α-Synuclein sedimentation assay
Arpine Sokratian
Abstract
The protocol describes step-by-step instructions on how to set a sedimentation assay to monitor the aggregation of alpha-synuclein over time. The protocol allows for precise quantification of fibril formation kinetics using SDS-PAGE. This methodology enhances our understanding of the underlying mechanisms involved in protein misfolding and offers a reliable tool for investigating potential therapeutic interventions targeting alpha-synuclein aggregation.
Steps
Amplification set-up
Thaw down a-syn monomer and/or sonicated fibril aliquots. Use water-bath.
Do not generate bubbles by pipetting or shaking
Measure monomer concentration via Nanodrop
Add Amount of 5x, 10x or 20x diluted aliquots in PBS onto nanodrop piedestal;
Parameters: other proteins; coefficient extinction:
5.98; MW: 14.4 kDA for human a-synuclein
7.45; MW: 14.4 kDA for mouse a-synuclein
Perform two measurements for each dilution and confirm <10% standard error between two measurements
For fibril preparations, dilute in 6M guanidine and follow the aforementioned instructions derscribed for monomeric protein
Calculate the volume of monomer need following: 0.3 mg/mL of monomer in a reaction (30 ul reaction/well).
Dilute the monomer preparation in PBS to 2,5 mg/mL in order to filter the protein preparation to avoid any high mol. weight contaminants
Use
to filter the diluted monomer concentration. Prior to adding the protein, wash the filter by adding PBS and centrifuging at high-speed.
Measure the size of proteins using DLS
Pour one zirconia bead into each well of the
Prepare serial dilutions of sonicated fibrils if needed, make sure to use protein-low-binding tubes and shake the preparation each time using vortex
Prepare reaction mix containing the monomer mix, PBS and if needed, sonicated fibrils.
Transfer reaction to the plate: start filling the plate with a standard curve samples (seal the portion with a sealing foil)
Proceed the experimental samples and cover the whole plate with a sealing foil. Spin down the plate at 500 g for 30 sec.
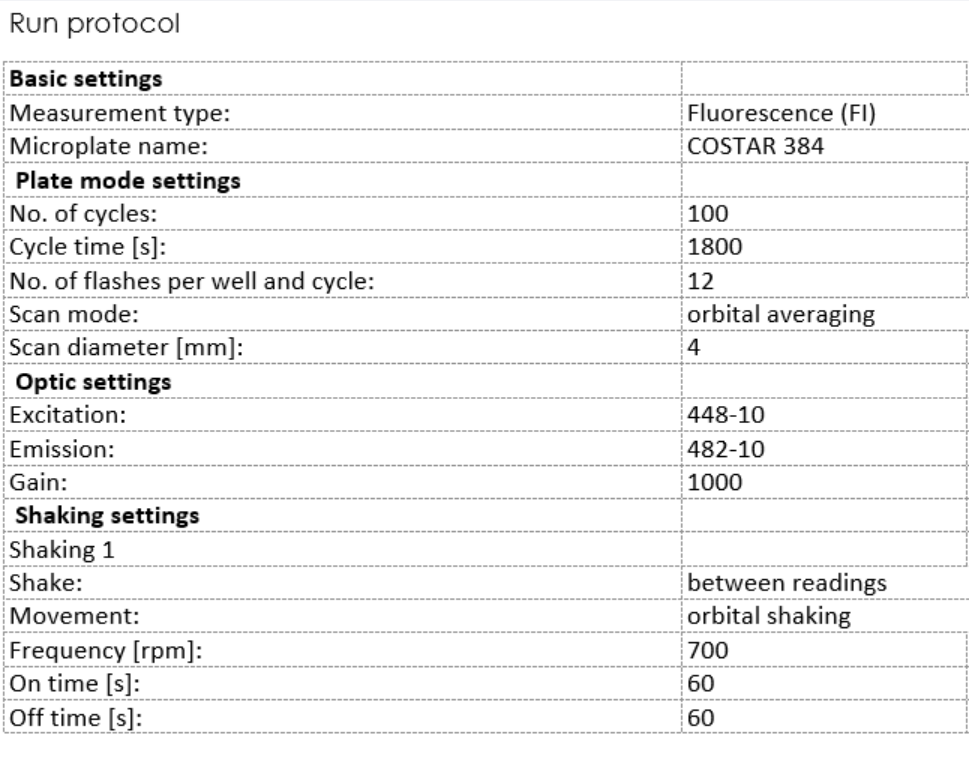
Set up the program on a plate reader
Equipment
| Value | Label |
|---|---|
| Omega - RT-QuIC / PRION Version - Fluorescence Base | NAME |
| BMG Fluostar plate reader | TYPE |
| BMG | BRAND |
| 0415-102P | SKU |
| https://www.bmglabtech.com/clariostar-plus/?utm_term=clariostar&utm_campaign=usa.roi.microplatereaders+products&utm_source=adwords&utm_medium&gclid=Cj0KCQjw4ImEBhDFARIsAGOTMj9ZLSjhHBhKWAli17cxj6V54U96GSg3qsfuFrfP1cETTLgLbn-kdUIaAkovEALw_wcB | LINK |
Sample collection and analysis
Collect replicates at the determined time-points (combine 3 replicates for 1 technical replicate, repeat three times)
Centrifuge at 20,000g for 10 min at 10C, collect 90 uL (the pellet should be visible).
Transfer the 50 ul of supernatant into a new epperdorf tube, remove soluble fraction from the centrifuged pellets and add 90 ul of fresh PBS
Add 4x Laemmli sample buffer to the collected reactions and freeze the samples untill further application
Run SDS-PAGE according the protocol described here, use
Stop the gel and cover it with 100mL of the fixing solution (50% Methanol, 10% Acetic acid) for 30 min
Stain the gel for 1 hr at room temperature with 100 mL of Coomassie stain (0.4g of Coomassie blue
Destain the gel using washing solution consisting of 10% Acetic acid
Image the resulting gels using
Equipment
| Value | Label |
|---|---|
| ChemiDoc™ MP Imaging System | NAME |
| Imaging System | TYPE |
| Bio-rad | BRAND |
| 12003154 | SKU |
| http://www.bio-rad.com/ | LINK |

