozen cell pellet remains.
9.Gently aspirate astrocyte suspension from the cryovial using a 1000-μl pipette, and transfer to an empty 15-ml centrifuge tube.
10.Slowly, drop by drop, add 5 ml prewarmed DMEM/F12 to the cell suspension. Gently rock 15-ml centrifuge tube containing the cell suspension while adding medium to minimize osmotic shock to the cells.
11.Centrifuge cell suspension 5 min at 340 × g , RT.
12.After centrifugation, aspirate supernatant with a suction needle or 10-ml serological pipette. Gently resuspend pellet in 1 ml prewarmed astrocyte medium using a 1000-μl pipette.
13.Using a 1000-μl pipette, plate astrocytes by adding the cell mixture drop-wise to Geltrex-coated 6-well plate containing astrocyte medium.
14.Transfer plate to a 37°C humidified incubator, 20% O2 and 5% CO2. In the incubator, move plate in several back-and-forth and side-to-side motions to evenly distribute cells across the surface of the wells. Do not disturb plate for 48 hr.
15.After 2 days, add 1 ml astrocyte medium. Return plate to a 37°C humidified incubator, 20% O2 and 5% CO2.
16.From 4 days after thawing, refresh two-thirds of the medium twice a week by removing 2 ml spent medium and adding 2 ml freshly prepared astrocyte medium (total volume 3 ml). Proceed to passaging (step 17) or co-culturing (Basic Protocol) once astrocyte cultures reach confluence.
Passage astrocytes
17.Before passaging astrocytes, coat a 6-well plate with Geltrex (see step 1).
18.Let Accutase come to RT.
19.Prepare astrocyte medium. Prepare 3 ml per well of a 6-well plate for plating and 1 ml for resuspension (prewarmed). Also prepare 5 ml DMEM/F12 for centrifugation (prewarmed) and a sterile 1.5-ml microcentrifuge tube with 10 μl trypan blue.
20.Remove Geltrex coating solution from the wells in the 6-well plate. Add 3 ml astrocyte medium per well using a 1000-μl pipette or serological pipette. Transfer plate to a 37°C humidified incubator, 20% O2 and 5% CO2.
21.Aspirate spent medium of human astrocytes with a suction needle or 5-ml serological pipette.
22.Gently add 1 ml Accutase per well of a 6-well plate. Return plate to a 37°C humidified incubator, 20% O2 and 5% CO2.
23.Detach astrocytes by flushing with Accutase. Gently make single cells by pipetting up and down using a 1000-μl pipette against the border of the well. Avoid bubble formation.
24.Collect cell suspension into a 15-ml tube with 5 ml prewarmed DMEM/F12 to neutralize the Accutase. Centrifuge 7 min at 340 × g , RT.
25.After centrifugation, aspirate supernatant with a suction needle or 10-ml serological pipette. Resuspend pellet in 1 ml prewarmed astrocyte medium.
26.Mix 10 μl cell suspension in 10 μl trypan blue, and count viable astrocytes in a hemocytometer following the manufacturer's instruction.
27.Calculate volume needed to reach 1–2.5 × 105 cells per well. Add this volume of cell suspension to one Geltrex-coated well of a 6-well plate containing astrocyte medium.
28.Transfer plate to a 37°C humidified incubator, 5% O2 and 5% CO2. In the incubator, move plate in several back-and-forth and side-to-side motions to evenly distribute cells across the surface of the wells. Do not disturb plate for 24 hr.
29.After 2 days, refresh two-thirds of the medium twice a week by removing 2 ml spent medium and adding 2 ml freshly prepared astrocyte medium (total volume 3 ml). Passage astrocytes or proceed to co-culture (Basic Protocol) once astrocyte cultures reach confluence.
Cryopreserve astrocytes
We recommend cryopreserving astrocytes at or below passage number five for stock-keeping.
30.Let Accutase come to RT, and cool freezing container to 4°C.
31.Per confluent well of a 6-well plate, prepare 2 ml astrocyte medium containing 10% (v/v) DMSO. Cool to 4°C. Also prepare 5 ml DMEM/F12 for centrifugation (prewarmed).
32.Aspirate spent medium of human astrocytes with a suction needle or 5-ml serological pipette.
33.Gently add 1 ml Accutase per well of a 6-well plate. Return plate to a 37°C humidified incubator, 20% O2 and 5% CO2.
34.Detach astrocytes by flushing with Accutase. Gently make single cells by pipetting up and down using a 1000-μl pipette against the border of the well. Avoid bubble formation.
35.Collect cell suspension into a 15-ml tube with 5 ml prewarmed DMEM/F12 to neutralize the Accutase. Centrifuge 7 min at 340 × g , RT.
36.After centrifugation, aspirate supernatant with a suction needle or 10-ml serological pipette. Resuspend pellet in 2 ml prechilled astrocyte medium with 10% (v/v) DMSO.
37.Split 2 ml astrocyte suspension into two appropriately labelled cryovials using a 1000-μl pipette.
38.Immediately place cryovials into a prechilled freezing container, and transfer freezing container to a –80°C freezer.
39.The next day, transfer cryovials to a liquid nitrogen cell storage container or −150°C deep freezer.
REAGENTS AND SOLUTIONS
AraC, 4 mM
Reconstitute AraC powder (e.g., Millipore Sigma, cat. no. 251010) in Dulbecco's PBS without calcium or magnesium (DPBS–; e.g., Thermo Fisher Scientific, cat. no. 14190250) containing 0.1% (w/v) bovine serum albumin (BSA), sterile filtered (e.g., Sigma-Aldrich, cat. no. A9418), to generate a 4 mM stock. Divide into aliquots, and store at –20°C for up to 1 year.
Use at 1:1000 for a 4 µM final concentration.
Astrocyte medium
- 480 ml astrocyte medium (ScienCell, cat. no. 1801-b)
- 10 ml FBS (e.g., ScienCell, cat. no. 0010)
- 5 ml astrocyte growth supplement (Sciencell, cat. no. 1852)
- 5 ml penicillin-streptomycin (e.g., Sciencell, cat. no. 0503)
- Store complete astrocyte medium at 4°C for up to 1 month
Basal astrocyte medium arrives at room temperate, and the supplements arrive on dry ice. Store basal medium at 4°C and supplements at –20°C for up to 1 year. Thaw supplements at 4°C before use.
Basal astrocyte medium and supplements are sterile. Prepare complete astrocyte medium in a laminar flow hood using standard aseptic technique.
BDNF, 20 µg/ml
Reconstitute 500 µg BDNF powder (e.g., Prospec, cat. no. Cyt 207) in 200 μl sterile distilled water, and dilute with 2.3 ml DPBS– (e.g., Thermo Fisher Scientific, cat. no. 14190250) containing 0.1% (w/v) BSA, sterile filtered (e.g., Sigma-Aldrich, cat. no. A9418), to generate a 200 µg/ml stock for long-term storage. Divide into aliquots, and store at –80°C for up to 1 year until further dilution. Dilute 200 μg/ml stock solution 1:10 in DPBS– (e.g., Thermo Fisher Scientific, cat. no. 14190250) containing 0.1% (w/v) BSA, sterile filtered (e.g., Sigma-Aldrich, cat. no. A9418), to generate a 20 µg/ml stock. Divide into aliquots, and store at –20°C for up to 3 months.
Use at 1:2000 for a 10 ng/ml final concentration.
Blocking buffer
- 94.7 ml PBS (see recipe)
- 6 ml normal goat serum (e.g., Life Technologies, cat. no. 16210-064)
- 0.1 g BSA (e.g., Sigma-Aldrich, cat. no. A9418)
- 300 μl Triton X-100 (e.g., Sigma-Aldrich, cat. no. T8787)
- Store 15-ml aliquots at –20°C for up to 1 year
Thawed blocking buffer should be used within 1 week when stored at 4°C.
DAPI, 50 mg/ml
Dilute DAPI powder (e.g., Sigma-Aldrich, cat. no. D9542) in filtered PBS (see recipe) to generate a 50 mg/ml stock. Divide into aliquots, and store at –20°C for up to 1 year.
Use at 1:2000 for a 2.5 µg/ml final concentration.
DMEM
- 48.5 ml DMEM/F12 without L-glutamine (e.g., Life Technologies, cat. no. 21331020)
- 500 μl N2 supplement (e.g., Life Technologies, cat. no. 17502048)
- 500 μl minimal essential medium nonessential amino acid (MEM-NEAA; e.g., Life Technologies, cat. no. 11140035)
- 500 μl penicillin-streptomycin (e.g., Life Technologies, cat. no. 15140122)
- Store complete DMEM at 4°C for up to 1 month
Basal DMEM/F12 and MEM-NEAA arrive at room temperate, and the other supplements arrive on dry ice. Store basal DMEM/F12 and MEM-NEAA at 4°C and other supplements at –20°C for up to 1 year. Thaw supplements at 4°C before use.
Basal DMEM/F12 and supplements are sterile. Prepare complete DMEM in a laminar flow hood using standard aseptic technique.
Doxycycline, 40 mg/ml
Dilute doxycycline powder (e.g., Sigma-Aldrich, cat. no. D98195) in DMSO (e.g., Sigma-Aldrich, cat. no. D2650) to generate a 40 mg/ml stock. Divide into aliquots, and store at –20°C for up to 1 year.
Use at 1:10.000 for a 4 µg/ml final concentration.
Geltrex
Dilute ∼15 mg/ml Geltrex 1:2 with cold DMEM/F12 (e.g., Life Technologies, cat. no. 11320033). Divide into aliquots, and store at –80°C for up to 3 years. Thaw at 4°C, and use 1:50 in cold DMEM/F12 for a 1:100 final dilution.
Neurobasal medium
- 48 ml neurobasal medium (e.g., Life Technologies, cat. no. 21103049)
- 1 ml B27 with vitamin A (e.g., Life Technologies, cat. no. 17504001)
- 500 μl Glutamax (e.g., Life Technologies, cat. no. 35050038)
- 500 μl penicillin-streptomycin (e.g., Life Technologies, cat. no. 15140122)
- Store complete neurobasal medium at 4°C for up to 1 month
Basal neurobasal medium and Glutamax arrive at room temperate, and the other supplements arrive on dry ice. Store Glutamax at RT for up to 2 years, and store basal neurobasal medium and other supplements at 4°C and –20°C, respectively, for up to 1 year. Thaw supplements at 4°C before use.
Basal neurobasal medium and supplements are sterile. Prepare complete neurobasal medium in a laminar flow hood using standard aseptic technique.
Neurobasal medium with FBS
- 47.75 ml neurobasal medium (e.g., Life Technologies, cat. no. 21103049)
- 1 ml B27 with vitamin A (e.g., Life Technologies, cat. no. 17504001)
- 500 μl Glutamax (e.g., Life Technologies, cat. no. 35050038)
- 500 μl penicillin-streptomycin (e.g., Life Technologies, cat. no. 15140122)
- 250 μl FBS (e.g., Thermo Fisher, cat. no. 16140063)
- Store at 4°C for up to 1 month
Basal neurobasal medium and Glutamax arrive at room temperate, and the other supplements arrive on dry ice. Store Glutamax at RT for up to 2 years, and store basal neurobasal medium and other supplements at 4°C and –20°C, respectively, for up to 1 year. Thaw supplements at 4°C before use.
Basal neurobasal medium and supplements are sterile. Prepare complete neurobasal medium in a laminar flow hood using standard aseptic technique.
NT3, 10 µg/ml
Reconstitute 100 µg NT3 powder (e.g., PeproTech, cat. no. 450-03) in 100 μl sterile distilled water, and dilute with 900 μl DPBS– (e.g., Thermo Fisher Scientific, cat. no. 14190250) containing 0.1% (w/v) BSA, sterile filtered (e.g., Sigma-Aldrich, cat. no. A9418), to generate a 100 µg/ml stock for long-term storage. Divide into aliquots, and store at –80°C for up to 1 year until further dilution. Dilute 1:10 in DPBS– (e.g., Thermo Fisher Scientific, cat. no. 14190250) containing 0.1% (w/v) BSA, sterile filtered (e.g., Sigma-Aldrich, cat. no. A9418), to generate a 10 µg/ml stock. Divide into aliquots, and store at –20°C for up to 3 months.
Use at 1:1000 for a 10 ng/ml final concentration.
PBS, pH 7.4, sterile or filtered
- 8 g NaCl (e.g., VWR, cat. no. S9888)
- 200 mg KCl (e.g., Sigma-Aldrich, cat. no. P5405)
- 1.44 g Na2HPO4 (e.g., Sigma-Aldrich, cat. no. S7907)
- 245 mg KH2PO4 (e.g., Sigma-Aldrich, cat. no. P5379)
- 800 ml ultrapure water (e.g., MilliQ water)
- Adjust pH to 7.4 with HCl
- Sterilize by autoclaving for 30 min at 120°C or filter through a 0.22-μm filter
- Store at RT for up to 6 months
RI, 10 mM
Reconstitute 50 µg Y-27632 dihydrochloride powder (e.g., Selleckchem, cat. no. S1049) in 1561 μl DMSO (e.g., Sigma-Aldrich, cat. no. D2650) to generate a 100 mM stock for long-term storage. Divide into aliquots, and store aliquots at –80°C for up to 3 years until further dilution. Dilute 1:10 in DMSO to generate a 10 mM stock. Divide into aliquots and store at –20°C for up to 6 months.
Use at 1:1000 for a 10 µM final concentration.
TesR-E8 medium
- 20.5 ml TesR-E8 medium (e.g., StemCell Technologies, cat. no. 05991)
- 20 ml TesR-E8 supplement (e.g., StemCell Technologies, cat. no. 05992)
- 2.5 ml penicillin-streptomycin (e.g., Life Technologies, cat. no. 15140122)
- Store complete TesR-E8 medium at 4°C for up to 1 month.
Basal TesR-E8 medium arrives at room temperate, and the other supplements arrive on dry ice. Store basal TesR-E8 medium at 4°C and other supplements at –20°C for up to 1 year. Thaw supplements at 4°C before use.
Basal TesR-E8 medium and supplements are sterile. Prepare complete TesR-E8 medium in a laminar flow hood using standard aseptic technique.
COMMENTARY
Background Information
Human iPSC-derived co-cultures are a powerful tool to investigate cellular mechanisms of neuron/astrocyte communication in physiological and pathological contexts, such as tau aggregation. Thus far, these models face technical challenges such as lengthy and variable neuronal differentiation, cell clustering, and culture detachment. In addition, inefficient tau aggregation currently limits the broad implementation of iPSC-derived neurons for the study of tau pathology, and co-culture with human astrocytes is required to further improve their translational value.
To address these shortcomings, we report the first step-by-step procedure to generate neuron/astrocyte co-cultures consisting of human iPSC-derived neurons and primary human astrocytes. We employed a previously generated, clonal human iPSC line with doxycycline-inducible Ngn2 (Frega et al., 2017) to facilitate the efficient and homogeneous generation of human iPSC-derived neurons (Thoma et al., 2012; Zhang et al., 2013). In Support Protocol 1, we report the step-by-step procedures on thawing, medium refreshment, passaging, and freezing of this human iPSCs line. Our data in the Basic Protocol show that neurons in the co-culture system have formed pre- and postsynaptic compartments at 4 weeks. In addition, Geltrex coating greatly prevented cell clustering and culture detachment to facilitate downstream microscopy analyses at 4 weeks. The stability of co-cultures at 8 weeks has been verified in the primary research paper (Batenburg et al., 2022), showing that the model can also be used for studies that require prolonged culturing.
We used commercially available primary human cortical astrocytes that were previously shown to support human iPSC-derived neurons in a similar format (Kuijlaars et al., 2016). We report the culture procedures for human primary astrocytes in Support Protocol 2. The use of human primary astrocytes circumvented the long differentiation time and possible heterogeneity that may be associated with human iPSC-derived astrocytes. Human primary astrocytes harvested from the embryonic human brain are not a renewable source and may evoke legal and ethical controversy. To overcome these limitations, future efforts should prioritize the inclusion of human iPSC-derived astrocytes in the co-culture model.
In addition to the Geltrex coating, a low percentage (0.5%) of serum is present to support the viability of astrocytes; therefore, the co-culture model is not completely animal free. Development of specific synthetic medium to allow serum-free culturing and synthetic coating to replace Geltrex would remove a source of variation and further reduce the use of experimental animals. In addition, this may resolve another limitation of the current system, which is that only a small population of astrocytes display immunoreactivity toward canonical astrocyte immunomarkers such as glial fibrillary acidic protein (GFAP) after 4 weeks of co-culturing. Indeed, an in vivo –like astrocyte state is modulated by contact with other cell types including neurons and the presence of secreted factors in the medium, and it is negatively affected by the presence of serum (Patani et al., 2023).
In this article, we included a time window—1 day after neural induction but before co-culture—for the lentiviral transduction of Ngn2-neural progenitors to enable neuron-specific expression of FTDtau variants. Overexpression of the single-mutated FTDtau1 variant and subsequent treatment with tau PFFs induced the formation of MeOH-insoluble, pathologically folded, and phosphorylated tau aggregates. In agreement with previous studies on rodent primary (Calafate et al., 2015; Jorge-Oliva et al., 2022; Wiersma et al., 2019) and human iPSC-derived neurons (Verheyen et al., 2015), neurons expressing FTDtau1 in our system did not develop intraneuronal tau aggregates in the absence of seeds. In contrast, expression of double mutated FTDtau1+2 led to the formation of tau aggregates that did not require seeding. These data, as well the data presented in our primary research paper (Batenburg et al., 2022), demonstrate the feasibility of FTDtau1+2 to model tau aggregation without the use of seeds that have thus far been a requirement for the induction of tau aggregation in human iPSC-derived neurons (Verheyen et al., 2015). In addition, we have showed that expression of FTDtau1+2 induces tau aggregation in a three-dimensional neuron/astrocyte co-culture (Batenburg et al., 2023), highlighting the versatility of this approach to induce spontaneous tau aggregation in more advanced cell culture models. Although seeds may be heterogeneous and directly affect neurons and astrocytes independent of intraneuronal tau pathology, their use represents an established paradigm in cellular models and may be of interest for specific research questions.
Because FTDtau1 and FTDtau1+2 are both mutated tau variants that differ in tau aggregation, FTDtau1 is a feasible control for overexpression of mutant tau to ascertain that cellular effects in the co-culture are due to tau aggregation. This, in combination with a high-content microscopy approach that was previously validated in our laboratory (van Ziel et al., 2020; Wolzak et al., 2022), enabled us to quantify and target the timing, extent, and cell (non)autonomous effects of spontaneous intraneuronal tau aggregation in a standardized manner in thousands of cells (Batenburg et al., 2022). This indicates that this rapid, miniaturized, and standardized model is a powerful tool for the preclinical screening of tau-targeting therapeutics.
Critical Parameters
Cell attachment at room temperature
Uniform cell density within the wells is of vital importance for microscopy analysis. We and others (Lundholt et al., 2003) have observed that plating at RT reduces the edge effect (i.e., higher density of cells along the borders of the well, which is most pronounced in the outer wells). Therefore, medium and the culture plate should be at RT during cell attachment to ensure homogeneous cell distribution within the well.
Removal of viral particles
To prevent lentiviral transduction of human astrocytes in the co-culture, all medium should be removed, and all wells should be incubated with fresh medium before co-culture. Ensure all medium is removed before astrocyte plating.
PFA prefixation
Prefixation by addition of PFA to the culture medium prevents culture detachment that may result from complete medium removal. Thus, PFA fixation should be performed in two steps: in 2% by addition of 4% PFA to the same volume of medium and then final fixation with 4% PFA.
MeOH fixation
Ensure all medium is removed before addition of ice-cold MeOH because residual medium in the well will impair fixation and incomplete removal of soluble proteins.
Troubleshooting
See Table 2 for a list of possible problems, causes, and solutions
| Problem | Possible cause | Solution |
|---|---|---|
| Cell clumping | Incomplete cell dissociation | After detaching human iPSCs on day –2, ensure they are indeed single cells; mix cell suspension right before adding to wells |
| Poor astrocyte viability or number | Check viability and proliferation of astrocytes before co-culture; ensure passage number is ≤7 and plating density is accurate | |
| Prepare fresh astrocyte medium | ||
| Cells too sparse | Ensure cells are completely dissociated before counting, and mix suspension right before adding to wells (see above); ensure plating density is accurate | |
| Poor Geltrex coating | Ensure Geltrex is thawed at 4°C and does not contain visible clumps; store Geltrex-coated plates at 4°C for up to 2 weeks | |
| Cell death | iPSC or astrocyte overgrowth | Before experiments, passage cells routinely, and/or feed astrocytes more frequently |
| Harsh dissociation | Handle cells gently during dissociation; gently make single cells by pipetting up and down against the border of the well, avoiding bubble formation | |
| Reduce incubation time with dissociation reagent | ||
| Infection | Exclude infection in culture wells and wells filled with PBS; check for discoloration of phenol red | |
| Cells detach during culture | Poor Geltrex coating | See above; supplement Geltrex in culture medium (1:100 final dilution) |
| Pipette tip touches culture | During medium refreshments, do not allow tips to touch culture | |
| Harsh medium refreshment | Gently add medium to wells during refreshment to prevent flushing cells | |
| Cells detach during fixation | Poor Geltrex coating | See above; refresh cultures with medium supplemented with Geltrex (1:100 final dilution) 2 days before fixation |
| Harsh fixative addition | Gently add PFA or MeOH to wells | |
| Inefficient tau aggregation | Inefficient seeding of FTDtau1 | Ensure final concentration and sonication of PFFs is accurate; increase sonication time, or use another batch of PFFs |
| Use lipofectamine delivery to increase seeding capacity of PFFs | ||
| Low transduction efficiency or expression of FTDtau1+2 | Ensure viral particle concentration is correcta | |
| Prepare another batch of viral particles | ||
| Spotty immunofluorescent background | Particles in antibody solution | Centrifuge primary and secondary antibody solutions to pellet antibody aggregates |
| Cell debris/death | See solutions for infection and harsh dissociation |
- iPSC, induced pluripotent stem cell; MeOH, methanol; Ngn2, neurogenin 2; PBS, phosphate-buffered saline; PFA, paraformaldehyde; PFF, preformed fibril.
- aAlthough the concentration described here (4 μl/ml) should be appropriate for most Ngn2 iPSC-lines, we suggest optimizing the concentration using a series of increasing virus concentrations.
Understanding Results
Statistical analysis
To allow statistical analysis, experiments should be performed with a minimum of three independent experiments. We recommend including four wells per condition per independent experiment. The statistical test will vary depending on the experimental design. To account for the dependency of replicates within a single experiment (nestedness), we recommend the use of nested tests for statistical analyses of two (nested t -test) or more than two (nested one-way analysis of variance) groups.
High-content screening
Co-cultures are compatible with high-content screening on automated platforms such as the CellIInsight CX7 HCS platform. Neuronal complexity and (pre)synaptic terminals, as well as tau aggregation and its cell-specific effects on neurons and astrocytes, can be quantified using automated software packages, including but not limited to Columbus analysis software (v2.5.2; PerkinElmer). See Batenburg et al. (2022) for how we analyzed co-cultures with high-content microscopy. In addition, because the 96-well plates used in this study contain a thin film bottom with glass-like optical properties, cultures are compatible with confocal and live imaging techniques with high signal-to-noise ratio. Images can be adapted for publication using ImageJ software (National Institutes of Health) and Adobe Illustrator.
Visualization of neurons and astrocytes
Neurons and astrocytes can be visualized by double immunofluorescent staining of neuronal MAP2 (e.g., Millipore, cat. no. AB5543) and glial membrane marker CD44 (e.g., Developmental Studies Hybridoma Bank, cat. no. H4C4; as described in Batenburg et al., 2022) or astrocytic GFAP (e.g., Dako, cat. no. Z0334; as described in Batenburg et al., 2022, 2023).
We observed that most astrocytes gradually lose GFAP immunoreactivity at high passage number (>7) and at 4 weeks of co-culturing. See Background Information for a discussion on GFAP immunoreactivity.
Neuron-specific expression of FTDtau
Neuron-specific expression of FTDtau can be confirmed using the direct fluorescence of fluorescently tagged FTDtau variants and its overlap with neuronal MAP2 but not glial/astrocytic markers (see Batenburg et al., 2022).
Synapse formation
Formation of synapses can be assessed using immunolabeling of presynaptic synaptophysin 1 (SYP1) and/or presynaptic vesicular glutamate transporters such as vGLUT2 (Fig. 4, left panel). The overlap of presynaptic SYP1 and postsynaptic density protein 95 (PSD95) can be used as a proxy for the formation of a morphologically mature synapse pattern (Fig. 4, right panel).
![Details are in the caption following the image Formation of pre- and postsynaptic terminals in neurogenin 2 human induced pluripotent stem cell–derived neurons in co-culture. After 4 weeks in co-culture, immunostaining was performed for dendrites (microtubule-associated protein-2 [MAP2], blue) and presynaptic markers (synaptophysin 1 [SYP1], green; vGlut2, magenta; left) and pre- (SYP1; green) and postsynaptic markers (postsynaptic density [PSD95], magenta). Merged images are shown.](https://static.yanyin.tech/literature_test/cpz1900-fig-0004-m.jpg)
Tau pathology
Accumulation of tau aggregates can be demonstrated by PFA fixation and subsequent MC1 immunolabeling to detect pathologically folded tau. In addition, co-cultures can be fixed with MeOH to remove soluble proteins. Because tau phosphorylation is developmentally regulated, iPSC-derived neurons generally express tau that is highly phosphorylated at several epitopes, including those associated with disease (Cheng et al., 2021). We therefore recommend MeOH fixation to remove phosphorylated but soluble tau in order to increase detection of tau aggregates using phospho-specific antibodies such as AT8 (pSer202 and pThr205) or AT100 (pSer212 and pSer214). Typical examples of these analyses for seeded aggregation of FTDtau1 and spontaneous aggregation of FTDtau1+2 are shown in Figure 5A,B and Figure 5C,D, respectively.
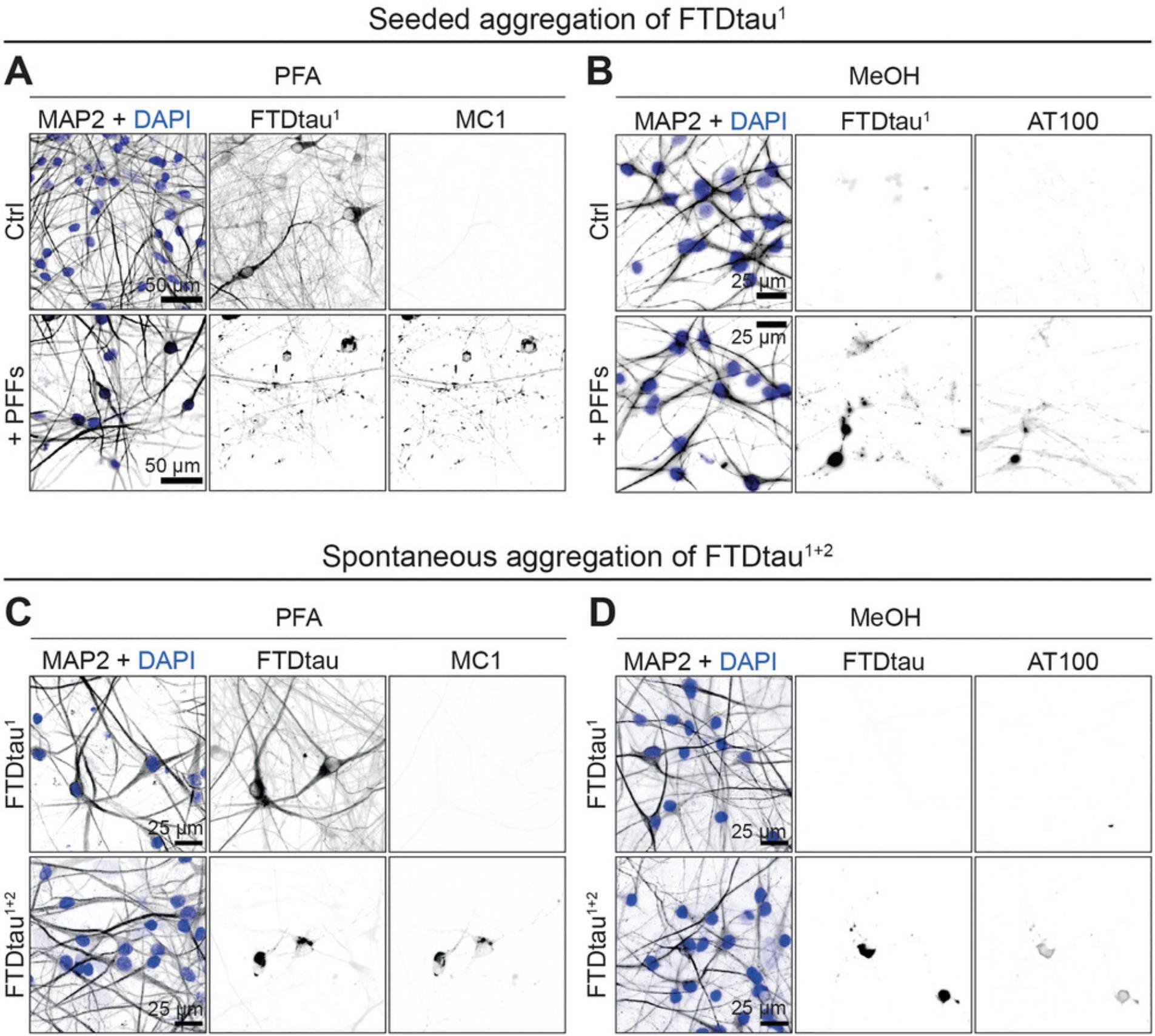
We have also validated that tau pathology can be addressed using the fluorescence intensity standard deviation of fluorescently labelled FTDtau (Batenburg et al., 2022). Thiophene dyes such as HS169 (Shirani et al., 2015) can also be used to label stacked beta sheets of pathological FTDtau (Batenburg et al., 2022, 2023). Finally, biochemical extraction and subsequent western blot analysis can be performed to confirm the formation of sarkosyl-insoluble tau aggregates. Examples of such analyses can be found in the primary research papers (Batenburg et al., 2022, 2023).
Time Considerations
From coating of culture plates to microscopy analysis of co-cultures, about 5 weeks needs to be scheduled. Only the first week of (co-)culture requires daily hands-on work: 2 hr for neural induction (day –2) and plating of human primary astrocytes (day 0) and 30 min for medium replacement on days –1, 1, and 2. For medium refreshment from week 1 until 4, only 30 min per week needs to be scheduled. With careful maintenance, cultures can be maintained for ≥8 weeks.
Acknowledgments
This study was supported by ZonMW and Stichting Proefdiervrij (MKMD no. 114022506 to WS) and co-funded by the PPP Allowance made available by Health∼Holland, Top Sector Life Sciences & Health, to stimulate public-private partnerships (no. LSHM17014 to VMH and LSHM18024 to WS).
The authors thank Nael Nadif Kasri for providing the Ngn2 human iPSC line; Lisa Gasparotto for advice and expert technical assistance on human iPSC cultures; and Robbert Zalm, Ingrid Saarloos, and Joost Hoetjes for preparation of constructs and viral particles. The authors also thank Peter Davies (Albert Einstein College of Medicine, New York, New York) for providing the MC1 antibody. The authors thank Vera Wiersma for expert advice on seeded FTDtau1 aggregation and members of the Molecular Neurodegeneration group (Functional Genomics, Vrije Universiteit, Amsterdam, Netherlands) and the Stem Cell Biology group (Complex Trait Genetics, Vrije Universiteit, Amsterdam, Netherlands) for stimulating discussions.
Author Contributions
Kevin Llewelyn Batenburg : conceptualization; data curation; formal analysis; investigation; methodology; project administration; validation; visualization; writing—original draft, review, and editing. Susan Karijn Rohde : investigation; validation; Paulien Cornelissen-Steijger : investigation; methodology. Nicole Breeuwsma : investigation; validation. Vivi Majella Heine : conceptualization; funding acquisition; supervision; writing–review and editing. Wiep Scheper : conceptualization; funding acquisition; supervision; writing–original draft, review, and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed.
Literature Cited
- Batenburg, K. L., Kasri, N. N., Heine, V. M., & Scheper, W. (2022). Intraneuronal tau aggregation induces the integrated stress response in astrocytes. Journal of Molecular Cell Biology , 14(10), mjac071. https://doi.org/10.1093/jmcb/mjac071
- Batenburg, K. L., Sestito, C., Cornelissen-Steijger, P., van Weering, J. R. T., Price, L. S., Heine, V. M., & Scheper, W. (2023). A 3D human co-culture to model neuron-astrocyte interactions in tauopathies. Biological Procedures Online , 25, 4. https://doi.org/10.1186/s12575-023-00194-2
- Calafate, S., Buist, A., Miskiewicz, K., Vijayan, V., Daneels, G., de Strooper, B., de Wit, J., Verstreken, P., & Moechars, D. (2015). Synaptic contacts enhance cell-to-cell tau pathology propagation. Cell Reports , 11(8), 1176–1183. https://doi.org/10.1016/j.celrep.2015.04.043
- Cheng, C., Reis, S. A., Adams, E. T., Fass, D. M., Angus, S. P., Stuhlmiller, T. J., Richardson, J., Olafson, H., Wang, E. T., Patnaik, D., Beauchamp, R. L., Feldman, D. A., Silva, M. C., Sur, M., Johnson, G. L., Ramesh, V., Miller, B. L., Temple, S., Kosik, K. S., … Haggarty, S. J. (2021). High-content image-based analysis and proteomic profiling identifies Tau phosphorylation inhibitors in a human iPSC-derived glutamatergic neuronal model of tauopathy. Scientific Reports , 11, 17029. https://doi.org/10.1038/S41598-021-96227-5
- Ehrlich, M., Hallmann, A. L., Reinhardt, P., Araúzo-Bravo, M. J., Korr, S., Röpke, A., Psathaki, O. E., Ehling, P., Meuth, S. G., Oblak, A. L., Murrell, J. R., Ghetti, B., Zaehres, H., Schöler, H. R., Sterneckert, J., Kuhlmann, T., & Hargus, G. (2015). Distinct neurodegenerative changes in an induced pluripotent stem cell model of frontotemporal dementia linked to mutant tau protein. Stem Cell Reports , 5(1), 83–96. https://doi.org/10.1016/j.stemcr.2015.06.001
- Fenyi, A., Coens, A., Bellande, T., Melki, R., & Bousset, L. (2018). Assessment of the efficacy of different procedures that remove and disassemble alpha-synuclein, tau and A-beta fibrils from laboratory material and surfaces. Scientific Reports , 8, 10788. https://doi.org/10.1038/s41598-018-28856-2
- Frega, M., van Gestel, S. H., Linda, K., van der Raadt, J., Keller, J., van Rhijn, J. R., Schubert, D., Albers, C. A., & Nadif Kasri, N. (2017). Rapid neuronal differentiation of induced pluripotent stem cells for measuring network activity on micro-electrode arrays. Journal of Visualized Experiments , 119, e54900. https://doi.org/10.3791/54900
- García-León, J. A., Cabrera-Socorro, A., Eggermont, K., Swijsen, A., Terryn, J., Fazal, R., Nami, F., Ordovás, L., Quiles, A., Lluis, F., Serneels, L., Wierda, K., Sierksma, A., Kreir, M., Pestana, F., van Damme, P., de Strooper, B., Thorrez, L., Ebneth, A., & Verfaillie, C. M. (2018). Generation of a human induced pluripotent stem cell–based model for tauopathies combining three microtubule-associated protein TAU mutations which displays several phenotypes linked to neurodegeneration. Alzheimer's and Dementia , 14(10), 1261–1280. https://doi.org/10.1016/j.jalz.2018.05.007
- Guo, J. L., Narasimhan, S., Changolkar, L., He, Z., Stieber, A., Zhang, B., Gathagan, R. J., Iba, M., McBride, J. D., Trojanowski, J. Q., & Lee, V. M. (2016). Unique pathological tau conformers from Alzheimer's brains transmit tau pathology in nontransgenic mice. Journal of Experimental Medicine , 213(12), 2635–2654. https://doi.org/10.1084/jem.20160833
- Hedegaard, A., Monzón-Sandoval, J., Newey, S. E., Whiteley, E. S., Webber, C., & Akerman, C. J. (2020). Pro-maturational effects of human iPSC-derived cortical astrocytes upon iPSC-derived cortical neurons. Stem Cell Reports , 15(1), 38–51. https://doi.org/10.1016/j.stemcr.2020.05.003
- Hodge, R. D., Bakken, T. E., Miller, J. A., Smith, K. A., Barkan, E. R., Graybuck, L. T., Close, J. L., Long, B., Johansen, N., Penn, O., Yao, Z., Eggermont, J., Höllt, T., Levi, B. P., Shehata, S. I., Aevermann, B., Beller, A., Bertagnolli, D., Brouner, K., … Lein, E. S. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature , 573(7772), 61–68. https://doi.org/10.1038/s41586-019-1506-7
- Iovino, M., Agathou, S., González-Rueda, A., Del Castillo Velasco-Herrera, M., Borroni, B., Alberici, A., Lynch, T., O'Dowd, S., Geti, I., Gaffney, D., Vallier, L., Paulsen, O., Káradóttir, R. T., & Spillantini, M. G. (2015). Early maturation and distinct tau pathology in induced pluripotent stem cell-derived neurons from patients with MAPT mutations. Brain , 138(11), 3345–3359. https://doi.org/10.1093/brain/awv222
- Johnson, M. A., Weick, J. P., Pearce, R. A., & Zhang, S. C. (2007). Functional neural development from human embryonic stem cells: Accelerated synaptic activity via astrocyte coculture. Journal of Neuroscience , 27(12), 3069–3077. https://doi.org/10.1523/jneurosci.4562-06.2007
- Jorge-Oliva, M., Smits, J. F. M., Wiersma, V. I., Hoozemans, J. J. M., & Scheper, W. (2022). Granulovacuolar degeneration bodies are independently induced by tau and α-synuclein pathology. Alzheimer's Research and Therapy , 14, 187. https://doi.org/10.1186/s13195-022-01128-y
- Kelley, K. W., Nakao-Inoue, H., Molofsky, A. V., & Oldham, M. C. (2018). Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nature Neuroscience , 21(9), 1171–1184. https://doi.org/10.1038/s41593-018-0216-z
- Kuijlaars, J., Oyelami, T., Diels, A., Rohrbacher, J., Versweyveld, S., Meneghello, G., Tuefferd, M., Verstraelen, P., Detrez, J. R., Verschuuren, M., de Vos, W. H., Meert, T., Peeters, P. J., Cik, M., Nuydens, R., Brône, B., & Verheyen, A. (2016). Sustained synchronized neuronal network activity in a human astrocyte co-culture system. Scientific Reports , 6, 36529. https://doi.org/10.1038/srep36529
- Leyns, C. E. G., & Holtzman, D. M. (2017). Glial contributions to neurodegeneration in tauopathies. Molecular Neurodegeneration , 12, 50. https://doi.org/10.1186/s13024-017-0192-x
- Lundholt, B. K., Scudder, K. M., & Pagliaro, L. (2003). A simple technique for reducing edge effect in cell-based assays. Journal of Biomolecular Screening , 8(5), 566–570. https://doi.org/10.1177/1087057103256465
- Moro, A., Hoogstraaten, R. I., Persoon, C. M., Verhage, M., & Toonen, R. F. (2021). Quantitative analysis of dense-core vesicle fusion in rodent CNS neurons. STAR Protocols , 2(1), 100325. https://doi.org/10.1016/J.XPRO.2021.100325
- Muratore, C. R., Srikanth, P., Callahan, D. G., & Young-Pearse, T. L. (2014). Comparison and optimization of hiPSC forebrain cortical differentiation protocols. PLoS One , 9(8), e105807. https://doi.org/10.1371/journal.pone.0105807
- Naldini, L., Blömer, U., Gallay, P., Ory, D., Mulligan, R., Gage, F. H., Verma, I. M., & Trono, D. (1996). In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science , 272(5259), 263–267. https://doi.org/10.1126/science.272.5259.263
- Oberheim, N. A., Takano, T., Han, X., He, W., Lin, J. H., Wang, F., Xu, Q., Wyatt, J. D., Pilcher, W., Ojemann, J. G., Ransom, B. R., Goldman, S. A., & Nedergaard, M. (2009). Uniquely hominid features of adult human astrocytes. Journal of Neuroscience , 29(10), 3276–3287. https://doi.org/10.1523/jneurosci.4707-08.2009
- Patani, R., Hardingham, G. E., & Liddelow, S. A. (2023). Functional roles of reactive astrocytes in neuroinflammation and neurodegeneration. Nature Reviews Neurology , 19, 395–409. https://doi.org/10.1038/S41582-023-00822-1
- Peeraer, E., Bottelbergs, A., van Kolen, K., Stancu, I. C., Vasconcelos, B., Mahieu, M., Duytschaever, H., Ver Donck, L., Torremans, A., Sluydts, E., van Acker, N., Kemp, J. A., Mercken, M., Brunden, K. R., Trojanowski, J. Q., Dewachter, I., Lee, V. M., & Moechars, D. (2015). Intracerebral injection of preformed synthetic tau fibrils initiates widespread tauopathy and neuronal loss in the brains of tau transgenic mice. Neurobiology of Disease , 73, 83–95. https://doi.org/10.1016/j.nbd.2014.08.032
- Reid, M. J., Beltran-Lobo, P., Johnson, L., Perez-Nievas, B. G., & Noble, W. (2020). Astrocytes in tauopathies. Frontiers in Neurology , 11, 572850. https://doi.org/10.3389/fneur.2020.572850
- Rhee, H. J., Shaib, A. H., Rehbach, K., Lee, C., Seif, P., Thomas, C., Gideons, E., Guenther, A., Krutenko, T., Hebisch, M., Peitz, M., Brose, N., Brüstle, O., & Rhee, J. S. (2019). An autaptic culture system for standardized analyses of iPSC-derived human neurons. Cell Reports , 27(7), 2212–2228. https://doi.org/10.1016/j.celrep.2019.04.059
- Schmid, B., Holst, B., Poulsen, U., Jørring, I., Clausen, C., Rasmussen, M., Mau-Holzmann, U. A., Steeg, R., Nuthall, H., Ebneth, A., & Cabrera-Socorro, A. (2021). Generation of two gene edited iPSC-lines carrying a DOX-inducible NGN2 expression cassette with and without GFP in the AAVS1 locus. Stem Cell Research , 52, 102240. https://doi.org/10.1016/J.SCR.2021.102240
- Shirani, H., Linares, M., Sigurdson, C. J., Lindgren, M., Norman, P., & Nilsson, K. P. R. (2015). A palette of fluorescent thiophene-based ligands for the identification of protein aggregates. Chemistry, A European Journal , 21, 15133–15137. https://doi.org/10.1002/chem.201502999
- Sidoryk-Wegrzynowicz, M., Gerber, Y. N., Ries, M., Sastre, M., Tolkovsky, A. M., & Spillantini, M. G. (2017). Astrocytes in mouse models of tauopathies acquire early deficits and lose neurosupportive functions. Acta Neuropathologica Communications , 5, 89. https://doi.org/10.1186/s40478-017-0478-9
- Sidoryk-Węgrzynowicz, M., & Struzyńska, L. (2019). Astroglial contribution to tau-dependent neurodegeneration. Biochemical Journal , 476(22), 3493–3504. https://doi.org/10.1042/BCJ20190506
- Taga, A., Dastgheyb, R., Habela, C., Joseph, J., Richard, J. P., Gross, S. K., Lauria, G., Lee, G., Haughey, N., & Maragakis, N. J. (2019). Role of human-induced pluripotent stem cell-derived spinal cord astrocytes in the functional maturation of motor neurons in a multielectrode array system. Stem Cells Translational Medicine , 8(12), 1272–1285. https://doi.org/10.1002/sctm.19-0147
- Tang, X., Zhou, L., Wagner, A. M., Marchetto, M. C. N., Muotri, A. R., Gage, F. H., & Chen, G. (2013). Astroglial cells regulate the developmental timeline of human neurons differentiated from induced pluripotent stem cells. Stem Cell Research , 11(2), 743–757. https://doi.org/10.1016/j.scr.2013.05.002
- Tarassishin, L., Suh, H. S., & Lee, S. C. (2014). LPS and IL-1 differentially activate mouse and human astrocytes: Role of CD14. Glia , 62(6), 999–1013. https://doi.org/10.1002/glia.22657
- Thoma, E. C., Wischmeyer, E., Offen, N., Maurus, K., Sirén, A. L., Schartl, M., & Wagner, T. U. (2012). Ectopic expression of neurogenin 2 alone is sufficient to induce differentiation of embryonic stem cells into mature neurons. PLoS One , 7(6), e38651. https://doi.org/10.1371/journal.pone.0038651
- van Ziel, A. M., Wolzak, K., Nölle, A., Hoetjes, P. J., Berenjeno-Correa, E., van Anken, E., Struys, E. A., & Scheper, W. (2020). No evidence for cell-to-cell transmission of the unfolded protein response in cell culture. Journal of Neurochemistry , 152(2), 208–220. https://doi.org/10.1111/jnc.14856
- Verheyen, A., Diels, A., Dijkmans, J., Oyelami, T., Meneghello, G., Mertens, L., Versweyveld, S., Borgers, M., Buist, A., Peeters, P., & Cik, M. (2015). Using human iPSC-derived neurons to model TAU aggregation. PLoS One , 10(12), e146127. https://doi.org/10.1371/journal.pone.0146127
- Wiersma, V. I., van Ziel, A. M., Vazquez-Sanchez, S., Nölle, A., Berenjeno-Correa, E., Bonaterra-Pastra, A., Clavaguera, F., Tolnay, M., Musters, R. J. P., van Weering, J. R. T., Verhage, M., Hoozemans, J. J. M., & Scheper, W. (2019). Granulovacuolar degeneration bodies are neuron-selective lysosomal structures induced by intracellular tau pathology. Acta Neuropathologica , 138(6), 943–970. https://doi.org/10.1007/s00401-019-02046-4
- Wolzak, K., Nölle, A., Farina, M., Abbink, T. E., van der Knaap, M. S., Verhage, M., & Scheper, W. (2022). Neuron-specific translational control shift ensures proteostatic resilience during ER stress. The EMBO Journal , 41(16), e110501. https://doi.org/10.15252/embj.2021110501
- Zhang, Y., Sloan, S. A., Clarke, L. E., Caneda, C., Plaza, C. A., Blumenthal, P. D., Vogel, H., Steinberg, G. K., Edwards, M. S., Li, G., Duncan, J. A. 3rd., Cheshier, S. H., Shuer, L. M., Chang, E. F., Grant, G. A., Gephart, M. G., & Barres, B. A. (2016). Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron , 89(1), 37–53. https://doi.org/10.1016/j.neuron.2015.11.013
- Zhang, Y., Pak, C., Han, Y., Ahlenius, H., Zhang, Z., Chanda, S., Marro, S., Patzke, C., Acuna, C., Covy, J., Xu, W., Yang, N., Danko, T., Chen, L., Wernig, M., & Südhof, T. C. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron , 78(5), 785–798. https://doi.org/10.1016/j.neuron.2013.05.029

